Abstract
A DNA methylation (DNAm) signature (the “Horvath clock”) has been proposed as a measure of human chronological and biological age. We determined peripheral blood DNAm in patients with nonalcoholic steatohepatitis (NASH) and assessed whether accelerated aging occurs in these patients. DNAm signatures were obtained in patients with biopsy-proven NASH and stage 2–3 fibrosis. The DNAm profile from one test and two validation cohorts served as controls. Age acceleration was calculated as the difference between DNAm age and the predicted age based on the linear model derived from controls. Hepatic collagen content was assessed by quantitative morphometry. The Horvath clock accurately predicts the chronological age of the entire cohort. Age acceleration was observed among NASH subjects compared with control data sets and our test controls. Age acceleration in NASH subjects did not differ by fibrosis stage but correlated with hepatic collagen content. A set of 152 differentially methylated CpG islands between NASH subjects and controls identified gene set enrichment for transcription factors and developmental pathways. Patients with NASH exhibit epigenetic age acceleration that correlates with hepatic collagen content.
Keywords: Hepatology
Keywords: Hepatitis
A peripheral blood DNA methylation profile identifies accelerated aging in patients with nonalcoholic steatohepatitis.
Introduction
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver condition globally. Although the majority of patients present with steatosis alone, the presence of inflammation and hepatocellular injury in a subgroup of patients with nonalcoholic steatohepatitis (NASH) signifies a risk of progressive fibrosis. Natural history studies of NAFLD have demonstrated that fibrosis stage, regardless of other histologic features, best predicts liver-related mortality (1–3). The assessment of fibrosis, however, represents only a single snapshot of liver injury, without providing a contextual picture of the biological health of the organ as a whole. Novel surrogate markers that measure the age of the liver may provide additional prognostic information beyond staging of fibrosis alone.
Recently, a molecular prediction model based on epigenetic DNA methylation (DNAm) has been proposed as a measure of human chronological and biological age (4–9). The ability to measure human aging from DNAm permits an unbiased assessment of aging, with calculation of a predicted “normal” aging rate based on methylation pattern. A DNAm age above that predicted for a defined chronologic age would represent age acceleration, a condition reflecting a more active epigenetic “maintenance system” working to ensure genomic stability (5). One of these algorithms, the so-called “Horvath clock,” was derived from methylation levels of 353 discrete CpG dinucleotide sites (5). These 353 CpG sites represent an epigenetic profile of aging and are categorized into two sets of CpGs: 193 hypermethylated CpGs that are positively and 160 hypomethylated CpGs that are negatively associated with age. As an illustration of the time frame captured by the “clock,” the DNAm score of embryonic stem cells is approximately 0, increases rapidly during development, and then assumes an intrinsic rate during adulthood. The ability of this peripheral DNAm signature to gauge chronological age has been validated in multiple cohorts and demonstrates predictive capacity across different tissue sites, including the liver (5, 10, 11).
The intrinsic rate of the epigenetic clock can be altered by diseases and metabolic insults related to progressive liver disease. For instance, HIV and obesity are associated with increased liver fibrosis, and both accelerate the epigenetic clock more than would be expected compared with age-matched control specimens (11–13). Interestingly, obesity altered the epigenetic clock of the liver but not for any other tissue sites (11). Acceleration of this clock has functional implications, as age-accelerated DNAm correlates with poorer performance on physical and mental aptitude tests and higher overall mortality, even after adjusting for known risk factors (14, 15). Thus, the Horvath clock can be assayed from peripheral blood, demonstrates concordance with tissue DNAm, and can be used to calculate accelerated aging to provide prognostic information.
Previous studies of DNAm in NAFLD have examined gene-specific epigenetic modifications that may be involved in disease pathogenesis by targeting genes known to regulate fibrogenesis or metabolic function (16–20). For instance, Hardy et al. examined two specific CpG loci on the PPARγ promoter in circulating DNA and found differences in methylation between NAFLD patients with severe fibrosis and those with mild fibrosis (17). Comprehensive integration of genome-wide methylation and gene expression studies has also been performed on liver tissue and been used to identify differential targets between mild and severe NAFLD (18, 20). Limited data exist, however, on whether patients with NASH exhibit a global methylation profile consistent with accelerated aging compared with healthy controls. Therefore, we designed the current study to address the hypothesis that patients with NASH would exhibit accelerated aging compared with controls using the Horvath clock.
In the current study, we determined that the DNAm signature in NASH patients correlates with the severity of hepatic fibrosis. Using three control cohorts (two test, one validation), NASH patients with documented liver fibrosis demonstrated age acceleration compared with healthy controls. DNAm correlates significantly with hepatic collagen content quantified morphometrically and serum markers of fibrosis. We further define two distinct clusters of NASH patients with functional differences in cellular apoptosis and necrosis. These results identify a peripheral blood DNAm signature that links enhanced age acceleration of NASH patients with tissue injury.
Results
The study population included 44 patients with NASH and stage 2–3 fibrosis enrolled in a phase II, randomized, controlled trial of selonsertib (formerly GS-4997; ClinicalTrials.gov, NCT02466516). The median age of the NASH cohort was 56 years; the majority were female (66%) and had diabetes mellitus (70%). Other demographic and baseline characteristics of the NASH patients and the three control populations are included in Table 1. To validate the DNAm age prediction model, we calculated the biological age of the individuals in our cohort and compared it to their chronological age (Figure 1A). The correlation coefficient between DNAm-based and chronological age is 0.84, with a root-mean-square error of 4.7 years. The slope of this line represents the intrinsic rate of DNAm that occurs with aging — the rate of the epigenetic clock during adulthood (4, 5). We next aimed to determine if the NASH cohort is age accelerated (i.e., vertically shifted) compared with normal controls.
Table 1. Demographic and baseline characteristics of NASH patients and the three control populations.
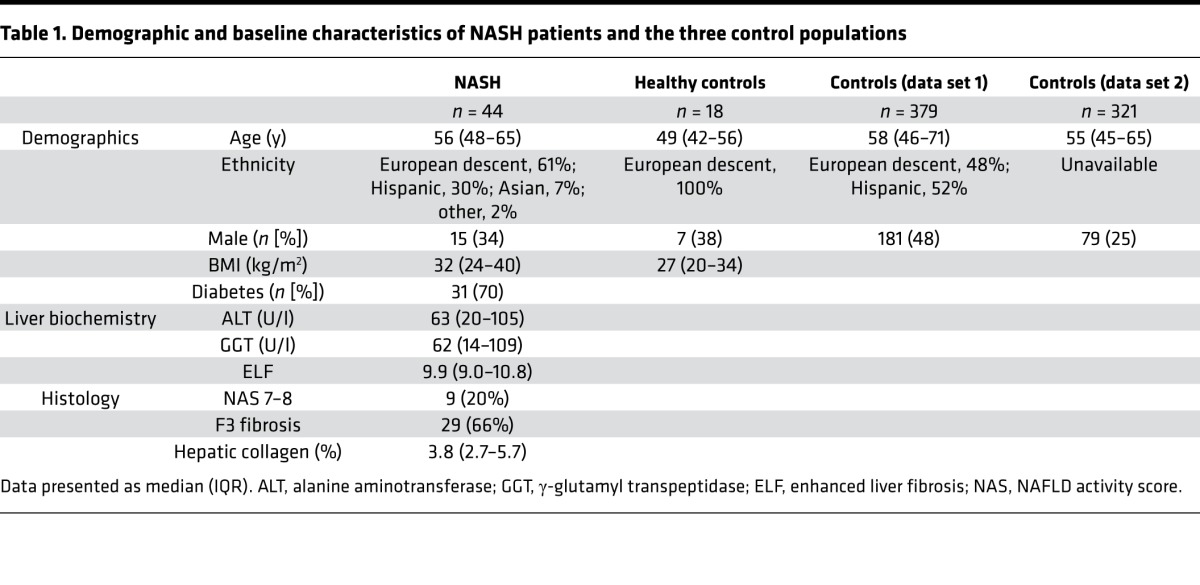
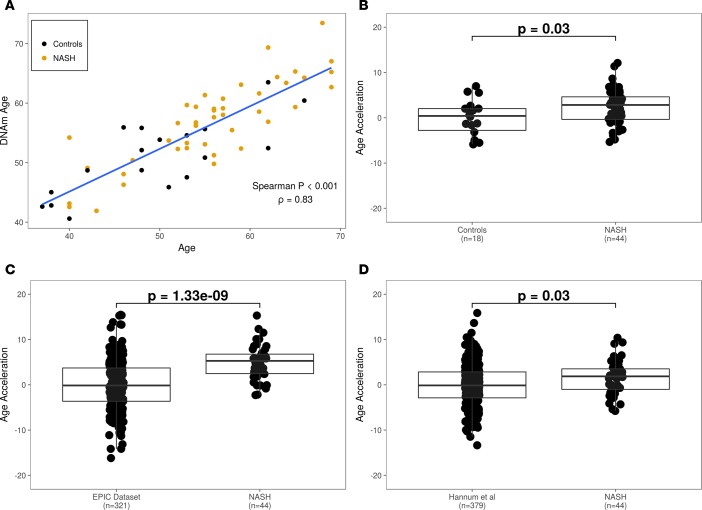
Figure 1. NASH patients are age accelerated compared with healthy controls.
(A) DNAm age as a function of chronological age. (B) Comparison of age acceleration between NASH and test control cohorts. (C) Acceleration of DNAm between NASH subjects and the validation control cohort derived from the EPIC data set (http://epic.iarc.fr/). (D) Comparison of age acceleration between NASH and control cohorts derived from the Hannum et al. data set (4). P values were computed using a 2-tailed Mann-Whitney U test.
We calculated age acceleration in our cohort by first deriving a linear regression model between DNAm age and chronological age in control samples and then comparing the distributions of residuals of NASH and control samples. The median age acceleration of NASH samples is 2.8 years (interquartile range [IQR]: –0.35–4.68), which is significantly greater than that observed among age- and sex-matched healthy controls (P = 0.03; Figure 1B). We then validated these findings using publically available DNAm data sets containing healthy controls: the European Prospective Investigation into Cancer and Nutrition (EPIC; http://epic.iarc.fr/) data set and the Hannum et al. (4) data set. We found that the NASH cohort was age accelerated compared with controls in the EPIC data set (median age acceleration = 5.3 years; P = 1.33 × 10–9; Figure 1C) and in the Hannum et al. data set (median age acceleration = 1.9 years; P = 0.03; Figure 1D). Taken together, these results show that biological age, based on DNAm profile, is highly correlated with chronological age. Furthermore, compared with three independent data sets of healthy controls, the NASH patient cohort demonstrates significant biological age acceleration.
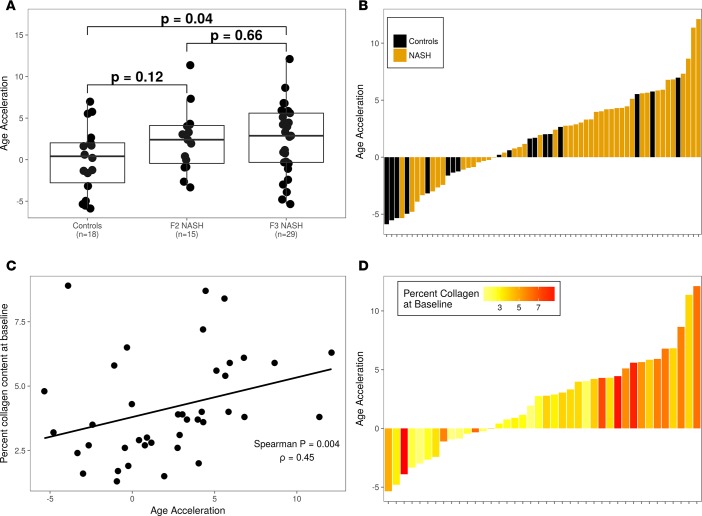
Since liver fibrosis is the defining feature that predicts mortality in NASH, we next evaluated whether age acceleration was associated with the severity of liver fibrosis. Compared with control subjects, DNAm from NASH patients with stage 2 fibrosis did not demonstrate increased age acceleration (P = 0.12), while those with stage 3 fibrosis demonstrated modestly increased age acceleration (P = 0.04; Figure 2A). Since the histological staging of liver fibrosis is limited by an ordinal scoring system, we evaluated the associations between age acceleration and two continuous measures of liver fibrosis, namely hepatic collagen content determined morphometrically and the enhanced liver fibrosis (ELF) score, a serum marker of fibrosis. As shown in Figure 2B, a greater proportion of patients with NASH exhibited age acceleration compared with controls in the test cohort (68% [30 of 44] vs. 55% [10 of 18]). In the NASH cohort, age acceleration was significantly associated with hepatic collagen content (ρ = 0.45; P = 0.004; Figure 2, C and D). Similarly, serum ELF levels were positively correlated with age acceleration (ρ = 0.35; P = 0.03; Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.96685DS1). Thus, these results identify an association between accelerated aging in NASH patients and liver fibrosis.
Figure 2. Age acceleration is associated with liver fibrosis and collagen content.
(A) Age acceleration in control and NASH data sets according to liver fibrosis score (F2 or F3). (B) The magnitude and direction of age acceleration by disease status show enrichment of NASH subjects in those with positive age acceleration. Each bar represents a sample in the cohort; color corresponds to disease status. (C) Relationship between percentage collagen at baseline (y axis) and age acceleration (x axis). Each dot represents a sample in the NASH cohort. (D) The magnitude and direction of age acceleration by disease status. Each bar represents a sample in the cohort; color corresponds to disease status. P values were computed using a 2-tailed Mann-Whitney U test.
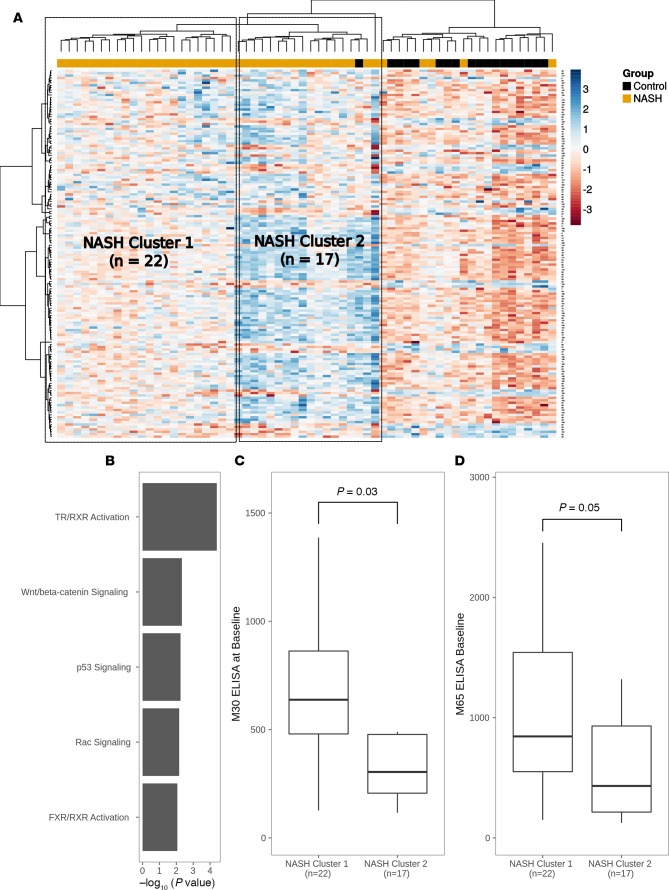
To determine whether NASH patients can be stratified based on a methylation signature, we performed genome-wide methylation analysis to examine differences in the status of CpG islands in controls versus NASH patients. After adjusting for multiple testing, this analysis identified 152 differentially methylated CpG islands, 120 of which exhibited greater methylation in NASH patients (Figure 3A). An enrichment analysis of the genes associated with the differentially methylated CpG islands revealed significant enrichment of developmental signaling pathways, including Wnt/β-catenin signaling and thyroid hormone/retinoid X receptor activation (Figure 3B). Hierarchical clustering of NASH and control samples based on the 152-CpG island signature identified two distinct clusters within the NASH cohort (Figure 3A). Since methylation of developmental pathways suggests functional differences in cell survival, we evaluated differences in cellular apoptosis/necrosis based on serum levels of cytokeratin-18 (CK-18) M30 and M65 fractions by ELISA. Levels of CK-18 M30 and M65 at baseline were significantly greater in NASH cluster 2 patients (Figure 3, C and D), indicating that a subset of NASH patients exists with increased cellular apoptosis/necrosis based on their DNAm pattern.
Figure 3. NASH patients have a distinct methylation profile.
(A) Heatmap of methylation values at CpG islands (rows) by samples (columns). Two distinct NASH sample clusters are annotated. (B) Pathway enrichment analysis of differentially methylated CpG islands between NASH and control cohorts. (C and D) Relationship between M30 and M65 ELISA values and NASH sample clusters 1 and 2, respectively. P values were computed using a 2-tailed Mann-Whitney U test.
Discussion
The ability to measure a surrogate marker of liver aging from a peripheral blood sample has broad implications for assessing clinically “silent” chronic diseases, such as NASH, and, potentially, their response to interventions. In the current study, we validate the utility of the Horvath clock in measuring age acceleration in a defined cohort of NASH patients with moderate to severe liver fibrosis. Using three different control data sets — two validation and one test cohort — the intrinsic rate of DNAm that occurs with aging was determined. We then ascertained that the DNAm signature of NASH patients was age accelerated approximately 5 years compared with public control data sets and 3 years versus our test control cohort. Further, based on two continuous measures of fibrosis, namely hepatic collagen content assessed morphometrically and the serum ELF test, we identified an association between NASH-related fibrosis and the degree of age acceleration. In sum, a peripheral DNAm signature in NASH patients identified age acceleration that reflected the extent of liver fibrosis.
The current study adds to the emerging literature examining the role of epigenetics and the NAFLD phenotype. Using liver biopsy tissue from NAFLD patients, Sookoian et al. identified methylation of promoter sites on PPARγ coactivator 1α (PPARGC1A) with markers of insulin resistance such as HOMA-IR and serum insulin (19). Other investigators have demonstrated that specific methylation sites on promoters of genes are linked to more severe liver fibrosis (17, 18). Although these reports establish a potential relationship between fibrosis and specific epigenetic modifications, targeted individual CpG sites may not accurately reflect the complex interaction between causal and compensatory measures in chronic diseases such as NASH. Thus, utilization of an aggregate DNAm profile obtained from peripheral blood that has been evaluated across different tissue types may provide a valuable measure of overall liver health. Here, we provide confirmation that the Horvath epigenetic clock, a measure of DNAm age, correlates with the degree of liver fibrosis in biopsy-proven NASH.
The results presented herein are consistent with studies of other systemic diseases that are characterized by age acceleration. For instance, HIV infection has been found to significantly accelerate age-related methylation by nearly 14 years in some studies (12, 21). These findings may have relevance to the current study, since HIV is a major determinant of liver fibrosis progression in chronic liver diseases, such as hepatitis C virus (HCV) infection. Prospective studies of patients with HCV and HIV-HCV coinfection confirm more rapid fibrosis progression in coinfected patients using paired liver biopsies (22, 23). Other conditions, such as obesity, also contribute to age acceleration but to a lesser extent (11). Therefore, DNAm age acceleration appears disease specific and modified by environmental factors. Given this complexity, a global signature of the health of the individual reflected in DNAm may serve as an indicator of the chronic injury found in liver diseases, such as NAFLD. Whether a DNAm signature captures those NASH patients at high risk of developing future complications, such as hepatocellular carcinoma, remains unknown and warrants investigation.
The findings in the current work are in contrast to an earlier study that found no association of DNAm with the NAFLD activity score or stage of liver fibrosis in patients with NASH (11). Importantly, that study assessed liver fibrosis based on conventional histological staging only, using the ordinal METAVIR classification. Similarly, we also found no difference in age acceleration between patients with stage 2 and 3 fibrosis according to the NASH Clinical Research Network (CRN) classification. On the contrary, by evaluating two continuous measures of fibrosis (hepatic collagen content by morphometry and the serum ELF test), which have a greater dynamic range than traditional histological staging, we found that patients with higher age acceleration have increased hepatic fibrosis. These findings illustrate the utility of continuous liver fibrosis scales that cannot be ascertained with traditional pathological staging. Whether DNAm is associated with other noninvasive measures of liver fibrosis is unknown and warrants investigation.
Limitations of our study are its relatively small sample size, its inclusion of predominately subjects of European descent, and the absence of specimens representing the full spectrum of liver fibrosis, early (F1) fibrosis and cirrhosis (F4). This study, however, included patients who were part of a clinical trial in which protocol-obtained biopsies were read by a central pathologist (ZG) and morphometric quantification of collagen standardized. Recent work indicates that the intrinsic pace of DNAm does vary by ethnicity (24). Thus, our study represents an important initial study in a population of mostly European descent with NASH, which identifies age acceleration and its correlation with fibrosis content. Future studies that examine the full spectrum of the NASH phenotype in different racial groups may identify specific DNAm aging patterns based on ethnicity.
In summary, a DNAm signature obtained from the peripheral blood of NASH patients identifies epigenetic age acceleration that correlates with increased liver fibrosis. These findings have potential prognostic implications and may serve as a surrogate marker of therapeutic efficacy in NASH.
Methods
Patients and control data sets.
The study population was derived from a phase II study of selonsertib in patients with biopsy-proven NASH and stage 2 or 3 fibrosis according to the NASH CRN classification (ClinicalTrials.gov, NCT02466516). Liver biopsies were obtained at baseline. Age- and sex-matched healthy control peripheral blood mononuclear cells (PBMCs) were acquired from Conversant Biologics.
The data sets for validating the control population were obtained from previously published work (4, 25). The data set by Hannum et al. (4) was downloaded from Gene Expression Omnibus (GSE40279) as β values using the GEOquery R package (26). The EPIC data set was downloaded from ArrayExpress (E-GEOD-51032) as IDAT files and processed in the same manner as data generated for this study (25). We restricted the subjects’ ages in the Hannum and EPIC data sets to between 40 and 69 years to reflect the age range of the NASH subjects in our cohort. For the EPIC data set, we excluded all subjects that have been annotated with a disease category.
Sample collection and methylation analysis.
DNA extracted from PBMCs was assayed for cytosine methylation using the Infinium Methylation Assay protocol for the Infinium Human Methylation450K BeadChip (Illumina). In brief, DNA was treated with sodium bisulfite to convert unmethylated cytosines to uracil, leaving methylated cytosines unchanged. The treated DNA sample was then denatured, neutralized, and isothermally amplified. The amplified DNA was fragmented, precipitated with isopropanol, and resuspended prior to hybridization onto BeadChips. The converted and nonconverted amplified DNAs were hybridized to their corresponding probes, and excess DNA was washed away. Hybridized DNAs then underwent single-base extension and staining for labeling, followed by scanning on an Illumina iScan instrument for detection. Scanned images were analyzed using Illumina Genome Studio software.
Data processing.
Data were processed using the minfi R Bioconductor package version 1.2 (27) and wateRmelon package version 1.18 (28).
Epigenetic age and determination of differential DNAm.
Epigenetic age, based on the Horvath model, was calculated from the methylation β values using the minfi’s agep function. Age acceleration was calculated as the difference between an individual’s DNAm age and one calculated from a linear model derived using the control samples (5). Differential DNAm analysis was restricted to genomic locations that do not overlap known SNPs and those that lie within CpG islands. Methylation β values were first quantile normalized and then converted to M values. CpG island methylation values were calculated using an average value of individual probes that span it. Tests for differential methylation were carried out using the limma R package version 3.3 (29). An FDR-adjusted P value of 0.05 was used to identify differentially methylated CpG islands. Associated between genes and CpG islands was done by genomic distance.
Ingenuity pathway analysis.
Genomic DNAm and the relationship between different patterns of gene methylation were analyzed using Ingenuity Pathway Analysis software (Ingenuity Systems). Networks of methylated genes were algorithmically generated based on their connectivity and assigned a score. Scores were used to rank signaling and developmental pathway networks according to their methylation content.
Hepatic collagen determination.
Sections of each liver biopsy stained with Picrosirius red were used for hepatic collagen content determination as previously described (30). A digital image of the entire stained section was acquired with an Aperio Scanscope CS (Aperio Technologies Inc.). The pixel count was obtained according to the manufacturer’s instructions to quantify the fraction of positively stained pixels. Results were expressed as a fraction of the entire biopsy specimen that was positive for the red staining. The central pathologist (ZG) reviewed biopsy specimens in a blinded fashion.
Serum markers of fibrosis.
The ELF panel was determined as previously described (31). Approximately 20 ml blood was obtained under fasting conditions. Procollagen III N-terminal peptide (PIIINP), hyaluronic acid (HA), and tissue inhibitor of metalloproteinase-1 (TIMP-1) were measured by an automated clinical immunochemistry analyzer that performs magnetic separation enzyme immunoassay tests (ADVIA Centaur, Siemens Healthcare Diagnostics). The ELF score was calculated using the algorithm [ELF = 2.278 + 0.851 ln(HA) + 0.751 ln(PIIINP) + 0.394 ln(TIMP-1)].
CK-18 M30 and M65 ELISA.
CK-18 M30 Apoptosense and M65 ELISA kits were obtained from PEVIVA AB and were performed according to the manufacturer’s instructions as previously described (32). Values are expressed in units, where 1 U/l is equivalent to 1.24 pM of a synthetic 21–amino acid fragment of CK18 containing both M30 (e.g., M30) and M65 (e.g., M5) epitopes. Patient plasma samples were run in duplicate, and values were plotted against standard curve.
Statistics.
A 2-tailed Mann-Whitney test was used to test the statistical significance of age acceleration in two groups. A P value of less than 0.05 was considered significant.
Study approval.
The study was approved by the institutional review boards of the University of California, San Diego, Gilead Sciences Inc., Texas Liver Institute, University of Virginia, Inova Fairfax Hospital, Intermountain Medical Center, Beth Israel Deaconess Medical Center, Harvard Medical School, and Duke Clinical Research Institute. Written informed consent was obtained from each individual prior to his or her participation in the study.
Author contributions
RL, GMS, RPM, and AMD conceived the project and designed experiments. YG, ZJ, CC, and RPM performed analysis. RL, YG, CC, RPM, and GMS wrote the manuscript. RL, YG, ZJ, EL, SC, CSD, RX, CC, RPM, GMS, ZG, MC, NHA, and AMD reviewed the data and the manuscript.
Supplementary Material
Acknowledgments
This work was supported by Gilead Sciences Inc. We thank Lakshmi Alaparthi and Fanny Monge for assistance with the morphometry analyses.
Version 1. 01/25/2018
Electronic publication
Footnotes
Conflict of interest: R. Loomba is on advisory committees for Arrowhead Pharmaceuticals, Conatus Pharmaceuticals, Galmed Pharmaceuticals, Gilead Sciences, Intercept Pharmaceuticals, NGM Bio, Nimbus Pharma, Octeta Therapeutics, and Tobira; has served as a member of NASH scientific advisory board; is a consultant for Alnylam Pharmaceuticals, Bird Rock Bio, Bristol-Myers Squibb, Boehringer Ingelheim, Celgene, Conatus Pharmaceuticals, DeuteRx, Eli Lilly and Co., Enanta Pharmaceuticals, FibroGen, GenKyoTex, Gilead Sciences, GRI Bio, ISIS Pharmaceuticals, Janssen Inc., Kyowa Kirin Pharmaceutical Development, Madrigal Pharmaceuticals, Metacrine, NGM Bio, Nitto Denko, Pfizer, Receptos, Roivant Sciences, RuiYi, Sanofi, Scholar Rock, Shire, Tasly, Viking Therapeutics, Yuhan Pharmaceuticals, and Zafgen; co-founded Liponexus Inc.; and has received research funding from Gilead Sciences.
Reference information: JCI Insight. 2017;2(24):e96685. https://doi.org/10.1172/jci.insight.96685.
Contributor Information
Rohit Loomba, Email: roloomba@ucsd.edu.
Yevgeniy Gindin, Email: Yevgeniy.Gindin@Gilead.com.
Zhaoshi Jiang, Email: Zhaoshi.Jiang@Gilead.com.
Eric Lawitz, Email: lawitz@txliver.com.
Stephen Caldwell, Email: shc5c@virginia.edu.
Ren Xu, Email: ren.xu@gilead.com.
Chuhan Chung, Email: chuhan.chung@gilead.com.
Zachary Goodman, Email: Zachary.Goodman@inova.org.
Nezam H. Afdhal, Email: nafdhal@bidmc.harvard.edu.
Anna Mae Diehl, Email: annamae.diehl@duke.edu.
References
- 1.Angulo P, et al. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149(2):389–97.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhala N, et al. The natural history of nonalcoholic fatty liver disease with advanced fibrosis or cirrhosis: an international collaborative study. Hepatology. 2011;54(4):1208–1216. doi: 10.1002/hep.24491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loomba R, Chalasani N. The hierarchical model of NAFLD: Prognostic significance of histologic features in NASH. Gastroenterology. 2015;149(2):278–281. doi: 10.1053/j.gastro.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 4.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch CM, Wagner W. Epigenetic-aging-signature to determine age in different tissues. Aging (Albany NY) 2011;3(10):1018–1027. doi: 10.18632/aging.100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Numata S, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90(2):260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakyan VK, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20(4):434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teschendorff AE, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20(4):440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen BH, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8(9):1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci USA. 2014;111(43):15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212(10):1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine AJ, et al. Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders. J Neurovirol. 2016;22(3):366–375. doi: 10.1007/s13365-015-0406-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christiansen L, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15(1):149–154. doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marioni RE, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44(4):1388–1396. doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahrens M, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Hardy T, et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. 2017;66(7):1321–1328. doi: 10.1136/gutjnl-2016-311526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy SK, et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145(5):1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sookoian S, et al. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology. 2010;52(6):1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 20.Kirchner H, et al. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol Metab. 2016;5(3):171–183. doi: 10.1016/j.molmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rickabaugh TM, et al. Acceleration of age-associated methylation patterns in HIV-1-infected adults. PLoS One. 2015;10(3):e0119201. doi: 10.1371/journal.pone.0119201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benhamou Y, et al. Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. The Multivirc Group. Hepatology. 1999;30(4):1054–1058. doi: 10.1002/hep.510300409. [DOI] [PubMed] [Google Scholar]
- 23.Mohsen AH, et al. Impact of human immunodeficiency virus (HIV) infection on the progression of liver fibrosis in hepatitis C virus infected patients. Gut. 2003;52(7):1035–1040. doi: 10.1136/gut.52.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath S, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17(1):171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riboli E, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5(6B):1113–1124. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 26.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 27.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McHutchison J, et al. Farglitazar lacks antifibrotic activity in patients with chronic hepatitis C infection. Gastroenterology. 2010;138(4):1365–1373. doi: 10.1053/j.gastro.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Crespo G, et al. ARFI, FibroScan, ELF, and their combinations in the assessment of liver fibrosis: a prospective study. J Hepatol. 2012;57(2):281–287. doi: 10.1016/j.jhep.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 32.Cummings J, et al. Evaluation of cell death mechanisms induced by the vascular disrupting agent OXi4503 during a phase I clinical trial. Br J Cancer. 2012;106(11):1766–1771. doi: 10.1038/bjc.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.