Abstract
Skeletal muscle mass is regulated by a complex array of signaling pathways. TGF-β–activated kinase 1 (TAK1) is an important signaling protein, which regulates context-dependent activation of multiple intracellular pathways. However, the role of TAK1 in the regulation of skeletal muscle mass remains unknown. Here, we report that inducible inactivation of TAK1 causes severe muscle wasting, leading to kyphosis, in both young and adult mice.. Inactivation of TAK1 inhibits protein synthesis and induces proteolysis, potentially through upregulating the activity of the ubiquitin-proteasome system and autophagy. Phosphorylation and enzymatic activity of AMPK are increased, whereas levels of phosphorylated mTOR and p38 MAPK are diminished upon inducible inactivation of TAK1 in skeletal muscle. In addition, targeted inactivation of TAK1 leads to the accumulation of dysfunctional mitochondria and oxidative stress in skeletal muscle of adult mice. Inhibition of TAK1 does not attenuate denervation-induced muscle wasting in adult mice. Finally, TAK1 activity is highly upregulated during overload-induced skeletal muscle growth, and inactivation of TAK1 prevents myofiber hypertrophy in response to functional overload. Overall, our study demonstrates that TAK1 is a key regulator of skeletal muscle mass and oxidative metabolism.
Keywords: Metabolism, Muscle Biology
Keywords: Mouse models, Skeletal muscle
Transforming growth factor β (TGF-β)-activated kinase 1 (TAK1) is essential for maintenance of skeletal muscle mass.
Introduction
Loss of skeletal muscle mass and strength leads to severe consequences, resulting in permanent disability and mortality in the elderly and in settings of functional denervation, as well as in many chronic disease states (1). Skeletal muscle mass is governed by a fine balance between the rate of protein synthesis and degradation (2). In addition, mitochondria play a pivotal role in the maintenance of skeletal muscle mass and metabolic function (3). Perturbation in mitochondrial dynamics and function is responsible for skeletal muscle wasting in multiple pathophysiological conditions (4, 5).
Skeletal muscle mass is regulated by a number of signaling pathways. For example, the activation of the Akt/mTOR signaling pathway induces skeletal muscle growth (6, 7) and prevents muscle protein degradation (6–9). While promoting mitochondrial biogenesis and metabolic adaptation, activation of AMPK inhibits protein synthesis in skeletal muscle through suppressing mTOR activity (10, 11). AMPK also induces protein degradation through stimulating the ubiquitin-proteasome system (UPS) and autophagy (12–14). Several inflammatory cytokines, microbial products, and tumor-derived factors induce muscle wasting through the activation of canonical NF-κB and p38 MAPK signaling pathways (15–17). Furthermore, TGF-β and its closely related family members, myostatin, activin, and growth and differentiation factor 11 (GDF11), cause muscle wasting through the activation of Smad2/3 transcription factors (2, 18). However, the proximal signaling events that regulate the activation of these pathways in skeletal muscle remain less understood.
TGF-β–activated kinase 1 (TAK1, also known as MAPK3K7) is a member of the MEK kinase (MAP3K) family that mediates context-dependent activation of distinct signaling pathways. TAK1 forms a complex with TAK1-binding protein 1 (TAB1) and with either TAB2 or TAB3 (19). Upon cytokine stimulation, the TAK1 complex is activated by K63-linked polyubiquitination reactions catalyzed by the E2 enzyme complex Uev1A-Ubc13 and the RING finger E3 ligases TRAF2 or TRAF6. The specific interaction of the K63-Ub chains with the C-terminal domains of TAB2 and TAB3 induces a conformational change that leads to the autophosphorylation of TAK1 (20). Activated TAK1 phosphorylates the specific MAPK kinases (MKK), which leads to the activation of JNK1/2 and p38 MAPK (21). TAK1 also phosphorylates inhibitor of κ B (IκB) kinase β (IKKβ), leading to the phosphorylation and degradation of IκB proteins, which results in the activation of NF-κB transcription factor (22, 23). It has been consistently observed that NF-κB is a negative regulator of skeletal muscle mass and function (2, 15, 16). However, the physiological role of TAK1 in the regulation of skeletal muscle remains unknown.
Because conventional Tak1-null mice are embryonically lethal (24, 25), for this study, we generated inducible skeletal muscle–specific Tak1-null mice. Our results demonstrate that inducible inactivation of TAK1 causes severe muscle wasting and kyphosis in mice without having any effect on the survival of myofibers. Deletion of TAK1 inhibits the rate of protein synthesis and stimulates the activation of proteolytic systems in skeletal muscle. Furthermore, targeted ablation of TAK1 induces oxidative stress and leads to the accumulation of dysfunctional mitochondria in the skeletal muscle of adult mice. Finally, we found that TAK1 is essential for overload-induced skeletal muscle hypertrophy in adult mice. Altogether, our study demonstrates a previously unrecognized role of TAK1 in the regulation of skeletal muscle mass and health.
Results
Inactivation of TAK1 reduces skeletal muscle mass in young mice.
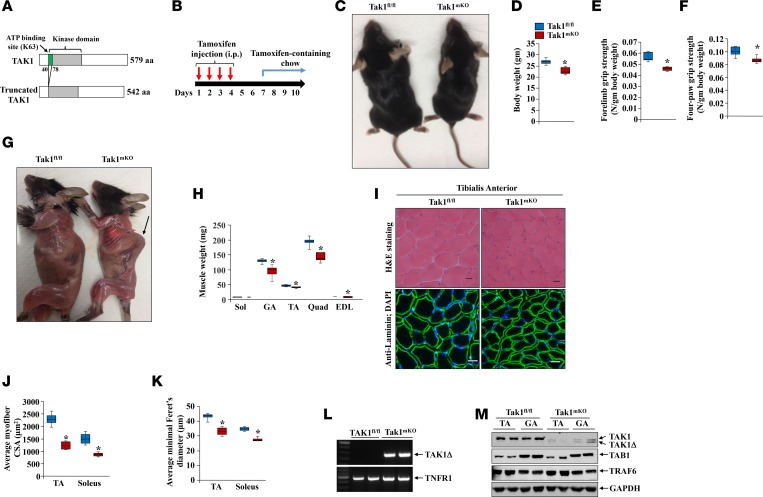
For our studies, we used mice in which the second exon of the TAK1 gene, which encodes the enzyme’s catalytic domain, is flanked with 2 loxP sites (24) (Figure 1A). Our attempts to generate skeletal muscle–specific Tak1-KO mice by crossing floxed Tak1 (i.e., Tak1fl/fl) mice with constitutive muscle creatine kinase-Cre (MCK-Cre) or myosin light chain 1-Cre (MLC1-Cre) mice, were futile. While muscle-specific Tak1-KO pups were born at expected frequencies, they did not survive beyond 2 days after birth. We then crossed Tak1fl/fl mice with a tamoxifen-inducible human α-skeletal actin-Cre (HSA-Cre) line (e.g., HSA-MerCreMer [MCM] as described; ref. 26) and generated inducible muscle–specific TAK1-KO (HSA-MCM;Tak1fl/fl, henceforth Tak1mKO) mice. To understand the role of TAK1 in the regulation of skeletal muscle mass in young animals, 6-week-old Tak1mKO mice were given i.p. injections of tamoxifen every day for 4 days to inactivate TAK1. Littermate Tak1fl/fl mice were also treated with tamoxifen and served as corresponding controls for Tak1mKO mice. The mice were maintained on a tamoxifen-containing chow for the entire duration of experimentation (Figure 1B). Intriguingly, both male and female Tak1mKO mice looked smaller in size (Figure 1C), and their body weight was significantly reduced compared with littermate Tak1fl/fl mice by 3 weeks after the initiation of tamoxifen injections (Figure 1D). There was a significant reduction in grip strength normalized by body weight of Tak1mKO mice compared with littermate Tak1fl/fl mice (Figure 1, E and F). We also observed that several Tak1mKO mice developed kyphosis after 3 weeks of inactivation of TAK1 (Figure 1G). Furthermore, wet weights of gastrocnemius (GA), tibialis anterior (TA), quadriceps (Quad), and extensor digitorum longus (EDL) muscle were significantly reduced in Tak1mKO compared with Tak1fl/fl mice (Figure 1H). We next generated transverse sections of TA and soleus muscles of Tak1fl/fl and Tak1mKO mice and performed H&E- or anti-laminin staining (Figure 1I). Quantitative analysis showed that average myofiber cross-sectional area (CSA) as well as average minimal Feret’s diameter were significantly reduced in the TA and soleus muscles of Tak1mKO mice compared with Tak1fl/fl mice (Figure 1, J and K). Deletion of TAK1 in the skeletal muscle of Tak1mKO mice was confirmed by performing PCR on genomic DNA (Figure 1L) using a primer set that detects the truncated form of TAK1 (27). Western blot analysis also showed that TAK1 was efficiently deleted in the skeletal muscle of Tak1mKO mice, but not in Tak1fl/fl mice, after treatment with tamoxifen. There was no difference in the levels of related proteins, TAB1 and TRAF6, in skeletal muscle of Tak1fl/fl and Tak1mKO mice (Figure 1M).
Figure 1. TAK1 is required for post-natal skeletal muscle growth in mice.
(A) Schematic representation of WT TAK1 and kinase domain-truncated (green) TAK1 protein. (B) Treatment protocol for tamoxifen-induced Cre recombination in Tak1mKO mice. Six-week-old Tak1fl/fl and Tak1mKO mice were given i.p. injections of tamoxifen, and 3 weeks later, the mice were analyzed. (C) Gross appearance of Tak1fl/fl and Tak1mKO mice. (D) Average body weight of Tak1fl/fl and Tak1mKO mice. (E) Average forelimb and (F) average 4-paw grip strength in Tak1fl/fl and Tak1mKO mice normalized by their body weight. (G) Representative images of skinned Tak1fl/fl and Tak1mKO mice. Arrow pointing to kyphotic phenotype in Tak1mKO mice. (H) Average weight of soleus (Sol), gastrocnemius (GA), tibialis anterior (TA), quadriceps (Quad), and extensor digitorum longus (EDL) muscles in Tak1fl/fl and Tak1mKO mice. (I) Representative photomicrographs of TA muscle sections of Tak1fl/fl and Tak1mKO mice after staining for H&E (top panel) and anti-laminin (lower panel). Scale bars: 20 μm. Quantification of (J) average myofiber CSA, and (K) mean minimal Feret’s diameter in TA and soleus muscle sections of Tak1fl/fl and Tak1mKO mice. (L) PCR analysis of genomic DNA from GA muscle of tamoxifen-treated Tak1fl/fl and Tak1mKO mice using primer sets that detect truncated TAK1 and TNF receptor 1 (TNFR1). Representative PCR gel images presented here demonstrate efficient deletion of the kinase domain of TAK1 in Tak1mKO mice following tamoxifen administration. (M) Immunoblots presented here demonstrate that the protein levels of TAK1, but not TAB1 or TRAF6, were diminished in GA muscle of Tak1mKO mice. n = 6 or 7 in each group. Error bars represent ± SEM. *P < 0.05, values significantly different from corresponding Tak1fl/fl mice by unpaired 2-tailed t test.
To understand the above phenotype at the molecular level, we performed a transcriptome analysis on the GA muscle from Tak1fl/fl and Tak1mKO mice. The analysis of the microarray data shows 163 upregulated and 168 downregulated genes at the threshold values of 1.5-fold and P < 0.05 (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.98441DS1). The gene ontology (GO) analysis of differentially expressed genes showed that many GO terms related to skeletal muscle tissue and organ development were highly affected in Tak1mKO mice (Supplemental Figure 1B). Indeed, mRNA levels of many skeletal muscle–related molecules and growth factors were significantly reduced in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Supplemental Figure 1, C and D).
Inactivation of TAK1 causes skeletal muscle wasting in adult mice.
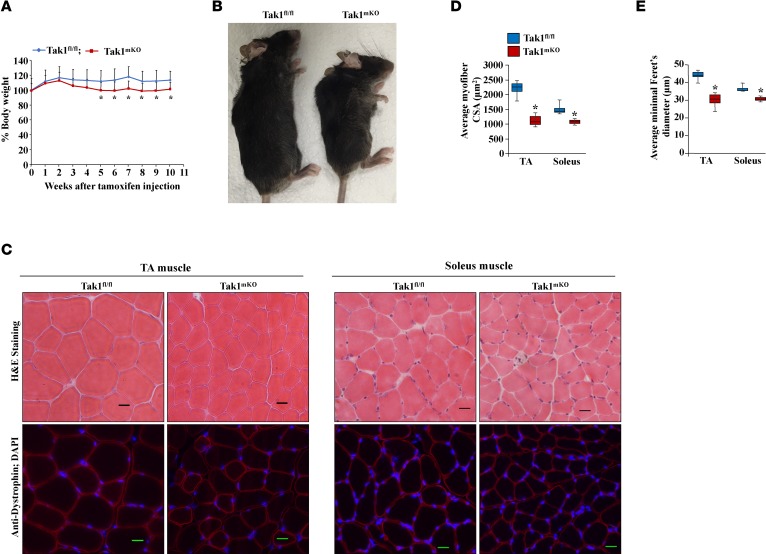
We next investigated the effect of muscle-specific inactivation of TAK1 in adult mice. For this experiment, 14-week-old littermate Tak1fl/fl and Tak1mKO mice were given i.p. injections of tamoxifen and further fed with a tamoxifen-containing chow, and their overall body weight was measured weekly. There was no significant difference in the body weight of Tak1fl/fl and Tak1mKO mice up to 4 weeks from the first injection of tamoxifen. However, by 5 weeks, the average body weight of Tak1mKO mice was significantly lower compared with Tak1fl/fl mice (Figure 2A). We also observed that Tak1mKO mice developed kyphosis by 10 weeks of inactivation of TAK1 (Figure 2B). Moreover, grip strength normalized with body weight was also significantly reduced in Tak1mKO mice compared with Tak1fl/fl mice (Supplemental Figure 2, A and B). After 10 weeks from the start of tamoxifen injections, we isolated hind limb muscles from the mice and performed H&E- and anti-dystrophin staining (Figure 2C), followed by morphometric analysis. Results showed that the average myofiber CSA and minimal Feret’s diameter of myofibers were drastically reduced in TA and soleus muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 2, D and E). While we observed a drastic reduction in average myofiber size, there was no overt sign of muscle pathology, such as centronucleation or infiltration of mononucleated cells in the skeletal muscle of Tak1mKO mice (Figure 2C). We also compared the number of myofibers in muscle sections. Our analysis showed that there was no significant difference in the total number of myofibers in TA muscle of Tak1fl/fl and Tak1mKO mice, suggesting that inactivation of TAK1 does not cause myofiber dropout (Supplemental Figure 2C). Published reports suggest that TAK1 is essential for the survival of multiple cell types, including muscle stem cells (19, 24, 28). By performing TUNEL staining, we investigated whether inactivation of TAK1 causes myonuclear apoptosis. However, we did not find any TUNEL+ nuclei in TA muscle sections of Tak1fl/fl or Tak1mKO mice (Supplemental Figure 3), suggesting that inactivation of TAK1 does not induce apoptosis in skeletal muscle.
Figure 2. TAK1 is required for maintenance of skeletal muscle mass in adult mice.
Fourteen-week-old Tak1fl/fl and Tak1mKO mice were given i.p. injections of tamoxifen, and their body weight was recorded weekly. (A) Representative line diagram of percentage change in body weight of Tak1fl/fl and Tak1mKO mice after initiation of tamoxifen injection. Error bars represent ± SD. (B) Gross appearance of Tak1fl/fl and Tak1mKO mice after 10 weeks of initiation of tamoxifen injections. (C) Representative photomicrographs of TA and soleus muscle sections of Tak1fl/fl and Tak1mKO mice after staining for H&E (top panel) and anti-dystrophin (lower panel). Scale bars: 20 μm. Quantification of average (D) myofiber CSA, and (E) minimal Feret’s diameter in TA and soleus muscle sections of Tak1fl/fl and Tak1mKO mice. n = 6 in each group. Error bars represent ± SEM. *P < 0.05 values significantly different from corresponding Tak1fl/fl mice by unpaired 2-tailed t test.
To understand whether TAK1 regulates skeletal muscle mass in a cell-autonomous or non–cell-autonomous manner, we investigated the effects of knockdown of TAK1 in fully differentiated cultured myotubes. Primary myotubes prepared from WT mice were transduced with adenovirus control (Ad.Control) shRNA or Ad.TAK1 shRNA. After 48 hours, the myotube cultures were fixed and the average myotube diameter was quantified. Interestingly, knockdown of TAK1 significantly reduced the average diameter of cultured myotubes (Supplemental Figure 4, A and B). Altogether, these results suggest that TAK1 is required for maintenance of skeletal muscle mass both in vivo and in vitro.
Inactivation of TAK1 reduces the rate of protein synthesis and induces proteolysis.
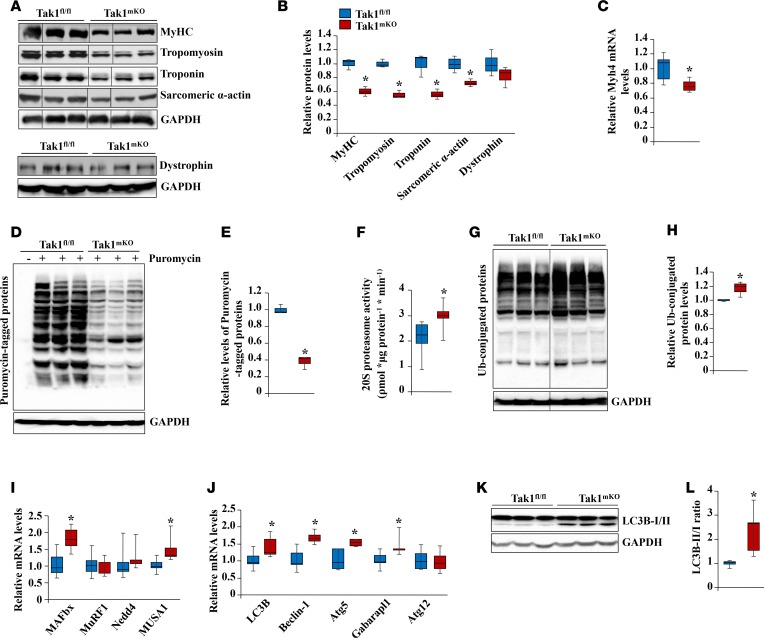
We next investigated the biochemical basis of skeletal muscle atrophy in Tak1mKO mice. We first measured relative levels of various thick- and thin-filament proteins in the GA muscle of Tak1fl/fl and Tak1mKO mice after 3 weeks of inactivation of TAK1. Results showed that protein levels of myosin heavy chain (MyHC), tropomyosin, troponin, and sarcomeric α-actin — but not dystrophin — were significantly reduced in Tak1mKO mice compared with Tak1fl/fl mice (Figure 3, A and B). There was also a significant decrease in the mRNA levels of MyHC fast-type (gene name, Myh4) in the skeletal muscle of Tak1mKO mice (Figure 3C). By performing surface sensing of translation (SUnSET) assay, we next investigated whether inactivation of TAK1 affects the rate of protein synthesis in skeletal muscle of mice. Intriguingly, a sharp reduction in puromycin-tagged protein was noticeable in the skeletal muscles of Tak1mKO mice compared with Tak1fl/fl mice (Figure 3, D and E). Consistent with in vivo results, we found that knockdown of TAK1 reduced the rate of protein synthesis in cultured myotubes, as well (Supplemental Figure 4C).
Figure 3. Inactivation of TAK1 perturbs the rate of protein synthesis and protein degradation in skeletal muscle.
Six-week-old Tak1fl/fl and Tak1mKO mice were given i.p. injections of tamoxifen, and 3 weeks later, skeletal muscle were collected and processed for biochemical analysis. (A) Immunoblots demonstrating the protein levels of MyHC fast-type, tropomyosin, troponin, sarcomeric α-actin, dystrophin, and unrelated protein GAPDH in GA muscle of Tak1fl/fl and Tak1mKO mice. Dystrophin was run on a separate gel with loading control GAPDH. (B) Densitometry quantification of relative protein levels of MyHC fast-type, tropomyosin, troponin, sarcomeric α-actin, and dystrophin in GA muscle of Tak1fl/fl and Tak1mKO mice. (C) Relative mRNA levels of Myh4 in GA muscle of Tak1fl/fl and Tak1mKO mice assayed by qPCR. (D) Tak1fl/fl and Tak1mKO mice were given i.p. injections of puromycin (0.04 μmol/g body weight). The mice were euthanized 30 minutes later, and GA muscle was collected and processed by Western blot. Immunoblots of puromycin-tagged protein and unrelated protein GAPDH in GA muscle of Tak1fl/fl and Tak1mKO mice. (E) Densitometry quantification of relative levels of puromycin-tagged proteins in GA muscle of Tak1fl/fl and Tak1mKO mice. (F) Fold change in 20S proteasome activity in GA muscle of Tak1fl/fl and Tak1mKO mice measured using a commercially available kit (Catalog # K245, BioVision). (G) Immunoblots showing relative amounts of ubiquitin-conjugated protein and unrelated protein GAPDH in skeletal muscle of Tak1fl/fl and Tak1mKO mice. (H) Densitometry quantification of relative levels of ubiquitin-conjugated proteins. (I) Relative mRNA levels of MAFbx/Atrogin-1, MuRF1, Nedd4, and MUSA1 in skeletal muscle of Tak1fl/fl and Tak1mKO mice. (J) Relative mRNA levels of autophagy related genes LC3B, Beclin-1, Atg5, Gabarapl1, and Atg12 in skeletal muscle of Tak1fl/fl and Tak1mKO mice. (K) Representative immunoblots of LC3B-I/II and unrelated protein GAPDH in skeletal muscle of Tak1fl/fl and Tak1mKO mice. (L) Densitometry quantification of the ratio of LC3B-II vs. LC3B-I in skeletal muscle of Tak1fl/fl and Tak1mKO mice. n = 4 or 5 in each group. Error bars represent ± SEM. *P < 0.05 values significantly different from corresponding Tak1fl/fl mice by unpaired 2-tailed t test.
Recent studies have provided evidence that perturbation in ribosomal content can also contribute to the reduced rate of protein synthesis in skeletal muscle (29). Surprisingly, analysis of microarray data showed that the gene expression of a number of molecules involved in ribosomal biogenesis was significantly increased in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Supplemental Figure 5A). Moreover, our Western blot analysis showed that levels of ribosomal protein L7 (RPL7), RPL22, and regulators of ribosomal biogenesis (e.g., c-Myc, Wnt3a, and β-catenin) were significantly increased in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Supplemental Figure 5, B and C). These results suggest that the translational capacity of skeletal muscle is increased in Tak1mKO mice, which could be a compensatory mechanism attempting to overcome the inhibition in translational efficiency (i.e., the rate of protein synthesis).
The UPS and autophagy are 2 major mechanisms for protein degradation in skeletal muscle in atrophy conditions (1, 14). Results showed that there was a significant increase in 20S proteasome activity in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 3F). Moreover, there was also a significant increase in the levels of ubiquitinated proteins in skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 3, G and H). We also compared mRNA levels of various E3 ubiquitin ligases that mediate protein degradation in skeletal muscle. Results showed that mRNA levels of MAFbx (also known as Atrogin-1) and MUSA1, but not MuRF1 or Nedd4, were significantly increased in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 3I). Consistent with in vivo results, we found that the level of MAFbx was increased in cultured myotubes upon knockdown of TAK1 (Supplemental Figure 4D). We next compared the expression of autophagy markers in the skeletal muscle of Tak1fl/fl and Tak1mKO mice. There was a significant increase in the mRNA levels of LC3B, Beclin-1, Atg5, and Gabarapl1 in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 3J). Moreover, the ratio of LC3B-II/I protein was found to be significantly increased in the skeletal muscle of Tak1mKO mice (Figure 3, K and L), as well as in TAK1-knockdown myotubes (Supplemental Figure 4D) compared with their corresponding controls, further suggesting that the inactivation of TAK1 induces autophagy in skeletal muscle. Collectively, these results suggest that inactivation of TAK1 inhibits protein synthesis and induces the activity of the UPS and autophagy in skeletal muscle.
TAK1 regulates the activation of multiple signaling pathways in skeletal muscle.
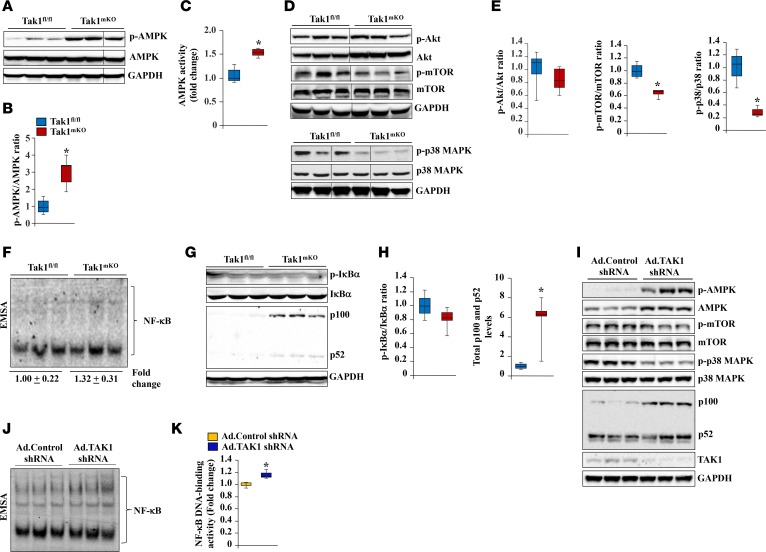
AMPK is a major signaling protein that represses the rate of protein synthesis and increases the activation of the UPS and autophagy (5, 13, 30). Our analysis showed that the level of phosphorylated AMPK was significantly increased in the skeletal muscle of Tak1mKO mice (Figure 4, A and B). There was also a significant increase in the enzymatic activity of AMPK in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 4C). The PI3K/Akt/mTOR pathway is an important regulator of protein synthesis and skeletal muscle mass (2, 6). While there was no significant effect on the phosphorylation of Akt, the level of phosphorylated mTOR was found to be significantly reduced in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 4, D and E). TAK1 is known to activate p38 MAPK in different cell types in response to stimulation by cytokines (19, 25). We found a significant reduction in the levels of phosphorylated p38 MAPK in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 4, D and E).
Figure 4. TAK1 regulates the activation of multiple signaling pathways in skeletal muscle.
(A) Representative immunoblots demonstrating the levels of phosphorylated and total AMPK protein in TA muscle of Tak1fl/fl and Tak1mKO mice. (B) Densitometry quantification of the ratio of phosphorylated and total AMPK protein. (C) Fold change in enzymatic activity of AMPK in TA muscle of Tak1fl/fl and Tak1mKO mice. (D) Representative immunoblots demonstrating the levels of phosphorylated and total Akt, mTOR, p38 MAPK, and unrelated protein GAPDH in TA muscle of Tak1fl/fl and Tak1mKO mice. Phosphorylated and total p38 MAPK and GAPDH were run on a separate gel. (E) Densitometry quantification of the ratio of phosphorylated vs. total protein levels of Akt, mTOR, and p38 MAPK. n = 6 in each group. (F) Representative EMSA gel demonstrating DNA-binding activity of NF-κB in quadriceps muscle of Tak1fl/fl and Tak1mKO mice. (G) Immunoblots presented here show levels of phosphorylated and total IκBα, total p100, and total p52 protein in TA muscle of Tak1fl/fl and Tak1mKO mice. (H) Densitometry quantification of the ratio of phosphorylated vs. total IκBα and total p100 and p52 bands in immunoblots. n = 4 or 5 in each group. Fully differentiated mouse primary myotubes were transduced with Ad.Control shRNA or Ad.TAK1 shRNA at MOI 1:50. After 48 hours, the cells were collected and analyzed. (I) Representative immunoblots demonstrating the levels of phosphorylated and total AMPK, mTOR, and p38 MAPK protein and levels of p100 and p52, TAK1, and unrelated protein GAPDH. (J) EMSA gel demonstrating DNA-binding activity of NF-κB in control and TAK1-knockdown myotube cultures. (K) Densitometry quantification of DNA-binding activity in Ad.TAK1 shRNA and Ad.Control shRNA myotube cultures. Error bars represent ± SEM. *P < 0.05, values significantly different from corresponding Tak1fl/fl mice or control myotubes by unpaired 2-tailed t test.
In response to receptor-mediated events, TAK1 phosphorylates IKKβ, leading to activation of the canonical NF-κB signaling pathway (19, 22, 25). We next sought to determine whether inactivation of TAK1 affects the activation of NF-κB in skeletal muscle of mice. We first performed electrophoretic mobility shift assay (EMSA) to measure the DNA-binding activity of NF-κB. Although there was a trend toward increased DNA-binding activity of NF-κB in skeletal muscle of Tak1mKO mice, it was not markedly different from Tak1fl/fl mice (Figure 4F). By performing Western blot, we also measured the levels of phosphorylated and total IκBα protein, the markers of activation for the canonical NF-κB pathway (16). There was no significant difference in the phosphorylation or total levels of IκBα in the skeletal muscle of Tak1fl/fl and Tak1mKO mice (Figure 4, G and H). Interestingly, we found that the levels of p100 and p52, the components of the noncanonical NF-κB pathway, were significantly elevated in the skeletal muscle of Tak1mKO mice (Figure 4, G and H).
Consistent with in vivo results, we found that the level of phosphorylated AMPK was drastically increased in TAK1-knockdown myotubes compared with control cultures. Moreover, the levels of phosphorylated mTOR and p38 MAPK were reduced, while the levels of p100 and p52 proteins were increased in TAK1 shRNA–expressing cultures compared with those expressing scrambled shRNA (Figure 4I). Furthermore, there was a significant increase in the DNA-binding activity of NF-κB in TAK1-knockdown myotubes compared with controls (Figure 4, J and K), which could be attributed to the activation of noncanonical NF-κB signaling. Taken together, these results suggest that inactivation of TAK1 disrupts multiple signaling pathways, which may ultimately lead to the loss of skeletal muscle mass.
Ablation of TAK1 increases the proportion of fast-type oxidative fibers in skeletal muscle of mice.
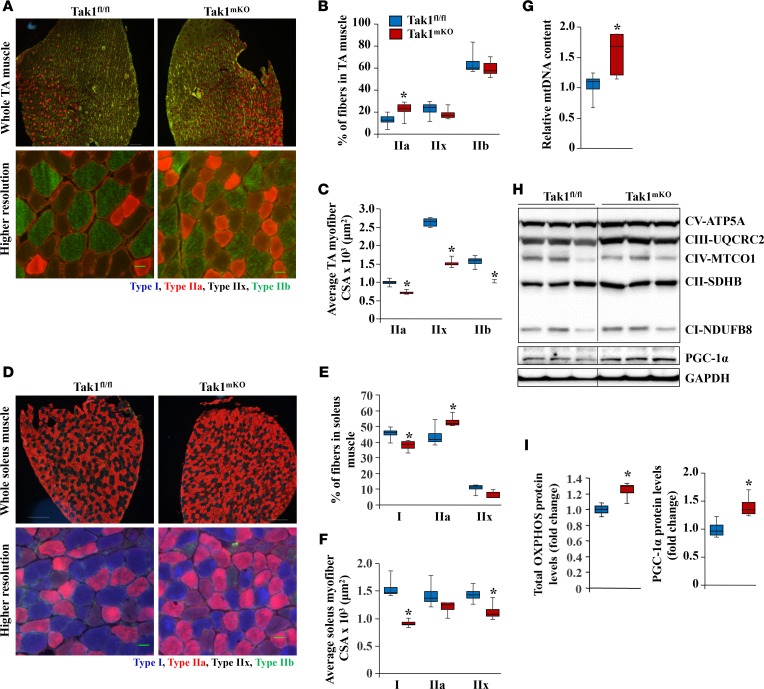
Skeletal muscle contains a mixture of fibers with diverse contractile and metabolic characteristics attributed to the expression of specific MyHC isoforms (31). Type I fibers are “slow-twitch,” rich in mitochondria, and use predominantly oxidative phosphorylation for energy production. In contrast, type IIb fibers are “fast-twitch,” contain comparatively low numbers of mitochondria, and rely more heavily on anaerobic glycolysis for energy production. Type IIa and type IIx fibers have characteristics between type I and type IIb fibers in that they have intermediate numbers of mitochondria and oxidative potential (31). The TA muscle of adult mice predominantly contains type IIa, -IIx, and -IIb myofibers, whereas the soleus muscle contains type I, -IIa, and -IIx myofibers (31, 32). To understand the effect of TAK1 inactivation on the composition of different types of myofibers, 14-week-old littermate Tak1fl/fl and Tak1mKO mice were given i.p. injections of tamoxifen. After 10 weeks, the TA and soleus muscles were isolated and analyzed by performing immunostaining with antibodies against MyHC I, -IIa, and -IIb protein. Intriguingly, we found that there was a significant increase in the proportion of type IIa fibers in the TA muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 5, A and B). An increased number of oxidative fibers in TA muscle was also evidenced by performing succinate dehydrogenase (SDH) staining (Supplemental Figure 6, A and B). While the soleus muscle contains ~45% slow-type oxidative fibers (e.g., type I), we found that the proportion of type IIa myofibers, but not type I, was significantly increased in the soleus muscle of Tak1mKO mice compared with Tak1fl/fl mice. Indeed, there was a concomitant decrease in the proportion of type I fibers in the soleus muscle of Tak1mKO mice (Figure 5, D and E). To understand whether loss in myofiber CSA observed in Tak1mKO mice is due to changes in fiber type composition or atrophy, we also measured the CSA of individual fiber types. We found that the average CSA of all types of myofibers was significantly reduced in both the TA and soleus muscles of Tak1mKO mice compared with Tak1fl/fl mice (Figure 5, C and F), suggesting that even though there is an increase in fast-type oxidative fibers, all types of myofibers undergo atrophy after inactivation of TAK1.
Figure 5. Inactivation of TAK1 favors a shift toward oxidative fast-twitch fibers in skeletal muscle of adult mice.
Fourteen-week-old Tak1fl/fl and Tak1mKO mice were given i.p. injections of tamoxifen, and 10 weeks later, the mice were euthanized and skeletal muscles were collected and processed for histological and biochemical analysis. (A) Muscle sections prepared from TA and soleus muscles of Tak1fl/fl and Tak1mKO mice were subjected to triple immunostaining against MyHC I, -IIa, and -IIb proteins. Representative photomicrographs of triple-stained sections of TA muscle. Scale bars: 50 μm. Quantification of (B) percentage of each fiber type, and (C) average CSA of type IIa, -IIx, and -IIb fibers in TA muscle of Tak1fl/fl and Tak1mKO mice. (D) Representative photomicrographs of triple-stained sections of soleus muscle. Scale bars: 50 μm. Quantification of (E) percentage of each fiber type, and (F) average CSA of type I, -IIa, -IIx fibers in soleus muscle of Tak1fl/fl and Tak1mKO mice. n = 6 in each group for A–F. (G) Mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) ratio measured by performing quantitative real-time PCR. (H) Representative immunoblots demonstrating levels of OXPHOS complex proteins, PGC-1α, and unrelated protein GAPDH in TA muscle of Tak1fl/fl and Tak1mKO mice. (I) Densitometry quantification of relative protein levels of OXPHOS complexes and PGC-1α in TA muscle of Tak1fl/fl and Tak1mKO mice. n = 4 or 5 in each group. Error bars represent SEM. *P < 0.05, values significantly different from corresponding Tak1fl/fl mice by unpaired 2-tailed t test.
We also compared mitochondrial content in the skeletal muscle of Tak1fl/fl and Tak1mKO mice. Quantitative PCR (qPCR) analysis of DNA showed that the relative abundance of mitochondrial DNA (mtDNA) normalized to nuclear DNA (nDNA) was increased in soleus muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 5G). Moreover, Western blot analysis using OXPHOS rodent antibody cocktail showed a significant increase in various subunits of the mitochondrial electron transport chain complexes in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 5, H and I). PGC-1α is a major regulator of mitochondrial biogenesis, and its gene expression is induced upon activation of AMPK (33). Consistent with the increase in activation of AMPK, we found a significant increase in the level of PGC-1α protein in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 5, H and I). Collectively, these results suggest that inactivation of TAK1 increases mitochondrial content, which may be responsible for an increased proportion of fast oxidative myofibers in skeletal muscle of mice.
Inactivation of TAK1 disrupts mitochondria structure and function in the skeletal muscle.
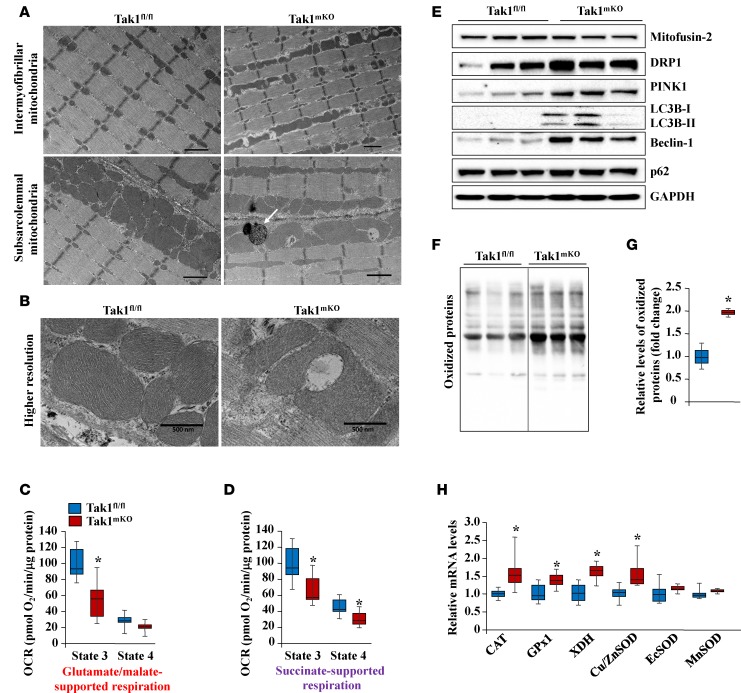
Since type IIa myofibers and overall mitochondrial content were increased in the skeletal muscle of Tak1mKO mice, we next sought to determine whether mitochondria are functional in the skeletal muscle of Tak1mKO mice. We first analyzed mitochondrial structure by performing transmission electron microscopy (TEM). Consistent with preceding results, we found that the abundance of both intermyofibrillar and subsarcolemmal mitochondria was increased in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 6A). Interestingly, we found that mitochondrial structure and organization were disrupted in the skeletal muscle of Tak1mKO mice. Many mitochondria were abnormally enlarged, and there was an increased proportion of vacuolated mitochondria. Mitochondrial cristae morphology was also disrupted in the skeletal muscle of Tak1mKO mice (Figure 6, A and B). To specifically investigate the impact of TAK1 inactivation on mitochondrial function, we isolated mitochondria from the GA muscles of Tak1fl/fl and Tak1mKO mice, and their function was examined by respirometry. Results showed that state 3 respiration fueled by glutamate and malate was significantly lower in mitochondria from Tak1mKO mice compared with Tak1fl/fl mice (Figure 6C). Similar results were obtained when succinate was used as a substrate (Figure 6D). State 4 respiration, induced by addition of oligomycin, was also significantly lower in succinate-fueled mitochondria from Tak1mKO mice, but it was not different in glutamate/malate-fueled mitochondria. These results suggest that ablation of TAK1 diminishes mitochondrial oxidative capacity in skeletal muscle.
Figure 6. TAK1 is required for mitochondrial homeostasis in skeletal muscle.
Fourteen-week-old Tak1fl/fl and Tak1mKO mice were given i.p. injections of tamoxifen. After 10 weeks, the mice were euthanized and skeletal muscles were collected and processed for histological and biochemical analysis. (A) Longitudinal muscle sections prepared from soleus muscle of Tak1fl/fl and Tak1mKO mice were processed for transmission electron microscopy (TEM) analysis. Representative EM images featuring intermyofibrillar (top panel) and subsarcolemmal (lower panel) mitochondria are presented here. Scale bars: 1 μm. (B) Higher-magnification EM images depicting vacuolated mitochondria in soleus muscle of Tak1mKO mice. Scale bars: 0.5 μm. Mitochondria were isolated from GA muscle of Tak1fl/fl and Tak1mKO mice, and state 3 and state 4 respiration was measured by respirometry . Oxygen consumption rate (OCR) was measured in the presence of (C) glutamate/malate and (D) succinate. n = 6 in each group from A–D. Error bars represent ± SD. *P < 0.05 values significantly different from corresponding Tak1fl/fl mice by unpaired t test. (E) Representative immunoblots of Mitofusin-2, Drp1, Pink1, LC3B-I/II, Beclin-1, p62, and unrelated protein GAPDH in mitochondrial fraction of GA muscle of Tak1fl/fl and Tak1mKO mice. (F) Representative immunoblot depicting the levels of irreversibly oxidized (carbonylated) protein in GA muscle of Tak1fl/fl and Tak1mKO mice. (G) Densitometry quantification of oxidized proteins. (H) Relative mRNA levels of oxidative stress related genes catalase (CAT), glutathione peroxidase 1 (GPx1), xanthine dehydrogenase (XDH), copper- and zinc–containing SOD (Cu/ZnSOD), extracellular SOD (EcSOD), and manganese SOD (MnSOD) by qPCR in skeletal muscle of Tak1fl/fl and Tak1mKO mice. n = 5 or 6 in each group. Error bars represent ± SEM. *P < 0.05, values significantly different from corresponding Tak1fl/fl mice by unpaired 2-tailed t test.
Mitophagy/autophagy is a major mechanism by which dysfunctional mitochondria are eliminated in mammalian cells, including skeletal muscle (34, 35). To determine whether inhibition of TAK1 also inhibits mitophagy in skeletal muscle, we next measured the levels of mitophagy markers in the mitochondrial fractions of skeletal muscle. Interestingly, there was a significant increase in levels of Drp1, Pink1, LC3B-I/II, Beclin-1, and p62 in mitochondrial fractions of the skeletal muscle of Tak1mKO compared with Tak1fl/fl mice (Figure 6E). Consistent with biochemical analysis, we also found the presence of electron dense autophagosomes in the skeletal muscle of Tak1mKO mice by TEM analysis (Figure 6A).
Oxidative stress plays a prominent role in skeletal muscle atrophy and weakness (36), and an increase in oxidative stress is one of the important mechanisms contributing to the accumulation of dysfunctional mitochondria in skeletal muscle (3, 4). Thus, we next investigated whether inactivation of TAK1 causes oxidative stress in skeletal muscle. Interestingly, the amount of irreversibly oxidized (carbonylated) proteins was significantly increased in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 6, F and G). Furthermore, mRNA levels of several antioxidant molecules were significantly increased in the skeletal muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 6H), further suggesting that inactivation of TAK1 disrupts redox homeostasis in the skeletal muscle.
We also investigated the effect of inactivation of TAK1 on oxidative stress in cultured myotubes. Intriguingly, the levels of oxidized proteins were significantly increased in cultured myotubes within 48 hours of TAK1 knockdown (Supplemental Figure 4E). However, there was no significant change in the levels of OXPHOS proteins in control and TAK1-knockdown cultures (Supplemental Figure 4F). These results suggest that oxidative stress may be a potential trigger for the loss of mitochondrial function in the skeletal muscle upon inactivation of TAK1.
TAK1 does not affect denervation-induced skeletal muscle atrophy.
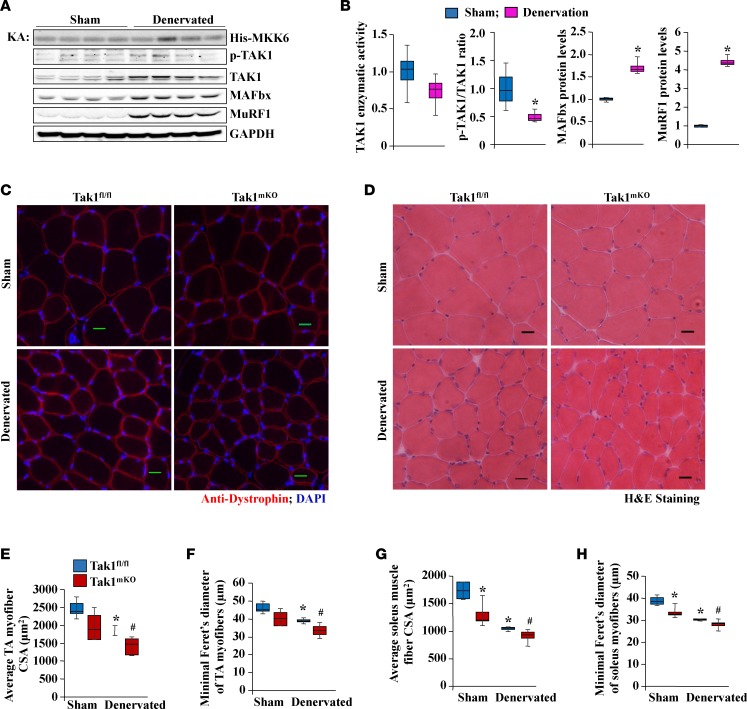
Many inflammatory cytokines induce canonical NF-κB signaling through the activation of TAK1 signalosomes (22, 23). Increased activation of NF-κB causes muscle wasting in many conditions, including in response to denervation (16, 37, 38). We next sought to determine the effect of inactivation of TAK1 on denervation-induced skeletal muscle atrophy. We first studied whether the activity of TAK1 changes in the skeletal muscle of mice in response to denervation. The right hind limb muscles of 4-month-old WT mice were denervated by transecting the sciatic nerve, while the left hind limb was sham-operated. After 7 days, the TA muscle was isolated, and the enzymatic activity — as well as levels of phosphorylated and total TAK1 protein — were measured. We observed a small increase in the levels of total TAK1 protein (Figure 7A). However, no significant increase in the enzymatic activity of TAK1 was noticeable (Figure 7, A and B). Indeed, the ratio of phosphorylated vs. total TAK1 protein was significantly reduced in denervated muscle compared with contralateral undenervated muscle (Figure 7B). Denervated TA muscle showed increased levels of MAFbx and MuRF1 proteins, suggesting underlying atrophy in this muscle (Figure 7, A and B).
Figure 7. Role of TAK1 in denervation-induced skeletal muscle atrophy in adult mice.
Right hind limb of WT mice were subjected to denervation by transecting the sciatic nerve, whereas the left hind limb was sham operated. After 7 days, TA muscle was isolated and processed for biochemical analysis. (A) Representative gel showing phosphorylated MKK6 protein in in vitro kinase assay (KA) in undenervated and denervated muscle. Immunoblots showing levels of phosphorylated TAK1 and total TAK1, MAFbx, MuRF1, and GAPDH protein. (B) Densitometry quantification of TAK1 enzymatic activity, phosphorylated vs. total TAK1 levels, MAFbx, and MuRF1 protein in skeletal muscle of Tak1fl/fl and Tak1mKO mice. n = 4 in each group. Error bars represent ± SD. *P < 0.05 values significantly different from sham-operated muscle by unpaired t test. In a separate experiment, 14-week-old Tak1fl/fl and Tak1mKO mice were given i.p. injections of tamoxifen, and immediately thereafter, the sciatic nerve of the right hind limb was transected to induce muscle atrophy, while the left side was sham operated. Fourteen days after denervation surgery, the mice were euthanized and TA and soleus muscles were isolated and processed for histological analysis. Representative photomicrographs of TA muscle sections of Tak1fl/fl and Tak1mKO mice after (C) anti-dystrophin staining and (D) H&E staining. Scale bars: 20 μm. Quantification of (E) average myofiber CSA, and (F) average minimal Feret’s diameter in anti-dystrophin-stained TA muscle sections. Quantification of (G) average myofiber CSA, and (H) average minimal Feret’s diameter in anti-dystrophin-stained soleus muscle sections of Tak1fl/fl and Tak1mKO. n = 4 or 5 in each group. Error bars represent ± SEM. *P < 0.05, values significantly different from sham-operated muscle of Tak1fl/fl mice by 1-way ANOVA with post hoc Bonferroni’s multiple comparison test. #P < 0.05, values significantly different from denervated muscle of Tak1fl/fl mice by 1-way ANOVA with post hoc Bonferroni’s multiple comparison test.
We next investigated whether inactivation of TAK1 inhibits denervation-induced skeletal muscle atrophy in adult mice. For this experiment, 4-month-old Tak1fl/fl and Tak1mKO mice were treated with tamoxifen, followed by transecting sciatic nerve of the right hind limb. After 14 days, the TA and soleus muscles were isolated and analyzed by performing anti-dystrophin staining and H&E staining. Both TA and soleus muscles of Tak1fl/fl mice showed a significant reduction in average myofiber CSA and minimal Feret’s diameter at 14 days after denervation (Figure 7, C–H, Supplemental Figure 7, A and B). Although the myofiber CSA was significantly reduced even in sham-operated TA and soleus muscle of Tak1mKO mice compared with corresponding Tak1fl/fl mice, there was further loss in myofiber CSA in Tak1mKO mice following denervation (Figure 7, C–H). Our analysis also showed that the percentage loss in myofiber CSA upon denervation was similar in Tak1fl/fl and Tak1mKO mice (Supplemental Figure 7C). Apart from the reduction in myofiber CSA, there was no sign of myopathy in the denervated muscle of Tak1mKO mice (Figure 7, C and D). These results suggest that, even though TAK1 is an upstream activator of NF-κB in some signaling cascades, inhibition of TAK1 does not attenuate denervation-induced muscle loss.
TAK1 is required for overload-induced myofiber hypertrophy in adult mice.
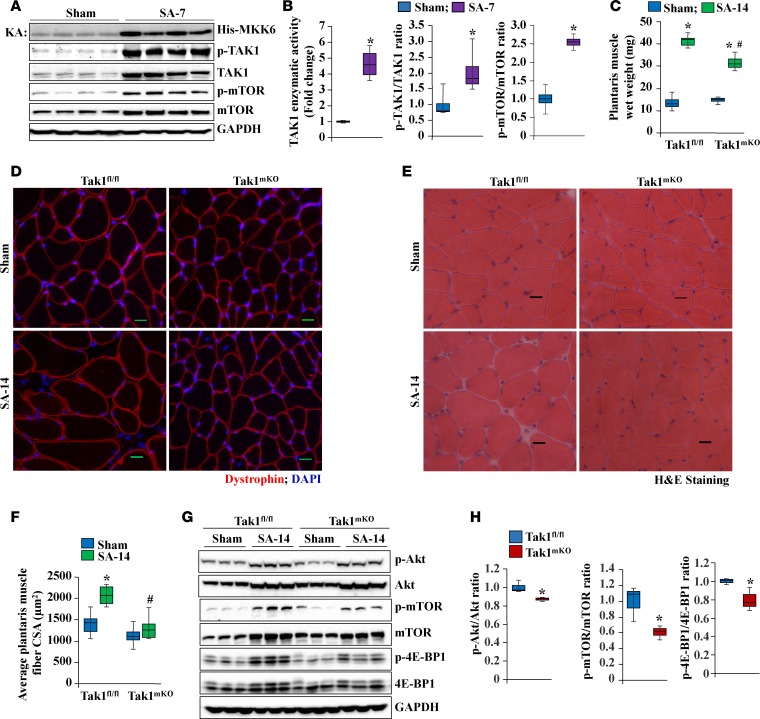
We first investigated how the level and activity of TAK1 are modulated during overload-induced muscle hypertrophy. For these experiments, we used the bilateral synergistic ablation (SA) model that has been widely used to induce hypertrophy of the plantaris muscle (39, 40). Interestingly, enzymatic activity as well as phosphorylation of TAK1 were significantly increased 7 days after performing the SA surgery (Figure 8, A and B). Consistent with published reports (6), there was also a significant upregulation in phosphorylated and total levels of mTOR in the plantaris muscle after 7 days from performing the SA surgery (Figure 8, A and B).
Figure 8. Role of TAK1 in overload-induced myofiber hypertrophy in adult mice.
(A) Four-month-old WT mice were subjected to bilateral synergistic ablation (SA) by surgical removal of GA and soleus muscles to induce overload hypertrophy in plantaris muscles. After 7 days, plantaris muscle was isolated and processed for biochemical analysis. Representative gel showing phosphorylated MKK6 protein in in vitro kinase assay (KA) in plantaris muscle. Immunoblots showing levels of phosphorylated and total TAK1 and mTOR protein. (B) Densitometry quantification of TAK1 enzymatic activity and phosphorylated vs. total levels of TAK1 and mTOR in plantaris muscle of Tak1fl/fl and Tak1mKO mice. n = 4 in each group. Error bars represent ± SEM. *P < 0.05, values significantly different from sham-operated muscle by unpaired t test. In another experiment, 14-week-old Tak1fl/fl and Tak1mKO mice were given i.p. injections of tamoxifen, and immediately thereafter, the mice were subjected to SA surgery. After 14 days, the mice were euthanized. One side plantaris muscle was collected and processed for biochemical examination, while the other side was processed for histological analysis. (C) Average wet weight of plantaris muscle of sham and SA-14 Tak1fl/fl and Tak1mKO mice. Representative photomicrographs of plantaris muscle sections of Tak1fl/fl and Tak1mKO mice after (D) anti-dystrophin staining and (E) H&E staining. Scale bars: 20 μm. (F) Quantification of average fiber CSA in dystrophin-stained plantaris muscle sections of Tak1fl/fl and Tak1mKO mice. n = 5–7 in each group. Error bars represent ± SEM. *P < 0.05 values significantly different from sham-operated muscle of Tak1fl/fl mice by 1-way ANOVA with post hoc Bonferroni’s multiple comparison test. #P < 0.05, values significantly different from SA-14 Tak1fl/fl mice by 1-way ANOVA with post hoc Bonferroni’s multiple comparison test. (G) Representative immunoblots of phosphorylated and total Akt, mTOR, and 4E-BP1 protein levels and unrelated protein GAPDH in plantaris muscle of Tak1fl/fl and Tak1mKO mice. (H) Densitometry quantification of the ratio of phosphorylated vs. total protein levels of Akt, mTOR, and 4E-BP1 in plantaris muscle of Tak1fl/fl and Tak1mKO mice 14 days after SA surgery. n = 3 in each group. Error bars represent ± SEM. *P < 0.05 values significantly different from Tak1fl/fl mice by unpaired 2-tailed t test.
To understand the role of TAK1 in overload-induced muscle hypertrophy, 4-month-old Tak1fl/fl and Tak1mKO mice were subjected to bilateral SA surgery. After 14 days, the plantaris muscle was isolated and weighed. There was a significant increase in wet weight of plantaris muscle 14 days after SA in Tak1fl/fl mice. Although the wet weight of plantaris muscle was also increased in Tak1mKO mice 14 days after SA surgery, it was significantly lower compared with corresponding Tak1fl/fl mice (Figure 8C). We next analyzed plantaris muscle by performing anti-dystrophin staining and H&E staining (Figure 8, D and E). We found that there was ~63% increase in the average myofiber CSA in the plantaris muscle of Tak1fl/fl mice after 2 weeks of performing SA surgery. By contrast, there was no significant increase in the myofiber CSA in the plantaris muscles of Tak1mKO mice (Figure 8F). Since activation of the Akt/mTOR pathway mediates protein synthesis during overload-induced skeletal muscle hypertrophy, we investigated whether inactivation of TAK1 affects the phosphorylation of components of this pathway during overload-induced hypertrophy. Results showed that an overload-induced increase in the levels of phosphorylated Akt, mTOR, and 4E-BP1 were significantly reduced in the plantaris muscle of Tak1mKO mice compared with Tak1fl/fl mice (Figure 8, G and H). Taken together, these results suggest that TAK1 plays a critical role in overload-induced myofiber hypertrophy in adult mice.
Discussion
TAK1 is an important intracellular signaling intermediate that regulates multiple signaling pathways, including canonical NF-κB and p38 MAPK (19, 25). Previous studies have shown that targeted inhibition of NF-κB or p38 MAPK does not affect skeletal muscle mass in naive conditions (15, 38, 41). In contrast, our results demonstrate that inactivation of TAK1 causes severe muscle wasting, mitochondrial abnormalities, and oxidative stress in naive conditions. A more crucial role of TAK1 in the regulation of skeletal muscle mass could be attributed to the fact that it is an upstream signaling kinase, which — in addition to NF-κB and p38 MAPK — also regulates several other pathways, such as JNK, Notch, Wnt, and canonical and noncanonical TGF-β signaling that have been implicated in the regulation of muscle mass (1, 19). Moreover, TAK1 may also improve skeletal muscle mass through inducing the expression of certain growth factors, evidenced by our microarray analysis of Tak1mKO skeletal muscle (Supplemental Figure 1D).
Our results demonstrate that inactivation of TAK1 drastically reduces the rate of protein synthesis and augments protein degradation, potentially through the activation of the UPS and autophagy (Figure 3). Previous studies have shown that TAK1 causes the activation of AMPK in cardiomyocytes and liver cells (42, 43). Contrary to these reports, we found that AMPK is highly activated upon deletion of TAK1 in skeletal muscle, suggesting that different mechanisms might be involved in the regulation of AMPK activation in skeletal muscle (Figure 4, A–C). AMPK has diverse functions in skeletal muscle. While AMPK activation improves mitochondrial biogenesis and metabolic function (33, 44), it also inhibits the activity of mTOR (TORC1/Raptor) through activation of the TSC1-TSC2 complex (45) or through direct phosphorylation of Raptor (46). Furthermore, AMPK can also induce proteolytic degradation through augmenting autophagy and enhancing the expression of components of the UPS (1, 45–47). Consistent with an increased activation of AMPK, we found a significant reduction in mTOR phosphorylation, as well as an upregulation in proteasome activity and autophagy in the skeletal muscle of Tak1mKO mice (Figure 3).
Muscle wasting is generally accompanied by slow- to fast-type fiber transition (14, 47). Fast-type glycolytic myofibers (e.g., type IIx and -IIb) undergo atrophy at a faster rate when compared with slow-type oxidative myofibers in catabolic conditions (1). Intriguingly, we observed that inactivation of TAK1 favors a type IIa myofiber phenotype, which is a fast type but uses oxidative metabolism for energy production (Figure 5). The increased number of type IIa myofibers in Tak1mKO mice could be attributed to the increased activation of AMPK, which stimulates mitochondrial biogenesis through increasing the expression and phosphorylation of PGC-1α (12, 33). The activity of noncanonical NF-κB signaling is also enhanced in skeletal muscle upon inhibition of TAK1 (Figure 4). Since noncanonical NF-κB signaling promotes oxidative metabolism (48, 49), activation of this pathway may also contribute to an increased proportion of type IIa myofibers and mitochondrial content in the skeletal muscle of Tak1mKO mice. Intriguingly, we noticed that the average CSA of both glycolytic and oxidative myofibers was significantly reduced in Tak1mKO mice compared with Tak1fl/fl mice (Figure 5, C and F). While there is an increase in type IIa myofibers, it is noteworthy that the mitochondria are dysfunctional and oxidative stress is increased, both of which can contribute to enhanced catabolism in both slow- and fast-type myofibers in Tak1mKO mice (Figure 6).
An important observation of the present investigation is that the deletion of TAK1 leads to the accumulation of enlarged or highly interconnected mitochondria in skeletal muscle of adult mice (Figure 6A). Enlarged mitochondria have been consistently observed in the skeletal muscle of the elderly (50). Indeed, myofiber atrophy, development of kyphosis, and concomitant accumulation of enlarged and dysfunctional mitochondria observed in Tak1mKO mice suggest that deletion of TAK1 accelerates an aging phenotype in skeletal muscle. One of the important mechanisms for the clearance of dysfunctional mitochondria is autophagy. Recent studies have shown that a basal level of autophagy is essential for the maintenance of skeletal muscle health (34, 35). Autophagy is also induced in multiple conditions, such as endurance exercise that improves skeletal muscle metabolic function (51). Previously, it was reported that TAK1 mediates the activation of the LKB1/AMPK/ULK1 signaling axis and autophagy in hepatocytes (43). However, we found that ablation of TAK1 stimulates the activation of AMPK in skeletal muscle. Moreover, we found that several markers of autophagy are upregulated in the skeletal muscle of Tak1mKO mice (Figure 3, J–L, and Figure 6E), suggesting that the initial activation of autophagy is enhanced, which could be an attempt to clear the dysfunctional mitochondria. Nevertheless, it is possible that this level of autophagy is not sufficient or some later steps of autophagy are impaired, preventing efficient clearance of defunct mitochondria in the skeletal muscle of Tak1mKO mice. Alternatively, deletion of TAK1 may alter other processes that are also essential to maintaining healthy mitochondria (50).
Although the exact mechanisms by which deletion of TAK1 activates AMPK in skeletal muscle remains unknown, reduced mitochondrial oxidative phosphorylation capacity could be one of the potential explanations. Reduction in mitochondrial function leads to the elevation in the AMP/ATP ratio, which activates AMPK. We found that deletion of TAK1 significantly diminished mitochondrial respiratory function in skeletal muscle (Figure 6, C and D). Importantly, our TEM analysis showed that the mitochondrial matrix of the skeletal muscle of Tak1mKO mice contains lipid-like inclusions or vacuolation (Figure 6, A and B). Abnormal aggregates of mitochondria with vacuolation have been consistently observed in skeletal muscle and motor neurons of the SOD1 mutant mouse model of amyotrophic lateral sclerosis (ALS), in which oxidative stress plays a major role in pathogenesis (52). Intriguingly, we also observed a significant increase in oxidative stress in the skeletal muscle of Tak1mKO mice (Figure 6, F–H). Mitochondria are one of the most important sources of oxidative stress in mammalian cells. Moreover, increased oxidative stress has also been found to be a cause of mitochondrial dysfunction in skeletal muscle in multiple conditions (4, 50). Similar to in vivo studies, we also noticed a significant increase in levels of carbonylated proteins within 48 hours of knockdown of TAK1 in cultured myotubes (Supplemental Figure 4E). These results are in agreement with published reports also suggesting that deletion of TAK1 leads to increased production of ROS in keratinocytes and skin tumors (27, 53). However, the cellular source of oxidative stress and whether dysfunctional mitochondria are the cause or consequence of disrupted redox homeostasis in skeletal muscle of Tak1mKO mice remains unknown. Future studies will determine whether targeted inhibition of oxidative stress in mitochondria or from nonmitochondrial sources can improve mitochondrial health and attenuate atrophy in skeletal muscle of Tak1mKO mice.
Further support for a role of TAK1 in the regulation of skeletal muscle mass came from the models of muscle atrophy and hypertrophy employed in our study. Denervation-induced muscle atrophy was not attenuated in Tak1mKO mice, suggesting that even though TAK1 is an upstream activator of NF-κB, it does not prevent denervation-induced atrophy (Figure 7). In response to TLR and IL-1R signaling, TRAF6 acts as an upstream activator of TAK1 and NF-κB (21). Interestingly, targeted deletion of TRAF6 inhibits muscle wasting in multiple catabolic conditions, including denervation (38, 54, 55). The exact reasons why the inhibition of TRAF6, but not TAK1, attenuates denervation-induced muscle atrophy remain unknown. However, it is well known that in response to some stimuli, such as TNFα, TRAF2 mediates the activation of TAK1 and NF-κB (56). There are also published reports suggesting that TRAF6 and TAK1 can activate distinct intracellular pathways to regulate tissue homeostasis (19, 21, 57). Finally, NF-κB can also be activated in a cell-autonomous manner due to increased oxidative stress that is commonly observed in atrophying skeletal muscle (1, 16).
In contrast to denervation, we found that the overload-induced myofiber hypertrophy was inhibited in Tak1mKO mice (Figure 8). Overload-induced myofiber hypertrophy involves an increased rate of protein synthesis, which is mediated through the activation of the Akt/mTOR pathway (6). We found that the overload-induced activation of the Akt/mTOR pathway is inhibited in the skeletal muscle of Tak1mKO mice (Figure 8). These results suggest that similar to young animals, TAK1 is required for overload-induced myofiber growth in adult animals.
In summary, our study provides initial evidence that TAK1 plays an important role in the regulation of skeletal muscle mass and mitochondrial health. Future studies will examine how the levels and activity of TAK1 are regulated in other conditions of skeletal muscle wasting, such as sarcopenia, cancer cachexia, type II diabetes, and ALS, and whether the modulation of TAK1 activity can improve skeletal muscle mass and function.
Methods
Animals.
Tak1mKO were generated by crossing HSA-MCM mice (The Jackson Laboratory, Tg[ACTA1-cre/Esr1*]2Kesr/J) with Tak1fl/fl mice as described (24). All mice were in the C57BL/6 background, and their genotype was determined by PCR from tail DNA. For inactivation of TAK1, mice were given i.p. injections of tamoxifen (75 mg/kg body weight) every day for 4 days, and the mice were fed with a chow containing tamoxifen (250 mg/kg) for the entire duration of the experiment. For denervation surgery, mice were anesthetized with an i.p. injection of tribromoethanol (MilliporeSigma) followed by shaving the right and left hind quarters, making a ~0.5-cm incision proximal to the knee on the lateral side of the right leg, separating the muscles at the fascia and lifting out the sciatic nerve with a surgical hook or forceps, and removing a 2 mm piece of sciatic nerve. Left-side hind limb was subjected to same procedure, except that the sciatic nerve was not removed. For bilateral SA surgery, the mice were anesthetized using tribromoethanol, and the soleus and ~60% of the GA muscles were surgically excised. A sham surgery was performed for controls following the same procedures, except that GA and soleus muscles were not excised. Finally, the incisions were closed with surgical sutures. At various time points, hind limb muscles were collected from euthanized mice for biochemical and histology studies.
Grip strength measurements.
Forelimb and total 4-paw grip strength of mice were measured using a digital grip-strength meter (Columbus Instruments) and normalized by body weight as described (58).
Histology and morphometric analysis.
TA, soleus, or plantaris muscle of mice was removed, frozen in liquid nitrogen, and sectioned in a microtome cryostat. For the assessment of tissue morphology, 10 μm–thick transverse sections of muscles were prepared and stained with H&E dye as described (59). The sections were examined under an inverted microscope (Nikon Eclipse TE 2000-U) using a Plan 10x, NA 0.25 PH1 DL, or Plan-Fluor ELWD 20x, NA 0.45 Ph1 DM, objective lens; a digital camera (Digital Sight DS-Fi1); and NIS Elements BR 3.00 software (Nikon). Images of H&E- or anti-dystrophin-stained TA or plantaris muscle sections were analyzed by measuring myofiber CSA. The distribution of myofiber CSA was calculated by analyzing ~500 myofibers per field using the NIS Elements BR 3.00 software (Nikon).
IHC.
Frozen TA, soleus, or plantaris muscle sections were fixed in acetone or 4% paraformaldehyde (PFA) in PBS and blocked in 2% BSA in PBS for 60 minutes. The sections were then incubated with rabbit anti-dystrophin or rabbit anti-laminin, in blocking solution at 4°C overnight under humidified conditions. The sections were washed briefly with PBS and then incubated with goat anti–rabbit Alexa Fluor 468 or goat anti–rabbit Alexa Fluor 488 secondary antibody for 60 minutes at room temperature. TUNEL staining was performed following a protocol from manufacturer (in situ Cell Death Detection Kit, MilliporeSigma). Briefly, muscle sections were fixed in 4% PFA and permeabilized with 0.1% Triton X-100 in 0.1% sodium citrate and incubated in TUNEL reaction mixture for 60 minutes at 37°C. Finally, the sections were washed 3 times for 15 minutes with PBS. Nuclei were counterstained with DAPI. The slides were mounted using fluorescence medium (Vector Laboratories) and visualized at room temperature on Nikon Eclipse TE 2000-U microscope (Nikon). Image levels were equally adjusted using Photoshop CS6 software (Adobe).
Fiber typing and SDH staining.
The composition of different types of myofibers in the soleus and TA muscle of mice was determined following a protocol as described (60). We used the entire TA or soleus muscle sections to quantify the percentage of type I, -IIa, -IIx, and -IIb fibers. SDH staining was performed on TA muscle sections using same protocol as described (61). At least 500 fibers were counted to determine SDH+ fibers in each section in a blinded fashion. The percentage of SDH-stained fibers was then determined based on a criteria using integrated optical density.
Relative mtDNA measurements.
Mitochondrial abundance was estimated by measuring mtDNA abundance relative to nDNA. Total DNA was isolated from soleus muscle of mice using a DNAeasy Blood & Tissue Kit (Qiagen). Following isolation, relative amounts of mtDNA and nDNA were compared using qPCR, using 2 ng of the isolated DNA. Primers for cytochrome b (mtDNA) and β-actin (nDNA) were used; the sequences are cytochrome b, 5′-TTG GGT TGT TTG ATC CTG TTT CG-3′ and 5′-CTT CGC TTT CCA CTT CAT CTT ACC-3′; and β-actin, 5′-CAG GAT GCC TCT CTT GCT CT-3′ and 5′-CGT CTT CCC CTC CAT CGT-3′.
Microarray data analysis.
GeneChipMouse Gene 2.0 ST Arrays (Affymetrix) were utilized according to the manufacturer’s protocol and were scanned. The CEL files were analyzed through R using the oligo package to normalize the data using the Robust Multi-array Average (RMA) normalization method, followed by the application of moderated t-statistics via the limma package (62) for statistical analysis. The differentially expressed genes were selected based on the 1.5-fold expression changes between Tak1fl/fl and Tak1mKO GA muscle with P value of less than 0.05. The CEL files have been deposited in the Gene Expression Omnibus (GEO GSE104586). The GO analysis was performed using the DAVID bioinformatics tool. The heatmap was created using the MeV bioinformatics tool. The network analysis was performed via the NetworkAnalyst (63).
Preparation and culturing of mouse primary myotubes.
Myoblasts were isolated from hind limb of 8-week-old mice using a method as described (64). To induce differentiation, the cells were incubated in differentiation medium (DM; 2% horse serum in DMEM) for 48 hours.
Mitochondrial functional assay.
Mitochondrial oxidative capacity was measured in isolated mitochondria using a Seahorse Bioscience XF24 Extracellular Flux Analyzer (Billerica). For measurements of isolated mitochondria function, tissue from the limb muscle (approximately 50 mg) was isolated and homogenized in 1 ml of isolation buffer (220 mM mannitol, 70 mM sucrose, 5 mM 3-[N-morpholino] propanesulfonic acid [MOPS], 1 mM EGTA, 0.3% fatty acid-free BSA, pH 7.2) using a Potter Elvehjem tube and a Teflon pestle. The homogenate was centrifuged at 500 g for 5 minutes at 4°C. The supernatant containing mitochondria was centrifuged at 10,000 g for 5 minutes. After 2 wash-centrifugation steps in BSA-free isolation buffer, the mitochondria were suspended in respiration buffer (120 mM KCl, 25 mM sucrose, 10 mM 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid [HEPES], 1 mM MgCl2, 5 mM KH2PO4, pH 7.2). Protein in the mitochondrial suspension was estimated using the Lowry DC assay (Bio-Rad), and 5 μg of mitochondrial protein was sedimented in XF culture plates as described (61). State 3 respiratory activity was determined by measuring the oxygen consumption rate (OCR) after injection of glutamate (5 mM), malate (2.5 mM), and ADP (1 mM). To determine state 3 respiratory activity supported by Complex II, succinate (10 mM), rotenone (1 μM) and ADP (1 mM) were provided. The OCR after exposure of mitochondria to oligomycin (1 μg/ml) was used to measure state 4 respiratory activity under both substrate conditions. Data are expressed as pmol O2/min/μg protein.
Generation of TAK1 shRNA adenovirus.
Adenoviral vector expressing mouse Tak1 shRNA was generated using the same protocol as described (65). Target sequences for Tak1 was 5′-GGT GCT GAA CCA TTG CCT TAC-3′. We used adenovirus at MOI 1:50.
AMPK assay.
The enzymatic activity of AMPK was measured using a commercially available kit following a procedure suggested by the manufacturer (CycLex Co.).
Proteasome activity assay.
The activity of 20S proteasome in muscle extracts was determined using a commercially available kit following recommended protocol by the manufacturer (MilliporeSigma).
Western blot.
The relative amount of various proteins was determined by performing Western blot as described (28, 59). Black lines on the immunoblots indicate that intervening lanes have been spliced out. Antibody sources and dilutions used are provided in Supplemental Table 1.
Carbonylation assays.
The carbonyl groups in the protein side chains were derivatized to 2,4-dinitrophenylhydrazone by reaction with 2,4-dinitrophenylhydrazine using OxyBlot Protein Oxidation Detection Kit (catalog S7150, MilliporeSigma). The carbonylated/oxidatively modified proteins were detected by performing Western blot using anti–2,4-dinitrophenyl hydrazone antibodies.
RNA isolation and qPCR.
Total RNA was isolated from skeletal muscle tissues of mice or cultured myotubes and subjected to qPCR analysis similar to as described (28, 59). The sequence of the primers are presented in Supplemental Table 2.
SUnSET assay.
In vivo, the mice were anesthetized and given i.p. injection of 0.04 μm of puromycin per gram of body weight. The mice were euthanized exactly 30 minutes after injection of puromycin, and hind limb muscle was isolated and snap-frozen. For cultured myotubes, 1 μM puromycin was added in the culture medium for 30 minutes. Finally, protein extracts were made from muscles or cultured myotubes, and newly synthesized protein was detected by performing immunoblotting using primary antibody anti-puromycin (1:1,000; catalog MABE343, MilliporeSigma).
Immunoprecipitation and in vitro kinase assay.
Skeletal muscle of mice were homogenized prepared in lysis buffer (50 mM Tris pH 7.5, 100 mM NaCl, 0.1% Triton X 100, 1 mM dithiothreitol, 1 mM sodium orthovanadate and protease inhibitors). Equal amounts of protein were immunoprecipitated using 1 μg TAK1 (clone D94D7) rabbit monoclonal antibody (Cell Signaling Technology; catalog 5206) followed by in vitro kinase assay using His-MKK6 protein as substrate as described (57). The radioactive bands were visualized by exposing to a PhosphorImager screen and quantified using ImageJ (NIH) software.
TEM.
Mitochondrial ultrastructure in soleus muscle was analyzed by TEM following a protocol as described (38). Images were stored as TIFF files, and image levels were equally adjusted using Photoshop CS2 software (Adobe).
Statistics.
Results are expressed as mean ± SEM. Statistical analyses used 2-tailed Student’s t test to compare quantitative data populations with normal distribution and equal variance. For experiments involving more than 2 groups, we used 1-way ANOVA with post hoc Bonferroni’s multiple comparison test. A value of P < 0.05 was considered statistically significant, unless otherwise specified.
Study approval.
All animal procedures were conducted in strict accordance with the institutional guidelines and were approved by the IACUC and Institutional Biosafety Committee of the University of Louisville (IACUC nos. 13097 and 16663).
Author contributions
AK conceived the study and designed the work. SMH and AK wrote the manuscript, and all authors edited the manuscript. SMH, SS, KRB, JDM, YSG, and GX performed all the experiments. BGH helped with mitochondrial functional assays. SU performed analysis of microarray dataset and contributed to statistical analysis of results.
Supplementary Material
Acknowledgments
We thank S. Akira (Osaka University, Osaka, Japan) for providing floxed TAK1 mice. This work was supported by funding from NIH grants AR059810, AR068313, and AG029623 (to AK); NIH grant AR069985 (to SH); and NIH grants HL130174 and HL078825, and the American Diabetes Association Pathway to Stop Diabetes grant 1-16-JDF-041 (to BH).
Version 1. 02/08/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(3):e98441. https://doi.org/10.1172/jci.insight.98441.
Contributor Information
Shuichi Sato, Email: sxs9151@louisiana.edu.
Guangyan Xiong, Email: guangyan.xiong@louisville.edu.
Kyle R. Bohnert, Email: krbohn02@louisville.edu.
Andrew A. Gibb, Email: aagibb03@louisville.edu.
Joseph D. McMillan, Email: jdmcmi02@louisville.edu.
Shizuka Uchida, Email: shizuka.uchida@louisville.edu.
Ashok Kumar, Email: ashok.kumar@louisville.edu.
References
- 1.Bonaldo P, Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech. 2013;6(1):25–39. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol. 2014;49(1):59–68. doi: 10.3109/10409238.2013.857291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romanello V, Sandri M. Mitochondrial Quality Control and Muscle Mass Maintenance. Front Physiol. 2015;6:422. doi: 10.3389/fphys.2015.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnard C, et al. Mitochondrial dysfunction results from oxidative stress in the skeletal muscle of diet-induced insulin-resistant mice. J Clin Invest. 2008;118(2):789–800. doi: 10.1172/JCI32601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romanello V, et al. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29(10):1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodine SC, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3(11):1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 7.Rommel C, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3(11):1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 8.Latres E, et al. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem. 2005;280(4):2737–2744. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- 9.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem. 2002;277(27):23977–23980. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- 11.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–590. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 12.Bujak AL, et al. AMPK activation of muscle autophagy prevents fasting-induced hypoglycemia and myopathy during aging. Cell Metab. 2015;21(6):883–890. doi: 10.1016/j.cmet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krawiec BJ, Nystrom GJ, Frost RA, Jefferson LS, Lang CH. AMP-activated protein kinase agonists increase mRNA content of the muscle-specific ubiquitin ligases MAFbx and MuRF1 in C2C12 cells. Am J Physiol Endocrinol Metab. 2007;292(6):E1555–E1567. doi: 10.1152/ajpendo.00622.2006. [DOI] [PubMed] [Google Scholar]
- 14.Sandri M. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int J Biochem Cell Biol. 2013;45(10):2121–2129. doi: 10.1016/j.biocel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai D, et al. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119(2):285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Malhotra S, Kumar A. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J Mol Med. 2008;86(10):1113–1126. doi: 10.1007/s00109-008-0373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang G, Jin B, Li YP. C/EBPβ mediates tumour-induced ubiquitin ligase atrogin1/MAFbx upregulation and muscle wasting. EMBO J. 2011;30(20):4323–4335. doi: 10.1038/emboj.2011.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egerman MA, et al. GDF11 Increases with Age and Inhibits Skeletal Muscle Regeneration. Cell Metab. 2015;22(1):164–174. doi: 10.1016/j.cmet.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mihaly SR, Ninomiya-Tsuji J, Morioka S. TAK1 control of cell death. Cell Death Differ. 2014;21(11):1667–1676. doi: 10.1038/cdd.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanayama A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15(4):535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Walsh MC, Lee J, Choi Y. Tumor necrosis factor receptor- associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol Rev. 2015;266(1):72–92. doi: 10.1111/imr.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 23.Smit L, Baas A, Kuipers J, Korswagen H, van de Wetering M, Clevers H. Wnt activates the Tak1/Nemo-like kinase pathway. J Biol Chem. 2004;279(17):17232–17240. doi: 10.1074/jbc.M307801200. [DOI] [PubMed] [Google Scholar]
- 24.Sato S, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6(11):1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 25.Shim JH, et al. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28(14):2028–2041. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy JJ, Srikuea R, Kirby TJ, Peterson CA, Esser KA. Inducible Cre transgenic mouse strain for skeletal muscle-specific gene targeting. Skelet Muscle. 2012;2(1):8. doi: 10.1186/2044-5040-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omori E, Morioka S, Matsumoto K, Ninomiya-Tsuji J. TAK1 regulates reactive oxygen species and cell death in keratinocytes, which is essential for skin integrity. J Biol Chem. 2008;283(38):26161–26168. doi: 10.1074/jbc.M804513200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogura Y, Hindi SM, Sato S, Xiong G, Akira S, Kumar A. TAK1 modulates satellite stem cell homeostasis and skeletal muscle repair. Nat Commun. 2015;6:10123. doi: 10.1038/ncomms10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaillou T, Kirby TJ, McCarthy JJ. Ribosome biogenesis: emerging evidence for a central role in the regulation of skeletal muscle mass. J Cell Physiol. 2014;229(11):1584–1594. doi: 10.1002/jcp.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez AM, et al. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2012;113(2):695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 31.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 32.Mittal A, et al. The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice. J Cell Biol. 2010;188(6):833–849. doi: 10.1083/jcb.200909117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jäger S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci USA. 2007;104(29):12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carnio S, et al. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep. 2014;8(5):1509–1521. doi: 10.1016/j.celrep.2014.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masiero E, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10(6):507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Reid MB, Moylan JS. Beyond atrophy: redox mechanisms of muscle dysfunction in chronic inflammatory disease. J Physiol (Lond) 2011;589(Pt 9):2171–2179. doi: 10.1113/jphysiol.2010.203356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mourkioti F, et al. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J Clin Invest. 2006;116(11):2945–2954. doi: 10.1172/JCI28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul PK, et al. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol. 2010;191(7):1395–1411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyazaki M, McCarthy JJ, Fedele MJ, Esser KA. Early activation of mTORC1 signalling in response to mechanical overload is independent of phosphoinositide 3-kinase/Akt signalling. J Physiol (Lond) 2011;589(Pt 7):1831–1846. doi: 10.1113/jphysiol.2011.205658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armstrong DD, Esser KA. Wnt/beta-catenin signaling activates growth-control genes during overload-induced skeletal muscle hypertrophy. Am J Physiol, Cell Physiol. 2005;289(4):C853–C859. doi: 10.1152/ajpcell.00093.2005. [DOI] [PubMed] [Google Scholar]
- 41.Pogozelski AR, et al. p38gamma mitogen-activated protein kinase is a key regulator in skeletal muscle metabolic adaptation in mice. PLoS One. 2009;4(11):e7934. doi: 10.1371/journal.pone.0007934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie M, et al. A pivotal role for endogenous TGF-beta-activated kinase-1 in the LKB1/AMP-activated protein kinase energy-sensor pathway. Proc Natl Acad Sci USA. 2006;103(46):17378–17383. doi: 10.1073/pnas.0604708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inokuchi-Shimizu S, et al. TAK1-mediated autophagy and fatty acid oxidation prevent hepatosteatosis and tumorigenesis. J Clin Invest. 2014;124(8):3566–3578. doi: 10.1172/JCI74068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zong H, et al. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci USA. 2002;99(25):15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond MJ, Dreyer HC, Fry CS, Glynn EL, Rasmussen BB. Nutritional and contractile regulation of human skeletal muscle protein synthesis and mTORC1 signaling. J Appl Physiol. 2009;106(4):1374–1384. doi: 10.1152/japplphysiol.91397.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- 48.Bakkar N, et al. IKKα and alternative NF-κB regulate PGC-1β to promote oxidative muscle metabolism. J Cell Biol. 2012;196(4):497–511. doi: 10.1083/jcb.201108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shintaku J, et al. MyoD Regulates Skeletal Muscle Oxidative Metabolism Cooperatively with Alternative NF-κB. Cell Rep. 2016;17(2):514–526. doi: 10.1016/j.celrep.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo AY, Joseph AM, Dutta D, Hwang JC, Aris JP, Leeuwenburgh C. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J Cell Sci. 2010;123(Pt 15):2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lira VA, et al. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J. 2013;27(10):4184–4193. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi P, Gal J, Kwinter DM, Liu X, Zhu H. Mitochondrial dysfunction in amyotrophic lateral sclerosis. Biochim Biophys Acta. 2010;1802(1):45–51. doi: 10.1016/j.bbadis.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Omori E, Inagaki M, Mishina Y, Matsumoto K, Ninomiya-Tsuji J. Epithelial transforming growth factor β-activated kinase 1 (TAK1) is activated through two independent mechanisms and regulates reactive oxygen species. Proc Natl Acad Sci USA. 2012;109(9):3365–3370. doi: 10.1073/pnas.1116188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paul PK, et al. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol. 2012;32(7):1248–1259. doi: 10.1128/MCB.06351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paul PK, Kumar A. TRAF6 coordinates the activation of autophagy and ubiquitin-proteasome systems in atrophying skeletal muscle. Autophagy. 2011;7(5):555–556. doi: 10.4161/auto.7.5.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26(3):203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gallot YS, et al. Distinct roles of TRAF6 and TAK1 in the regulation of adipocyte survival, thermogenesis program, and high-fat diet-induced obesity. Oncotarget. 2017;8(68):112565–112583. doi: 10.18632/oncotarget.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hindi SM, Sato S, Choi Y, Kumar A. Distinct roles of TRAF6 at early and late stages of muscle pathology in the mdx model of Duchenne muscular dystrophy. Hum Mol Genet. 2014;23(6):1492–1505. doi: 10.1093/hmg/ddt536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hindi SM, Kumar A. TRAF6 regulates satellite stem cell self-renewal and function during regenerative myogenesis. J Clin Invest. 2016;126(1):151–168. doi: 10.1172/JCI81655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bohnert KR, Gallot YS, Sato S, Xiong G, Hindi SM, Kumar A. Inhibition of ER stress and unfolding protein response pathways causes skeletal muscle wasting during cancer cachexia. FASEB J. 2016;30(9):3053–3068. doi: 10.1096/fj.201600250RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato S, et al. TWEAK promotes exercise intolerance by decreasing skeletal muscle oxidative phosphorylation capacity. Skelet Muscle. 2013;3(1):18. doi: 10.1186/2044-5040-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc. 2015;10(6):823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 64.Hindi SM, et al. Reciprocal interaction between TRAF6 and notch signaling regulates adult myofiber regeneration upon injury. Mol Cell Biol. 2012;32(23):4833–4845. doi: 10.1128/MCB.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiong G, et al. The PERK arm of the unfolded protein response regulates satellite cell-mediated skeletal muscle regeneration. Elife. 2017;6:e22871. doi: 10.7554/eLife.22871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.