Abstract
The bacterium Staphylococcus aureus produces an array of anti-inflammatory molecules that prevent the innate immune system from recognizing it as a pathogen and clearing it from the host. In the acute phase of inflammation, our immune system relies on neutrophils to clear invading bacteria. Recently, novel classes of secreted proteins from S. aureus, including the Extracellular Adherence Protein (EAP) family (Stapels et al. 2014) and the Staphylococcal Peroxidase Inhibitor (SPIN), (unpublished work) have been identified as highly selective inhibitors acting on Neutrophil Serine Proteases (NSPs) and myeloperoxidase (MPO) respectively.
SPIN is a protein found only in Staphylococci, with no sequence homology to any known proteins. Solution NMR structural studies of SPIN are therefore expected to provide a deeper understanding of its interaction with MPO. In this study, we report the backbone and side-chain 1H, 15N, and 13C resonance assignments of SPIN. Furthermore, using the chemical shifts of these resonances, we predicted the secondary structure of SPIN in solution via the TALOS-N server. The assignment data has been deposited in the BMRB data bank under accession number 27069.
Keywords: NMR assignment, Staphylococcus aureus, virulence factor, Staphylococcal Peroxidase Inhibitor (SPIN)
Biological context
Staphylococcus aureus is a pathogenic bacterium known for its ability to cause a variety of infections such as endocarditis, skin abscesses and bacteremia (Archer 1998; Lowy 1998). Previous studies have demonstrated that S. aureus produces an array of virulence proteins that disrupt normal function of the human innate immune system (Foster 2005). The combined activities of these proteins are believed to prolong bacterial survival within the host, allowing for infections to take hold.
Neutrophils are the most abundant type of white blood cells in humans and the most prominent cellular component of the innate immune system. They can be rapidly activated by various biochemical stimuli during the earliest phase of bacterial invasion and recruited into the sites of infection (Kruger et al. 2015). Neutrophils contain subcellular granules that are replete with vital anti-bacterial enzymes, including a series of chymotrypsin-like Neutrophil Serine Proteases (NSPs) as well as a heme-containing Myeloperoxidase (MPO) (Amulic et al. 2012; Kruger et al. 2015). Novel classes of S. aureus secreted proteins, such as the Extracellular Adherence Protein (EAP) family (Stapels et al. 2014) and Staphylococcal Peroxidase Inhibitor (SPIN) proteins (unpublished work) have been identified as inhibitors of NSPs and MPO, respectively. Our understanding of the mechanisms used by neutrophils to eliminate pathogens continues to expand. While it has been long believed that neutrophils eliminate pathogens through phagocytosis alone, more recent work has shown that neutrophils can employ additional killing mechanisms such as degranulation and NETosis (Amulic et al. 2012; Kolaczkowska and Kubes 2013; Kruger et al. 2015). These mechanisms, particularly the release of digestive and oxidative granular enzymes into the inflammatory environment, can expose otherwise healthy host cells and extracellular matrices to substantial damage. In the case of recurring infections, the immune response can lead to scar tissue and in some cases, even malignancy (Shacter and Weitzman 2002).
Since MPO is a vital component of neutrophil’s anti-bacterial arsenal and is a major component of neutrophil granules, release of MPO into the extracellular space can result in production of damaging hypohalous acids. Characterization of novel classes of enzymatic inhibitors that target key enzymes within the neutrophil anti-bacterial repertoire may therefore open new directions for development of anti-inflammatory therapies. Our previous studies have identified SPIN as the first described innate immune evasion protein that acts as an exclusively specific inhibitor of MPO. SPIN binds MPO with nanomolar affinity (KD ~10 nM) and a crystal structure of the complex has been obtained (unpublished work). While the structure of SPIN in bound form is predominantly characterized by a three helix bundle, an ~10 residue region at its N-terminus adopts a peculiar beta hairpin conformation that inserts into the MPO active site channel. Preliminary analyses have revealed that loss of this N-terminal region dramatically reduces SPIN’s ability to inhibit MPO without any notable effect its binding affinity. As a prelude to detailed structure/function studies, we investigated herein the secondary structural features of SPIN in the free form in solution. After assigning the backbone and side-chain 1H, 15N, and 13C resonances of SPIN, we were able to predict the secondary structure using TALOS-N server using the observed chemical shifts.
Methods and Experiments
Protein expression and purification
SPIN was overexpressed using the method described by Geisbrecht et al. (2006)
gBlocks Gene Fragments (Integrated DNA Technologies, USA) encoding the S. aureus SPIN sequence were subcloned into the Sal1 and EcoRI sites of a modified prokaryotic expression vector pT7HMT. The fragment inserted into the vector was verified by sequencing and uniformly 15N and 13C/15N double-labeled SPIN protein was expressed in Escherichia coli BL21(DE3) cells, grown in minimal medium (M9) enriched with 15NH4Cl and 13C-glucose as described in the protocol by Woehl et al. (2016). The vector used encoded for an N-terminal affinity His-tag which was used for Ni-affinity chromatography purification of the protein and then removed by digestion with tabacco etch virus (TEV) protease. Therefore, this version of the recombinant SPIN protein contains an additional “Gly-Ser-Thr” sequence at the N-terminus.
The purified protein yield from 1 L of E. coli culture was in the range of 7–9 mg for both 15N and 13C/15N double-labeled SPIN. The samples for NMR experiments contained 0.75 – 1.0 mM uniformly 15N or 13C/15N double-labeled SPIN protein in 50 mM sodium phosphate buffer (pH 6.5) containing 5 % (v/v) D2O (used as a lock solvent). The mass and purity of the protein were verified using mass spectrometry (Ultra Flex III TOF, Bruker Daltonics) prior to acquisition of the NMR data to ensure the protein integrity.
NMR experiments
NMR measurements were performed at 25 °C on a Varian 500VNMR System (Agilent Technologies) equipped with a 5-mm triple-resonance inverse detection pulse gradient cold probe operating at 499.84 MHz for 1H frequency, and on a Bruker Avance 700 MHz spectrometer equipped with a TXI cryo-probe. The NMR data recorded on uniformly 15N-labeled SPIN protein included sensitivity enhanced 2D 1H-15N HSQC using water-flip-back for minimizing water saturation. The triple resonance NMR experiments performed on uniformly 13C/15N double-labeled SPIN samples were 3D HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, and HNCO. The following NMR spectra were collected for side chain assignments: 2D 13C HSQC and 3D HCCONH, CCCONH, HCCH-TOCSY, and 15N-edited NOESY (tm =100 ms). The validation of the assignment was also conducted with a 15N HSQC-TOCSY (tm=80 ms) spectrum. The 1H chemical shifts were referenced to the external standard 2,2-dimethyl-2-silapentane-5-sulphonic acid (DSS) at 25 °C. However, 13C and 15N chemical shifts were referenced indirectly from DSS.
NMR data were processed using NMRPipe (Delaglio et al. 1995) and the analysis and visualization of the spectra were performed in CARA (http://www.nmr.ch; Keller 2004). The secondary structure of SPIN was predicted using the TALOS-N platform (Shen and Bax 2013).
Results
Extent of the assignments and data deposition
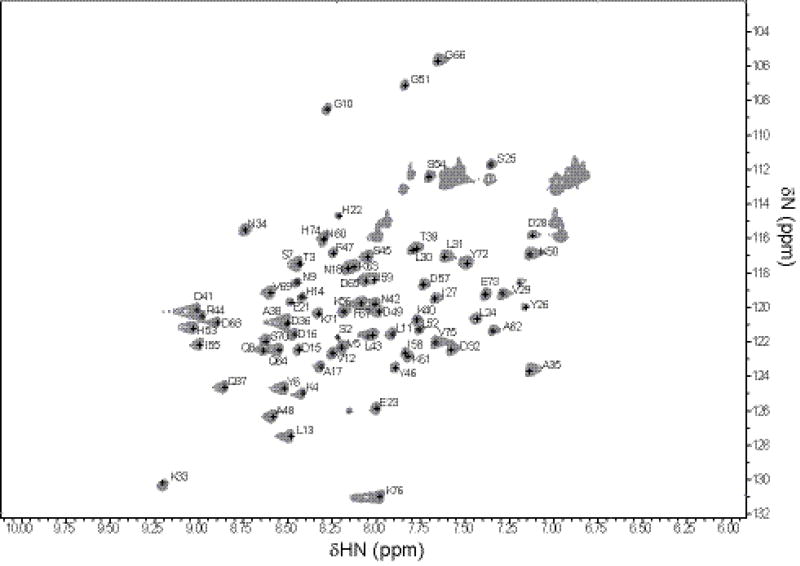
As illustrated in Fig. 1, the 2D 1H–15N HSQC measurement of SPIN resulted in a well-dispersed spectrum. The unlabeled peaks in this figure belong to the side chains of Asn and Gln residues. Amino acid numbering is based on the SPIN sequence, including the N-terminal “Gly-Ser-Thr”. The sequence specific backbone resonance assignments for nearly all 1H, 15N and 13C spins were completed (97% 1H/15N assignments) using the NMR experiments described in the Methods and Experiments section. The backbone amide protons that could not be assigned include F19 and L20. Based on secondary structure prediction, these unassigned residues are located on the C-terminus of a dynamic region (1–20AA) connecting to an alpha-helix (21–32AA) and probably could not be identified due overlap or line-broadening of the relevant Cα and Cβ resonances.
Fig. 1.
The 2D 1H - 15N HSQC spectrum of 1 mM of 13C/15N-labeled SPIN recorded at 25°C on Bruker Avance 700 MHz spectrometer
Spin resonances for HN, N, Cα, Cβ, Hα, Hβ and C were assigned for >95% of the residues. The analysis of HN(CO)CA, HNCACB, CBCA(CO)NH, and HNCO resulted in the assignment of 95% of 13Cα resonances, 92% of 13Cβ resonances, and 76% of all 13C resonances. The chemical shift assignments have been deposited in BioMagResBank (www.bmrb.wisc.edu) under the accession number 27069.
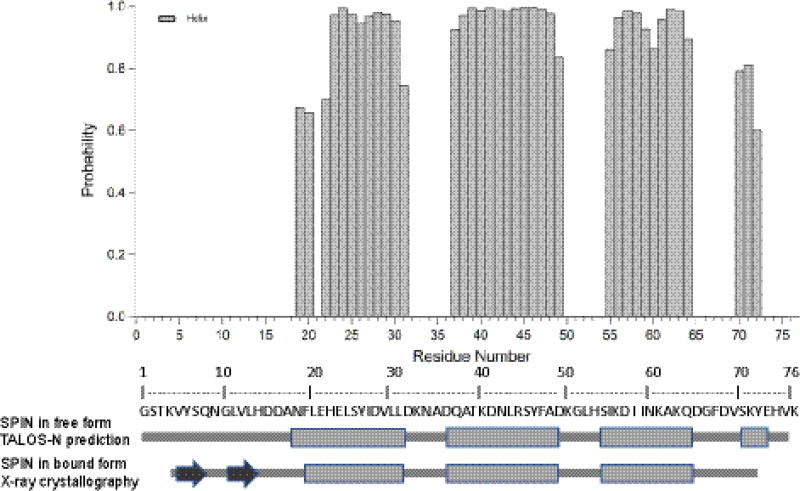
The secondary structure of SPIN was predicted by TALOS-N program (Shen and Bax 2013) using the resonance assignments of 13Cα, 13Cβ, and 13C, NH and 1H (Fig. 2). We compared this predicted secondary structure of SPIN in the free form in solution with the one determined by X-ray crystallography for SPIN bound to recombinant human MPO (unpublished work). Although the SPIN-MPO co-crystal structure shows that N-terminal region of SPIN adopts a beta-hairpin, TALOS-N prediction of the free SPIN in solution suggests that this region is not similarly structured in the unbound protein. Additional studies are underway to further investigate the structure/function consequences of this discrepancy.
Fig. 2.
Secondary structure prediction for SPIN based on TALOS-N program with obtained chemical shift values of 13Cα, 13Cβ, and 13C, NH and 1H. Alpha helix probabilities are given by the shaded columns and positive values. Below is represented a schematic of secondary structure (rectangle = alpha helix, arrow =beta strand) of the TALOS-N prediction of SPIN in free form and of the crystal structure of SPIN in bound form.
Acknowledgments
This research was funded by awards from the National Institutes of Health to B.V.G. (GM121511 and AI111203) and for this study we made use of the NMR facility at University of Arkansas, Fayetteville, AR.
Footnotes
Ethical standards
The experiments described in this paper comply with the current laws of USA.
Conflict of interest
The authors declare no conflict of interest.
References
- Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–89. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- Archer GL. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Foster TJ. Immune evasion by staphylococci. Nature Reviews Microbiology. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- Geisbrecht BV, Bouyan S, Pop M. An optimized system for expression and purification of secreted bacterial proteins. Protein Expr. Purif. 2006;46:23–32. doi: 10.1016/j.pep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Keller R. The Computer Aided Resonance Assignment Tutorial. 2004. [Google Scholar]
- Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–75. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- Kruger P, Saffarzadeh M, Weber ANR, Rieber N, Radsak M, von Bernuth H, Benarafa C, Roos D, Skokowa J, Harti D. Neutrophils: Between host defence, immune modulation, and tissue injury. Plos Pathogens. 2015;11:e1004651. doi: 10.1371/journal.ppat.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy FD. Staphylococcus aureus Infections. N. Engl. J. Med. 1998;339:550–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Shacter E, Weitzman SA. Chronic Inflammation and Cancer. Oncology Journal. 2002;16:217. [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapels DA, Ramyar KX, Bischoff M, von Köckritz-Blickwede M, Milder FJ, Ruyken M, Eisenbeis J, McWhorter WJ, Herrmann M, van Kessel KP, Geisbrecht BV, Rooijakkers SH. Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proc Natl Acad Sci. 2014;111:13187–92. doi: 10.1073/pnas.1407616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehl JL, Takahashi D, Herrera AI, Geisbrecht BV, Prakash O. 1H, 15N, and 13C resonance assignments of Staphylococcus aureus extracellular adherence protein domain 4. Biomol NMR Assign. 2016;10:301–5. doi: 10.1007/s12104-016-9688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]