Abstract
Efferocytosis, the phagocytic clearance of apoptotic cells, can provide host protection against certain types of viruses by mediating phagocytic clearance of infected cells undergoing apoptosis. It is known that HIV-1 induces apoptosis and HIV-1-infected cells are efferocytosed by macrophages, although its molecular mechanisms are unknown. To elucidate the roles that efferocytosis of HIV-1-infected cells play in clearance of infected cells, we sought to identify molecules that mediate these processes. We found that protein S, present in human serum, and its homologue, Gas6, can mediate phagocytosis of HIV-1-infected cells by bridging receptor tyrosine kinase Mer, expressed on macrophages, to phosphatidylserine exposed on infected cells. Efferocytosis of live infected cells was less efficient than dead infected cells; however, a significant fraction of live infected cells were phagocytosed over 12 h. Our results suggest that efferocytosis not only removes dead cells, but may also contribute to macrophage removal of live virus producing cells.
Keywords: HIV-1, Phagocytosis, Efferocytosis, Protein S, Gas6, Mer, Phosphatidylserine
1. Introduction
Phagocytosis of dying or dead cells by phagocytes such as macrophages is known as efferocytosis (Birge et al., 2016; Zent and Elliott, 2017). Removal of dead or dying cells by efferocytosis is critical for maintaining homeostasis and preventing inflammation and autoimmune reactions (Elliott and Ravichandran, 2016). Because infection by certain viruses induces massive cell death, efferocytosis plays an important role in maintaining host homeostasis during viral infection (Jorgensen et al., 2017; Nainu et al., 2017). Viral infection induces changes of cell surface molecules related to cell death, and macrophages can recognize these changes and engulf the affected cells. It has been reported that macrophages perform efferocytosis of cells infected with the influenza (Fujimoto et al., 2000; Hashimoto et al., 2007; Shiratsuchi et al., 2000; Watanabe et al., 2005, 2002, 2004; Mukherjee et al., 2017), papilloma (Hermetet et al., 2016), and Drosophila C viruses (Nainu et al., 2015; Nonaka et al., 2017), as well as Lymphocytic choriomeningitis virus (LCMV) (Alatery et al., 2010), and Human Immunodeficiency Virus Type 1 (HIV-1) (Akbar et al., 1994).
Efferocytosis can also play a role in protecting the host from pathogens (Martin et al., 2014; Nainu et al., 2015; Tufail et al., 2017). When cells are infected with certain types of viruses, the cells undergo apoptosis to prevent pathogens from exploiting the host cell machinery. Efferocytosis then mediates phagocytic clearance of the infected cells and prevents further production of viruses.
The roles of efferocytosis in protection against pathogens have been well-studied for influenza virus infection (Fujimoto et al., 2000; Hashimoto et al., 2007; Shiratsuchi et al., 2000; Watanabe et al., 2005, 2002, 2004). When cells are infected with influenza virus, they start exposing phosphatidylserine (PtdSer), which is mainly present in the inner layers of the cell membrane but becomes exposed when cells undergo apoptosis. Macrophages and neutrophils efferocytose infected cells by recognizing exposed PtdSer. By clearing the infected cells, efferocytosis inhibits virus spread.
HIV-1 infection is known to induce exposure of PtdSer by inducing cell death (apoptosis or pyroptosis) (Terai et al., 1991; Doitsh et al., 2014). Analysis of tissue sections from HIV-1-infected patients has shown that macrophages engulf HIV-1-infected T-cells by efferocytosis (Akbar et al., 1994). In vitro studies showed that macrophages can recognize and engulf HIV-1-infected cells by a mechanism that is independent of viral envelope proteins and antiviral antibodies (Baxter et al., 2014). Although the molecular mechanisms by which macrophages selectively capture and engulf apoptotic HIV-1-infected cells are not known, it is likely that recognition of PtdSer plays a role.
Phagocytes recognize PtdSer on dead cells by various molecular mechanisms that can be largely categorized as mediated by soluble molecules that bridge dead cells and phagocytes, including protein S, Gas6, and MFG-E8 (Hafizi and Dahlback, 2006; Hanayama et al., 2002), or mediated by the receptors that directly bind PtdSer, including TIM-1, -3, and -4, CD300a, BAI-1, RAGE, and Stabilin 1 and 2 (DeKruyff et al., 2010; Friggeri et al., 2011; He et al., 2011; Kobayashi et al., 2007; Miyanishi et al., 2007; Nakahashi-Oda et al., 2012; Park et al., 2007, 2009; Simhadri et al., 2012). In this study, we found that protein S/Gas6 can mediate phagocytosis of HIV-1-infected cells by bridging PtdSer exposed on the infected cells to one type of receptor tyrosine kinase, Mer, which is expressed on macrophages. We investigated whether this efferocytosis mechanism can inhibit virus production by engulfment of infected cells producing virus.
2. Results
2.1. HIV-1 infection induces PtdSer exposure
Because HIV-1 infection is known to induce exposure of PtdSer on infected cells, we hypothesized that macrophages capture infected cells by recognizing exposed PtdSer, similar to how they recognize influenza virus-infected cells (Fujimoto et al., 2000; Hashimoto et al., 2007; Shiratsuchi et al., 2000; Watanabe et al., 2005; Watanabe et al., 2002; Watanabe et al., 2004).
We first investigated the time-course of Gag (HIV-1 p24) expression, Env expression, PtdSer exposure, cell death, and virus production to determine whether exposed PtdSer can be a marker for phagocytes to recognize HIV-1-infected cells (Fig. 1A). For target cells, we used MT4CCR5, a CD4+ T-cell line ectopically expressing CCR5. Since nearly 100% of MT4CCR5 cells become infected within two days post-infection (Fig. 1A), this cell line provides an ideal model for experiments to investigate the molecular mechanisms of efferocytosis of HIV-1-infected cells.
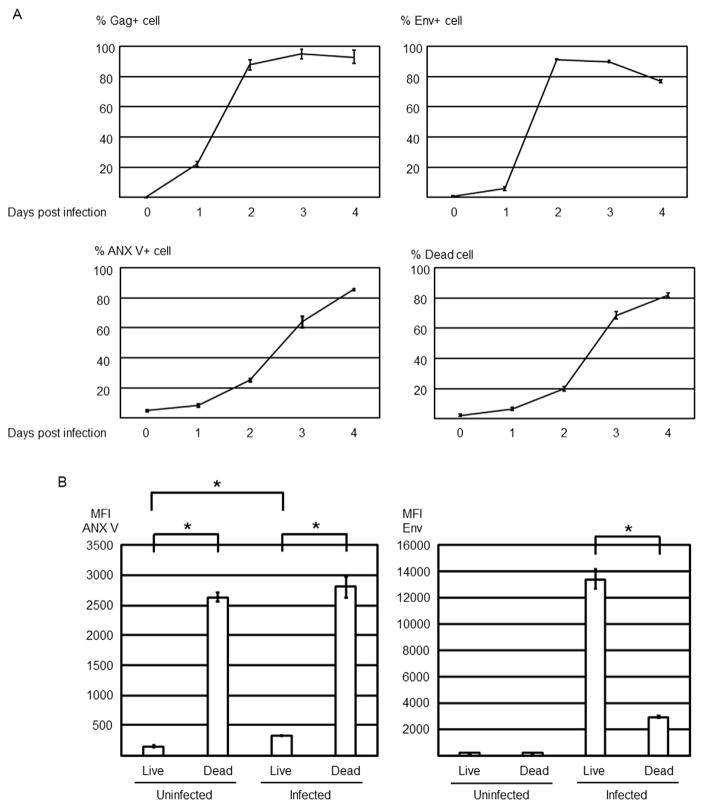
Fig. 1.
HIV-1 infection induces PtdSer exposure. MT4CCR5 cells were infected with HIV-1NL4-3 at MOI 5. Infected cells were analyzed by flow cytometry for expression of Gag and Env, exposure of PtdSer, and cell death, and virus production was quantified by ELISA and titration for up to four days post-infection. This experiment was repeated twice in singlicate and once in triplicate as independent experiments (A–C). The results shown are averages and standard deviations of the triplicate experiment. (A) HIV-1 Gag expression was quantitated by intracellular staining of cells with FITC-conjugated anti-HIV-1 p24 antibody. Env expression and cell death were quantitated by staining with Alexa 647-conjugated anti-HIV-1 gp120 antibody and Ghost Dye Violet 450. PtdSer exposure and cell death were quantitated by staining with APC-conjugated ANX V and Ghost Dye Violet 450. (B) Mean fluorescence intensity of ANX V and Env staining of live and dead populations of uninfected and infected cells. The infected cells were analyzed at 3 days post-infection. (C) Supernatants of infected cells were harvested every 24 h after infection for 4 days, and virus production was quantitated by measuring amounts of Gag by ELISA and titration of virus, using GHOST (3) CXCR4+CCR5+ cells. (D) Expression levels of TIM-1, TIM-4, Axl, TYRO3, and Mer were analyzed by staining cells with specific PE-conjugated antibodies against each molecule (red lines). The black line represents staining with PE-conjugated isotype control antibody.
MT4CCR5 cells were infected with X4-tropic strain of HIV-1 (HIV-1NL4-3). Cells infected with virus expressed Gag and Env at low levels one day post-infection and then at drastically increased levels two days post-infection (Fig. 1A). PtdSer exposure, which was analyzed by Annexin V (ANX V) staining, started two days post-infection and increased until four days post-infection. The cells infected with heat-inactivated virus do not expose PtdSer (Fig. S1), indicating that exposed PtdSer can be a marker for macrophages to recognize HIV-1-infected cells. Cell death also started at two days post-infection (~20%) and drastically increased at 3 days post-infection (~65%). When cell death and Env expression of infected cells were analyzed together (Fig. 1B and Fig. S1), Env expression was lower in dead cells than live cells (Fig. 1B). When cell death and PtdSer exposure were analyzed together, PtdSer exposure was higher on dead cells than live cells (Fig. 1B).
Progeny virus production started between one and two days post-infection, peaked between two and three days post-infection, and drastically decreased between three and four days post-infection (Fig. 1C). Time course analyses of cell death and virus production suggested that high viability is required for efficient production of infectious virus.
2.2. Expression of PtdSer receptors on macrophages
We next investigated whether macrophages express PtdSer receptors. We obtained macrophages by differentiating peripheral blood monocytes. To avoid the effects of protein S present in serum, we depleted bovine protein S from fetal calf serum used in this study by barium chloride precipitation (Bhattacharyya et al., 2013).
We recently investigated the PtdSer recognition mechanisms that can efficiently recognize exposed PtdSer (Morizono and Chen, 2014; Morizono et al., 2011). Our results showed that protein S/Gas6, which bridges PtdSer to TAM receptors (TYRO 3, Axl, and Mer) on phagocytes, and TIM-1 and -4, which bind PtdSer directly, bind PtdSer most efficiently. To investigate whether any of these molecular mechanisms can mediate phagocytosis of HIV-1-infected cells by macrophages, we first analyzed expression of these receptors on macrophages and found only Mer was expressed on macrophages (Fig. 1D). We also analyzed expression of those five receptors on macrophages differentiated under various culture conditions, including use of granulocyte-macrophage colony-stimulating factor (GM-CSF), macrophage colony-stimulating factor (M-CSF), or no cytokines, or using medium supplemented with normal human or fetal bovine serum or serum-free medium, or in the presence and absence of Dexamethasone. Of the five receptors, only Mer was expressed on macrophages under any conditions tested (data not shown).
2.3. Human serum induces phagocytosis of infected MT4CCR5 cells
Because Mer can mediate efferocytosis through an interaction with protein S in serum, we investigated whether human serum can induce phagocytosis of HIV-1-infected cells. We used a pH-sensitive fluorescent dye, pHrodo Red, to analyze phagocytosis of cells, as previously described (Aziz et al., 2013). We labeled MT4CCR5 cells or the cells infected with HIV-1NL4-3 (MT4CCR5/NL4-3) with pHrodo Red so that they emit fluorescent signals upon exposure to the low-pH environment in the phagosomes of phagocytes. We used MT4CCR5/NL4-3 cells at three days post-infection to investigate phagocytic mechanisms. We co-cultured macrophages with pHrodo Red-labeled MT4CCR5 or MT4CCR5/NL4-3 cells in the absence or presence of human serum, using macrophages obtained from four donors. The gating strategy to quantitate phagocytosis by flow cytometry analysis is shown in Fig. 2A. In the absence of serum, percentages of pHrodo Red signal-positive macrophages co-cultured with MT4CCR5/NL4-3 were higher than those co-cultured with MT4CCR5 (Fig. 2B), indicating that macrophages can preferentially engulf HIV-1-infected cells independently of serum, albeit at low efficiency. Human serum enhanced phagocytosis of MT4CCR5/NL4-3 more efficiently than for MT4CCR5 cells (Fig. 2B), demonstrating that human serum preferentially induces phagocytosis of HIV-1-infected cells by macrophages. We confirmed that four lots of human AB serum could induce phagocytosis of MT4CCR5/NL4-3 cells by macrophages (data not shown). After confirmation, we used the same lot throughout this study.
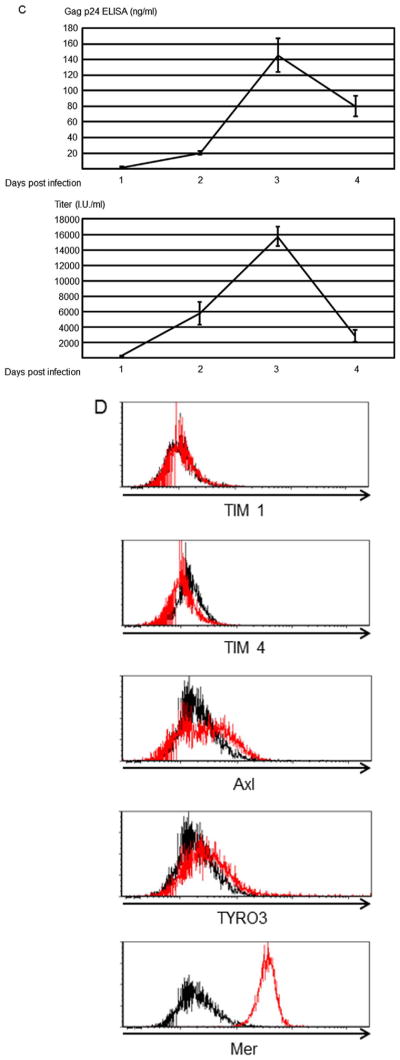
Fig. 2.
Human serum induces efferocytosis of HIV-1-infected MT4CCR5 cells. (A) pHrodo Red-labeled MT4CCR5 and MT4CCR5/NL4-3 cells were co-cultured with equivalent numbers of macrophages in the absence or presence of 3% human serum for 90 min. The cells were then stained with APC-conjugated anti-CD14 antibody, and pHrodo Red signals in CD14-positive population were analyzed by flow cytometry. (B) Experiments to examine the effects of human serum on phagocytosis were repeated four times as independent experiments, using cells from different donors for each independent experiment. The results shown are averages and standard deviations of the all experiments. Significance was calculated by a one-sided Mann- Whitney U test. * p=0.05. (C) MT4CCR5 and MT4CCR5/NL4-3 cells were labeled with CellTrace Violet and co-cultured with the same numbers of macrophages in the absence or presence of 3% human serum for 90 min. Cells were then stained with APC-conjugated anti-CD14 antibody and subjected to imaging flow cytometry. Binding of CD14-positive and CellTrace violet-positive populations were analyzed first. The CD14- and CellTrace violet double-positive populations were examined to score internalization CellTrace violet-positive cells into CD14-positive cells. (D) Percentages of macrophages containing CellTrace violet-labeled cells were calculated via imaging flow cytometry. A threshold internalization score of 1.5 was used to define internalized CellTrace violet-labeled cells. (E) Macrophages labeled with CellTrace Far Red (red color) and MT4CCR5/NL4-3 labeled with CFSE were co-cultured in the absence or presence 3% human AB serum for 90 min, followed by nuclear staining with DAPI. Phagocytosis was analyzed by confocal microscopy. Four slice images of each condition were examined, and percentages of macrophages containing MT4CCR5/NL4-3 cells were calculated. More than 120 macrophages were counted for each condition. The results shown are averages and standard deviations, and significance was calculated by a one-sided Mann-Whitney U test. * p=0.05.
We next confirmed the flow cytometric analysis of phagocytosis, using quantitative analysis of microscopic images by imaging flow cytometry. To analyze binding of macrophages to prey cells, we tracked MT4CCR5 and MT4CCR5/NL4-3 cells by labeling them with CellTrace Violet fluorescent dye, co-cultured the cells with macrophages in the presence or absence of human serum, then stained cells with APC-conjugated antibody against CD14 to identify macrophages. In the absence of serum, 1% and 3.6% of macrophages bound MT4CCR5 and MT4CCR5/NL4-3 cells, respectively (Fig. 2C), demonstrating that macrophages can recognize HIV-1-infected cells independently of serum, which is consistent with the results using pHrod Red labeling and flow cytometry. Human serum enhanced binding of macrophages to MT4CCR5/NL4-3 (from 3.6% to 40.7%), but only minimally to MT4CCR5 (from 1% to 1.1%) (Fig. 2C). To distinguish macrophage internalization of prey cells from macrophage:prey cell binding, we employed imaging flow cytometry analysis software, IDEAS 6.2, which can quantitate internalization of violet signals into macrophage APC signals as internalization scores. Events with internalization scores of less than 0 indicate that MT4CCR5/NL4-3 cells (violet color) bound the surfaces of macrophages (red color) (Fig. 2C), and those with internalization scores greater than 0 indicate internalization of the MT4CCR5/NL4-3 signal into macrophages (Fig. 2C). We used an internalization score of 1.5 as a threshold to stringently identify engulfment of cells, and calculated percentages of macrophages that had internalized MT4CCR5 or MT4CCR5/NL4-3 cells. Consistent with the results of flow cytometric analysis of pHrodo Red-labeled cells, imaging flow cytometry demonstrated that human serum drastically enhanced internalization of MT4CCR5/NL4-3 into macrophages (from 1.33% to 18.8%), but only minimally enhanced phagocytosis of uninfected cells (from 0.69% to 0.92%) (Fig. 2D).
We also confirmed internalization of MT4CCR5/NL4-3 into macrophages, using confocal microscopy. Macrophages were labeled with CellTrace Far Red, and MT4CCR5/NL4-3 cells were labeled with CSFE. Both cell types were co-cultured in the absence or presence of 3% human serum. After staining the nuclei with DAPI, co-cultured cells were visualized by confocal microscopy. We counted macrophages that contained both cytoplasm and nuclei of MT4CCR5/NL4-3 cells (Fig. S2A and B). Addition of human serum increased the percentages of macrophages containing MT4CCR5 cells from 4.2% to 14.5% (Fig. 2E).
The results of imaging flow cytometry and confocal microscopy visually confirmed the results of conventional flow cytometry, which showed that human serum induces engulfment of MT4CCR5/NL4-3 cells.
2.4. Human serum mediates phagocytosis of HIV-1-infected cells through a PtdSer- and Mer-dependent mechanism
We next determined whether exposed PtdSer plays a role in serum-induced phagocytosis. We investigated the role of exposed PtdSer by blocking PtdSer on MT4CCR5/NL4-3 cells using MFG-E8 mutants. Wild-type MFG-E8 binds to PtdSer with its discoidin domain, C2, and also binds integrins αVβ3 and αVβ5 at its RGD motif in EGF domain 2 (E2) (Hanayama et al., 2002) (Fig. 3A). Dr. Hanayama et al. generated a mutant MFG-E8 (D89E) that contains a mutation within its integrin-binding RGD motif (Asano et al., 2004; Hanayama et al., 2002). D89E has been used as a dominant-negative mutant to block PtdSer-dependent phagocytosis of dead cells (Asano et al., 2004; Miyanishi et al., 2007; Toda et al., 2012; Yoshida et al., 2005) because it can bind PtdSer, whereas its binding to integrins is impaired. We also generated other types of dominant-negative mutants of MFG-E8 by deleting the entire E2 domain of MFG-E8, designated ΔE2 MFG-E8 (Fig. 3A), which can no longer bind integrins on phagocytes but can still bind to PtdSer. We attempted to block serum-induced phagocytosis by incubating MT4CCR5/NL4-3 cells with D89E or ΔE2 MFG-E8 prior to incubation with macrophages.
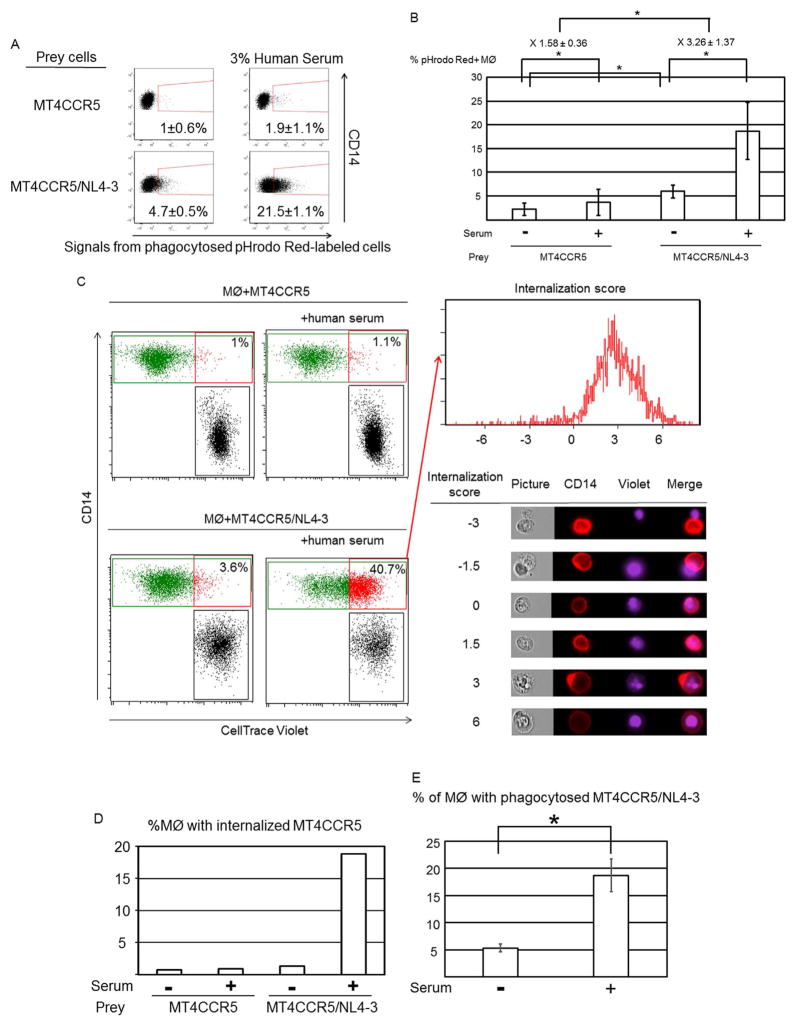
Fig. 3.
Human serum-induced phagocytosis of HIV-1-infected cells is dependent on PtdSer exposed on infected cells and Mer expressed on macrophages. (A) Schematic representation of recombinant MFG-E8 and two types of mutants of MFG-E8 (D89E and ΔE2 MFG-E8) that bind PtdSer but not integrins. E1 and E2, epidermal growth factor-like domains 1 and 2; P/T, proline/threonine-enriched motif; C1 and C2, discoidin domains 1 and 2. The C2 domain of MFG-E8 binds PtdSer, and the RGD motif in the E2 domain of MFG-E8 binds integrins. (B) pHrodo red-labeled MT4CCR5/NL4-3 cells were incubated with or without 20 μg/ml of D89E, ΔE2 MFG-E8 or sCD4, followed by co-culture with macrophages in the presence or absence of 3% human serum. Efferocytosis of MT4CCR5/NL4-3 cells was analyzed by flow cytometry. This experiment was repeated twice in triplicate as independent experiments, using cells from different donors for each independent experiment. The results shown are averages and standard deviations. Significance was calculated by comparing the values of the samples with 3% serum and no blocking reagents to the values of the samples with 3% serum with the blocking reagents indicated in the figures, using a one-sided Mann-Whitney U test. * p=0.05. (C) MT4CCR5 cells were infected with NLΔBgl (VSV-G) or HIV-1Ada at MOI 5. Three days post-infection, Gag and Env expression and PtdSer exposure were analyzed by flow cytometry. This experiment was repeated 5 times in singlicate as independent experiments. The representative results are shown. (D) pHrodo Red-labeled MT4CCR5/NLΔBgl (VSV-G) cells were co-cultured with equivalent numbers of macrophages in the absence or presence of 3% human serum for 90 min. The cells were then stained with APC-conjugated anti-CD14 antibody, and pHrodo Red signals in CD14-positive population were analyzed by flow cytometry. This experiment was repeated three times in triplicate, using cells from different donors, and the results shown are averages and standard deviations of the all experiments. (E) Macrophages were incubated with or without anti-Axl or anti-Mer antibodies (10 μg/ml), followed by co-culture with pHrodo Red-labeled MT4CCR5/NL4-3 cells in the presence or absence of 3% human serum. Efferocytosis of MT4CCR5/NL4-3 cells was analyzed by flow cytometry. This experiment was repeated once in singlicate and twice in triplicate as independent experiments, using cells from different donors for each independent experiment. The results shown are averages and standard deviations of the triplicate experiments. Significance was calculated by comparing the value of the samples with 3% serum and no blocking reagents to the value of the samples with 3% serum with the blocking reagents indicated in the figures, using a one-sided Mann-Whitney U test. * p=0.05.
We incubated pHrodo red-labeled MT4CCR5/NL4-3 cells with ΔE2 MFG-E8, D89E, or control soluble CD4 (sCD4), followed by co-culture with macrophages and human serum. ΔE2 MFG-E8 and D89E inhibited serum-induced engulfment, while sCD4 did not significantly inhibit phagocytosis (Fig. 3B), demonstrating that phagocytosis of HIV-1-infected cells is mediated by exposed PtdSer.
Because sCD4 did not inhibit phagocytosis of the infected cells, it is unlikely that Env plays a significant role in phagocytosis of infected cells. To investigate whether Env is necessary for human serum-induced phagocytosis, we used Env-deleted HIV-1 (Pang et al., 2000) pseudotyped with VSV-G [NLΔBgl (VSV-G)] to infect MT4CCR5 cells, which resulted in Gag expression and PtdSer exposure without expression of Env (Fig. 3C). Phagocytosis of pHrodo Red-labeled MT4CCR5/NLΔBgl (VSV-G) was enhanced by human serum, indicating that Env is not required for human serum-induced phagocytosis (Fig. 3D).
We next investigated whether Mer is involved in this phagocytosis mechanism. We incubated macrophages with either anti-Mer antibody or control anti-Axl antibody before addition of human serum. Anti-Mer inhibited serum induced phagocytosis of MT4CCR5/NL4-3 to the level of phagocytosis activity occurring without serum (Fig. 3E), demonstrating that Mer plays a role in human serum-induced phagocytosis of HIV-1-infected cells.
2.5. Protein S and Gas6 facilitate phagocytosis of MT4CCR5/NL4-3 by macrophages
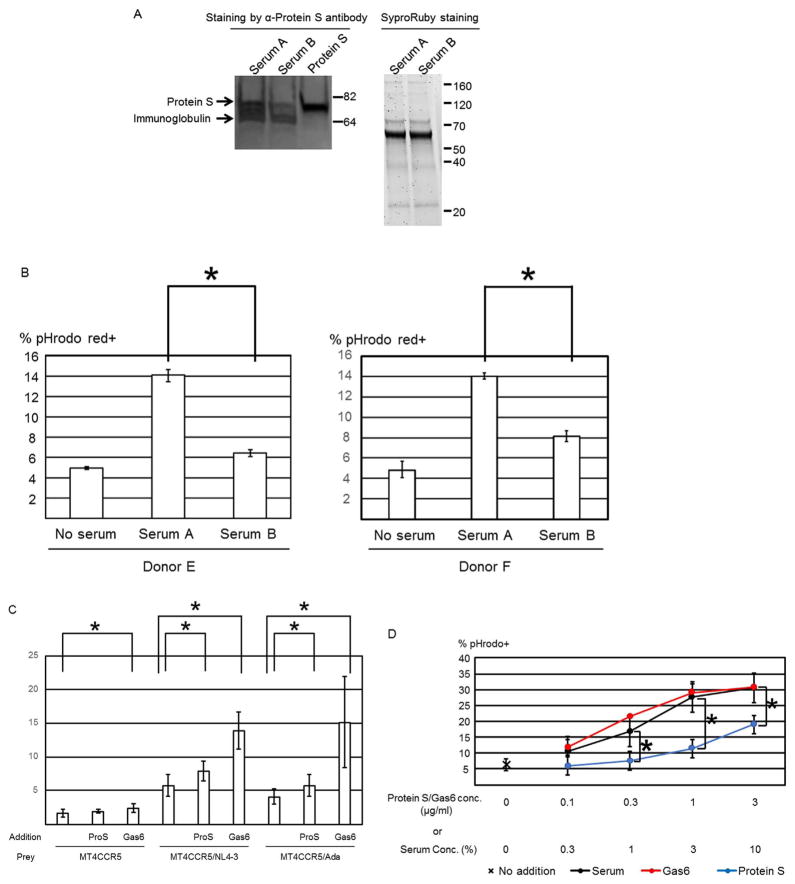
We next investigated whether protein S in human serum mediates phagocytosis of MT4CCR5/NL4-3. We depleted protein S from serum using anti-protein S antibody-conjugated microbeads (Serum B). We also treated serum with microbeads conjugated with an isotype control antibody (Serum A). Anti-protein S antibody, but not the control, depleted approximately 70% of protein S in serum (Fig. 4A). Depletion of protein S inhibited human serum-mediated induction of efferocytosis of MT4CCR5/NL4-3 cells (Fig. 4B), demonstrating that protein S mediates human serum-induced efferocytosis of infected cells.
Fig. 4.
Protein S and Gas6 mediate phagocytosis of HIV-1-infected MT4CCR5 cells. (A) Human serum was incubated with microbeads conjugated with isotype control antibody (serum A) or anti-protein S monoclonal antibody (serum B). After removal of the microbeads, 15 μl of sera A and B (1.5%) and recombinant protein S (1 μg/ml) were subjected to SDS-PAGE and western blotting analysis, using anti-protein S antibody. As the controls for the amounts of protein subjected, the entire proteins in the samples were analyzed by SDS-PAGE and SyproRuby staining. (B) Macrophages and pHrodo Red-labeled MT4CCR5/NL4-3 cells were co-cultured in the presence or absence of 3% serum A or B. The cells were then stained with APC-conjugated anti-CD14 antibody, and pHrodo Red signals in the CD14-positive population were analyzed by flow cytometry. This experiment was repeated twice in triplicate, using cells from two donors as independent experiments. Donors of cells differed in each independent experiment. The results shown are averages and standard deviations. Significance of these triplicated experiments was calculated by a one-sided Mann-Whitney U test. * p=0.05. (C) Macrophages and pHrodo Red-labeled MT4CCR5, MT4CCR5/NL4-3, or MT4CCR5/Ada cells were co-cultured in the presence or absence of recombinant protein S or Gas6 (1 μg/ml). Phagocytosis was analyzed by flow cytometry. This experiment was repeated twice in singlicate and three times in triplicate, using cells from different donors for each experiment, and the results shown are averages and standard deviations of the all triplicate experiments. Significance was calculated by comparing the value of the samples with no additional protein to the value of the samples with protein S or Gas6, using a one-sided Mann-Whitney U test. *p=0.05. (D) Macrophages and pHrodo Red-labeled MT4CCR5/NL4-3 cells were co-cultured in the presence or absence of various concentrations of human serum, recombinant protein S, or Gas6. The cells were then stained with APC-conjugated anti-CD14 antibody, followed by flow cytometry analysis. This experiment was repeated three times as independent experiments, using cells from different donors for each independent experiment. Significance of these triplicate experiments was calculated by a one-sided Mann-Whitney U test. * p=0.05.
We next investigated whether purified recombinant protein S and its homologue, Gas6, can induce phagocytosis of infected cells. Addition of protein S and Gas6 enhanced engulfment of MT4CCR5/NL4-3 cells by macrophages from 5.8% to 7.9% and 13.9%, respectively (Fig. 4C). Gas6 also enhanced phagocytosis of uninfected MT4CCR5 cells from 1.7% to 2.4%, which was likely due to phagocytosis of small PtdSer-exposing populations present in uninfected cells.
We compared the abilities of Gas6, protein S, and serum to induce phagocytosis at various concentrations. The serum concentration of protein S was approximately 30 μg/ml (Hafizi and Dahlback, 2006; Anderson et al., 2003). When comparing recombinant protein S and serum at the concentration containing equivalent protein S (i.e., 1% human AB serum vs 0.3 μg/ml), recombinant protein S induced phagocytosis less efficiently than serum (Fig. 4D). We compared other lots of human serum and consistently observed that recombinant protein S induced phagocytosis less efficiently than serum (data not shown).
In the human body, Gas6 can be produced by various cell types, including kidney cells (Nagai et al., 2003) which are mimicked in the HEK-293T kidney fibroblast cell line used to produce recombinant Gas6, but protein S is mainly produced in the liver (Fernandez-Fernandez et al., 2008; Hafizi and Dahlback, 2006). Since recombinant protein S may lack liver-specific post-translational modification necessary for its function, including γ-carboxylation (McCann and Ames, 2009) and oxidization (Uehara and Shacter, 2008), we used recombinant Gas6 and its mutants for further investigation of the molecular mechanisms of Gas6/protein S-mediated phagocytosis of HIV-1- infected cells and its effects on virus production and macrophage infection.
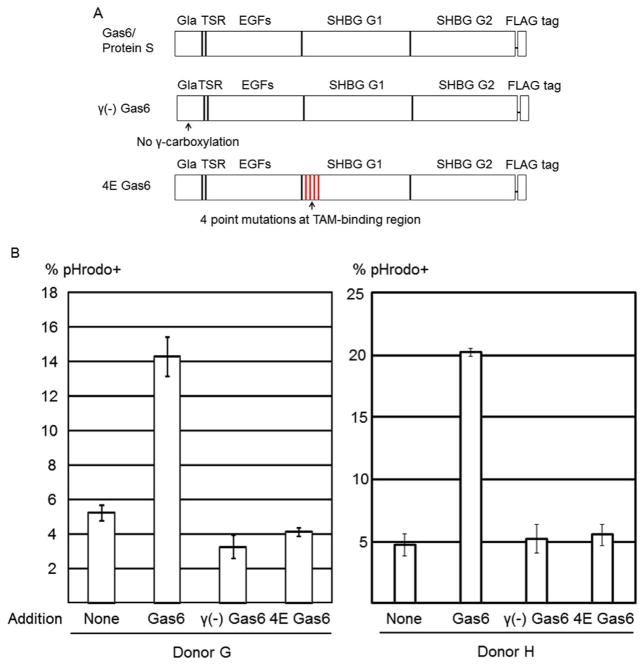
To confirm the roles of PtdSer and Mer in efferocytosis induction by Gas6, we used two types of mutant Gas6 recombinant proteins that lack divalent binding activity. The Gla domain of the Gas6/protein S domain undergoes post-translational modification of glutamic acid residues in a vitamin K-dependent manner (γ-carboxylation), which is necessary for binding to PtdSer (Morizono et al., 2011) (Fig. 5A). We produced recombinant Gas6 in the absence of vitamin K in the culture medium, designated as γ(−) Gas6, which cannot bind PtdSer due to the lack of γ-carboxylation. We also generated a Gas6 variant that has four TAM receptor-binding amino acids mutated, designated as 4E Gas6. A previous study by other research groups showed that these mutations abrogate binding of Gas6 to TAM receptors (Sasaki et al., 2006). Neither of these two mutant proteins could induce efferocytosis of HIV-1-infected cells (Fig. 5B), demonstrating that binding to both Mer and PtdSer is required for Gas6-induced efferocytosis of infected cells.
Fig. 5.
Efferocytosis by Gas6 mutants. (A) Schematic representation of Gas6, γ(−) Gas6 (a mutant of Gas that cannot bind PtdSer), and 4E Gas6 (a mutant Gas6 that cannot bind TAM receptors). Gla, γ-carboxyglutamic acid domain; SHBG, sex hormone-binding globulin domain. The Gla domain of Gas6 binds PtdSer. The SHBG G1 domain of Gas6 binds TAM receptors. (B) Macrophages and pHrodo Red-labeled MT4CCR5/NL4-3 cells were co-cultured in the presence or absence of various concentrations of Gas6, γ(−) Gas6, or 4E Gas6. Phagocytosis was analyzed by flow cytometry. This experiment was repeated once in singlicate and twice in triplicate, using cells from different donors for each experiment.
Because efferocytosis of infected cells is PtdSer-dependent, it should occur regardless of the tropism of HIV-1, as long as the virus induces PtdSer exposure on infected cells. We next investigated whether recombinant protein S and Gas6 could induce phagocytosis of MT4CCR5 infected with the R5- and macrophage-tropic HIV-1 strain, HIV-1Ada (MT4CCR5/Ada). HIV-1Ada infection induced PtdSer exposure (Fig. 3C), and protein S and Gas6 induced efferocytosis of MT4CCR5/Ada although induction by protein S was less efficient than human serum and Gas6 (Fig. 4C). These results confirmed that protein S and Gas6 mediate phagocytosis of infected MT4CCR5 regardless of HIV-1 tropism.
We considered protein S/Gas6-induced phagocytosis of HIV-1-infected cells as efferocytosis because of the indispensable role of exposed PtdSer.
2.6. Efferocytosis of live populations of infected cells
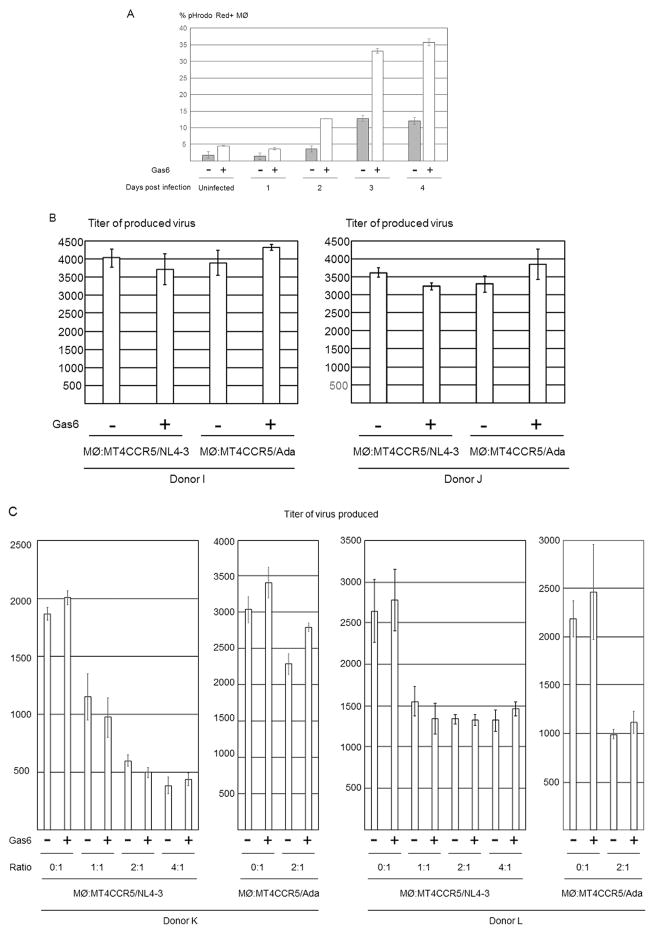
We investigated whether Gas6 can mediate efferocytosis of live HIV-1-infected cells, which can vigorously produce infectious virus. Fig. 1C shows that infectious virus is produced abundantly at 2 days post-infection, and production ends at 3 days-post infection. Since the cells at 2 days-post infection do not expose PtdSer at high levels, we investigated whether cells at 2 days post-infection can be engulfed. Consistent with PtdSer exposure levels and viability, both Gas6-dependent and -independent engulfment of MT4CCR5/NL4-3 occurred at two days post-infection, albeit at lower levels compared to three and four days post-infection (Fig. 6A).
Fig. 6.
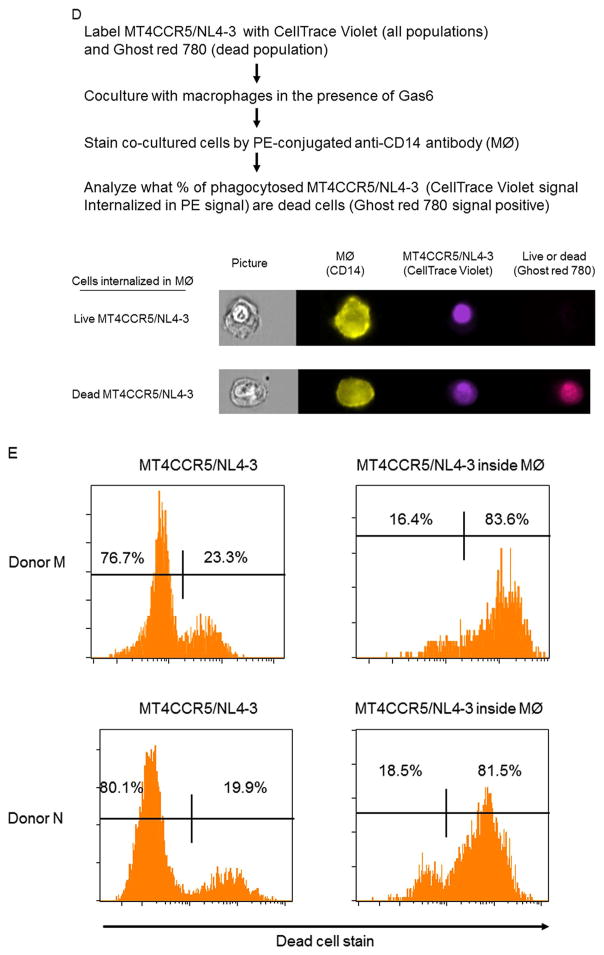
The effects of Gas6-mediated efferocytosis on viral production from infected MT4CCR5 cells. (A) pHrodo Red-labeled MT4CCR5 and MT4CCR5/NL4-3 (2×105 cells) 1, 2, 3, and 4 days post-infection were co-cultured with macrophages (2×105) for 90 min in the absence and presence of Gas6 (1 μg/ml). The cells stained by APC-conjugated anti-CD14 antibody and pHrodo Red signals in the CD14-positive population were analyzed by flow cytometry. This experiment was repeated twice in triplicate, using cells from different donors for each experiment, and the results shown are averages and standard deviations of one representative triplicate experiment. (B) MT4CCR5/NL4-3 and MT4CCR5/Ada (1×105 cells) were co-cultured with the same numbers of macrophages. The supernatants of the co-cultured cells were harvested 15 h later, and the titers of virus in the supernatants were titrated by GHOST (3) CXCR4+CCR5+ cells. The results shown are averages and standard deviations of two triplicate experiments, using macrophages from different donors. (C) MT4CCR5/NL4-3 and MT4CCR5/Ada (1×105 cells) were co-cultured with the different cell numbers of macrophages. The supernatants of the co-cultured cells were harvested 15 h later, and the titers of virus in the supernatants were titrated by GHOST (3) CXCR4+CCR5+ cells. The results shown are averages and standard deviations of two triplicate experiments, using macrophages from donors K and L. (D) MT4CCR5/NL4-3 cells were labeled with CellTrace Violet and Ghost Red 780 2 days post-infection. The labeled cells (5×105) were co-cultured with macrophages (5×105) for 90 min in the presence of Gas6 (1 μg/ml). The cells were then stained with PE-conjugated anti-CD14 antibody and subjected to imaging flow cytometry. The representative images of live and dead MT4CCR5/NL4-3 cells internalized in macrophages are shown. (E) The results of Ghost Red 780 staining of the total population and the population internalized in macrophages of MT4CCR5/NL4-3 are shown as histograms. This experiment was repeated twice using cells from different donors.
We next investigated whether Gas6 can inhibit virus production by inducing phagocytosis of MT4CCR5/NL4-3 cells. We co-cultured MT4CCR5/NL4-3 cells at two days post-infection with macrophages in the absence or presence of Gas6. The supernatants of the co-cultured cells were harvested 15 h after initiation of co-culture, and the virus titers of the supernatants were quantitated. Gas6 did not significantly decrease the viral titers produced in the supernatant (Fig. 6B). Inhibitory effects of Gas6 on virus production were not observed when increasing the numbers of macrophages co-cultured with MT4CCR5/NL4-3 (Fig. 6C). Gas6 had no inhibitory effects on virus production from MT4CCR5/Ada cells co-cultured with macrophages (Fig. 6B and C). These results indicated that the majority of engulfed cells detected in the experiment shown in Fig. 6A is comprised of dead cells that cannot produce infectious virus.
Since the experimental setting of Fig. 6B and C measures virus production from entire cultures, it was difficult to detect statistically significant reductions in virus production after engulfment of a relatively small population(s) of live infected cells. Thus, we investigated whether live populations of MT4CCR5/NL4-3 cells can be efferocytosed by directly visualizing viability of cells engulfed in macrophages. We labeled total populations of MT4CCR5/NL4-3 cells with CellTrace Violet and the dead populations with Ghost Red 780. We then co-cultured labeled cells with macrophages, followed by staining with PE-conjugated anti-CD14 antibody (Fig. 6D). Imaging flow cytometry showed that ~17% of MT4CCR5/NL4-3 cells internalized into macrophages were Ghost Red 780-negative (Fig. 6E). These results demonstrate that while Gas6 preferentially induces engulfment of dead populations of MT4CCR5/NL4-3, live populations of infected cells can also be engulfed, albeit less efficiently than dead cells.
3. Discussion
Our study showed that protein S/Gas6 can mediate efferocytosis of HIV-1-infected cells by bridging Mer, expressed on macrophages, and PtdSer, exposed on infected cells. We also observed basal levels of phagocytosis of HIV-1-infected cells without addition of protein S/Gas6, and this is not blocked by MFG-E8 mutants. Because MFG-E8 mutants can universally block binding of PtdSer to PtdSer-recognizing molecules (Morizono and Chen, 2014), including TIM-1, TIM-4, MFG-E8, protein S, and Gas6, it is likely that basal level phagocytosis is PtdSer-independent. In addition to PtdSer, several types of molecules, including calreticulin (Ogden et al., 2001) and DD1α (Yoon et al., 2015), are known to be expressed on apoptotic cells and to stimulate phagocytosis. These molecules might also be involved in efferocytosis of HIV-1-infected cells in the absence of protein S and Gas6.
Our recombinant protein S does not induce efferocytosis as efficiently as human serum. One possible explanation of the relatively low activity of recombinant protein S could be the differences in posttranslational modification, as described in the Results section. Another possibility is that efferocytosis by protein S requires a molecule(s) in serum that supports binding between macrophages and apoptotic cells, since the affinity of protein S for TAM receptors is not as high as that of Gas6 (Hafizi and Dahlback, 2006). Although the normal human serum concentrations of Gas6 are approximately 1000-fold less than protein S (20–30 ng/ml) (Uehara et al., 2009), protein S would need the synergistic effects of Gas6 for efficient efferocytosis. Recent studies showed that normal human serum contains poly-active antibodies that bind apoptotic but not healthy cells and mediate phagocytosis of apoptotic HIV-1-infected cells (Zhou et al., 2013). Protein S might need the support of such molecules in serum to efficiently induce efferocytosis.
Phagocytosis of simian immunodeficiency virus (SIV)-infected cells was observed in vivo in experimental infection of monkeys (Calantone et al., 2014). Based on mathematical analysis, this study suggested that SIV DNA detected in the macrophage population is not the result of de novo infection of macrophages, but instead from engulfment of SIV-infected T-cells. Several research groups have reported phagocytosis of HIV-1-infected cells by macrophages in vivo (Sips et al., 2016; Orenstein, 2000; Akbar et al., 1994). Some of this phagocytosis is likely mediated by efferocytosis (Akbar et al., 1994). Because Mer has been shown to be expressed on human monocytes and tissue macrophages, including alveolar macrophages, macrophages in the liver and in mesenteric lymph nodes, and microglia (Bernsmeier et al., 2015; Zhu et al., 2014; Hodge et al., 2017; Kazeros et al., 2008; Healy et al., 2016), it would be of interest to investigate whether PtdSer/protein S/Mer mediates efferocytosis of SIV and/or HIV-1-infected cells and contributes to the amounts of viral DNA and proteins detected in macrophages in vivo.
It is known that one HIV-1 accessory protein, Nef, inhibits efferocytosis (Torre et al., 2002). Because both endogenously expressed Nef and exogenously added soluble Nef can inhibit efferocytosis, it is possible that HIV-1 can systemically inhibit efferocytosis by both infected and uninfected macrophages. HIV-1 infection is known to induce autoimmune diseases (Huang et al., 2016). It is possible that accumulation of dead cells by HIV-1 replication and Nef-mediated inhibition of efferocytosis play a role in development of autoimmunity against self-antigens.
Our results indicate that macrophages can efferocytose live infected cells (approximately 17% in the total engulfed cells) over 12 h albeit the level of efferocytosis is less efficient than dead cells. Since our experimental setting measures virus production from entire cultures, it was difficult to detect statistically significant reductions in virus production in supernatant after engulfment of a relatively small population (s) of live infected cells. It is likely that efferocytosis inhibits virus production from live infected cells at the single cell level. In addition, the cumulative effects of efferocytosis on removal of infected cells over time could be more significant over long-term in vivo settings.
Hermetet et al. reported that Macrophages engulf the late but not the early apoptotic populations of papilloma virus-infected cells (Hermetet et al., 2016). In this study, the levels of PtdSer exposure were almost the same in both the early and late apoptotic populations, suggesting involvement of other types of molecules associated with cell death in efferocytosis.
It is known that expression of an additional molecule(s) associated with cell death and/or down-regulation of the “don’t eat-me signal” is necessary for efferocytosis (Oldenborg et al., 2000; Majeti et al., 2009; Jaiswal et al., 2009), in addition to PtdSer exposure. Gas6/protein S might need to synergize with additional signals for efficient phagocytosis of live HIV-1-infected cells. Because broadly neutralizing antibodies against HIV-1 can provide signals for phagocytosis via the Fcreceptor (Musich et al., 2017), it would be of interest to determine whether the broadly neutralizing antibodies can synergize with Gas6/protein S to clear infected cells by phagocytosis/efferocytosis.
In previous studies of influenza virus, efferocytosis could clear cells actively producing virus, which inhibits virus replication. Efferocytosis of influenza virus is stimulated not only by PtdSer exposed on infected cells, but also by desialylation by influenza virus neuraminidase of cell surface proteins of infected cells and macrophages (Watanabe et al., 2002, 2004). The additional stimulation signal by neuraminidase might enable macrophages to engulf the live infected cells actively producing progeny viruses.
The release of many types of enveloped viruses need intact cell membranes of live cells for budding; thus, efferocytosis of dead cells does not efficiently reduce virus production of the envelope viruses. However, many types of non-enveloped viruses require cell death (lysis) before their egress (Fields et al., 2013). Thus, efferocytosis of dead or late apoptotic virus-infected cells will be able to inhibit production of such non-enveloped viruses. Indeed, efferocytosis has been shown to inhibit replication of a non-enveloped virus, Drosophila C, both in vitro and in vivo (Nainu et al., 2015; Nonaka et al., 2017).
Cells transduced with adenovirus vectors, which expose PtdSer due to the toxicity of viral transduction, were recently shown to be removed by efferocytosis by the interactions between PtdSer, protein S/Gas6, and TAM receptors, resulting in decreased transgene expression of adenoviral vectors (Tufail et al., 2017). Because adenoviruses require cell death for egress (Fields et al., 2013), it is of interest to determine whether protein S/Gas6-mediated efferocytosis can inhibit release of replication-competent adenovirus from infected cells.
The molecular mechanisms of efferocytosis were previously utilized for enhancement of antibody-mediated removal of viral particles of HIV-1. One research group conjugated anti-gp120 monoclonal antibodies with PtdSer and investigated whether macrophages can eliminate HIV-1 by engulfing virus bound to the anti-gp120 antibody-PtdSer conjugate (Gramatica et al., 2014). Their data showed that anti-gp120 antibody-PtdSer conjugate mediates phagocytic removal of HIV-1 more efficiently than anti-gp120 antibody, suggesting that efferocytosis can induce engulfment of viruses more efficiently than Fc receptor-mediated phagocytosis. It would be of interest to see whether fusion proteins between antibodies and the TAM-binding regions (SHBG) of Gas6 can induce efficient phagocytosis of virus and virus-infected cells.
In addition to removal of virus-producing cells, efferocytosis is known to play a role in induction of acquired immunity against viruses. Macrophages can present virus-derived peptides on MHC class I molecules and stimulate T-cells expressing virus-specific T-cell receptors after efferocytosis of LCMV-infected cells (Alatery et al., 2010). Thus, it is possible that enhancement of efferocytosis can inhibit replication of both enveloped and non-enveloped viruses by eliciting acquired immunity against viral antigens.
Pulmonary delivery of Bacillus Calmette-Guerinn (BCG) was previously shown to protect mice from influenza pneumonia by enhancing efferocytosis (Mukherjee et al., 2017). BCG enhances efferocytosis by increasing expression of the PtdSer receptor, TIM-4, on alveolar macrophages. Because human alveolar macrophages are shown to express Mer, administration of Gas6/protein S would also be able to enhance efferocytosis and help protect a host from viral infection by clearing infected cells, suppressing inflammatory responses by clearing apoptotic cells, and facilitating antigen presentation of viral proteins.
Further studies of the roles of efferocytosis in virus infection will lead to a better understanding of the pathogenesis of viruses and establish basic information for developing novel therapeutic approaches.
4. Materials and methods
4.1. Plasmids
A Flag-tagged Gas6 and protein S expression vectors were constructed by inserting Flag-tagged Gas 6 and protein S with synthetic gene fragment (Integrated DNA Technologies, Coralville, IA) into the 3′-end of Gas6 cDNA of the Gas6 expression vector obtained from OriGene (Rockville, MD). The expression vector of 4E Gas6 was generated by introducing 4 mutations into the TAM receptor-binding region of Gas6 (R299E, R308E, K310E, and R312E) by site-directed mutagenesis of Flag-tagged Gas6 expression vector. Expression vectors of MFG-E8 and its D89E mutant were provided by Dr. Shigekazu Nagata (Osaka University, Japan) (Hanayama et al., 2002). Expression vector ΔE2 MFG-E8 was constructed by deleting the E2 domain of a mouse MFG-E8 expression vector.
4.2. Proteins
Protein S, Gas6, and 4E Gas6 were produced by transfecting 293T cells with expression vectors of protein S, Gas6, and 4E Gas6. The cells were cultured in IMDM supplemented with 10% Fetal Bovine Serum (FBS), antibiotics, and 10 μg/ml vitamin K1 (Sigma-Aldrich, St. Louis, MO). γ(−) Gas6 was produced by transfecting 293T cells with expression vectors of Gas6. The cells were cultured in IMDM supplemented with 10% FBS, antibiotics, and 1 μM warfarin (UCLA Pharmacy, Los Angeles, CA). D89E and ΔE2 MFG-E8 were produced by transfecting 293T cells with expression vectors of ΔE2 MFG-E8 and culturing cells in IMDM supplemented with 10% FBS and antibiotics. The culture supernatants of transfected cells were harvested 5 days post-transfection, filtered, and incubated overnight at 4 °C with anti-Flag antibody-conjugated microbeads. The microbeads were collected by centrifugation, and proteins were eluted using the FLAG M Purification Kit (Sigma-Aldrich) according to the manufacturer’s protocol. The fractions containing target proteins were consolidated and dialyzed into Hanks balanced buffer (Life Technologies, Carlsbad, CA). The purity of the proteins was confirmed by SDS-PAGE and SyproRuby staining (Life Technologies), and the amounts of proteins were measured by OD280.
4.3. Serum
Four different lots of human AB serum were obtained from Sigma-Aldrich and MP Biomedicals (Burlingame, CA). After confirming that all four lots could induce efferocytosis of HIV-1-infected cells, we used the same one lot for all the experiments. FBS was purchased from Sigma-Aldrich. FBS used in this study was subjected to BaCl2 precipitation to remove bovine protein S (Bhattacharyya et al., 2013). Briefly, FBS was incubated with saturating amounts of BaCl2 at 4 °C for one hour, followed by ultracentrifugation to pellet down the precipitate. Supernatant of ultra-centrifuged serum was dialyzed in 0.15 M NaCl and filtered.
Anti-human protein S monoclonal antibody (clone PS7) (Santa Cruz Biotechnology, Dallas, TX) and its isotype control antibody (rat IgG2a) (Biolegend, San Diego, CA) were used for experiments to analyze the function of protein S in human AB serum by depletion of protein S. Anti-human protein S and isotype control antibodies were individually conjugated to protein A/G microbeads (Thermo Fisher Scientific, Canoga Park, CA) by cross-linking the antibodies and microbeads according the manufacturer’s protocol. Human AB serum was incubated overnight at 4 °C with microbeads conjugated with anti-human protein S or isotype control antibody. The serum was then centrifuged and filtered to remove the microbeads. Depletion of protein S was confirmed by SDS-PAGE and western blotting, using goat anti-human protein S antibody (R&D Systems, Minneapolis, MN). Recombinant protein S was used as a positive control for western blotting analysis. The amounts of protein S were analyzed by densitometry, using GelQuant NET software (BiochemLab Solutions, CA). The amounts of the proteins in the samples were confirmed by SDS-PAGE, SyproRuby staining, and imaging and analyses were carried out using an FX imager (BioRad, Hercules, CA).
4.4. Cells and viruses
MT4CCR5 cells were generated by transducing a CD4+ T cell line with lentiviral vector expressing human CCR5 and blasticidin-resistant genes under the control of the SFFV promoter and IRES. The stably transduced cells were selected using 10 μg/ml Blasticidin (InvivoGen, San Diego, CA). MT4CCR5 cells were cultured in X-VIVO 15 medium (Lonza, Walkersville, MD) supplemented with 5% BaCl2-precipitated FBS and antibiotics before efferocytosis assays.
GHOST (3) CXCR4+CCR5+ cells were obtained from the NIH AIDS Reagent program and cultured according to the instructions provided by NIH AIDS reagent program (Morner et al., 1999).
Flasks and plates used for monocyte and macrophage cultures were coated with rat collagen type I (TREVIGEN, Gaithersburg, MD) before use. Human peripheral blood mononuclear cells (PBMC) were obtained from the UCLA CFAR Virology Core. Monocytes were isolated from PBMC using the Pan Monocyte Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s protocol. Monocytes were differentiated to macrophages by culturing the cells in X-VIVO 15 medium supplemented with 5% BaCl2-precipitated FBS, 30 μg/ml human recombinant M-CSF (Shenandoah Biotechnology, Warwick, PA), and 1 nM Dexamethasone (Selectchem.com, Houston, TX) for 7 days.
HIV-1NL4-3 was prepared from supernatant of MT4CCR5 cells infected with HIV-1NL4-3 that were produced by transfection of 293T cells with pNL4-3. HIV-1Ada was prepared from supernatant of MT4CCR5 cells that were infected with HIV-1Ada obtained from the NIH AIDS Reagent program. The virus supernatant was filtered and frozen at −80 °C. Heat inactivation of HIV-1NL4-3 was done by incubating the virus at 56 °C for 2 h. Titer of the virus was analyzed by infecting MT4CCR5 cells. The infected cells were cultured for 2 days in the presence of 10 μg/ml T20 (NIH AIDS Reagent program), then fixed with fixation buffer (Life Technologies) and stained with FITC-conjugated anti-HIV p24 antibody (Beckman Coulter, Brea, CA) in permeabilization buffer (Life Technologies). Gag expression levels were analyzed by flow cytometry, and the titer of virus was calculated by percentages of cells expressing Gag, the number of infected cells, and dilution of virus that yielded 5–20% of infection.
Env-deleted HIV-1 pseudotyped with VSV-G [NLΔBgl (VSV-G)] was generated by transfecting 293T cells with an expression vector of VSV-G and a molecular clone of NL4-3 lacking the Env region (NL4-3 ΔBglII) (Pang et al., 2000).
4.5. Flow cytometry analysis of expression of cell surface molecules on macrophages
PtdSer receptor expression was analyzed by staining with PE-conjugated anti-TIM-1 (Biolegend), TIM-4 (Biolegend), Axl (R&D Systems), TYRO3 (R&D Systems), or Mer (R&D Systems). APC-conjugated IgG2a (BD Bioscience, Flanklin Lakes, NJ), PE-conjugated IgG1 (R&D Systems) and IgG2a (BD Bioscience) were used as isotype controls when appropriate. An LSRFortessa flow cytometer (BD Bioscience) was used for conventional flow cytometry, and FCS Express Version 5 (De Novo Software, Glendale, CA) was used for data analysis.
4.6. Analysis of Env and Gag expression, PtdSer exposure, cell death, and virus production
For analysis of Env expression, the anti-HIV gp120 monoclonal antibody, 2G12 (NIH AIDS Reagent program) (Buchacher et al., 1994; Trkola et al., 1996), was conjugated with Alexa 647 (Life Technologies) according to the manufacturer’s protocol. MT4CCR5 cells were incubated with virus at MOI 5 for 2 h, washed 3 times with medium after infection, and harvested on the day of analysis. For analysis of Env expression, the cells were stained with Alexa 647-conjugated 2G12 and fixed with 2% paraformaldehyde. For analysis of Gag expression, the cells were fixed with fix buffer and stained with FITC-conjugated anti-HIV p24 antibody in permeabilization buffer. For analysis of PtdSer exposure, the cells were stained with APC-conjugated ANX V (eBiosicence, San Diego, CA) in ANX V staining buffer (140 mM NaCl, 4 mM KCl, 0.75 mM MgCl2, 1.5 mM CaCl2, and 10 mM HEPES), then resuspended and fixed in ANX V staining buffer containing 2% paraformaldehyde. Cell death was analyzed by Ghost Red Dye 780 or Ghost Violet Dye 450 (Tonbo Biosciences, San Diego, CA) according to the manufacturer’s protocol.
The supernatant of infected cells was harvested every 24 h after infection, filtered, and analyzed for virus production. HIV p24 production was analyzed with ELISA by the UCLA CFAR Virology Core. The titers of harvested virus were measured by GHOST (3) CXCR4+CCR5+ assay according to the protocol provided by the NIH AIDS Reagent program.
4.7. Imaging flow cytometry
An ImageStream MkII (EMD Millipore, Billerica, MA) at the UCLA JCCC/CFAR Flow Cytometry Core was used for imaging flow cytometry. It is equipped with four lasers (405, 488, 658, and 758 nm), one CCD camera, and three objective lenses (X20, 40, and 60). Data was acquired and analyzed using INSPIRE and IDEAS software, respectively. All data acquisition was done using a 40-fold objective lens and slow flow speed. For all imaging flow cytometric analyses, each event was first analyzed by gradient RMS values of bright field images. The events with more than 60 gradient RMS were further analyzed as focused images. The images with multiple events and images of debris were excluded by analyzing sizes and aspect ratios (height vs width) of the image.
4.8. Efferocytosis assay
MT4CCR5 cells were infected with NLΔBgl (VSV-G), HIV-1NL4-3, or HIV-1Ada 3 days prior to co-culture with macrophages at MOI 5. Macrophages were washed three times with MACS buffer (Miltenyi Biotec) and seeded in collagen-coated plates one day before co-culture. On the day of co-culture, we confirmed that infected MT4CCR5 cells expressed HIV-1 Gag and exposed PtdSer using flow cytometry. For flow cytometric analysis of efferocytosis of MT4CCR5 cells, infected and uninfected MT4CCR5 cells were labeled with pHrodo Red according to the manufacturer’s protocol. The labeled cells (2×105) were co-cultured with macrophages (2×105) in 24-well plates in the presence or absence of human AB serum, human AB serum treated with anti-protein S antibody- or isotype control antibody-conjugated microbeads, protein S, Gas6, γ(−) Gas6, or 4E Gas6 for 90 min at 37 °C. Co-cultured cells were harvested, stained with APC-conjugated anti-CD14 antibody (eBioscience), and resuspended in pH 9 flow cytometry buffer containing 2% paraformaldehyde. CD14 expression was used to gate macrophages, and pHrodo Red signals in CD14-positive populations were analyzed to assess efferocytosis of MT4CCR5 cells by macrophages. The labeled MT4CCR5 cells were also resuspended in pH 5.5 flow cytometry buffer and used as a positive control of fluorescent signal induction in a low-pH environment.
For imaging flow cytometry analysis of phagocytosis of MT4CCR5 cells, MT4CCR5 and MT4CCR5/NL4-3 cells were labeled with CellTrace Violet (Life Technologies) according to the manufacturer’s protocol. The labeled cells (5×105) were co-cultured with macrophages (5×105) in 12-well plates in the presence or absence of human AB serum for 90 min at 37 °C. Co-cultured cells were harvested and stained with APC-conjugated anti-CD14 antibody. The data of 104 cells/sample were acquired by Imagestream MKII, and first analyzed to calculate percentages of CD14+ macrophages that bound to CellTrace Violet-positive cells. Those bound cells were then analyzed for internalization into CD14+ macrophages, using the IDEAS wizard that calculates internalization scores. Percentages of macrophages containing internalized CellTrace Violet-positive cells were calculated using an internalization score of 1.5 as a threshold to identify internalized CellTrace Violet-positive cells.
To investigate the role of Mer in efferocytosis, macrophages were incubated with goat anti-Mer or Axl antibodies (R&D Systems) 30 min before co-culture with MT4CCR5/NL4-3. To investigate the role of PtdSer, MT4CCR5/NL4-3 cells were incubated with ΔE2 MFG-E8, D89E, or soluble CD4 (NIH AIDS Reagent program) 30 min before co-culture with macrophages. Inhibitory or control antibodies and proteins were present during the co-culture period.
For phagocytosis analysis by imaging flow cytometry, co-culture experiments were done in the presence of T20 (10 μg/ml) to avoid internalization of Gag into macrophage by Env-dependent fusion.
4.9. Analysis of phagocytosis by confocal microscopy
Macrophages were labeled with CellTrace Far Red (Life Technologies) according to the manufacturer’s protocol, then seeded in collagen-coated 24-well plates (1×105/well) one day prior to co-culture with MT4CCR5/NL4-3. On the day of co-culture, MT4CCR5/NL4-3 cells were labeled with CFSE (Biolegend) according to the manufacturer’s protocol. The labeled MT4CCR5/NL4-3 cells (1×105/well) were cultured with macrophages in the presence or absence of 3% human serum for 90 min. Co-cultured cells were harvested and fixed with 3.2% paraformaldehyde. The thin layer of cells on slides were generated by Cytospin 4 (Thermo Fisher Scientific). The nuclei were stained by ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific). Image slices were obtained with a LSM 780 confocal microscope (Carl Zeiss, Jena, Germany), and the images were analyzed by Zen software (Carl Zeiss). Image slices were analyzed for numbers of entire macrophages and macrophages engulfing MT4CCR5/NL4-3. Several engulfment events were analyzed by 3D imaging with Z-stack projection to further confirm internalization of MT4CCR5/NL4-3 cells into macrophages.
4.10. Quantitation of virus production from infected MT4CCR5 cells co-cultured with macrophages
pHrodo Red-labeled MT4CCR5 and MT4CCR5/NL4-3 cells 1, 2, 3, and 4 days post-infection were cultured with macrophages in the absence and presence of Gas6 (1 μg/ml) for 90 min. Phagocytosis was analyzed by flow cytometry as previously described. MT4CCR5/NL4-3 and MT4CCR5/Ada cells (1×105 cells) at 2 days post-infection were co-cultured with various numbers of macrophages (0 – 4×105 cells) in the absence and presence of Gas6 (1 μg/ml) for 15 h. The supernatants of the cells were filtered and subjected to titration of HIV-1 using GHOST (3) CXCR4+CCR5+ cells. To investigate viability of cells engulfed in macrophages, MT4CCR5/NL4-3 cells at 2 days post-infection were labeled by CellTrace Violet and Ghost Red Dye 780. The labeled cells were co-cultured with macrophages for 90 min. The cells were stained by PE-conjugated anti-CD14 antibody (BD Biosciences), followed by analysis by imaging flow cytometry. The internalization of CellTrace Violet signal into CD14-positive masks were first analyzed. The cells above internalization score 1.5 were analyzed for the signals of Ghost Red Dye 780 staining.
Acknowledgments
We thank NIH AIDS Reagent Program for providing GHOST (3) CXCR4+CCR5+ Cells (deposited by Drs. Vineet N. KewalRamani and Dan R. Littman), 2G12, soluble CD4, and T20. We thank Drs. Irvin Chen, Airi, Harui, Takeshi Miyamoto, Matthew Marsden, Olivier Pernet, and Gene-Errol Ringpis for discussion, and Ms. Wendy Aft for proof-reading of the manuscript. We thank the UCLA CFAR Virology Core Laboratory (supported by NIH 5P30AI28697 grant) for the p24 ELISA and UCLA JCCC flow cytometry core (supported by NIH P30 CA016042 and 5P30 AI02869 grants) for imaging flow cytometry, and UCLA CFAR Gene and Cell Therapy core (supported by NIH AI028697) for autoMACS usage.
This work was supported by the UCLA AIDS Institute and the UCLA Center for AIDS Research, and U.S. National Institute of Health grants R21AI095004 (K.M) and R01AI108400 (K. M.).
We have no financial conflicts of interest to declare.
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2017.12.025.
Footnotes
Declarations of interest
None.
References
- Akbar AN, Savill J, Gombert W, Bofill M, Borthwick NJ, Whitelaw F, Grundy J, Janossy G, Salmon M. The specific recognition by macrophages of CD8+,CD45RO+ T cells undergoing apoptosis: a mechanism for T cell clearance during resolution of viral infections. J Exp Med. 1994;180(5):1943–1947. doi: 10.1084/jem.180.5.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alatery A, Siddiqui S, Chan M, Kus A, Petrof EO, Basta S. Cross, but not direct, presentation of cell-associated virus antigens by spleen macrophages is influenced by their differentiation state. Immunol Cell Biol. 2010;88(1):3–12. doi: 10.1038/icb.2009.90. [DOI] [PubMed] [Google Scholar]
- Anderson HA, Maylock CA, Williams JA, Paweletz CP, Shu H, Shacter E. Serum-derived protein S binds to phosphatidylserine and stimulates the phagocytosis of apoptotic cells. Nat Immunol. 2003;4(1):87–91. doi: 10.1038/ni871. [DOI] [PubMed] [Google Scholar]
- Asano K, Miwa M, Miwa K, Hanayama R, Nagase H, Nagata S, Tanaka M. Masking of phosphatidylserine inhibits apoptotic cell engulfment and induces auto-antibody production in mice. J Exp Med. 2004;200(4):459–467. doi: 10.1084/jem.20040342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz M, Yang WL, Wang P. Measurement of phagocytic engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. Curr Protoc Immunol. 2013;Chapter 14(Unit 14):31. doi: 10.1002/0471142735.im1431s100. [DOI] [PubMed] [Google Scholar]
- Baxter AE, Russell RA, Duncan CJ, Moore MD, Willberg CB, Pablos JL, Finzi A, Kaufmann DE, Ochsenbauer C, Kappes JC, Groot F, Sattentau QJ. Macrophage infection via selective capture of HIV-1-infected CD4+ T cells. Cell Host Microbe. 2014;16(6):711–721. doi: 10.1016/j.chom.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsmeier C, Pop OT, Singanayagam A, Triantafyllou E, Patel VC, Weston CJ, Curbishley S, Sadiq F, Vergis N, Khamri W, Bernal W, Auzinger G, Heneghan M, Ma Y, Jassem W, Heaton ND, Adams DH, Quaglia A, Thursz MR, Wendon J, Antoniades CG. Patients with acute-on-chronic liver failure have increased numbers of regulatory immune cells expressing the receptor tyrosine kinase MERTK. Gastroenterology. 2015;148(3):603–615. e14. doi: 10.1053/j.gastro.2014.11.045. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Zagorska A, Lew ED, Shrestha B, Rothlin CV, Naughton J, Diamond MS, Lemke G, Young JA. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host Microbe. 2013;14(2):136–147. doi: 10.1016/j.chom.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge RB, Boeltz S, Kumar S, Carlson J, Wanderley J, Calianese D, Barcinski M, Brekken RA, Huang X, Hutchins JT, Freimark B, Empig C, Mercer J, Schroit AJ, Schett G, Herrmann M. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 2016;23(6):962–978. doi: 10.1038/cdd.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retrovir. 1994;10(4):359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Calantone N, Wu F, Klase Z, Deleage C, Perkins M, Matsuda K, Thompson EA, Ortiz AM, Vinton CL, Ourmanov I, Lore K, Douek DC, Estes JD, Hirsch VM, Brenchley JM. Tissue myeloid cells in SIV-infected primates acquire viral DNA through phagocytosis of infected T cells. Immunity. 2014;41(3):493–502. doi: 10.1016/j.immuni.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKruyff RH, Bu X, Ballesteros A, Santiago C, Chim YL, Lee HH, Karisola P, Pichavant M, Kaplan GG, Umetsu DT, Freeman GJ, Casasnovas JM. T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J Immunol. 2010;184(4):1918–1930. doi: 10.4049/jimmunol.0903059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, Greene WC. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505(7484):509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott MR, Ravichandran KS. The Dynamics of Apoptotic Cell Clearance. Dev Cell. 2016;38(2):147–160. doi: 10.1016/j.devcel.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fernandez L, Bellido-Martin L, Garcia de Frutos P. Growth arrest-specific gene 6 (GAS6). An outline of its role in haemostasis and inflammation. Thromb Haemost. 2008;100(4):604–610. doi: 10.1160/th08-04-0253. [DOI] [PubMed] [Google Scholar]
- Fields BN, Knipe DM, Howley PM. Fields virology. 6. Vol. 2. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2013. [Google Scholar]
- Friggeri A, Banerjee S, Biswas S, de Freitas A, Liu G, Bierhaus A, Abraham E. Participation of the receptor for advanced glycation end products in efferocytosis. J Immunol. 2011;186(11):6191–6198. doi: 10.4049/jimmunol.1004134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto I, Pan J, Takizawa T, Nakanishi Y. Virus clearance through apoptosis- dependent phagocytosis of influenza A virus-infected cells by macrophages. J Virol. 2000;74(7):3399–3403. doi: 10.1128/jvi.74.7.3399-3403.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramatica A, Petazzi RA, Lehmann MJ, Ziomkowska J, Herrmann A, Chiantia S. alphaEnv-decorated phosphatidylserine liposomes trigger phagocytosis of HIV-virus-like particles in macrophages. Nanomedicine. 2014;10(5):981–989. doi: 10.1016/j.nano.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Hafizi S, Dahlback B. Gas6 and protein S. Vitamin K-dependent ligands for the Axl receptor tyrosine kinase subfamily. FEBS J. 2006;273(23):5231–5244. doi: 10.1111/j.1742-4658.2006.05529.x. [DOI] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417(6885):182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Moki T, Takizawa T, Shiratsuchi A, Nakanishi Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J Immunol. 2007;178(4):2448–2457. doi: 10.4049/jimmunol.178.4.2448. [DOI] [PubMed] [Google Scholar]
- He M, Kubo H, Morimoto K, Fujino N, Suzuki T, Takahasi T, Yamada M, Yamaya M, Maekawa T, Yamamoto Y, Yamamoto H. Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep. 2011;12(4):358–364. doi: 10.1038/embor.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy LM, Perron G, Won SY, Michell-Robinson MA, Rezk A, Ludwin SK, Moore CS, Hall JA, Bar-Or A, Antel JP. MerTK is a functional regulator of myelin phagocytosis by human myeloid cells. J Immunol. 2016;196(8):3375–3384. doi: 10.4049/jimmunol.1502562. [DOI] [PubMed] [Google Scholar]
- Hermetet F, Jacquin E, Launay S, Gaiffe E, Couturier M, Hirchaud F, Sandoz P, Pretet JL, Mougin C. Efferocytosis of apoptotic human papillomavirus-positive cervical cancer cells by human primary fibroblasts. Biol Cell. 2016;108(7):189–204. doi: 10.1111/boc.201500090. [DOI] [PubMed] [Google Scholar]
- Hodge S, Tran HB, Hamon R, Roscioli E, Hodge G, Jersmann H, Ween M, Reynolds PN, Yeung A, Treiberg J, Wilbert S. Nonantibiotic macrolides restore airway macrophage phagocytic function with potential anti-inflammatory effects in chronic lung diseases. Am J Physiol Lung Cell Mol Physiol. 2017;312(5):L678–L687. doi: 10.1152/ajplung.00518.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YM, Hong XZ, Xu JH, Luo JX, Mo HY, Zhao HL. Autoimmunity and dysmetabolism of human acquired immunodeficiency syndrome. Immunol Res. 2016;64(3):641–652. doi: 10.1007/s12026-015-8767-5. [DOI] [PubMed] [Google Scholar]
- Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, Weissman IL. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138(2):271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, Rayamajhi M, Miao EA. Programmed cell death as a defence against infection. Nat Rev Immunol. 2017;17(3):151–164. doi: 10.1038/nri.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazeros A, Harvey BG, Carolan BJ, Vanni H, Krause A, Crystal RG. Overexpression of apoptotic cell removal receptor MERTK in alveolar macrophages of cigarette smokers. Am J Respir Cell Mol Biol. 2008;39(6):747–757. doi: 10.1165/rcmb.2007-0306OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Karisola P, Pena-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, Butte MJ, Nagumo H, Chernova I, Zhu B, Sharpe AH, Ito S, Dranoff G, Kaplan GG, Casasnovas JM, Umetsu DT, Dekruyff RH, Freeman GJ. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 2007;27(6):927–940. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, Jr, van Rooijen N, Weissman IL. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CJ, Peters KN, Behar SM. Macrophages clean up: efferocytosis and microbial control. Curr Opin Microbiol. 2014;17:17–23. doi: 10.1016/j.mib.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann JC, Ames BN. Vitamin K, an example of triage theory: is micronutrient inadequacy linked to diseases of aging? Am J Clin Nutr. 2009;90(4):889–907. doi: 10.3945/ajcn.2009.27930. [DOI] [PubMed] [Google Scholar]
- Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450(7168):435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- Morizono K, Chen IS. Role of phosphatidylserine receptors in enveloped virus infection. J Virol. 2014;88(8):4275–4290. doi: 10.1128/JVI.03287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, Xie Y, Olafsen T, Lee B, Dasgupta A, Wu AM, Chen IS. The soluble serum protein Gas6 bridges virion envelope phosphatidylserine to the TAM receptor tyrosine kinase Axl to mediate viral entry. Cell Host Microbe. 2011;9(4):286–298. doi: 10.1016/j.chom.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morner A, Bjorndal A, Albert J, Kewalramani VN, Littman DR, Inoue R, Thorstensson R, Fenyo EM, Bjorling E. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol. 1999;73(3):2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Subramaniam R, Chen H, Smith A, Keshava S, Shams H. Boosting efferocytosis in alveolar space using BCG vaccine to protect host against influenza pneumonia. PLoS One. 2017;12(7):e0180143. doi: 10.1371/journal.pone.0180143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musich T, Li L, Liu L, Zolla-Pazner S, Robert-Guroff M, Gorny MK. Monoclonal antibodies specific for the V2, V3, CD4-binding site, and gp41 of HIV-1 mediate phagocytosis in a dose-dependent manner. J Virol. 2017;91(8) doi: 10.1128/JVI.02325-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Arai H, Yanagita M, Matsubara T, Kanamori H, Nakano T, Iehara N, Fukatsu A, Kita T, Doi T. Growth arrest-specific gene 6 is involved in glomerular hypertrophy in the early stage of diabetic nephropathy. J Biol Chem. 2003;278(20):18229–18234. doi: 10.1074/jbc.M213266200. [DOI] [PubMed] [Google Scholar]
- Nainu F, Shiratsuchi A, Nakanishi Y. Induction of apoptosis and subsequent phagocytosis of virus-infected cells as an antiviral mechanism. Front Immunol. 2017;8:1220. doi: 10.3389/fimmu.2017.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nainu F, Tanaka Y, Shiratsuchi A, Nakanishi Y. Protection of Insects against Viral Infection by Apoptosis-Dependent Phagocytosis. J Immunol. 2015;195(12):5696–5706. doi: 10.4049/jimmunol.1500613. [DOI] [PubMed] [Google Scholar]
- Nakahashi-Oda C, Tahara-Hanaoka S, Honda S, Shibuya K, Shibuya A. Identification of phosphatidylserine as a ligand for the CD300a immunoreceptor. Biochem Biophys Res Commun. 2012;417(1):646–650. doi: 10.1016/j.bbrc.2011.12.025. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Ando Y, Kanetani T, Hoshi C, Nakai Y, Nainu F, Nagaosa K, Shiratsuchi A, Nakanishi Y. Signaling pathway for phagocyte priming upon encounter with apoptotic cells. J Biol Chem. 2017;292(19):8059–8072. doi: 10.1074/jbc.M116.769745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CA, deCathelineau A, Hoffmann PR, Bratton D, Ghebrehiwet B, Fadok VA, Henson PM. C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J Exp Med. 2001;194(6):781–795. doi: 10.1084/jem.194.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science. 2000;288(5473):2051–2054. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- Orenstein JM. In vivo cytolysis and fusion of human immunodeficiency virus type 1-infected lymphocytes in lymphoid tissue. J Infect Dis. 2000;182(1):338–342. doi: 10.1086/315640. [DOI] [PubMed] [Google Scholar]
- Pang S, Yu D, An DS, Baldwin GC, Xie Y, Poon B, Chow YH, Park NH, Chen IS. Human immunodeficiency virus Env-independent infection of human CD4(−) cells. J Virol. 2000;74(23):10994–11000. doi: 10.1128/jvi.74.23.10994-11000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450(7168):430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- Park SY, Jung MY, Lee SJ, Kang KB, Gratchev A, Riabov V, Kzhyshkowska J, Kim IS. Stabilin-1 mediates phosphatidylserine-dependent clearance of cell corpses in alternatively activated macrophages. J Cell Sci. 2009;122(Pt 18):3365–3373. doi: 10.1242/jcs.049569. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Knyazev PG, Clout NJ, Cheburkin Y, Gohring W, Ullrich A, Timpl R, Hohenester E. Structural basis for Gas6-Axl signalling. EMBO J. 2006;25(1):80–87. doi: 10.1038/sj.emboj.7600912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratsuchi A, Kaido M, Takizawa T, Nakanishi Y. Phosphatidylserine-mediated phagocytosis of influenza A virus-infected cells by mouse peritoneal macrophages. J Virol. 2000;74(19):9240–9244. doi: 10.1128/jvi.74.19.9240-9244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simhadri VR, Andersen JF, Calvo E, Choi SC, Coligan JE, Borrego F. Human CD300a binds to phosphatidylethanolamine and phosphatidylserine, and modulates the phagocytosis of dead cells. Blood. 2012;119(12):2799–2809. doi: 10.1182/blood-2011-08-372425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sips M, Krykbaeva M, Diefenbach TJ, Ghebremichael M, Bowman BA, Dugast AS, Boesch AW, Streeck H, Kwon DS, Ackerman ME, Suscovich TJ, Brouckaert P, Schacker TW, Alter G. Fc receptor-mediated phagocytosis in tissues as a potent mechanism for preventive and therapeutic HIV vaccine strategies. Mucosal Immunol. 2016;9(6):1584–1595. doi: 10.1038/mi.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai C, Kornbluth RS, Pauza CD, Richman DD, Carson DA. Apoptosis as a mechanism of cell death in cultured T lymphoblasts acutely infected with HIV-1. J Clin Invest. 1991;87(5):1710–1715. doi: 10.1172/JCI115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S, Hanayama R, Nagata S. Two-step engulfment of apoptotic cells. Mol Cell Biol. 2012;32(1):118–125. doi: 10.1128/MCB.05993-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre D, Gennero L, Baccino FM, Speranza F, Biondi G, Pugliese A. Impaired macrophage phagocytosis of apoptotic neutrophils in patients with human immunodeficiency virus type 1 infection. Clin Diagn Lab Immunol. 2002;9(5):983–986. doi: 10.1128/CDLI.9.5.983-986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufail Y, Cook D, Fourgeaud L, Powers CJ, Merten K, Clark CL, Hoffman E, Ngo A, Sekiguchi KJ, O’Shea CC, Lemke G, Nimmerjahn A. Phosphatidylserine exposure controls viral innate immune responses by microglia. Neuron. 2017;93(3):574–586. e8. doi: 10.1016/j.neuron.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara H, Shacter E. Auto-oxidation and oligomerization of protein S on the apoptotic cell surface is required for Mer tyrosine kinase-mediated phagocytosis of apoptotic cells. J Immunol. 2008;180(4):2522–2530. doi: 10.4049/jimmunol.180.4.2522. [DOI] [PubMed] [Google Scholar]
- Uehara S, Handa H, Gotoh K, Tomita H, Sennshuu M. Plasma concentrations of growth arrest-specific protein 6 and protein S in patients with acute pancreatitis. J Gastroenterol Hepatol. 2009;24(9):1567–1573. doi: 10.1111/j.1440-1746.2009.05875.x. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Hashimoto Y, Shiratsuchi A, Takizawa T, Nakanishi Y. Augmentation of fatality of influenza in mice by inhibition of phagocytosis. Biochem Biophys Res Commun. 2005;337(3):881–886. doi: 10.1016/j.bbrc.2005.09.133. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Shiratsuchi A, Shimizu K, Takizawa T, Nakanishi Y. Role of phosphatidylserine exposure and sugar chain desialylation at the surface of influenza virus-infected cells in efficient phagocytosis by macrophages. J Biol Chem. 2002;277(20):18222–18228. doi: 10.1074/jbc.M201074200. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Shiratsuchi A, Shimizu K, Takizawa T, Nakanishi Y. Stimulation of phagocytosis of influenza virus-infected cells through surface desialylation of macrophages by viral neuraminidase. Microbiol Immunol. 2004;48(11):875–881. doi: 10.1111/j.1348-0421.2004.tb03619.x. [DOI] [PubMed] [Google Scholar]
- Yoon KW, Byun S, Kwon E, Hwang SY, Chu K, Hiraki M, Jo SH, Weins A, Hakroush S, Cebulla A, Sykes DB, Greka A, Mundel P, Fisher DE, Mandinova A, Lee SW. Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science. 2015;349(6247):1261669. doi: 10.1126/science.1261669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Kawane K, Koike M, Mori Y, Uchiyama Y, Nagata S. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature. 2005;437(7059):754–758. doi: 10.1038/nature03964. [DOI] [PubMed] [Google Scholar]
- Zent CS, Elliott MR. Maxed out macs: physiologic cell clearance as a function of macrophage phagocytic capacity. FEBS J. 2017;284(7):1021–1039. doi: 10.1111/febs.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Wild T, Xiong Y, Sylvers P, Zhang Y, Zhang L, Wahl L, Wahl SM, Kozlowski S, Notkins AL. Polyreactive antibodies plus complement enhance the phagocytosis of cells made apoptotic by UV-light or HIV. Sci Rep. 2013;3:2271. doi: 10.1038/srep02271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Sun X, Zhu L, Hu F, Shi L, Li Z, Su Y. The expression and clinical significance of different forms of Mer receptor tyrosine kinase in systemic lupus erythematosus. J Immunol Res. 2014;2014:431896. doi: 10.1155/2014/431896. [DOI] [PMC free article] [PubMed] [Google Scholar]