Abstract
Precision oncology involves identifying drugs that will effectively treat a tumor and then prescribing an optimal clinical treatment regimen. However, most first-line chemotherapy drugs do not have biomarkers to guide their application. For molecularly targeted drugs, using the genomic status of a drug target as a therapeutic indicator has limitations. In this study, machine learning methods (e.g., deep learning) were used to identify informative features from genome scale omics data and to train classifiers for predicting the effectiveness of drugs in cancer cell lines. The methodology introduced here can accurately predict the efficacy of drugs, regardless of whether they are molecularly targeted or non-specific chemotherapy drugs. This approach, on a per-drug basis, can identify sensitive cancer cells with an average sensitivity of 0.82 and specificity of 0.82; on a per-cell line basis, it can identify effective drugs with an average sensitivity of 0.80 and specificity of 0.82. This report describes a data-driven precision medicine approach that is not only generalizable but also optimizes therapeutic efficacy. The framework detailed herein, when successfully translated to clinical environments, could significantly broaden the scope of precision oncology beyond targeted therapies, benefiting an expanded proportion of cancer patients.
Introduction
Precision oncology aims to detect and target tumor-specific aberrations with effective therapies(1,2). In the current practice of precision oncology, the prescription of molecularly targeted drugs is mainly based on the genomic status of a drug-target gene as a therapeutic indicator(2,3). However, this approach only benefits a small percentage of patients(4,5). Nonspecific cytotoxic drugs lack well-established biomarkers to guide their usage, yet they remain first-line chemotherapy for many patients(6), despite recent advances in molecularly targeted therapy and immunotherapy. Therefore, there exists a need for data driven approaches to improve therapeutics.
Recent large-scale pharmacogenomics screening on cancer cell lines(7,8) and patient-derived xenografts(9) (PDXs) have demonstrated that almost every cancer cell line or PDX is sensitive to one or more targeted or non-targeted drugs, but current approaches cannot accurately match sensitive drug-cancer pairs. For nonspecific cytotoxic medications, few data driven models exist. In the case of molecularly targeted drugs, genomic markers are not accurate indicators. Translated into a clinical setting, this indicates that many patients are treated with an ineffective first line of chemotherapy due to the lack of accurate prognostic predictors. On the other hand, for most molecularly targeted drugs, the majority of sensitive cancers do not host genomic alterations in the targeted gene. The clinical implication is that there exist patients who could benefit from molecularly targeted medications but they are being missed due to the inaccuracy of genomic markers. Accurately identifying these groups of patients would maximize the therapeutic usefulness of existing anti-cancer drugs for improved treatment outcomes.
Recently, pharmacogenomics experiments have collected genomic and transcriptomic data on a large number of cancer cell lines and PDXs, together with drug sensitivity data. Typically, these studies have attempted to uncover associations between omics features and drug sensitivity measurements, such as IC50(10). Different studies have explored the use of current state-of-the-art classification models, such as ridge regression and support vector machines(11–13) to train predictive models for IC50 using genome-scale omics data as input features. However, the performance of current computational models is far from adequate. Difficulty arises from the high dimensionality of omics data and the relatively small number of training cases available, which often leads to overfitting. Thus, learning novel informative features from omics data is a critical step in model-based prediction of drug sensitivity.
In this study, we investigate the utility of combining genome-scale omics data with contemporary machine learning techniques to develop predictive models that can be applied to both molecularly targeted and conventional chemotherapy drugs. The models developed in this study are trained to predict discretized effectiveness measures for drug-cell line pairs. This novel systematic production of classification models holds advantages over drug concentration regression models in both flexibility and ease of clinical translation. We addressed the dimensionality challenge by concentrating on feature selection and deep learning based feature learning techniques to extract informative features that reflect the activation states of drug-target proteins and cell-signaling pathways. We show that deep learning models can learn novel representations of cellular signaling systems (14). Using these informative features, we trained a classification model for each anti-cancer drug to predict the sensitivity of cancer cell lines to that drug (Figure 1A). The results indicate that informative features derived from deep learning can significantly enhance the accuracy of prediction models. We further show that our predictions can significantly expand the therapeutic scope of molecularly targeted drugs and reduce ineffective administration of nonspecific first-line drugs. If these results are reproduced (even partially) in clinical settings, they have the potential to significantly improve the practice of precision oncology.
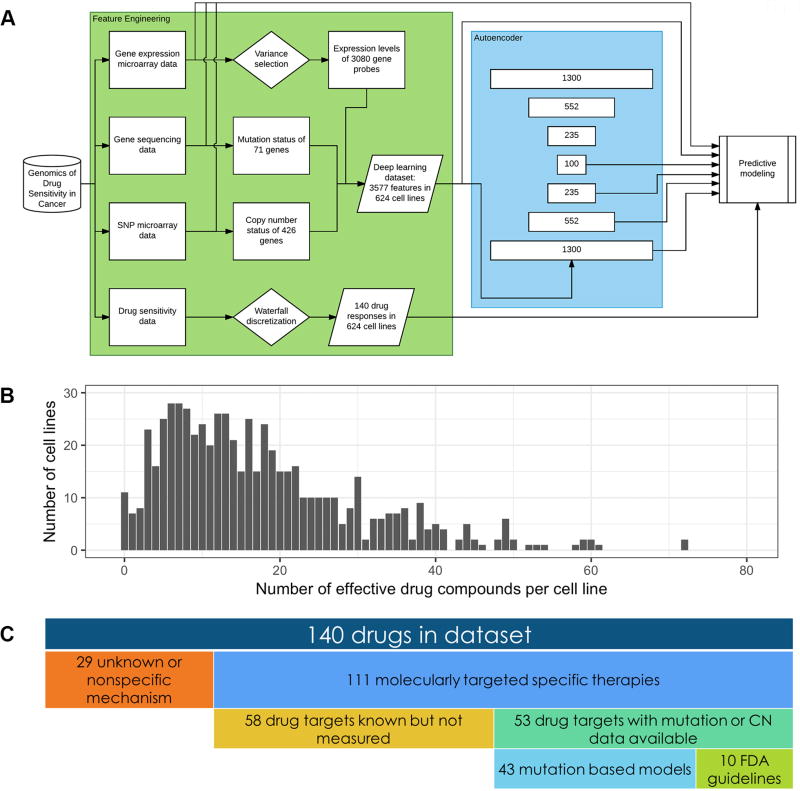
Figure 1. Drug sensitivity prediction workflow utilizing GDSC data.
A, Workflow for the training and optimization of drug sensitivity prediction models. There are three main steps. First is the feature engineering of omics data from the Genomics of Drug Sensitivity in Cancer Project. Second is feature construction via a deep neural network autoencoder. Third is training of machine learning models to predict drug sensitivity response using various feature sets as inputs. B, Histogram of the number of effective drug compounds for any given cell line in the GDSC pharmacogenomics study (median = 14.5). C, Descriptive breakdown of drugs tested in the GDSC pharmacogenomics study.
Materials and Methods
Data retrieval
We retrieved and utilized data from two large pharmacogenomics studies, the Genomics of Drug Sensitivity in Cancer Project (GDSC), and the Cancer Cell Line Encyclopedia (CCLE).
GDSC gene expression data were downloaded from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-783/) in the form of raw Affymetrix CEL files. Drug sensitivity measurements, copy number variation data, mutation data, and cell line annotations were downloaded from the GDSC website (http://www.cancerrxgene.org/downloads).
CCLE gene expression data, copy number variation data, mutation data, and cell line annotations were downloaded from the CCLE website (http://www.broadinstitute.org/ccle). Drug sensitivity measurements were obtained from the associated publication(7).
Feature engineering
GDSC gene expression data in the form of Affymetrix CEL files were normalized using Robust Multi-Array Averaging. For replicate experiments, expression values were averaged. After removal of spike control probes, this procedure generated an array of 22,215 probe expression measurements in 727 cell lines.
The normalized gene expression data were filtered using three different variance metrics. We applied Hartigans’ dip test for unimodality to select for genes with multimodal distributions(15). The outlier sum method was used to select for genes that had largely unimodal distributions with significant outlier populations(16). Finally, median absolute deviation was used to select for genes with a high variance across samples regardless of distribution shape. We attempted to keep the approximately 1500 most variant gene probes selected by each metric. However, for the dip test, far fewer probes had statistically significant scores for multimodality, and only 664 were retained. The union of the three methods resulted in the selection of 3080 gene expression measurements out of 22,215, a retention of approximately 14%. After variance selection, a mixture of two normal distributions was fitted to each gene’s expression profile. A t-test was performed to verify statistical significance between the two groups for each gene. These groups were then used to determine a cutoff to discretize the expression levels of each gene into low and high values. After this procedure, the dataset contains discretized gene expression data of 3080 genes in 727 cell lines.
GDSC copy number and mutation data for 624 cell lines were extracted from gdsc_en_input_w5.csv available on the GDSC website. Copy number variation data ranging from 0 to 10 were normalized to real values between 0 and 1. Copy number variation data above this range were set to 1. Mutation data was already encoded in a binary form and required no further processing. 426 genes were characterized for copy number variation and 71 genes were characterized for mutations. These two feature sets were relatively low-dimensional compared to the gene expression data, and it was determined that selective preprocessing would introduce a high risk of discarding important predictive variables for the sake of relatively insignificant improvements in computational efficiency.
Cell line annotations were used to combine the three feature sets (gene expression, copy number variation, and mutation data) into a single array of data containing information on 3577 features in 624 cell lines. Cell lines for which all three data types were not available were excluded from analysis.
Deep learning
Code for training a deep autoencoder as described by Hinton and Salakhutdinov was obtained from Hinton’s website (http://www.cs.toronto.edu/~hinton/MatlabForSciencePaper.html) and modified to utilize the feature selected GDSC dataset for unsupervised representation learning. To train the autoencoder, the 624 cell lines in the dataset were randomly split into training and testing datasets of 520 and 104 samples, respectively. The training dataset is used to train the weights of the model via conjugate gradient descent. During training, the current performance of the model is periodically evaluated on the testing dataset, enabling application of the early stopping rule to prevent overfitting. Using a batch size of 26, an autoencoder with hidden layers of size 1300, 552, 235, and 100 was trained. This model was learned using 50 epochs of pretraining a stacked restricted Boltzmann machine and 400 epochs of backpropagation.
Drug sensitivity data
GDSC drug sensitivity measurements in the form of activity area values were extracted from gdsc_manova_input_w5.csv available on the GDSC website. These were discretized into sensitive and resistant categories by applying the waterfall method to each drug(7). The waterfall method is summarized as follows: Drug sensitivity measurements for all cell lines are sorted in increasing order to generate a waterfall distribution. A linear regression is fitted to this distribution. A Pearson correlation is calculated to determine goodness of fit for the linear regression equation. If the Pearson correlation coefficient is less than 0.95, the major inflection point is estimated to be the point on the sensitivity curve with the maximal distance to a line drawn between the start and end points of the waterfall distribution. If the Pearson correlation coefficient is greater than 0.95, the median value is used instead. This value is then determined to be the cutoff for separating sensitive and resistant cell lines for each drug.
Elastic net regression
We used elastic net regression to generate logistic models for drug sensitivity prediction. Elastic net regression is a form of logistic regression with a hybrid regularization term that combines lasso and ridge regularization(17). The elastic net contains two hyperparameters, alpha and lambda. Alpha defines the relative weight of the lasso and ridge penalization terms. Lambda determines the overall size of the regularization penalty. We fixed alpha at 0.5 and optimized for predictive performance over a range of lambdas7. Regression was performed with 25-fold cross validation using the glmnet R package.
For each model, the target vector consists of the discretized sensitivity data for that particular drug. For each drug, six models were built, with input vectors of varying size. These input vectors were: the original unprocessed genomic features, the feature selected dataset, and each of the four layers of latent variables from the deep learning autoencoder.
Support Vector Machine
We also used a support vector machine (SVM) with a Gaussian kernel to predict drug sensitivity. Although SVM is ordinarily a linear classifier, the custom kernel maps the input into high-dimensional feature spaces, allowing for the modeling of non-linear classifications. SVM training was performed with 25-fold cross validation using the e1071 R interface to the libsvm C++ implementation.
As with elastic net regression, the target vector consists of the discretized sensitivity data for that particular drug. For each drug, two SVM models were built. One used the original unprocessed genomic features as input, while the other used the feature selected dataset.
Consensus clustering
Consensus clustering was performed with 50 repetitions and a sample probability of 0.8 using the ConsensusClusterPlus algorithm(18). During each repetition, 80% of the cell lines in the dataset were randomly selected to be clustered via agglomerative hierarchical clustering. Samples that consistently clustered together during these repetitions were subsequently assigned to the same cluster upon compilation of the repetitions. Enrichment of drug sensitivity in specific clusters was determined by Fisher’s exact test, Bonferroni corrected for the number of clusters.
Tissue type modeling
In addition to omics data, the GDSC provides tissue type information describing the cell line samples studied in the experiment. The 624 cell lines originate from one of 19 different tissues. To sufficiently power statistical tests, we chose to analyze those tissue categories with more than 30 cell lines. The 9 largest tissue categories are lymphoma, breast, large intestine, skin, aerodigestive tract, nervous system, leukemia, urogenital system, and non-small cell lung carcinomas. Enrichment of drug sensitivity in specific tissue categories was determined by chi-square test, yielding 89 drug-tissue pairs.
External validation
CCLE gene expression data in the form of Affymetrix CEL files were normalized using Robust Multi-Array Averaging. This procedure generated an array of 54,675 probe expression measurements in 1067 cell lines. A mixture of two normal distributions was fitted to each gene’s expression profile, and these groups were used to determine a cutoff to discretize the expression levels of each gene into low and high values.
CCLE copy number data were extracted from CCLE_copynumber_byGene_2013-12-03.txt available on the CCLE website. These values are HapMap normalized. To make them comparable to the estimated copy number counts used in GDSC, we assumed a base frequency of two copies per gene to estimate raw copy number. We then normalized values from 0 to 10 to values between 0 and 1. Copy number variation above this range were set to 1.
CCLE mutation data were collected using two methods: Oncomap 3.0 and hybrid capture analysis. These data were extracted from CCLE_Oncomap3_2012-04-09.maf, and CCLE_hybrid_capture1650_hg19_NoCommonSNPs_NoNeutralVariants_CDS_2012.05.07.maf respectively, both available on the CCLE website. These annotations were classified as mutated or not based on The Cancer Genome Atlas specification for the Mutation Annotation Format.
After this extraction and processing, values for the 3577 features in the GDSC selected dataset were extracted and combined to create a CCLE selected dataset. Information on all 3577 features were available in CCLE, with the exception of mutation data for four genes. These missing values were filled in with uninformative average values derived from the GDSC dataset. After this procedure, the CCLE dataset consists of 3577 features measured in 1067 cell lines.
CCLE drug sensitivity data in the form of activity area values was extracted from CCLE_NP24.2009_Drug_data_2015.02.24.csv available on the CCLE website. These were discretized into sensitive and resistant categories by applying the waterfall method as described previously.
The GDSC autoencoder was used to encode the CCLE selected dataset, and the subsequent encoding was used as inputs to make drug sensitivity predictions using GDSC elastic net models for 15 drugs shared by the two datasets. These predictions were evaluated against the waterfall discretized CCLE sensitivity data for those drugs.
Results
Limited predictive capability of genome status markers
We collected data from the Genomics of Drug Sensitivity in Cancer (GDSC)(8) pharmacogenomics study and systematically analyzed the utility of using genome-status markers as therapeutic indicators for molecularly targeted drugs. The GDSC dataset contains the results of drug sensitivity experiments of 140 drugs in 624 cell lines. Each cell line is characterized by the following genomic features: somatic copy number alteration (SCNA) status of 426 genes, mutation status of 71 genes, and gene expression values of 22,215 gene probes.
Each drug was tested on a median of n = 586 cell lines. In these experiments, only 11 cell lines were found to be not responsive to any drugs. The remaining 613 cell lines, comprising 98.2% of the dataset, were typically responsive to a median of 14.5 drug compounds (Figure 1B). These results suggest the existence of effective therapies for the majority of cancer cells investigated in the dataset.
Out of the 140 drugs in the dataset, 29 have unknown or nonspecific mechanisms of action, leaving 111 molecularly targeted specific therapies (Supplementary Tables S1, S2). Of these 111 drugs, some combination of mutation or SCNA information is available for 53 drug targets, whereas the genomic status of the target genes of the remaining 58 drugs were not measured. For the 53 drugs, we built a rule-based classifier for each drug to predict drug sensitivity. Among these, 10 drugs have an FDA-approved genomic testing indication for their clinical use, and our rule-based classifier mirrors these predefined indications. These 10 drugs are comprised of PARP and tyrosine kinase inhibitors, with genetic tests for BRCA1/2, EGFR, ERBB2, ALK, and BCR-ABL mutations. In the cases of the remaining 43 drugs, the rule-based classifier consists of one simple rule: If the genomic status of the target protein of a drug is abnormal – either mutated or copy number amplified – the cell line should be sensitive to the drug (Figure 1C).
Some of these models appear to perform well. For example, responsiveness of cell lines to the BCR-ABL inhibitor GNF-2 is well correlated with the presence of the BCR-ABL fusion (Supplementary Figure S1A). However, there are still cell lines that may be responsive to GNF-2 that do not have the gene fusion. In other cases, the rule-based models perform significantly worse. For example, it is difficult to associate the responsiveness of cell lines to the EGFR inhibitor Gefitinib with the genomic status of EGFR, if such a correlation exists at all (Supplementary Figure S1B).
We evaluated the performance of these rule-based classifiers using several metrics. First, sensitivity for a drug rule or model is the proportion of cell lines responsive to the drug that are correctly identified by the rule or model. Second, specificity is the proportion of cell lines that are not responsive to the drug that are correctly identified as such. Third, positive predictive value (PPV) refers to the proportion of cell lines predicted to be responsive to the drug that are in fact responsive.
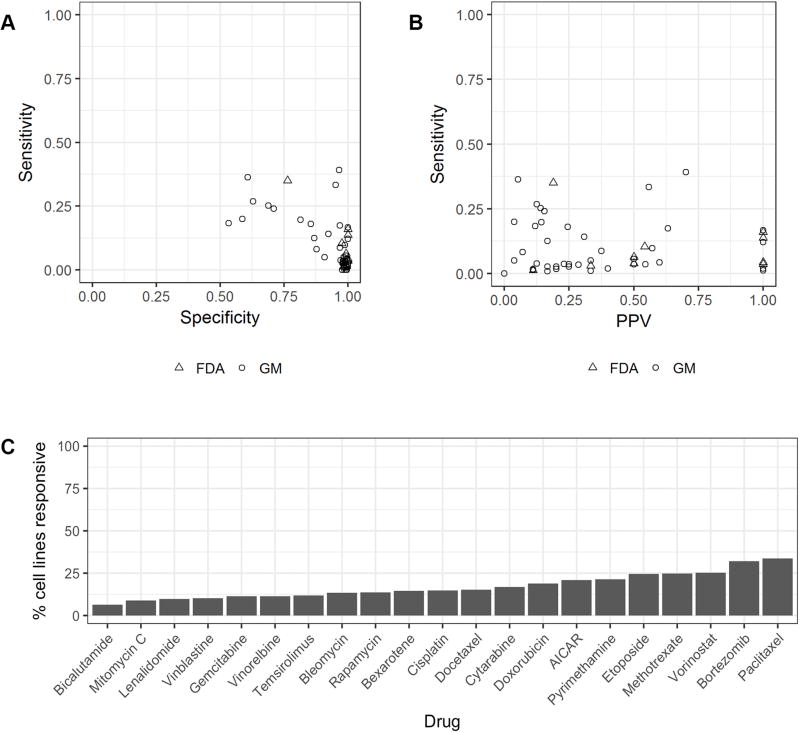
The average sensitivity of the 53 rule-based models is 0.10 and the average specificity is 0.93, indicating genomic markers fail to identify the vast majority of cancer cells sensitive to the molecularly targeted drugs (Figure 2A). Most cell lines are insensitive to molecularly targeted therapies. The majority of cell lines were predicted by each rule as being insensitive, because only a few cell lines host the specific genomic markers required to be present by the rule. Thus, these rules are generally characterized by high specificity. Under these circumstances, sensitivity and PPV are better indicators of the accuracy of a classifier. The 53 models share an average PPV of 0.38 (Figure 2B), indicating that the majority of cell lines hosting a genomic marker are actually resistant to the drugs.
Figure 2. Limited predictive capability of genomic markers.
A, Sensitivity and specificity of 43 genomic marker rule based models (GM) and 10 FDA genomic guideline clinical indications (FDA). B, Sensitivity and positive predictive value of 43 genomic marker rule based models (GM) and 10 FDA genomic guideline clinical indications (FDA). C, Drug sensitivity of 21 nonspecific FDA-approved medications.
Meanwhile, there are 21 drugs in the dataset with FDA approved guidelines that do not involve genetic testing. Most of these are nonspecific cytotoxic agents, and many of them are indicated as first line treatment for a variety of cancers. When applied indiscriminately across the cell lines in the dataset (equivalent to predicting every cell line as sensitive to the drug), these drugs achieve an average PPV of 0.17 (Figure 2C). The actual administration of a first line treatment often takes into account tumor size, stage, and other factors, but these features do not apply to cell lines, so we have calculated a reasonable lower bound on the PPV.
These results indicate two areas with opportunity for meaningful improvement. First, better prediction of the effectiveness of targeted therapies can expand the application of those drugs to patients with cancer cells that are receptive despite lacking the related genomic marker. Second, accurate prediction of the effectiveness of nonspecific treatments can reduce the prescription of those drugs to patients for whom they will not be effective, leading to improvements in cost and quality of life for patients who would otherwise only suffer through a toxic, ineffective first line regimen. To this end, we set out to investigate whether combining state-of-the-art machine learning methods and genome-scale omics data can derive more accurate therapeutic indicators.
Omics data contain information for predicting drug sensitivity
We first investigated whether current state-of-the-art classification models trained with omics data as input features could predict the drug sensitivity of cell lines. We tested two classification methods with proven ability to handle high-dimensional data: elastic net regression(17) and support vector machines(19) (SVMs). Since we are no longer restricted by the availability of known and measured genomic markers, we were able to train a model for each of the 140 drugs in the dataset.
The elastic net models trained with all omics features achieved an average sensitivity of 0.75 and an average specificity of 0.78. The corresponding SVM models achieved an average sensitivity of 0.59 and an average specificity of 0.56. PPV averaged 0.43 for the elastic net models and 0.18 for the SVM models. The performance of these classification models is better than that of the genomic markers for molecularly targeted drugs (Supplementary Figure S2).
We also evaluated the area under the receiving operator curve (AUROC) as a summary statistic for how well the model performs across various sensitivity and specificity values. The elastic net models have an average AUROC of 0.81. In contrast, the SVM models have an average AUROC of 0.55 (Supplementary Figure S2). These results indicate that omics data contain useful predictive information that can be captured by different classification models, and that machine learning algorithms outperform the rule-based method. Since elastic net models appear to outperform SVMs, we hereafter only present results derived using elastic net models.
Although the elastic net model has intrinsic feature selection capability, most classification algorithms suffer from an overfitting problem when the dimensionality of input features is very large. We sought to determine whether additional feature selection techniques could be applied to enhance the performance of these classifiers. We applied a variance-based mixture-fitting feature selection scheme, since features that lack significant variation across samples should have low predictive value(20). We found that, on the level of individual models, some drugs are better predicted with feature selection than without. However, feature selection did not enhance the overall aggregate performance of the elastic net, likely due to the loss of some useful information during feature selection (Figure 3A).
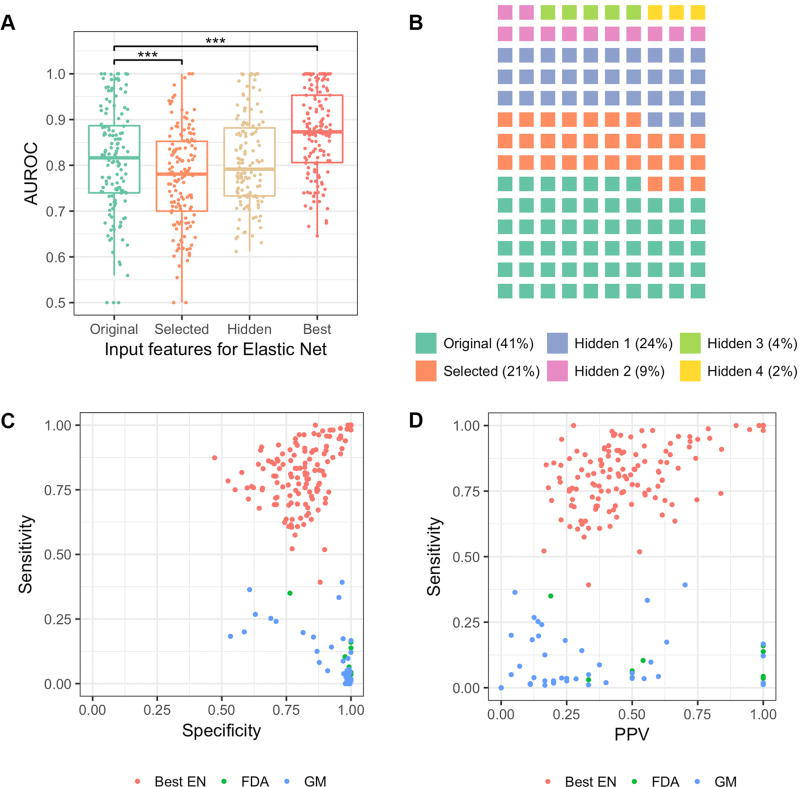
Figure 3. Learning cellular state features using deep learning.
A, Predictive performance of elastic net models relative to predictive features used as inputs. B, Proportion of best models from each category of input feature. C, Sensitivity and specificity of 140 best elastic net models (Best EN) compared to 43 genomic marker rule based models (GM) and 10 FDA genomic guideline clinical indications (FDA). D, Sensitivity and positive predictive value of 140 best elastic net models (Best EN) compared to 43 genomic marker rule based models (GM) and 10 FDA genomic guideline clinical indications (FDA) *** p < 10e-3
Learning cellular state features using deep learning
For certain drugs, neither elastic net nor SVM perform well using the original or the selected features as predictive inputs. We hypothesized that the signals of some cellular pathways are embedded as complex statistical structures in the omics data, which cannot be detected and fully utilized by elastic net and SVM. This problem may be addressed by models designed to reveal such complex statistical structures. Recently, our team reported that deep learning algorithms may be able to capture the signals of biological entities in cell signaling systems(14,21). In this study, we applied an autoencoder(22), a type of unsupervised deep neural network, to learn features that are potentially reflective of the cellular state.
The autoencoder aims to learn new representations of a vector of observed variables using multiple hidden layers of hierarchically organized latent variables. In each hidden layer, the input data are transformed using a set of weights and then propagated to the next layer. In this manner, the statistical distributions underlying the omics data are compositionally encoded by latent variables in each of the hidden layers. We learn one such model for each drug. Once learned, we can apply the model to infer the expected states of latent variables for each cell line, providing a set of new representations, potentially reflecting the state of signaling pathways in these cells. These latent variables become additional features in learning a drug-specific elastic-net model that predicts a response of each cell-line to the drug.
Since different layers of an autoencoder capture information with differing degrees of abstraction, we represented each cell line using the states of latent variables within specific layers and then trained an elastic net classifier for each drug. Although aggregate performance does not improve when using latent variables as predictive features, some drugs modeled poorly by original or selected omics features are significantly better predicted by hidden layer models (Figure 3A). For a given drug, the performance often varies when latent variables from different layers are used as predictive features. 57 drugs are best predicted using the original omics features, 30 are best predicted using selected features, and 53 drugs are best predicted by deep-learning-derived hidden layer features. These findings indicate that useful complex relationships are uncovered by deep learning (Figure 3B).
As a group, the best models have an average AUROC of 0.87 (Figure 3A). Average sensitivities, specificities, and positive predictive values are 0.82, 0.82, and 0.51, respectively (Figure 3C, D). Exceptional performance was achieved for 15 drugs, in which sensitivity and specificity values were greater than 0.98, positive predictive value was greater than 0.94, and AUROC values were greater than 0.99 (Supplementary Table S3).
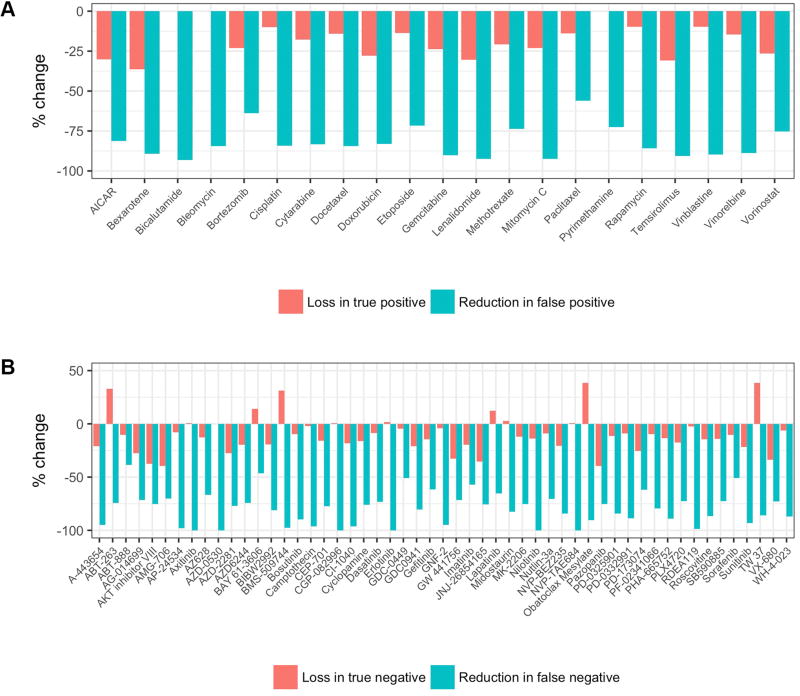
In the 53 rule-based models for molecularly targeted drugs, the opportunity for improvement lies in the expansion of therapeutic use. When compared to these rule-based models, the best models reduced the rate of false negatives (recovering missed therapeutic opportunities) by an average of 80%, at the cost of reducing the rate of true negatives (introducing ineffective therapies) by just 9% (Figure 4A). Conversely, for the 21 FDA approved nonspecific medications, the improvement potential is in the reduction of ineffective administration. In this group, use of the best models resulted in an 82% reduction in false positives (reducing ineffective therapies) while incurring a 18% reduction in true positives (missing some other therapeutic opportunities) (Figure 4B).
Figure 4. Improvement in predictive performance achieved with optimized models.
A, Percent change in true positives and false positives identified by optimized elastic net models relative to simply giving the drug to all patients for 21 FDA-approved nonspecific medications. B, Percent change in true negatives and false negatives identified by optimized elastic net models relative to genomic marker rule-based models for 53 targeted drugs.
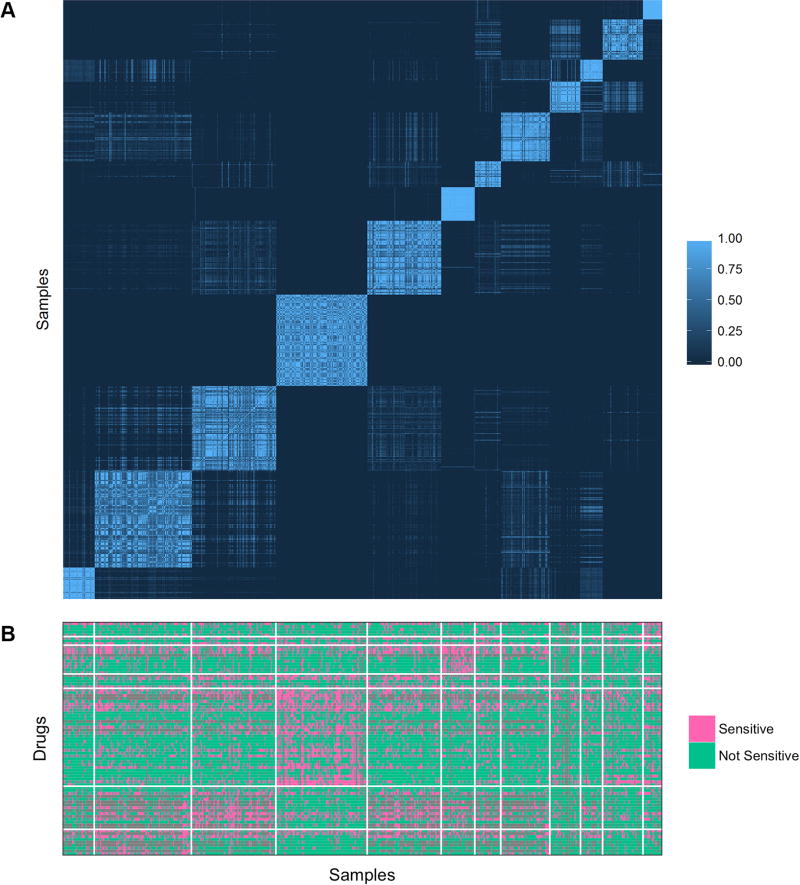
To investigate whether deep-learning-derived features performed well because they capture information relevant to the cellular signaling system, we hypothesized that cell lines with similar representations may present similar drug response profiles. We applied agglomerative hierarchical clustering to the GDSC cell lines based on the autoencoder’s first hidden layer of 1300 features. Consensus clustering into 12 groups recovered a stable partitioning (Figure 5A). We found that sensitivity to 74 drugs was significantly enriched (p < 0.05) in at least one group (Figure 5B). Altogether, this supports the idea that deep learning produces a novel representation from the input data, and that the deep learning representation is useful for predicting drug sensitivity.
Figure 5. Consensus clustering of GDSC tumor cell line samples.
A, consensus clustering of GDSC cell lines based on autoencoder constructed Hidden 1 features. The intensity of the plot indicates the relative frequency, or consensus, with which a pair of samples cluster together in repeated hierarchical clustering of subsamplings from the dataset. B, enrichment of sensitivity to drugs in autoencoder constructed Hidden 1 consensus clusters.
Tissue specific prediction of predictive models
In the GDSC, sensitivity to 72 of the 140 drugs studied was significantly enriched (p < 0.05) in at least one major tissue type (Supplementary Table S4). As cell lines from the same tissue often share similar gene expression and genomic sequence profiles, these occurrences of enrichment are potential instances of confounding. In these situations, a predictive model could theoretically achieve strong performance by simply learning to predict tissue type based on omics features. However, such a model would have limited utility, because tissue type is usually a known variable that does not require predicting. Although the 89 drug-tissue pairs in which enrichment occurs represents only 7% of all drug-tissue combinations, we investigated the performance of our predictive models in these specific instances to explore the possibility of tissue type confounding.
For any particular drug, a model based on tissue type would assign a sensitive prediction to samples originating from tissue with enriched sensitivity. This method results in an average accuracy of 0.47 for drugs in enriched tissue. In the same samples, the corresponding best elastic net models achieve a significantly higher (p < 10e-7) average accuracy of 0.62 (Supplementary Figure S3). This advantage in performance indicates that the deep learning representations are not merely recapitulating the tissue type of the input sample. Additional information regarding the cellular state is being encoded and utilized to predict drug sensitivity. Training tissue specific models is currently not feasible, but may be interesting as more experimental samples become available.
External validity of predictive models
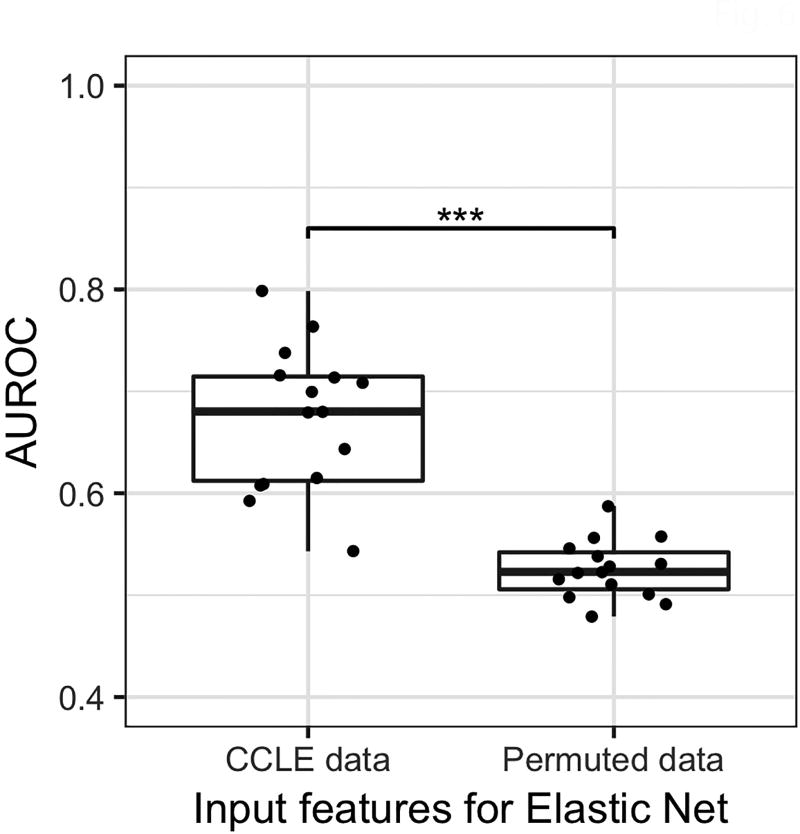
To further investigate the validity of our predictive models, we sought to evaluate them using data from a different study. A total of 15 of the drugs studied in the GDSC are also investigated in the Cancer Cell Line Encyclopedia (CCLE) pharmacogenomics study(7). Although the level of agreement between the two studies is unclear, the methods employed by the two groups are similar(23,24). We collected omics data from the CCLE, applied the autoencoder trained on GDSC data to derive features for the 1067 CCLE cell lines, and then applied the best elastic net models trained using GDSC to predict drug sensitivity for CCLE cell lines. We then evaluated these predictions against actual sensitivity calls from the CCLE experiment (Figure 6). The fifteen models achieved an average AUROC of 0.67. This is significantly higher than results obtained using randomly permuted input data (p < 10e-5), indicating that the relationships modeled by deep learning persist even under different experimental conditions.
Figure 6. External validity of predictive models.
AUROC values for fifteen elastic net models developed using GDSC omics data and autoencoder, evaluated using CCLE omics data, and randomly permuted data. *** p < 10e-6
DISCUSSION
In this study, we combined genome scale omics data and contemporary machine learning techniques to accurately predict the performance of a wide range of targeted and untargeted therapies on cancer cell lines. Our results indicate that data-driven approaches significantly outperform current rule-based methods using the genomic status of drug targets as therapeutic indicators. Although this represents a significant improvement, further refinements are possible. Individual models can be tuned for sensitivity or specificity based on the use case, and the performance of all models would be expected to improve with additional training data. The positive results reported here provide support for further investigating the extent to which the introduced methods can improve prediction of the sensitivity of patient tumors to currently available drugs. In addition, recent success using cell line studies to motivate the eventually successful clinical trials of cyclin D kinase 4/6 inhibitor palbociclib (25–27) indicates the value of cell line based drug screening.
Our study demonstrates that omics data contains information beyond genomic markers that are important and useful for the prediction of cancer drug sensitivity. As biotechnology advances and the cost of collecting omics data decreases, we anticipate that genomic, transcriptomic, proteomic, and metabolomics data may play a significant role in guiding data-driven precision medicine. In order for this to happen, the data must first be available. Therefore, systematic collection of molecular phenotypes must become standard clinical practice.
Precision oncology can and should be a practice of effectively utilizing all available treatments, including molecularly targeted, immunotherapy, and cytotoxic chemotherapies in a patient-specific manner. In the future, using procedures that build on the methods introduced here and elsewhere, we anticipate that an oncologist equipped with a computer-based decision support system will be able to select for any given patient an optimal regimen that maximizes therapeutic efficacy while minimizing the negative effects associated with ineffective treatments. We believe that such collaboration as well as the system itself will be key elements in realizing the promise of precision oncology.
Supplementary Material
Acknowledgments
Funding. Research reported in this publication was supported by grant R01LM012011 and 4T15LM007059-30 awarded by the National Library of Medicine. Funding was also provided by grant U54HG008540 awarded by the National Human Genome Research Institute through funds provided by the trans-NIH Big Data to Knowledge (BD2K) initiative (www.bd2k.nih.gov), and by grant #4100070287 awarded by the Pennsylvania Department of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Pennsylvania Department of Health.
List of abbreviations
- AUROC
area under receiving operator characteristic
- CCLE
the Cancer Cell Line Encyclopedia
- FDA
United States Food and Drug Administration
- GDSC
Genomics of Drug Sensitivity in Cancer
- PDX
Patient Derived Xenograft
- PPV
Positive Predictive Value
- SCNA
Somatic Copy Number Alteration
- SVM
Support Vector Machine
Footnotes
Conflict of interest. The authors state that they have no conflicts of interest to declare.
References
- 1.Fojo T. Precision oncology: a strategy we were not ready to deploy. Seminars in oncology. 2016;43(1):9–12. doi: 10.1053/j.seminoncol.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Prasad V, Fojo T, Brada M. Precision oncology: origins, optimism, and potential. The Lancet Oncology. 2016;17(2):e81–6. doi: 10.1016/S1470-2045(15)00620-8. [DOI] [PubMed] [Google Scholar]
- 3.Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. J Clin Oncol. 2013;31(15):1803–5. doi: 10.1200/JCO.2013.49.4799. [DOI] [PubMed] [Google Scholar]
- 4.Prasad V. Perspective: The precision-oncology illusion. Nature. 2016;537(7619):S63. doi: 10.1038/537S63a. [DOI] [PubMed] [Google Scholar]
- 5.Tannock IF, Hickman JA. Limits to Personalized Cancer Medicine. The New England journal of medicine. 2016;375(13):1289–94. doi: 10.1056/NEJMsb1607705. [DOI] [PubMed] [Google Scholar]
- 6.Rubio-Perez C, Tamborero D, Schroeder MP, Antolin AA, Deu-Pons J, Perez-Llamas C, et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer cell. 2015;27(3):382–96. doi: 10.1016/j.ccell.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nature medicine. 2015;21(11):1318–25. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 10.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166(3):740–54. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome biology. 2014;15(3):R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta S, Chaudhary K, Kumar R, Gautam A, Nanda JS, Dhanda SK, et al. Prioritization of anticancer drugs against a cancer using genomic features of cancer cells: A step towards personalized medicine. Scientific reports. 2016;6:23857. doi: 10.1038/srep23857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello JC, Heiser LM, Georgii E, Gonen M, Menden MP, Wang NJ, et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nature biotechnology. 2014;32(12):1202–12. doi: 10.1038/nbt.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Cai C, Chen V, Lu X. Learning a hierarchical representation of the yeast transcriptomic machinery using an autoencoder model. BMC bioinformatics. 2016;17(Suppl):1–9. doi: 10.1186/s12859-015-0852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartigan JAH, P M. The Dip Test of Unimodality. The Annals of Statistics. 1985;13(1):14. [Google Scholar]
- 16.Tibshirani R, Hastie T. Outlier sums for differential gene expression analysis. Biostatistics. 2007;8(1):2–8. doi: 10.1093/biostatistics/kxl005. [DOI] [PubMed] [Google Scholar]
- 17.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of statistical software. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortes C, Vapnik V. Support-Vector Networks. Machine Learning. 1995;20(3):273–97. doi: 10.1023/a:1022627411411. [DOI] [Google Scholar]
- 20.Hellwig B, Hengstler JG, Schmidt M, Gehrmann MC, Schormann W, Rahnenfuhrer J. Comparison of scores for bimodality of gene expression distributions and genome-wide evaluation of the prognostic relevance of high-scoring genes. BMC bioinformatics. 2010;11:276. doi: 10.1186/1471-2105-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Cai C, Chen V, Lu X. Trans-species learning of cellular signaling systems with bimodal deep belief networks. Bioinformatics. 2015;31(18):3008–15. doi: 10.1093/bioinformatics/btv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–7. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 23.Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504(7480):389–93. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Cell Line Encyclopedia C, Genomics of Drug Sensitivity in Cancer C. Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015;528(7580):84–7. doi: 10.1038/nature15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 26.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast cancer research : BCR. 2009;11(5):R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finn RS, Martin M, Rugo HS, Jones S, Im SA, Gelmon K, et al. Palbociclib and Letrozole in Advanced Breast Cancer. N Engl J Med. 2016;375(20):1925–36. doi: 10.1056/NEJMoa1607303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.