Abstract
Metagenomics and next generation sequencing were employed to discover new virus natural enemies of the fire ant, Solenopsis invicta Buren in its native range (i.e., Formosa, Argentina) with the ultimate goal of testing and releasing new viral pathogens into U.S. S. invicta populations to provide natural, sustainable control of this ant. RNA was purified from worker ants from 182 S. invicta colonies, which was pooled into 4 groups according to location. A library was created from each group and sequenced using Illumina Miseq technology. After a series of winnowing methods to remove S. invicta genes, known S. invicta virus genes, and all other non-virus gene sequences, 61,944 unique singletons were identified with virus identity. These were assembled de novo yielding 171 contiguous sequences with significant identity to non-plant virus genes. Fifteen contiguous sequences exhibited very high expression rates and were detected in all four gene libraries. One contig (Contig_29) exhibited the highest expression level overall and across all four gene libraries. Random amplification of cDNA ends analyses expanded this contiguous sequence yielding a complete virus genome, which we have provisionally named Solenopsis invicta virus 5 (SINV-5). SINV-5 is a positive-sense, single-stranded RNA virus with genome characteristics consistent with insect-infecting viruses from the family Dicistroviridae. Moreover, the replicative genome strand of SINV-5 was detected in worker ants indicating that S. invicta serves as host for the virus. Many additional sequences were identified that are likely of viral origin. These sequences await further investigation to determine their origins and relationship with S. invicta. This study expands knowledge of the RNA virome diversity found within S. invicta populations.
Introduction
The red imported fire ant, Solenopsis invicta Buren is an invasive species native to southern South America [1]. The ant was introduced into North America sometime in the 1930s [1], most likely from somewhere in Formosa Province, Argentina [2]. This ant is a very serious pest in the U.S., but generally not in its native range; although it is one of the most ecologically dominant ant species in northeastern Argentina [3, 4]. Damage and control efforts cost an estimated $6 billion annually in the U.S. [5]. Population studies on the two continents have shown that fire ant populations are 5–10 times greater in infested areas within the U.S. [6, 7]. These inter-continental disparities support the supposition that S. invicta likely escaped its natural enemies during U.S. founding events. Indeed, direct evaluations have shown a paucity of natural enemies in founding populations of S. invicta [8].
In the U.S., early eradication efforts were attempted [1], but eventually gave way to the implementation of quarantine [9] to limit the spread of the ant. Concomitantly, research focus shifted from eradication to the discovery, characterization, and release of natural enemies of S. invicta with the intention of providing sustainable control in the U.S. This effort led to the discovery of many pathogens and parasites of fire ants in their native and introduced ranges [10], some of which have been utilized and released as natural control agents against invasive fire ants in the U.S. [11–13]. Still, there remains a large discrepancy in both the abundance and the number of natural enemies found between populations of S. invicta in South and North America [14, 15] warranting continued efforts to identify new pathogens for use in providing natural control.
Despite the known usefulness of viruses to control insect pests [16], viruses have been only recently investigated for use against ants [11, 17]. Indeed, the first ant viruses discovered and characterized were from S. invicta [14]. To date, four RNA viruses and one DNA virus have been discovered from S. invicta. The RNA viruses include Solenopsis invicta virus 1 [18], Solenopsis invicta virus 2 [19], Solenopsis invicta virus 3 [20], and Solenopsis invicta virus 4 [21]. All of these viruses are present in both U.S. and Argentine populations of S. invicta. Solenopsis invicta queens infected with Solenopsis invicta virus 1 (SINV-1) have lower body weights that reduce the probability of successful colony founding [22]. Solenopsis invicta virus 2 (SINV-2) infections are associated with significant reductions in queen fecundity and other detrimental fitness effects including longer claustral periods and slower growth of incipient colonies [22]. Solenopsis invicta virus 3 (SINV-3) also reduces queen fecundity [23] and alters the feeding behavior exhibited by the worker caste, which results in colony starvation [24]. The impacts of Solenopsis invicta virus 4 (SINV-4) and the sole DNA virus, Solenopsis invicta densovirus (SiDNV), have not been established [25].
The objective of this research was to discover new virus natural enemies of S. invicta from the native range (i.e., Formosa, Argentina), with the ultimate goal of their release into introduced U.S. populations as self-sustaining biocontrol agents. Metagenomics and next generation sequencing were employed to achieve this objective and resulted in the discovery of one new virus and multiple high-probability target sequences of likely viral origin providing future leads to pursue.
Materials and methods
Solenopsis invicta collections
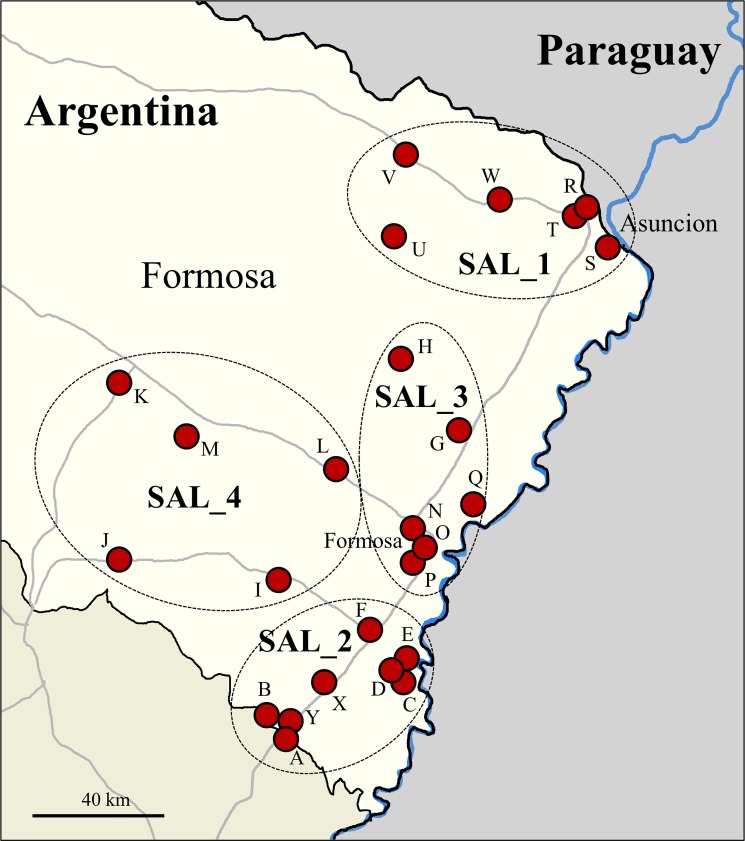
Samples of workers, brood, and sexuals were obtained from a total of 182 S. invicta colonies collected at 25 locations in the eastern portion of the Province of Formosa, Argentina (Fig 1). Collections were limited to the Province of Formosa by design because S. invicta in the U.S. has been shown to originate from this region [2]. Pathogen-matching based on genetic relatedness can have a profound influence on the ability of pathogens to infect a host [26]. Collection details, including specific location, date, and habitat are summarized in Table 1. After removal from the nest, live ants were held in 50 ml centrifuge tubes with a screened top and transported to the USDA quarantine facility in Gainesville, Florida, where they were placed in rearing trays and provided water and sugar water. We removed a sample of 15 workers 11–21 days after collection from each colony fragment for RNA extraction and any remaining individuals were frozen at -80 oC for future evaluations. Voucher specimens have been deposited in both the USDA-ARS, Center for Medical, Agricultural and Veterinary Entomology (CMAVE), Gainesville, Florida collection and the Fundación Para el Estudio de Especies Invasivas (FuEDEI), Hurlingham, Buenos Aires, Argentina collection.
Fig 1. Map of collection locations (capital letters) in Formosa Province, Argentina.
Dashed ovals show how locations were grouped for RNA library preparation. We sampled 2–9 colonies at each site. See Table 1 for colony numbers, GPS coordinates, and habitat descriptions.
Table 1. Summary of collection information for Solenopsis invicta colonies used in RNA library preparation.
Library designation represents South American Library (SAL).
| Site | Date | Latitude | Longitude | Habitat | Number of colonies | Library designation |
|---|---|---|---|---|---|---|
| R | 23-ix-14 | -25.2875 | -57.7054 | Dike along river | 7 | SAL_1 |
| S | 22-ix-14 | -25.3598 | -57.6631 | Dike | 5 | SAL_1 |
| T | 22-ix-14 | -25.2948 | -57.7445 | Road right of way | 8 | SAL_1 |
| U | 24-ix-14 | -25.3553 | -58.2796 | Low grass | 9 | SAL_1 |
| V | 24-ix-14 | -25.1432 | -58.2314 | Roadside | 9 | SAL_1 |
| W | 24-ix-14 | -25.2508 | -57.9907 | Roadside | 8 | SAL_1 |
| A | 16-ix-14 | -26.6609 | -58.6297 | River bank, Costanera park | 8 | SAL_2 |
| B | 16-ix-14 | -26.6222 | -58.6653 | Semi urban | 9 | SAL_2 |
| C | 17-ix-14 | -26.5173 | -58.2818 | Field adjacent to road | 9 | SAL_2 |
| D | 17-ix-14 | -26.4806 | -58.2737 | Marshy road area | 7 | SAL_2 |
| E | 17-ix-14 | -26.4795 | -58.3071 | City park, mowed grass |
9 | SAL_2 |
| F | 17-ix-14 | -26.3944 | -58.3442 | Gas station | 7 | SAL_2 |
| X | 25-ix-14 | -26.5412 | -58.5081 | Roadside | 1 | SAL_2 |
| Y | 25-ix-14 | -26.6474 | -58.6236 | Roadside | 3 | SAL_2 |
| G | 18-ix-14 | -25.8786 | -58.0872 | Abandoned parking area and periphery | 9 | SAL_3 |
| H | 18-ix-14 | -25.6733 | -58.2561 | Grassy roadside | 6 | SAL_3 |
| N | 20-ix-14 | -26.1182 | -58.2241 | Urban, highly disturbed | 5 | SAL_3 |
| O | 22-ix-14 | -26.1975 | -58.2144 | Adjacent to gas station | 7 | SAL_3 |
| P | 22-ix-14 | -26.2281 | -58.2405 | Abandoned picnic area | 9 | SAL_3 |
| Q | 22-ix-14 | -26.0416 | -58.0589 | Rural road side | 8 | SAL_3 |
| I | 19-ix-14 | -26.2391 | -58.6303 | Rough grassy area | 9 | SAL_4 |
| J | 19-ix-14 | -26.205 | -59.0708 | City park | 8 | SAL_4 |
| K | 19-ix-14 | -25.7411 | -59.1021 | Mowed road edge, west side | 9 | SAL_4 |
| L | 20-ix-14 | -25.9357 | -58.5124 | Road edge | 5 | SAL_4 |
| M | 20-ix-14 | -25.8637 | -58.8858 | Residential area | 8 | SAL_4 |
RNA preparation
Total RNA was extracted from a pooled group of 15 worker ants from each colony fragment using the Trizol method followed by the PureLink RNA Mini Purification Kit according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA). RNA quality of each preparation was assessed by microfluidic analysis on an Agilent 2100 Bioanalyzer (Agilent, Cary, NC) using the RNA 6000 Nano kit according to the manufacturer’s instructions. RNA samples were pooled from ant colonies into four groups according geographic region (Table 1; Fig 1). The four groups included South American Library _1 (SAL_1), collected north of 25.4o latitude; SAL_2, collected south of 26.3o latitude; SAL_3, collected east of 58.3o longitude; and SAL_4, collected west of 58.5o longitude. The total number of colonies pooled in each group was 46 (SAL_1), 53 (SAL_2), 44 (SAL_3), and 39 (SAL_4). Total RNA (10 μg per group) was submitted to GE Healthcare (Los Angeles, CA) for mRNA purification, library preparation, and Illumina RNA sequencing (MiSeq).
Library preparation and sequencing
Total RNA (200 ng) purified from each of the four pooled groups of worker ants (SAL_1, SAL_2, SAL_3, and SAL_4) was used for mRNA purification with the Illumina TruSeq Stranded mRNA Library Preparation Kit (Catalog # RS-122-2101). The low sample protocol was followed according to the manufacturer’s instructions. The RNA fragmentation step was omitted to maximize library insert length. Rather than fragmenting the RNA, the sealed plate was incubated at 80°C for 2 minutes to elute the primed mRNA from the RNA purification beads. This omission resulted in RNA fragmentation with an average final library size of 467 bp. Library sizes were determined empirically by microfluidic analysis on an Agilent 2100 Bioanalyzer and quantified using the Quant-iT dsDNA kit with broad range standards (ThermoFisher Scientific, Q-33130). Samples were pooled together at equimolar quantities and sequenced twice using the Illumina MiSeq (2x300) cycle kit with version 3 chemistry. Using the Illumina indices, the data were demultiplexed and the runs combined to assign the data to individual samples. All other procedures were followed according to the manufacturer’s instructions.
Bioinformatics analysis
Sequences were aligned to the Solenopsis invicta reference genome downloaded from http://www.ncbi.nlm.nih.gov/Traces/wgs/?val=AEAQ01#contigs using the Burrows-Wheeler Aligner (bwa-0.7.5a), a software package for mapping low-divergent sequences against a large reference genome [27]. The S. invicta unmapped reads were selected and converted to FASTA format using NextGENe-2.3.4 (SoftGenetics, State College, PA). Each read was then filtered and retained if the median score was ≥ 20 and base number ≥ 25. Unmapped and filtered individual MiSeq sequences were analyzed using BLASTX [28] against the curated Swiss Protein database (http://www.uniProt.org; download date 11/14/2014). Sequences returning an expectation score less than 10−5 were tabulated.
Based on the BLASTX results, each sequence was annotated and assorted taxonomically. The sequences were binned into the following groups: Animal, Plant, Fungi, Bacteria, Archaea, Phage, and Non-phage virus. Also at this stage, sequences exhibiting identity to Enterobacteria phage phiX174, an internal control for Illumina processing [29] were removed and not considered in subsequent analyses.
Virus sequences
Non-phage virus sequences from each library identified from the BLASTX analysis were assembled using the CAP3 algorithm [30] in the Vector NTI ContigExpress program (Invitrogen, Carlsbad, CA). Sequences from phage were not assembled because they infect bacteria and would not be expected to infect fire ant cells. Contiguous sequences (contigs) and remaining singletons were matched to the genomes of known fire ant viruses (Solenopsis invicta virus 1 [SINV-1, GCF_000854925.1], SINV-2 (GCF_000870805.1), SINV-3 (GCF_000881215.1), SINV-4 (MF_041808.1), and SiDNV (GCF_000912895.1)). Those sequences matching known fire ant viruses (≥ 95% identity) were binned according to virus species and excluded from further analysis. Virus unmatched sequences/contigs were re-analyzed by BLASTX and sequences returning an expectation score of less than 10−5 were tabulated.
Data availability
Raw sequence data from each library were deposited into the GenBank database as a Sequence Read Archive under accession number, SRP113235 (Bioproject PRJNA394996). Assembled sequences with viral identity (Table 2) have been deposited at DDBJ/EMBL/GenBank as a Transcriptome Shotgun Assembly project under the accession GFUG00000000. The version described in this paper is the first version, GFUG01000000. The SINV-5 annotated genome was deposited in GenBank under accession number MF593921.
Table 2. Contiguous sequences (Contigs) with significant viral identity by BLASTX analysis of the GenBank database and considered high likelihood viral prospects from RNA libraries created from Solenopsis invicta worker ants.
Contigs were first sorted in descending order based on the number of the sequences comprising it, followed by the libraries represented.
| Designation | Sequences comprising contig | Size (nt) | e-score | Query coverage:identity (%) | Sequence identity with | Number of singletons in | Genome | Virus family | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SAL_1 | SAL_2 | SAL_3 | SAL_4 | ||||||||
| Contig_29 (SINV-5) | 21294 | 9030 | 0 | 57: 67 | Israeli acute paralysis virus | 274 | 10312 | 4312 | 6396 | ssRNA | Dicistroviridae |
| Contig_66 | 13980 | 2626 | 2E-154 | 76: 39 | Aphid lethal paralysis virus | 3823 | 1704 | 5241 | 3212 | ssRNA | Dicistroviridae |
| Contig_30 | 6197 | 1869 | 1E-139 | 96: 40 | Aphid lethal paralysis virus | 2165 | 779 | 1833 | 1420 | ssRNA | Dicistroviridae |
| Contig_16 | 2104 | 2371 | 0 | 83:48 | Solenopsis invicta virus 1 | 2 | 133 | 1815 | 154 | ssRNA | Dicistroviridae |
| Contig_70 | 641 | 1068 | 6E-64 | 82: 43 | Alber virus | 117 | 180 | 141 | 203 | RNA | Unclassified |
| Contig_58 | 607 | 1453 | 1E-67 | 88: 34 | Nasonia vitripennis virus | 18 | 311 | 37 | 241 | RNA | Unclassified |
| Contig_13 | 510 | 1568 | 2E-102 | 87: 41 | Aphid lethal paralysis virus | 206 | 98 | 7 | 199 | ssRNA | Dicistroviridae |
| Contig_21 | 486 | 3211 | 0 | 85: 61 | Acute bee paralysis virus | 108 | 51 | 9 | 318 | ssRNA | Dicistroviridae |
| Contig_83 | 428 | 2623 | 0 | 97: 62 | Mosinovirus | 90 | 58 | 110 | 170 | RNA | Unclassified |
| Contig_55 | 202 | 2639 | 3E-147 | 91: 35 | Kashmir bee virus | 12 | 153 | 20 | 17 | ssRNA | Dicistroviridae |
| Contig_82 | 114 | 1203 | 0 | 82: 94 | Acute bee paralysis virus | 16 | 7 | 4 | 87 | ssRNA | Dicistroviridae |
| Contig_15 | 100 | 1042 | 0 | 92: 83 | Hubei orthoptera virus 1 | 28 | 14 | 3 | 55 | RNA | Unclassified |
| Contig_27 | 69 | 680 | 6E-135 | 91: 97 | Acute bee paralysis virus | 1 | 6 | 2 | 60 | ssRNA | Dicistroviridae |
| Contig_17 | 58 | 796 | 4E-162 | 99: 91 | Acute bee paralysis virus | 15 | 7 | 5 | 31 | ssRNA | Dicistroviridae |
| Contig_80 | 55 | 1173 | 0 | 98: 79 | Drosophila C virus | 44 | 4 | 3 | 4 | ssRNA | Dicistroviridae |
| Contig_75 | 36 | 589 | 6E-26 | 83: 39 | Hubei picorna-like virus 46 | 8 | 13 | 15 | 0 | RNA | Unclassified |
| Contig_47 | 734 | 1045 | 7E-82 | 89: 44 | Solenopsis invicta virus 2 | 0 | 667 | 6 | 61 | ssRNA | Unclassified |
| Contig_78 | 216 | 702 | 7E-111 | 99: 85 | Israeli acute paralysis virus | 153 | 42 | 21 | 0 | ssRNA | Dicistroviridae |
| Contig_57 | 146 | 1028 | 5E-116 | 93: 57 | Wuhan insect virus 11 | 0 | 93 | 28 | 25 | RNA | Unclassified |
| Contig_18 | 44 | 664 | 2E-84 | 97: 60 | Hubei Orthoptera virus 1 | 10 | 2 | 0 | 32 | RNA | Unclassified |
| Contig_88 | 44 | 1500 | 0 | 99: 87 | Rhopalosiphum padi virus | 12 | 0 | 7 | 25 | ssRNA | Dicistroviridae |
| Contig_53 | 39 | 522 | 1E-50 | 98: 52 | Wuhan insect virus 11 | 0 | 27 | 3 | 9 | RNA | Unclassified |
| Contig_9 | 9 | 471 | 3E-68 | 87: 95 | Acute bee paralysis virus | 3 | 0 | 3 | 3 | ssRNA | Dicistroviridae |
| Contig_85 | 8 | 542 | 2E-53 | 91: 56 | Solenopsis invicta virus 1 | 4 | 1 | 3 | 0 | ssRNA | Dicistroviridae |
| Contig_51 | 321 | 1465 | 0 | 99: 76 | Nodamura virus | 0 | 274 | 0 | 47 | ssRNA | Nodaviridae |
| Contig_49 | 182 | 1269 | 0 | 99: 95 | Aphid lethal paralysis virus | 0 | 153 | 29 | 0 | ssRNA | Dicistroviridae |
| Contig_12 | 95 | 438 | 2E-79 | 99: 77 | Big Sioux River virus | 49 | 0 | 0 | 46 | ssRNA | Unclassified |
| Contig_50 | 69 | 768 | 9E-68 | 99: 46 | Israeli acute paralysis virus | 0 | 68 | 0 | 1 | ssRNA | Dicistroviridae |
| Contig_60 | 69 | 926 | 0 | 99: 93 | Aphid lethal paralysis virus | 0 | 55 | 14 | 0 | ssRNA | Dicistroviridae |
| Contig_28 | 67 | 830 | 1E-46 | 97: 37 | Israeli acute paralysis virus | 0 | 0 | 13 | 54 | ssRNA | Dicistroviridae |
| Contig_52 | 66 | 356 | 1E-35 | 99: 94 | Shuangao insect virus 8 | 0 | 20 | 0 | 46 | RNA | Unclassified |
| Contig_25 | 58 | 869 | 8E-10 | 37: 40 | Hubei picorna-like virus 50 | 0 | 0 | 20 | 38 | RNA | Unclassified |
| Contig_141_3101 | 3101 | 2351 | 0 | 97: 55 | Solenopsis invicta virus 1 | 0 | 0 | 0 | 3101 | ssRNA | Dicistroviridae |
| Contig_19 | 1075 | 1080 | 2E-159 | 89: 71 | Solenopsis invicta virus 1 | 0 | 0 | 0 | 1075 | ssRNA | Dicistroviridae |
| Contig_19_480 | 480 | 623 | 8E-114 | 99: 88 | Solenopsis invicta virus 1 | 0 | 0 | 480 | 0 | ssRNA | Dicistroviridae |
| Contig_110_361 | 361 | 1214 | 0 | 99: 82 | Kashmir bee virus | 0 | 0 | 0 | 361 | ssRNA | Dicistroviridae |
| Contig_81_84 | 84 | 1103 | 6E-161 | 99: 67 | Wuhan arthropod virus 2 | 0 | 0 | 84 | 0 | RNA | Unclassified |
| Contig_108_79 | 79 | 715 | 5E-131 | 98: 83 | Shuangao insect virus 8 | 0 | 79 | 0 | 0 | RNA | Unclassified |
Virus genome re-sequencing
Contig_29 was unquestionably a near complete virus RNA genome. Therefore, this sequence was used as template for 5' and 3' RACE to acquire the entire genome sequence. For 3' RACE, cDNA was synthesized with the GeneRacer Oligo dT primer (Invitrogen, Carlsbad, CA). PCR was subsequently conducted with the GeneRacer 3' primer and gene-specific primer, P1604 (S1 Table). For 5' RACE, cDNA was synthesized with oligonucleotide primer P1601 and PCR conducted with P1601 and the GeneRacer Abridged Anchor Primer. Amplicons generated during RACE reactions were cloned into pCR4 vector and submitted for Sanger sequencing. After acquiring the genome termini, oligonucleotide primers were designed to provide complete, overlapping coverage of Contig_29. Amplicons were cloned and sequenced by the Sanger method and the genome assembled with CAP3 in Vector NTI (Life Technologies, Carlesbad, CA).
Preliminary RNA virus confirmation
In order to provide further evidence of whether a contig was of viral origin, or not, oligonucleotide primers (S1 Table) were designed to contigs with viral identity and expressed in all four libraries (i.e. the first 15 contigs in Table 2). RT-PCR was conducted with RNA pooled from all four libraries to establish the orientation of the template and to verify that cDNA synthesis was required for amplification. Once established, RT-PCR was conducted with RNA derived from S. invicta colonies (worker ants) field-collected from around Gainesville, Florida, (n = 27 from 3 locations) to determine whether the sequence was present in the U.S. field population and, if so, its prevalence. This experiment also provided additional confirmation of a potential viral origin. If amplification was observed in 100% of the samples, it was assumed that the sequence was of host origin and further experiments with the sequence were terminated. Viral infections rarely exhibit an incidence of 100% among field-collected arthropods [31].
Finally, to establish whether Contig_29 (SINV-5) was actively replicating in S. invicta, strand-specific RT-PCR was conducted to detect the replicative genome strand by the modified method of Craggs et al. [32]. Pooled total RNA (50 ng) used for library creation was mixed with 10 mM dNTPs, 1 μM of tagged reverse oligonucleotide primer p1600TAG and heated to 65°C for 5 minutes. First strand buffer and Superscript reverse transcriptase (Invitrogen, Carlsbad, CA) were then added and the reaction mixture was incubated at 55°C for 30 minutes before inactivating the RT at 70°C for 15 minutes. Unincorporated cDNA oligonucleotides were digested with 10 units of Exonuclease I (New England Biolabs, Ipswich, MA) at 37°C for 1 hour. The reaction was terminated by heating to 80°C for 20 minutes. PCR was subsequently conducted with minus-strand specific cDNA as template. The reaction was conducted in a 25 μl volume containing 2 mM MgCl2, 200 μM dNTP mix, 0.5 units of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), 0.2 μM of each oligonucleotide primer p1601 and TAG (S1 Table), and 5 μl of the cDNA preparation. The temperature cycling program was 1 cycle at 94° C for 2 minutes, 35 cycles of 94°C for 15 seconds, 59°C for 15 seconds, 68°C for 40 seconds, and 1 cycle of 68°C for 5 minutes. PCR products were separated on an agarose gel (1%) and visualized by SYBR-safe (Invitrogen, Carlsbad, CA) staining. Plus strand RT-PCR was included as a positive control as well as a non-template negative control.
Results
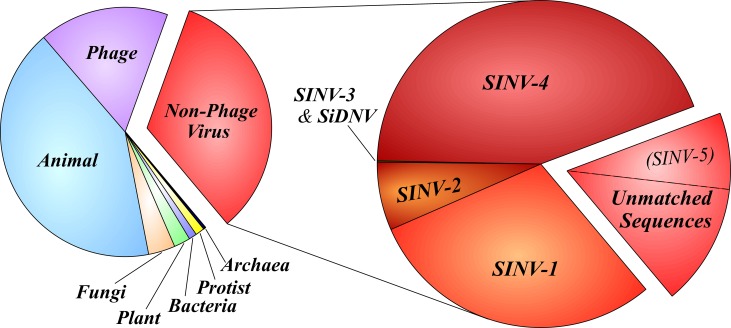
Fig 2 summarizes the annotation of the S. invicta-unmatched sequences. Phage and non-phage virus categories comprised 50.2% of the total number of non-S. invicta sequences (n = 876,528). The number of non-phage virus sequences was 292,499, or 33.4% of the total. Surprisingly, phage were well represented despite low numbers of bacterial sequences. However, those phage sequences detected exhibited high expression levels and were limited to the Microviridae (ssDNA). The remaining sequences were annotated to the following taxa: Animal (41.7%), Fungi (3.4%), Plant (2.1%), Protist (1.1%), Bacteria (1.0%), and Archaea (0.3%). The relative number of sequences across these broad taxonomic categories was fairly consistent among the four gene libraries (Fig 3A). Detection of a high percentage of animal-related sequences were largely fire ant sequences that were not filtered out during the fire ant matching phase.
Fig 2. Graphical summary of broad taxonomic assignments of RNA sequences from the four combined gene libraries (Left) which did not match the Solenopsis invicta fire ant genome.
These libraries were derived from 182 S. invicta colonies sampled from the 25 locations shown in Fig 1. Phage sequences were not of interest because they infect bacteria. The expanded pie chart (Right) shows the percentage of non-phage virus sequences matching known S. invicta viruses and those remaining sequences with significant viral identity. Note that both SINV-3 and SiDNV only accounted for a very small percentage of matched contigs. The dashed line in the unmatched section shows the proportion of sequences belonging to the new fire ant virus (SINV-5) described in this paper.
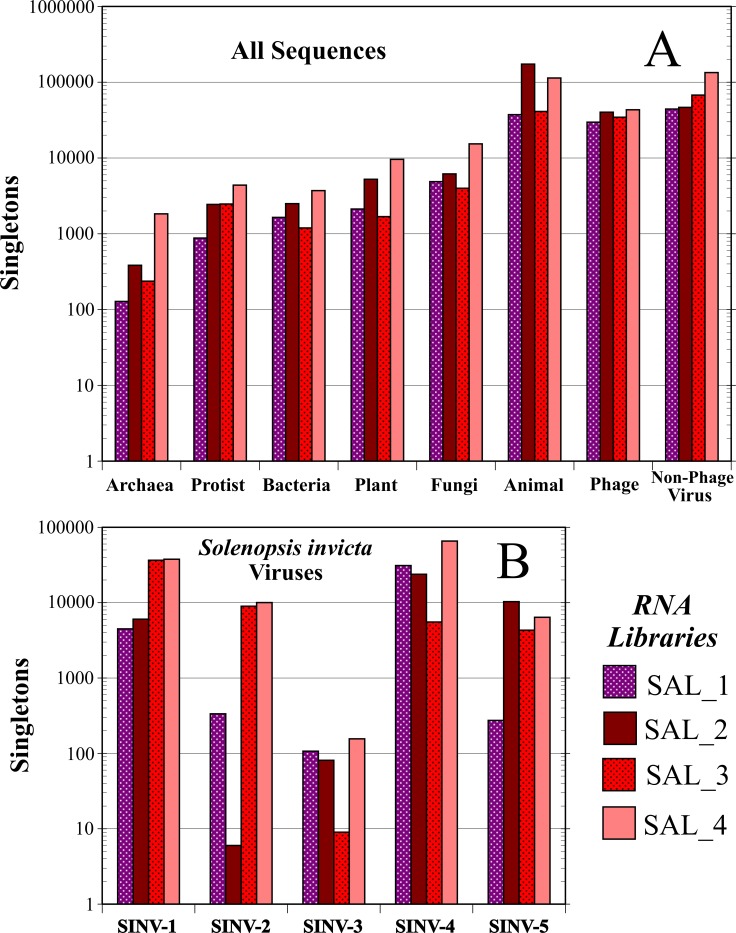
Fig 3. Geographic variation of singleton expression levels of sequences for each RNA library (see Fig 1 for map of collection locations and RNA library groupings).
Sequences were binned according to broader taxonomic categories (Top) and known or newly described Solenopsis invicta viruses (Bottom). Note that expression levels are shown in log scales.
Further winnowing was accomplished by removing the known S. invicta virus sequences from the non-phage virus pool (Fig 2). The total number of S. invicta virus-matched sequences was 230,555 (i.e., 78.8% of the non-phage sequences). SINV-4 (n = 126,086) represented the largest fraction of S. invicta known virus sequences in the pooled libraries, followed by SINV-1 (n = 84,769), SINV-2 (n = 19,338), SINV-3 (n = 353), and SiDNV (n = 9) (Fig 2). The small number of SiDNV sequences may be explained because SiDNV is a DNA virus which would normally only produce RNA while actively replicating. Conversely, many RNA viruses would be detected while replicating or not. Unlike the broader taxonomic assignments, the prevalence of S. invicta viruses varied by several orders of magnitude across each library/region (Fig 3B).
The S. invicta virus-unmatched sequences (n = 61,944) were assembled de novo yielding 171 contiguous sequences (comprised of 55,677 singletons) with significant identity to non-plant virus genes (Table 2 and S2 Table). An additional 30 contiguous sequences were assembled from 633 singletons, which showed significant identity to non-virus genes and 35 contiguous sequences (596 singletons) with significant plant virus identity, all of which were excluded from further examination. The remaining singletons and contiguous sequences (n = 5,038) were also excluded from further analysis because they either did not match any sequence in the GenBank database and/or were less than 100 nucleotides in length.
Among the 171 contiguous sequences with significant identity to non-plant virus genes, Table 2 summarizes those considered most likely to be of viral origin (n = 38) and using S. invicta as host. This assumption was based on the total sequence expression representation, representation across the libraries (i.e., by geography), and contig size. Expression representation has been used successfully to detect pathogens and is simply based on the fact that actively replicating genes (e.g., from viruses infecting a host) will be highly represented in non-normalized gene libraries [33]. Among the 38 sequences in Table 2 exhibiting significant identity with viral sequences from the GenBank database, fifteen (9%) were represented in all four gene libraries and were composed of 46,845 of the 55,677 singletons. Thus, these fifteen contiguous sequences alone accounted for 84% of the singletons assembled with non-plant virus identity and were considered high likelihood viral prospects. Nine contigs contained sequences detected in three of the four libraries and were composed of 1,276 singletons; eight contigs contained sequences detected in two of the four libraries and were composed of 927 singletons. Finally, six contigs contained sequences detected in a single library, but were highly represented within the single library (composed of 5,180 singletons). In total, these 38 contigs contained 54,228 singletons, or 97.4% of all sequences of those with non-plant virus identity. All of these sequences exhibited identity to viruses with RNA genomes, the majority of which to the Dicistroviridae (n = 23). Fourteen of the sequences exhibited identity with unclassified viruses and one with a virus in the Nodaviridae (i.e., Contig_51).
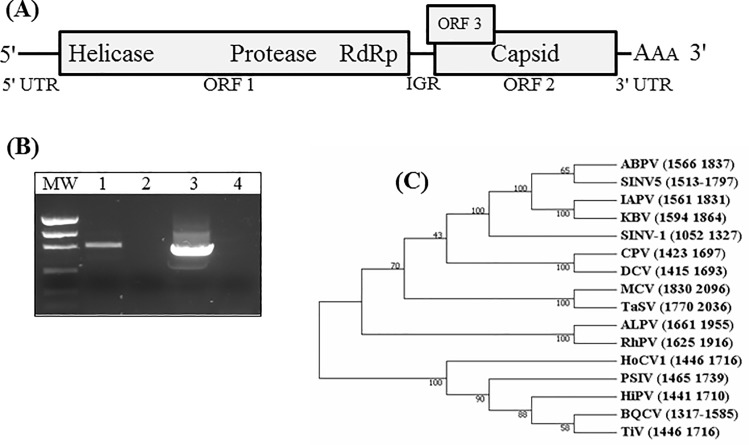
Sequences with the highest level of expression and represented in all four gene libraries (n = 15; Table 2) were considered the most likely prospects to be of viral origin (henceforth referred as high likelihood viral prospects). Therefore, we focused our effort on these fifteen contigs (Table 2). Contig_29 exhibited the highest expression level overall and across all four gene libraries; this sequence appeared to be a near complete virus genome. The large contig sequence was 9,030 nucleotides in length and BLASTX analysis [28] indicated that the sequence had significant identity to Israeli acute paralysis virus (IAPV) and other RNA viruses in the Dicistroviridae. Sanger re-sequencing and RACE reactions revealed a 9,313 nucleotide polyadenylated genome containing two, in-frame, open reading frames (ORFs) separated and flanked by untranslated regions, and a short, overlapping ORF at the 5' end of ORF 2 (Fig 4A). The 5'-proximal ORF contained domains with identity to RNA helicase (pfam00910), virus peptidase (pfam12381), and RNA-dependent RNA polymerase (pfam00680). The 3'-proximal ORF contained domains with identity to CRPV capid proteins (pfam08762) and the capsid protein, VP4, from dicistroviruses (pfam 11492). These characteristics indicate that this genome sequence represents a new dicistrovirus [34]. The virus is provisionally named, Solenopsis invicta virus 5 (SINV-5) and the sequence deposited in GenBank under accession number MF593921. Further evaluation of the SINV-5 genome revealed that the replicative strand was detected in S. invicta (from Argentinean colonies). The presence of a replicative genome strand and a high expression level of SINV-5 sequences detected in all four gene libraries indicate that S. invicta likely serves as host for the virus (Fig 4B). Phylogenetic analysis of the conserved RdRp region of ORF 1 of SINV-5 with the known Dicistroviridae species shows that SINV-5 assorts with dicistroviruses within the Aparavirus genus, near SINV-1 (Fig 4C). The short, overlapping ORF at the 5' end of the structural ORF (ORF 2) provides further support for the Aparavirus placement.
Fig 4.
SINV-5 genome architecture (A), detection of replicative genome strand (B), and phylogenetic analysis of the RNA-dependent RNA polyermase of SINV-5 and known dicistroviruses (C). (A) The SINV-5 genome is represented by the center black line and open reading frames by rectangles. Proteins with identity to an RNA helicase, protease, and RNA-dependent RNA polymerase (non-structural proteins) were detected within ORF 1 and those with identity to virus capsid proteins were detected in ORF 2. A short, overlapping ORF3 at the 5' end of ORF2 provides support for Aparavirus placement. (B) RT-PCR amplification of each SINV-5 genome strand. Replicative strand (lane 1), corresponding control (lane 2), positive strand (lane 3) and corresponding control (lane 4). (C) The evolutionary relationship of SINV-5 with known dicistroviruses by comparison of the RNA-dependent RNA polymerase from each virus inferred using the Neighbor-Joining method [43]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches [44]. Only the most conserved region of the RdRp was aligned and a total of 243 positions were included in the final dataset (exact positions for the translated ORF1 are indicated in the phylogenetic tree within parentheses by each taxa). Evolutionary analyses were conducted in MEGA7 [45]. Abbreviations and GenBank Accession numbers for regions of the translated ORF1 (shown in graph) used for analysis include SINV-5 (Solenopsis invicta virus 5; MF593921), ABPV (Acute bee paralysis virus; NC002548), IAPV (Israeli acute bee paralysis virus; NC009025), KBV (Kashmir bee virus; NC004807), SINV-1 (Solenopsis invicta virus 1; NC006559), CPV (Cricket paralysis virus; NC003924), DCV (Drosophila C virus; NC001834), MCV (Mud crab virus; NC014793), TaSV (Taura syndrome virus; NC003005), ALPV (Aphid lethal paralysis virus; NC004365), RhPV (Rhopalosiphum padi virus; NC001874), HoCV (Homalodisca coagulata virus 1; NC008029), PSIV (Plautia stali intestine virus; NC003779), HiPV (Himetobi P virus; NC003782), BQCV (Black queen cell virus; NC003784), TrV (Triatoma virus; NC003783).
Contiguous sequences (specifically, Contig_66, Contig_30 and Contig_16) also exhibited high expression levels and were represented in all four libraries. These contigs also exhibited significant identity to dicistrovirus non-structural and structural proteins and likely represent new virus species. The remaining 11 contigs, Contig_70 to Contig_80 (Table 2), had lower overall expression levels, but were considered high likelihood virus prospects because they were detected in all four libraries (geographic regions) and exhibited significant identity with viral genes.
Among the fifteen high likelihood viral sequences identified (Table 2), PCR amplification only occurred after reverse transcription (Table 3). No amplification was detected without reverse transcription confirming that these templates were RNA. A small number of field-collected S. invicta colonies from Florida (n = 27) were also examined by RT-PCR to determine whether any of the fifteen high likelihood viral sequences were present in the U.S. S. invicta population (Table 3). Five of the templates were apparently present in the U.S. population ranging in prevalence from 15 to 56%. However, the majority were not detected in U.S. S. invicta samples.
Table 3. gs composed of sequences from all four gene libraries evaluated by RT-PCR and PCR.
Conti Reverse transcription was required for successful amplification of all contigs. RT-PCR was also conducted with RNA obtained from S. invicta colonies (n = 27) collected in the U.S. to determine if the contig was present in the introduced range.
| Contig designation | PCR amplification with cDNA synthesis | PCR amplification without cDNA synthesis | Presence in U.S. (%) |
|---|---|---|---|
| Contig_29 (SINV-5) |
Yes | No | Not detected |
| Contig_66 | Yes | No | 56 |
| Contig_30 | Yes | No | 56 |
| Contig_16 | Yes | No | 56 |
| Contig_70 | Yes | No | 26 |
| Contig_58 | Yes | No | Not detected |
| Contig_13 | Yes | No | 15 |
| Contig_21 | Yes | No | Not detected |
| Contig_83 | Yes | No | Not detected |
| Contig_55 | Yes | No | Not detected |
| Contig_82 | Yes | No | Not detected |
| Contig_15 | Yes | No | Not detected |
| Contig_27 | Yes | No | Not detected |
| Contig_17 | Yes | No | Not detected |
| Contig_80 | Yes | No | Not detected |
Discussion
In an effort to discover new viral pathogens to possibly control S. invicta in the U.S., we collected samples from 182 nests from four distinct geographic areas across the Formosa region of Argentina, created gene libraries from each of these pooled groups, and sequenced each of them by the Illumina Miseq method. Through a series of winnowing methods, 171 contiguous sequences with significant viral identity were ultimately identified as viral candidates.
Among these 171 possibilities, we focused on 15 contigs because they exhibited the highest expression levels and were detected in all 4 geographic regions (Table 2). They were analyzed in an attempt to establish their origin—whether viral, host, or otherwise. Solenopsis invicta is an omnivorous insect, so viruses infecting prey or plant food items must be identified and excluded from consideration. We largely employed the step-by-step decision tree reported previously [35] as a general guide to determine the likelihood that a given sequence was of viral origin. Based on previous studies [18, 36], this winnowing method significantly improves virus identification and discovery. In addition to this decision tree, the relative prevalence of each contig/sequence was considered supporting evidence for viral replication and host status. Because no form of mRNA subtraction was conducted before library preparation, the representation of each transcript was expected to be relatively proportional to the actual number of sequences present in the sample. Basically, ingested viruses would likely be represented by fewer sequence copies. Conversely, replicating viruses would be indicated by higher sequence copy numbers. Furthermore, sequences found in more than a single region/library would provide further support that it was, in fact, of virus origin and infecting S. invicta.
However, low representation does not preclude the possibility that a sequence is not from a virus infecting S. invicta. Indeed, low gene/genome copy number has been reported for actively replicating viruses that have detrimental effects on their hosts [22]. Additionally, some RNA viruses, which can be virulent and highly represented, may occur in low numbers because of seasonal variation, low host infection rates, or other unknown factors [37, 38].
One nearly complete virus genome was assembled among the high likelihood virus contigs (Contig_29; Table 2). RACE reactions using this contig as template and subsequent Sanger re-sequencing resulted in a complete virus genome. We have provisionally named this virus sequence SINV-5. It exhibits characteristics consistent with dicistroviruses in the Aparavirus genus, including, a monopartite genome containing two, in-frame, ORFs that are flanked and separated by untranslated regions, a short, overlapping ORF at the 5' end of ORF 2, and a polyadenylated 3' terminus. The 5'-proximal ORF contained domains with identity to RNA helicase, protease, and RNA-dependent RNA polymerase, and the 3'-proximal ORF contained domains with identity to virus capsid proteins. SINV-5 was only detected in S. invicta colonies from Argentina; it was not detected in a limited number of S. invicta colonies from three locations in the U.S. In addition to the genomic architecture, phylogenetic analysis of the RdRp further supports a taxonomic assignment of SINV-5 in the Dicistroviridae (Fig 4C). Taken together, high expression levels, detection of a replicative strand, and phylogeny of SINV-5 indicate that S. invicta serves as the host for this novel virus (Fig 4B). Depending on the impact of this virus on fire ants and it host specificity, it may be a candidate for introduction into the U.S. as a classical biological control agent for S. invicta.
We also discovered what will likely be a second virus among the unidentified contigs in Table 2. Two contigs (Contig_66 and Contig_30) exhibited identity with Aphid lethal paralysis virus (ALPV) structural and non-structural proteins, respectively (Table 2). Because ALPV is a dicistrovirus, we postulated that these two fragments may have been part of the same genome, but were lacking sequence linking them. The number of sequences comprising each of these contigs further supported this notion. Notice that in every library (SAL-1, -2, -3, and -4), more sequences comprised Contig_66 than Contig_30 (Table 2). This relationship would be expected in a host in which a dicistrovirus was actively replicating. Specifically, there would be a molar excess of capsid proteins compared with non-structural proteins. RT-PCR with a reverse primer (P1617) specific for Contig_30 and forward primer (P1631) specific for Contig_66 produced an amplicon (~3 kbp), whose sequence linked the two contigs. The joined contig was 4,505 nucleotides in length. This sequence was detected in both Argentinean and U.S. S. invicta populations (Table 3) and likely represents another virus as the replicative strand of this genome was also detected in U.S. and Argentinean S. invicta colonies (Data not shown).
A majority of the contigs tested by RT-PCR (10/15; Table 3) were only detected in the RNA libraries from Argentina. This fits well with the hypothesis that most of the natural enemies of S. invicta were left behind in South America when it was accidently introduced into the U.S. [7]. Nevertheless, it is of interest that 4 of the 5 S. invicta viruses discovered to date, plus the likely one just mentioned above, plus 5 of 15 likely viral contigs tested (Table 3) have been found in both North and South American populations of S. invicta. Similarly, the microsporidian pathogen Kneallhazia solenopsae has been found infecting fire ants on both continents, but the microsporidian, Vairimorpha invictae is only found in South America [39]. This frequency of pathogens found on both continents supports the conclusion that multiple colonies of S. invicta were introduced into the U.S., perhaps over a period of years [1] because it is highly unlikely that a single invading colony or only a few colonies would carry this many pathogens [37]. It also appears that most of the viruses found in the U.S. were naturally common in native South American fire ant populations (Fig 3). Certainly, at least 3 of the viruses (SINV-1, SINV-2, SINV-3) can be seasonally abundant in U.S. populations [37]. Another possible explanation for viral pathogens on both continents is that some may be generalists with a wide host range that already occurred in the U.S. on other ant species, especially some of the native fire ants. However, the two viruses tested to date (SINV-1 and SINV-3) appear to have originated from South America and are specific for S. invicta [38, 40, 41].
While high expression levels are a logical place to start to discover new viruses from next generation sequencing data, low expression levels do not necessarily preclude a sequence from consideration. Indeed, SINV-2 was shown to exhibit comparatively low expression in S. invicta queens, yet had a profound impact on fecundity and gene expression during colony founding [22]. In fact, low copy sequences from gene libraries have previously resulted in virus discovery from the tawny crazy ant, Nylanderia fulva [35]. S2 Table contains 133 contiguous sequences with significant viral identity, all comprised of fewer than 50 singletons. Most of these sequences exhibited significant identity with RNA viral genomes, however, seven sequences showed identity to DNA viral genomes (5 single stranded and 2 double stranded). Thus, there are many possible virus leads resulting from this study that require additional investigation to establish their origin and relationship to S. invicta. All sequences have been deposited in Genbank to facilitate and encourage the discovery of additional viral pathogens of S. invicta in South America. Hopefully, new homologies can be discovered by employing new search algorithms and by periodically reanalyzing these libraries as new viral sequences are added to sequence databases.
In conclusion, our ongoing metagenomics/next generation sequencing efforts [14] have been very successful. In this study, we were able to match 79% of the non-phage virus contigs to known fire ant viruses (Fig 2). We also expanded the virome of S. invicta by discovery of a new S. invicta-infecting virus, provisionally named, SINV-5. SINV-5 is of particular interest because it does not appear to occur in the introduced U.S. range of S. invicta and therefore may be able to be released as a self-sustaining classical biological control agent for these invasive ants in the U.S. Another apparent virus sequence (Contigs_30 and 66; Table 2) appears to be a new virus of S. invicta, although the genome is incomplete. To date, we have discovered and sequenced the entire genomes of 6 viruses in South American fire ants (i.e., SINV-1, -2, -3, -4, -5 and SiDNV). By way of comparison, 31 viruses have been characterized from honey bees [42]. Future work with SINV-5 and other newly discovered fire ant viruses will focus on their pathogenicity, host specificity, and seasonality in order to assess their potential for use as self-sustaining biocontrol agents and/or biopesticides.
Supporting information
(DOCX)
Contigs were first sorted in descending order based on the number of the sequences comprising it, followed by the libraries represented.
(DOCX)
Acknowledgments
The use of trade, firm, and corporation names in this publication are for the information and convenience of the reader. Such use does not constitute an official endorsement or approval by the United States Department of Agriculture or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable. We thank Mariel Guala for her invaluable assistance with the field work. Ant collections were carried out under collecting permits issued by Subsecretaria de Recursos Naturales, Ordenamiento y Calidad Ambiental of the province of Formosa (Nota N° 108/13). Export permits were issued by the Secretaría de Ambiente y Desarrollo Sustentable (N° 24767) and the Servicio Nacional de Sanidad y Calidad Agroalimentaria (Nota DNPV N° 747) of Argentina.
Data Availability
Raw sequence data from each library were deposited into the GenBank database as a Sequence Read Archive under accession number, SRP113235 (Bioproject PRJNA394996). Assembled sequences with viral identity (Table 2) have been deposited at DDBJ/EMBL/GenBank as a Transcriptome Shotgun Assembly project under the accession GFUG00000000. The version described in this paper is the first version, GFUG01000000. The SINV-5 annotated genome was deposited in GenBank under accession number MF593921.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Tschinkel WR. The fire ants. Cambridge: The Belknap Press of Harvard University Press; 2006. 723 p. [Google Scholar]
- 2.Caldera EJ, Ross KG, DeHeer CJ, Shoemaker D. Putative native source of the invasive fire ant Solenopsis invicta in the USA. Biological Invasions. 2008;10(8):1457–79. [Google Scholar]
- 3.LeBrun EG, Tillberg CV, Suarez AV, Folgarait PJ, Smith CR, Holway DA. An experimental study of competition between fire ants and Argentine ants in their native range. Ecology. 2007;88(1):63–75. doi: 10.1890/0012-9658(2007)88[63:Aesocb]2.0.Co;2 PubMed PMID: WOS:000245668300010. [DOI] [PubMed] [Google Scholar]
- 4.Calcaterra LA, Livore JP, Delgado A, Briano JA. Ecological dominance of the red imported fire ant, Solenopsis invicta, in its native range. Oecologia. 2008;156(2):411–21. Epub 2008/02/29. doi: 10.1007/s00442-008-0997-y . [DOI] [PubMed] [Google Scholar]
- 5.Pereira RM. Areawide suppression of fire ant populations in pastures: project update. J Agric Urban Entomol. 2003;20(2):123–30. [Google Scholar]
- 6.Porter SD, Fowler HG, Mackay WP. Fire ant mound densities in the United States and Brazil (Hymenoptera: Formicidae). Journal of Economic Entomology. 1992;85(4):1154–61. [Google Scholar]
- 7.Porter SD, Williams DF, Patterson RS, Fowler HG. Intercontinental differences in the abundance of Solenopsis fire ants (Hymenoptera: Formicidae): escape from natural enemies? Environmental Entomology. 1997;26(2):373–84. [Google Scholar]
- 8.Yang CC, Yu YC, Valles SM, Oi DH, Chen YC, Shoemaker D, et al. Loss of microbial (pathogen) infections associated with recent invasions of the red imported fire ant Solenopsis invicta. Biological Invasions. 2010;12(9):3307–18. doi: 10.1007/s10530-010-9724-9 PubMed PMID: ISI:000280892600041. [Google Scholar]
- 9.Callcott AM, Collins HL. Invasion and range expansion of imported fire ants (Hymenoptera: Formicidae) in North America from 1918–1995. Florida Entomologist. 1996;79(2):240–51. [Google Scholar]
- 10.Schmid-Hempel P. Parasites in Social Insects. Princeton: Princeton University Press; 1998. 409 p. [Google Scholar]
- 11.Oi DH, Valles SM. Fire ant control with entomopathogens in the USA In: Hajek AE, Glare TR, O'Callaghan M, editors. Use of microbes for control and eradication of invasive arthropods. New York: Springer Science; 2009. p. 237–58. [Google Scholar]
- 12.Callcott AM, Porter SD, Weeks RD Jr., Graham LC, Johnson SJ, Gilbert LE. Fire ant decapitating fly cooperative release programs (1994–2008): Two Pseudacteon species (P. tricuspis, P. curvatus) rapidly expand across imported fire ant populations in the southeastern United States. Journal of Insect Science. 2010;11(19):1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter SD, Kumar V, Calcaterra LA, Briano JA, Seal DR. Release and establishment of the little decapitating fly Pseudacteon cultellatus (Diptera: Phoridae) on imported fire ants (Hymenoptera: Formicidae) in Florida. Florida Entomologist. 2013;96(4):1567–73. PubMed PMID: WOS:000329082500040. [Google Scholar]
- 14.Valles SM. Positive-strand RNA viruses infecting the red imported fire ant, Solenopsis invicta. Psyche. 2012;2012:1–14. doi: 10.1155/2012/821591 [Google Scholar]
- 15.Briano J, Calcaterra L, Varone L. Fire ants (Solenopsis spp.) and their natural enemies in southern South America. Psyche. 2012;2012:Article ID 198084, 1–19. [Google Scholar]
- 16.Lacey LA, Frutos R, Kaya HK, Vail P. Insect pathogens as biological control agents: Do they have a future? Biological Control. 2001;21(3):230–48. [Google Scholar]
- 17.Chen Y, Becnel JJ, Valles SM. RNA viruses infecting pest insects In: Vega F, Kaya HK, editors. Insect Pathology. 2nd ed. Amsterdam: Elsevier; 2012. p. 133–70. [Google Scholar]
- 18.Valles SM, Strong CA, Dang PM, Hunter WB, Pereira RM, Oi DH, et al. A picorna-like virus from the red imported fire ant, Solenopsis invicta: initial discovery, genome sequence, and characterization. Virology. 2004;328(1):151–7. Epub 2004/09/24. doi: 10.1016/j.virol.2004.07.016 S0042-6822(04)00491-X [pii]. . [DOI] [PubMed] [Google Scholar]
- 19.Valles SM, Strong CA, Hashimoto Y. A new positive-strand RNA virus with unique genome characteristics from the red imported fire ant, Solenopsis invicta. Virology. 2007;365(2):457–63. Epub 2007/05/05. doi: S0042-6822(07)00212-7 [pii] doi: 10.1016/j.virol.2007.03.043 . [DOI] [PubMed] [Google Scholar]
- 20.Valles SM, Hashimoto Y. Isolation and characterization of Solenopsis invicta virus 3, a new postive-strand RNA virus infecting the red imported fire ant, Solenopsis invicta. Virology. 2009;388(2):354–61. doi: 10.1016/j.virol.2009.03.028 [DOI] [PubMed] [Google Scholar]
- 21.Olendraite I, Lukhovitskaya NI, Porter SD, Valles SM, Firth AE. Polycipiviridae: a proposed new family of polycistronic picorna-like RNA viruses. J Gen Virol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manfredini F, Shoemaker D, Grozinger CM. Dynamic changes in host-virus interactions associated with colony founding and social environment in fire ant queens (Solenopsis invicta). Ecol Evol. 2016;6(1):233–44. doi: 10.1002/ece3.1843 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valles SM, Porter SD, Choi MY, Oi DH. Successful transmission of Solenopsis invicta virus 3 to Solenopsis invicta fire ant colonies in oil, sugar, and cricket bait formulations. Journal of Invertebrate Pathology. 2013;113(3):198–204. doi: 10.1016/j.jip.2013.04.003 PubMed PMID: WOS:000320633900002. [DOI] [PubMed] [Google Scholar]
- 24.Valles SM, Porter SD, Firth AE. Solenopsis invicta virus 3: pathogensis and stage specificity in red imported fire ants. Virology. 2014;461:66–71. [DOI] [PubMed] [Google Scholar]
- 25.Valles SM, Shoemaker D, Wurm Y, Strong CA, Varone L, Becnel JJ, et al. Discovery and molecular characterization of an ambisense densovirus from South American populations of Solenopsis invicta. Biological Control. 2013;67(3):431–9. doi: 10.1016/j.biocontrol.2013.09.015 PubMed PMID: ISI:000327720100016. [Google Scholar]
- 26.Gandon S, Van Zandt PA. Local adaptation and host–parasite interactions. Trends in Ecology & Evolution. 1998;13(6):214–6. http://dx.doi.org/10.1016/S0169-5347(98)01358-5. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–402. Epub 1997/09/01. doi: gka562 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee S, Huntemann M, Ivanova N, Kyrpides NC, Pati A. Large-scale contamination of microbial isolate genomes by Illumina PhiX control. Stand Genomic Sci. 2015;10:18 doi: 10.1186/1944-3277-10-18 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang X, Madan A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999;9(9):868–77. doi: 10.1101/gr.9.9.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuxa JR, Tanada Y. Epizootiology of insect diseases. New York: John Wiley and Sons; 1987. 555 p. [Google Scholar]
- 32.Craggs JK, Ball JK, Thomson BJ, Irving WL, Grabowska AM. Development of a strand-specific RT-PCR based assay to detect the replicative form of Hepatitis C virus RNA. Journal of Virological Methods. 2001;94(1–2):111–20. Epub 2001/05/05. doi: S0166093401002816 [pii]. . [DOI] [PubMed] [Google Scholar]
- 33.Johansson H, Dhaygude K, Lindstrom S, Helantera H, Sundstrom L, Trontti K. A metatranscriptomic approach to the identification of microbiota associated with the ant Formica exsecta. PLoS One. 2013;8(11):e79777 doi: 10.1371/journal.pone.0079777 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valles SM, Chen Y, Firth AE, Guerin DM, Hashimoto Y, Herrero S, et al. ICTV Virus Taxonomy Profile: Dicistroviridae. J Gen Virol. 2017;98(3):355–6. doi: 10.1099/jgv.0.000756 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valles SM, Oi DH, Yu F, Tan XX, Buss EA. Metatranscriptomics and pyrosequencing facilitate discovery of potential viral natural enemies of the invasive Caribbean crazy ant, Nylanderia pubens. PLoS ONE. 2012;7(2):e31828 Epub 2012/03/03. doi: 10.1371/journal.pone.0031828 PONE-D-11-23360 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valles SM, Strong CA, Hunter WB, Dang PM, Pereira RM, Oi DH, et al. Expressed sequence tags from the red imported fire ant, Solenopsis invicta: annotation and utilization for discovery of viruses. Journal of Invertebrate Pathology. 2008;99(1):74–81. Epub 2008/03/11. doi: S0022-2011(08)00018-9 [pii] doi: 10.1016/j.jip.2008.01.004 . [DOI] [PubMed] [Google Scholar]
- 37.Valles SM, Oi DH, Porter SD. Seasonal variation and the co-occurrence of four pathogens and a group of parasites among monogyne and polygyne fire ant colonies. Biological Control. 2010;54(3):342–8. [Google Scholar]
- 38.Valles SM, Strong CA, Oi DH, Porter SD, Pereira RM, Vander Meer RK, et al. Phenology, distribution, and host specificity of Solenopsis invicta virus-1. Journal of Invertebrate Pathology. 2007;96(1):18–27. Epub 2007/04/07. doi: S0022-2011(07)00038-9 [pii] doi: 10.1016/j.jip.2007.02.006 . [DOI] [PubMed] [Google Scholar]
- 39.Oi DH, Valles SM, Porter SD. The fire ant pathogen Vairimorpha invictae (Microsporidia: Burenellidae) not detected in Florida. Florida Entomologist. 2012;95(2):506–8. [Google Scholar]
- 40.Porter SD, Valles SM, Oi DH. Host specificity and colony impacts of Solenopsis invicta virus 3. Journal of Invertebrate Pathology. 2013;114:1–6. doi: 10.1016/j.jip.2013.04.013 [DOI] [PubMed] [Google Scholar]
- 41.Porter SD, Valles SM, Wild AL, Dieckmann R, Plowes NJR. Solenopsis invicta virus 3: further host-specificity tests with native Solenopsis ants (Hymenoptera: Formicidae). Florida Entomologist. 2015;98:122–5. [Google Scholar]
- 42.Remnant EJ, Shi M, Buchmann G, Blacquirere T, Holmes EC, Beekman M, et al. A diverse range of novel RNA viruses in geographically distinct honey bee populations. Journal of Virology. 2017;(In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–25. Epub 1987/07/01. doi: 10.1093/oxfordjournals.molbev.a040454 . [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39(4):783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x . [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. doi: 10.1093/molbev/msw054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Contigs were first sorted in descending order based on the number of the sequences comprising it, followed by the libraries represented.
(DOCX)
Data Availability Statement
Raw sequence data from each library were deposited into the GenBank database as a Sequence Read Archive under accession number, SRP113235 (Bioproject PRJNA394996). Assembled sequences with viral identity (Table 2) have been deposited at DDBJ/EMBL/GenBank as a Transcriptome Shotgun Assembly project under the accession GFUG00000000. The version described in this paper is the first version, GFUG01000000. The SINV-5 annotated genome was deposited in GenBank under accession number MF593921.
Raw sequence data from each library were deposited into the GenBank database as a Sequence Read Archive under accession number, SRP113235 (Bioproject PRJNA394996). Assembled sequences with viral identity (Table 2) have been deposited at DDBJ/EMBL/GenBank as a Transcriptome Shotgun Assembly project under the accession GFUG00000000. The version described in this paper is the first version, GFUG01000000. The SINV-5 annotated genome was deposited in GenBank under accession number MF593921.
Table 2. Contiguous sequences (Contigs) with significant viral identity by BLASTX analysis of the GenBank database and considered high likelihood viral prospects from RNA libraries created from Solenopsis invicta worker ants.
Contigs were first sorted in descending order based on the number of the sequences comprising it, followed by the libraries represented.
| Designation | Sequences comprising contig | Size (nt) | e-score | Query coverage:identity (%) | Sequence identity with | Number of singletons in | Genome | Virus family | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SAL_1 | SAL_2 | SAL_3 | SAL_4 | ||||||||
| Contig_29 (SINV-5) | 21294 | 9030 | 0 | 57: 67 | Israeli acute paralysis virus | 274 | 10312 | 4312 | 6396 | ssRNA | Dicistroviridae |
| Contig_66 | 13980 | 2626 | 2E-154 | 76: 39 | Aphid lethal paralysis virus | 3823 | 1704 | 5241 | 3212 | ssRNA | Dicistroviridae |
| Contig_30 | 6197 | 1869 | 1E-139 | 96: 40 | Aphid lethal paralysis virus | 2165 | 779 | 1833 | 1420 | ssRNA | Dicistroviridae |
| Contig_16 | 2104 | 2371 | 0 | 83:48 | Solenopsis invicta virus 1 | 2 | 133 | 1815 | 154 | ssRNA | Dicistroviridae |
| Contig_70 | 641 | 1068 | 6E-64 | 82: 43 | Alber virus | 117 | 180 | 141 | 203 | RNA | Unclassified |
| Contig_58 | 607 | 1453 | 1E-67 | 88: 34 | Nasonia vitripennis virus | 18 | 311 | 37 | 241 | RNA | Unclassified |
| Contig_13 | 510 | 1568 | 2E-102 | 87: 41 | Aphid lethal paralysis virus | 206 | 98 | 7 | 199 | ssRNA | Dicistroviridae |
| Contig_21 | 486 | 3211 | 0 | 85: 61 | Acute bee paralysis virus | 108 | 51 | 9 | 318 | ssRNA | Dicistroviridae |
| Contig_83 | 428 | 2623 | 0 | 97: 62 | Mosinovirus | 90 | 58 | 110 | 170 | RNA | Unclassified |
| Contig_55 | 202 | 2639 | 3E-147 | 91: 35 | Kashmir bee virus | 12 | 153 | 20 | 17 | ssRNA | Dicistroviridae |
| Contig_82 | 114 | 1203 | 0 | 82: 94 | Acute bee paralysis virus | 16 | 7 | 4 | 87 | ssRNA | Dicistroviridae |
| Contig_15 | 100 | 1042 | 0 | 92: 83 | Hubei orthoptera virus 1 | 28 | 14 | 3 | 55 | RNA | Unclassified |
| Contig_27 | 69 | 680 | 6E-135 | 91: 97 | Acute bee paralysis virus | 1 | 6 | 2 | 60 | ssRNA | Dicistroviridae |
| Contig_17 | 58 | 796 | 4E-162 | 99: 91 | Acute bee paralysis virus | 15 | 7 | 5 | 31 | ssRNA | Dicistroviridae |
| Contig_80 | 55 | 1173 | 0 | 98: 79 | Drosophila C virus | 44 | 4 | 3 | 4 | ssRNA | Dicistroviridae |
| Contig_75 | 36 | 589 | 6E-26 | 83: 39 | Hubei picorna-like virus 46 | 8 | 13 | 15 | 0 | RNA | Unclassified |
| Contig_47 | 734 | 1045 | 7E-82 | 89: 44 | Solenopsis invicta virus 2 | 0 | 667 | 6 | 61 | ssRNA | Unclassified |
| Contig_78 | 216 | 702 | 7E-111 | 99: 85 | Israeli acute paralysis virus | 153 | 42 | 21 | 0 | ssRNA | Dicistroviridae |
| Contig_57 | 146 | 1028 | 5E-116 | 93: 57 | Wuhan insect virus 11 | 0 | 93 | 28 | 25 | RNA | Unclassified |
| Contig_18 | 44 | 664 | 2E-84 | 97: 60 | Hubei Orthoptera virus 1 | 10 | 2 | 0 | 32 | RNA | Unclassified |
| Contig_88 | 44 | 1500 | 0 | 99: 87 | Rhopalosiphum padi virus | 12 | 0 | 7 | 25 | ssRNA | Dicistroviridae |
| Contig_53 | 39 | 522 | 1E-50 | 98: 52 | Wuhan insect virus 11 | 0 | 27 | 3 | 9 | RNA | Unclassified |
| Contig_9 | 9 | 471 | 3E-68 | 87: 95 | Acute bee paralysis virus | 3 | 0 | 3 | 3 | ssRNA | Dicistroviridae |
| Contig_85 | 8 | 542 | 2E-53 | 91: 56 | Solenopsis invicta virus 1 | 4 | 1 | 3 | 0 | ssRNA | Dicistroviridae |
| Contig_51 | 321 | 1465 | 0 | 99: 76 | Nodamura virus | 0 | 274 | 0 | 47 | ssRNA | Nodaviridae |
| Contig_49 | 182 | 1269 | 0 | 99: 95 | Aphid lethal paralysis virus | 0 | 153 | 29 | 0 | ssRNA | Dicistroviridae |
| Contig_12 | 95 | 438 | 2E-79 | 99: 77 | Big Sioux River virus | 49 | 0 | 0 | 46 | ssRNA | Unclassified |
| Contig_50 | 69 | 768 | 9E-68 | 99: 46 | Israeli acute paralysis virus | 0 | 68 | 0 | 1 | ssRNA | Dicistroviridae |
| Contig_60 | 69 | 926 | 0 | 99: 93 | Aphid lethal paralysis virus | 0 | 55 | 14 | 0 | ssRNA | Dicistroviridae |
| Contig_28 | 67 | 830 | 1E-46 | 97: 37 | Israeli acute paralysis virus | 0 | 0 | 13 | 54 | ssRNA | Dicistroviridae |
| Contig_52 | 66 | 356 | 1E-35 | 99: 94 | Shuangao insect virus 8 | 0 | 20 | 0 | 46 | RNA | Unclassified |
| Contig_25 | 58 | 869 | 8E-10 | 37: 40 | Hubei picorna-like virus 50 | 0 | 0 | 20 | 38 | RNA | Unclassified |
| Contig_141_3101 | 3101 | 2351 | 0 | 97: 55 | Solenopsis invicta virus 1 | 0 | 0 | 0 | 3101 | ssRNA | Dicistroviridae |
| Contig_19 | 1075 | 1080 | 2E-159 | 89: 71 | Solenopsis invicta virus 1 | 0 | 0 | 0 | 1075 | ssRNA | Dicistroviridae |
| Contig_19_480 | 480 | 623 | 8E-114 | 99: 88 | Solenopsis invicta virus 1 | 0 | 0 | 480 | 0 | ssRNA | Dicistroviridae |
| Contig_110_361 | 361 | 1214 | 0 | 99: 82 | Kashmir bee virus | 0 | 0 | 0 | 361 | ssRNA | Dicistroviridae |
| Contig_81_84 | 84 | 1103 | 6E-161 | 99: 67 | Wuhan arthropod virus 2 | 0 | 0 | 84 | 0 | RNA | Unclassified |
| Contig_108_79 | 79 | 715 | 5E-131 | 98: 83 | Shuangao insect virus 8 | 0 | 79 | 0 | 0 | RNA | Unclassified |