Abstract
Several transcription factors have been identified that activate an epithelial-to-mesenchymal transition (EMT), which endows cells with the capacity to break through basement membranes and migrate away from their site of origin. A key program in development, in recent years it has been shown to be a crucial driver of tumour invasion and metastasis. However, several of these EMT-inducing transcription factors are often expressed long before the initiation of the invasion-metastasis cascade as well as in non-invasive tumours. Increasing evidence suggests that they may promote primary tumour growth, but their precise role in this process remains to be elucidated. To investigate this issue we have focused our studies on two Drosophila transcription factors, the classic EMT inducer Snail and the Drosophila orthologue of hGATAs4/6, Serpent, which drives an alternative mechanism of EMT; both Snail and GATA are specifically expressed in a number of human cancers, particularly at the invasive front and in metastasis. Thus, we recreated conditions of Snail and of Serpent high expression in the fly imaginal wing disc and analysed their effect. While either Snail or Serpent induced a profound loss of epithelial polarity and tissue organisation, Serpent but not Snail also induced an increase in the size of wing discs. Furthermore, the Serpent-induced tumour-like tissues were able to grow extensively when transplanted into the abdomen of adult hosts. We found the differences between Snail and Serpent to correlate with the genetic program they elicit; while activation of either results in an increase in the expression of Yorki target genes, Serpent additionally activates the Ras signalling pathway. These results provide insight into how transcription factors that induce EMT can also promote primary tumour growth, and how in some cases such as GATA factors a ‘multi hit’ effect may be achieved through the aberrant activation of just a single gene.
Author summary
Many cancer cells acquire abnormal motility behaviour leading to metastasis, the main cause of cancer related deaths. In many cancers, transcription factors capable of inducing motile migratory cell behaviours, so-called EMT transcription factors, are found highly expressed. However, the expression of these genes is not restricted to metastatic invasive cancers; they are often found in benign tumours, or in tumours long before they show any sign of metastasis. This observation motivated us to ask if they may play a role in driving primary tumour growth. Our results show that the Drosophila EMT-inducers Snail and Serpent are both capable of driving overproliferation. However, Snail overproliferation is accompanied by a decrease in cell size as well as cell death, and consequently the tissue does not increase in size. Serpent also drives cell proliferation but this occurs together with an increase in cell size, but not cell death, thus having a profound effect on the overall size of the tissue. We show that both Snail and Serpent trigger activation of the Yorki pathway and in addition Serpent, but not Snail, also triggers activation of the Ras pathway. These results provide insight into how activation of some EMT-inducing genes can also promote primary tumour growth.
Introduction
Epithelial cells display a remarkable plasticity and while this property is vital for morphogenesis during normal embryonic development, there is now strong evidence supporting its critical role in tumour progression (reviewed in [1–3]). Several core inducers of the epithelial-to-mesenchymal transition (EMT) have been identified, such as the Snail/Slug family, Twist, SIP1, Zeb factors and Goosecoid, which effect EMT through the transcriptional repression of E-Cadherin. Our lab identified an alternative mechanism, whereby GATA factors affect cell plasticity through the modulation of apicobasal polarity [4, 5]. Similar to other EMT transcription factors, gain of function mutations in GATA factors have been found in a number of human cancers including breast, ovarian, pancreatic and colorectal cancer (CRC), and in metastases, such as liver metastasis from CRC patients [6–9].
Activating an EMT program promotes tumour invasion and metastasis, as it endows cells with the ability to break through basement membranes and migrate away from their site of origin. However, as well as promoting tumour dissemination, there is also evidence implicating EMT transcription factors in promoting primary tumour growth (reviewed in [10]). For example EMT transcription factors are often expressed in non-invasive tumours suggesting that they might promote oncogenic functions within the primary lesion, affecting tumour development long before the initiation of the invasion-metastasis cascade. Furthermore, Snail (Sna) interference in carcinoma cells dramatically decreases tumour growth, with tumours adopting a smaller size and having a decreased proliferation [11]. However, the effects of EMT transcription factors on primary tumour growth remain poorly characterised and many questions remain. Is activation of an EMT transcription factor alone sufficient to drive primary tumour growth, or just a step in the progression of the tumour? Are the mechanisms of driving growth shared by all EMT transcription factors?
To investigate this we have focused our studies on a classic EMT inducer Sna, and on Serpent (Srp), the Drosophila orthologue of hGATAs4/6, and modelled the effects of creating gain-of-function conditions in the wing imaginal discs of the fly. Due to the work of many laboratories, wing discs are now a well-established system for modelling epithelial tumours (see [12]), where the effects of proto-oncogenes can be analysed in a proliferating epithelium surrounded by a mature basement membrane. Here we show that activation of either Sna or Srp induces an increase in the proliferation of wing disc cells, together with tissue disorganisation and a loss of epithelial polarity. However, Sna-induced proliferation is accompanied by both a decrease in average cell size and extensive cell death, and consequently no overall increase in tissue size is observed. Conversely, Srp drives an increase in cell size as well as cell proliferation, leading to a large overall increase in the size of the wing disc. Furthermore, these Srp-induced tumor-like tissues are able to grow extensively when transplanted into the abdomen of adult hosts. We carried out a genetic screen of candidate genes to gain insight into the mechanisms underlying Srp-induced tissue growth and found that both the Yorki (Yki) and Ras pathways contribute to the phenotype. Our results show that while both Sna and Srp activation results in an increase in the expression of Yki target genes, Srp additionally activates Ras pathway signaling. Thus activation of an EMT transcription factor alone is sufficient to drive primary tumour growth, but this effect is transcription factor dependent. These results highlight the diverse consequences of reactivation of EMT transcription factors, which are often key developmental genes, and how a ‘multi hit’ effect may be achieved through the aberrant activation of just a single gene.
Results
Sna and Srp both drive EMT when ectopically expressed in the wing imaginal disc
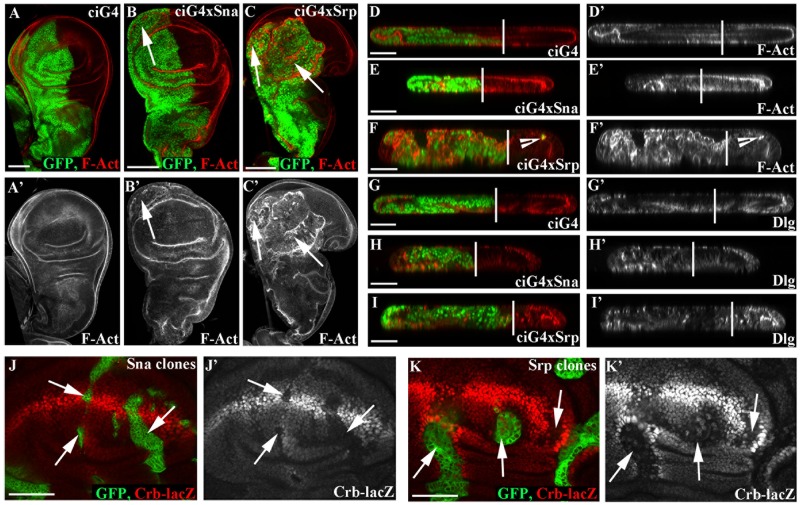
The Drosophila primordia of adult wing structures, the wing imaginal discs, are epithelial monolayers that actively proliferate during larval development to give rise to a 1,000-fold increase in the number of cells and tissue size. They are now widely used to study and dissect the basic mechanisms underlying different stages of epithelial tumour progression (for a review see [12]). We therefore decided to exploit this system to assess the consequences of generating gain-of-function conditions of the Sna and Srp transcription factors. To this end, we made use of the Gal4/UAS technique [13] and used Ci-Gal4 to drive expression of either sna or srp in the anterior wing compartments and compared these with the wild type posterior compartments in the same discs (Fig 1A–1C); or nub-Gal4, which drives expression throughout the wing pouch (S1 Fig). Regional expression of either sna or srp causes tissue disorganization characteristic of an EMT, including loss of cell polarity, multilayering and loss of epithelial architecture (Fig 1A–1I, S1A–S1G Fig). F-actin becomes completely disorganised, particularly for Srp (Fig 1B, 1C, 1E and 1F, arrows, S1E and S1G Fig) and cell polarity proteins such as Dlg lose their tight localisation (Fig 1H and 1I, S1D–S1G Fig). In addition we see small numbers of Srp expressing cells in the posterior compartment (Fig 1F, arrowhead), pointing to Srp-expressing cells gaining the migratory and invasive properties associated with EMT.
Fig 1. Both Srp and Sna drive an EMT in wing disc cells.
A-I Staining for polarity markers in control ci-Gal4, UAS-GFP; tub-Gal80TS discs (A, D, G); in ci-Gal4, UAS-GFP, UAS-sna;tub-Gal80TS discs (B, E, H) and in ci-Gal4, UAS-GFP, UAS-srp;tub-Gal80TS discs (C, F, I) 48 hours after shifting to 29°C, the permissive temperature. Staining for F-Act (A-F) and Dlg (G-I) in xy (A-C) and yz sections (D-I) shows that actin becomes delocalised in wing disc cells (arrows), and cells lose their regular columnar shape. Upon srp-overexpression some cells also appear in the posterior compartment (F, arrowhead). Vertical lines in D-I demarcate the anterior-posterior boundary J-K Clones of either UAS-sna (J) or UAS-srp (K) generated in a crb-LacZ reporter background. Clones were induced by a 30 min heat shock and fixed after 48 hours. Staining for LacZ showed that Crb is downregulated at the transcriptional level by both Sna and Srp (J, K, arrows). Scale bars A-C—100μm; D-K—50μm.
The effects of Sna and Srp on tissue organisation vary notably, depending on the region of the wing disc, with the effects of Sna relatively restricted to the peripodial membrane (Fig 1B, arrow) and of Srp more pronounced in both the wing disc proper and peripodial membrane (Fig 1C, arrows). Interestingly, a recent study highlighted a susceptibility of certain areas of the discs to particular tumorigenic stimuli [14], thus these observations suggest either that ectopic activation of Sna and Srp produce the same effects but at different strengths, or that Sna and Srp are affecting different pathways.
Both Sna and Srp have previously been shown to impinge on apicobasal polarity in the Drosophila embryo through the direct transcriptional repression of the key cell polarity regulator Crumbs (Crb) [4, 15]. To see if this is also the case in the wing disc, we next made clones of Sna or Srp expressing cells in a background where wing disc cells have a lacZ enhancer trap insertion in the crb regulatory region (crb-lacZ [16]). We find that crb transcription is repressed upon overexpression of either Sna or Srp (Fig 1J and 1K, arrows), indicating that, as in the embryo, Sna and Srp repress crb transcription. However, in contrast to the embryo, repression of crb alone in wing disc cells is not sufficient to perturb apico-basal polarity [17], and thus Sna and Srp likely affect the transcription of other genes regulating polarity in addition to crb, as seen during their embryonic roles in mesoderm [15] and midgut development [4].
Srp drives neoplastic-like growth when expressed in the wing disc
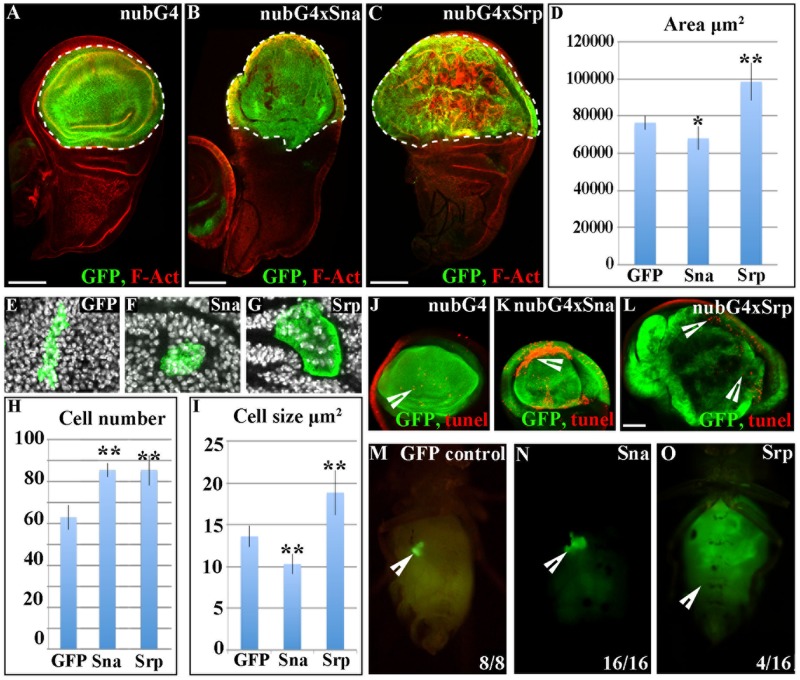
Loss of crb has previously been shown to affect growth of Drosophila imaginal cells [17, 18], thus these results indicated that EMT transcription factors might impinge on growth in addition to tissue organisation. To investigate this further we drove expression of GFP together with Sna or Srp throughout the wing disc pouch and distal hinge for 48 hours using Nub-Gal4, and then measured the size of the GFP expressing area in 3rd instar larvae. We found that overexpression of Srp induced a dramatic increase in the size of the tissue (Fig 2A–2D). Whereas, overexpression of Sna leads to a small, but significant decrease in tissue size (Fig 2A–2D).
Fig 2. Srp drives an overgrowth of the wing disc.
A-D, J-L Ectopic expression of either GFP alone (A, J), GFP and sna (B, K) or GFP and srp (C, L) in the wing disc; transgenes are under the control of nub-Gal4 driver and tub-Gal80TS and 3rd instar disc are fixed 48 hours after shifting to 29°C. Discs are stained for GFP (green) and F-act (red). Expression of sna results in a small, but significant decrease in wing disc size (B, D), whereas srp drives a large increase in the size of the GFP expressing compartment (C, D). E-G Clones of cells expressing either GFP alone (E), GFP and sna (F) or GFP and srp (G) were induced by a 30 min heatshock, and discs were fixed 48 hours after induction. H Counts of the number of cells in each clone showed that there is a significant increase in the number of cells in clones with either srp or sna expression. I Measuring the size of cells in clones of each condition revealed that in contrast to control clones, srp-expressing cells show an increase in both the magnitude and variability of their size. In contrast, sna expression shows a small but significant decrease in cell size. Data are presented as mean ± standard deviation (SD), *P<0.05, **P<0.005; paired t-test. J-L TUNEL staining reveals that Sna (K) but not Srp (L) causes a dramatic increase in cell death. M-O Adult fly micrographs taken 21 d after implantation of GFP-labeled larval wing tissue expressing GFP alone (M), GFP and sna (N) or GFP and srp (O). Ratios show the reproducibility of the phenotype are shown. Scale bars A-C—100μm; J-L—50μm.
Surprisingly, we found that Srp expression leads to heterogeneous effects, even within a single region of the wing disc, with a large number of cells appearing larger than in wild type (Fig 2C). As this suggested that tissue growth could be coming from an increase in cell size rather than overproliferation, we investigated this further by generating clones of cells expressing Sna or Srp and compared the number and size of cells within the clones with wild type clones generated concurrently (Fig 2E–2I). This analysis showed that Srp, and surprisingly also Sna, drives an increase in the average cell number within the clone (Fig 2H). In addition Srp drives a large increase in cell size in a very hetrogenous manner (Fig 2I). In contrast, Sna overproliferation is accompanied by a decrease in the average cell size (Fig 2I).
To further understand what is contributing to the phenotypes observed upon over-expression of Sna or Srp, we carried out TUNEL staining, to investigate a possible role for cell death. We found that overexpression of Sna causes massive cell death in certain regions of the wing disc proper (Fig 2K), whereas ectopic Srp does not cause a considerable increase in cell death (Fig 2L). Taken together, our experiments suggest that while Sna-induced cell proliferation occurs concurrently with cell death and a decrease in cell size, Srp induces both cell proliferation and cell growth in the absence of an increase of cell death. Thus the overall effect of ectopic Sna is a slight reduction in tissue size; in contrast overexpression of Srp leads to considerable tissue overgrowth.
To understand the contribution of cell death to the sna phenotype, we decided to overexpress sna together with the baculovirus p35 protein which blocks the action of a wide range of caspases and in inhibits cell death [19, 20]. To assess this we drove sna expression in the anterior wing compartment using ci-Gal4, and compared the size of this compartment with the wild type posterior compartment in the same disc. When sna alone is overexpressed, the A/P ratio is reduced from 1.6 to 1, and sna together with GFP, the A/P ratio is reduced to 1.2 (S2A and S2C Fig). When sna is overexpressed together with p35, which blocks cell death, we no longer see a decrease in tissue size, and the A/P ratio is 1.6, similar to wild type (S2B and S2C Fig). Our clonal analysis shows that Sna drives an increase in cell proliferation which is coupled to a decrease in average cell size. Taken together, our results suggest that when cell death is blocked, Sna-induced overproliferation is accompanied by a decrease in cell size which compensate for each other in terms of overall tissue size, and thus no overall change is observed. Furthermore, these data suggest that it is not differences in cell division that contribute to the differences in tissue size observed upon overexpression of Srp or Sna. Rather it is their contrasting effects on cell death and on cell size that lead to these pronounced differences.
To further understand the effect of Srp overexpression, we tried to follow the growth of srp expressing tissues for a longer period of time, however larvae bearing imaginal discs with ectopic srp expression died after two days, hence it was not possible to follow their growth for a longer period. Thus, we further characterized the growth potential of the tissue by allograft cultures [21, 22]. Wing tissue expressing GFP alone or GFP together with either sna or srp was transplanted into the abdomen of adult females and maintained for a period of 21 days. Tissue expressing GFP alone did not grow (Fig 2M), as shown previously [23, 24], nor did they grow when expressing GFP together with sna (Fig 2N). Conversely, when expressed together with srp the tissue grew many times larger than the original piece of transplanted tissue (Fig 2O) and could also grow when transplanted again into a new host. Altogether, these results indicate that srp expression can induce neoplastic tissue growth by triggering cells to proliferate and grow indefinitely.
Srp-induced overgrowth is partially rescued by reducing Ras and Yki pathway activity
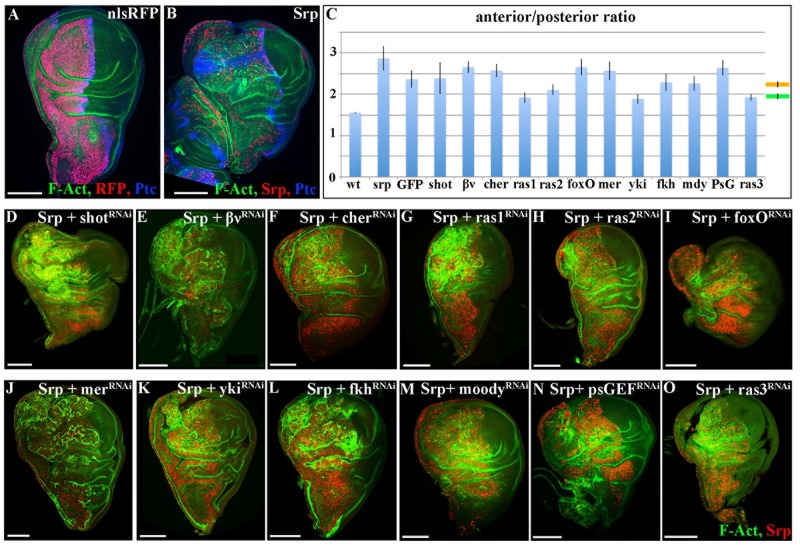
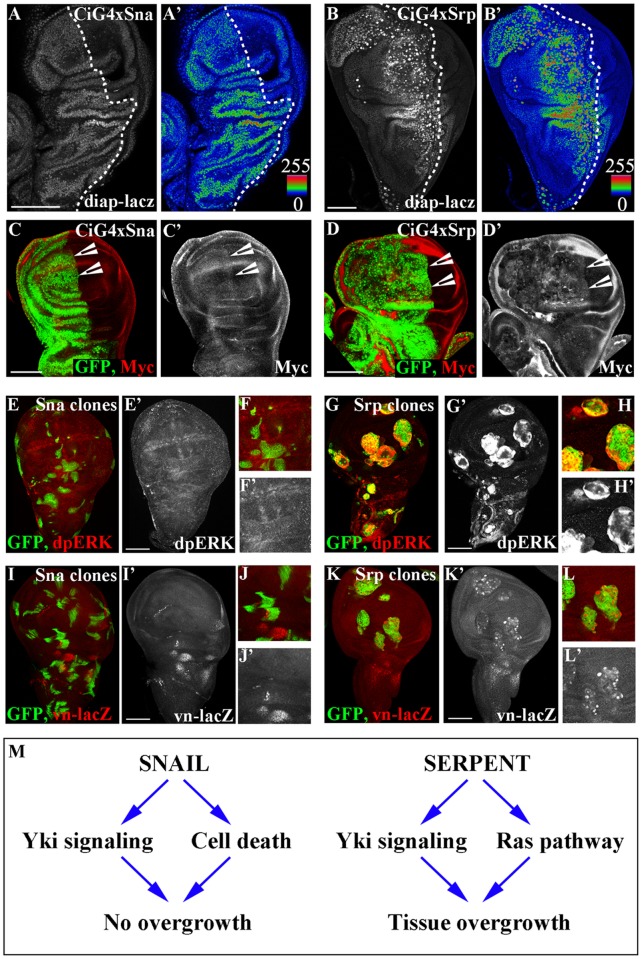
To uncover the mechanisms underlying Srp-induced overgrowth, we decided to set up conditions for carrying out a genetic modifier candidate screen for genes that rescue this aspect of the Srp-phenotype. We drove expression of either a wild type nuclear label alone (Fig 3A), or of srp (Fig 3B) in the anterior wing compartment using ci-Gal4, and compared the size of this compartment with the wild type posterior compartment in the same disc (Fig 3A and 3B). We quantified the sizes of the two compartments as a measure of the ratio between the width of the anterior and posterior compartments in the same discs; while this ratio is 1.6 in wild type discs, the ratio rises to 3 upon srp expression in the anterior compartment (Fig 3C). We used this system to carry out a series of epistatic experiments with RNAis for candidate genes selected either by their role in signalling pathways involved in cell proliferation, or with a role related to EMT. For a big majority of the UAS-RNAis we found a general decrease of the Srp-induced oversize, compatible with the presence of another UAS construct being activated by the same GAL4 driver (Fig 3C, orange line; Fig 3D, 3E, 3F, 3I, 3J, 3L, 3M and 3N); a similar result obtained with a control UAS-GFP construct supports this interpretation (Fig 3C). Interestingly, 4 UAS-RNAi constructs gave rise to a greater reduction of the Srp-induced overgrowth three targeting ras and one targeting yki, both coding for well-known inducers of cell growth/proliferation (reviewed in [25]) (Fig 3C, green line; Fig 3G, 3H, 3K and 3O). Indeed, we found that ectopic expression of either sna or srp in the wing disc activated expression of yki downstream targets, as revealed by staining for the diap1-lacZ reporter construct [26] (Fig 4A and 4B), and the cellular growth promoter Myc [27] (Fig 4C and 4D). Since both sna and srp repress transcription of crb, and crb loss can cause Hippo pathway repression and Yki-dependent overproliferation [17, 18], it is likely that repression of crb by Sna or Srp is activating, or at least contributing, to the increase in the transcription of Yki target genes.
Fig 3. srp-induced overgrowth is partially rescued by RNAis against ras and yki.
A, B, D-O 3rd instar wing discs from either control individuals expressing stingerRFP to mark anterior the anterior compartment (A) or individuals expressing the indicated transgenes in the anterior compartment (B, D-O). Discs are stained for F-Act (green), RFP (red) and Ptc (blue) (A); for F-Act (green), Srp (red) and Ptc (blue); and for F-Act (green) and Srp (red) (D-O). Ras1, ras2 and ras3 denote 3 independent RNAi transgenes against ras—see methods. Transgenes are under the control of the ci-Gal4 driver and tub-Gal80TS and the discs are fixed 48 hours after shifting to 29°C. Scale bars—100μm. C Histogram plotting the A/P width ratio of wing primordia expressing either nuclear RFP alone (wt), srp alone (Srp), srp together with GFP (GFP) or srp and dsRNA for the indicated genes under the control of ci-Gal4 driver. Data are presented as mean ± SD. The mean ratio of discs expressing srp and an additional transgene is 2.49 with a standard error of the mean (SEM) of 0.05 and is depicted by the orange line; the mean ratio of discs showing a partial rescue is 1.96 with a SEM of 0.04 and is depicted by the green line. Scale bars—100μm.
Fig 4. Srp activates both the Yki and Ras signalling pathways.
A-D CiGal4, Gal80ts was used to drive either sna (A, C) or srp (B, D) expression in the anterior compartment of discs containing the diap-lacZ reporter (A, B) or in a wildtype background together with GFP (C, D). Discs are stained for lacZ (A, B) or for GFP (green) and Myc (red) (C, D). E-K Clones of either UAS-sna (E, F, I, J) or UAS-srp (G, H, K, L) generated in either a wild type (E-H) or a vn-LacZ reporter background (I-L). Clones were induced by a 30 min heat shock and fixed after 48 hours. Staining for dpERK shows that Ras signalling is activated downstream of Srp (G, H), but not downstream of Sna (E, F). Staining for LacZ showed that vn is upregulated at the transcriptional level by Srp (K, L), but not by Sna (I, J). Scale bars A-K—100μm. (M) Model showing the effects of ectopic activity of Sna and Srp in the wing disc reported in this work (additional targets are likely to be triggered as well); Sna represses crumbs, which in turn activates Yki signalling; Srp represses crumbs, activating Yki signalling and also activates vein, which activates the RAS pathway via the EGFR.
Thus, while Yki activity might be necessary for Srp-induced overgrowth it is clearly not sufficient, as Yki is also activated by ectopic expression of sna, which does not induce tissue overgrowth. Hence, we focused on the Ras pathway, the other element identified by the RNAi screen. Ras is a transducer of a phosphorylation cascade that conveys the activity of Receptor Tyrosine Kinases (RTK) resulting in the di-phosphorylation and activation of the extracellular signal-regulated kinase (dpERK) [28]. Once active, Erk phosphorylates its substrates in the cytoplasm or in the nucleus [29]. Again, we found that srp expressing clones in the wing disc activate the Ras pathway as reported by dpERK staining (Fig 4G). Conversely, there was no dpERK staining in sna expressing clones in the wing disc (Fig 4E).
Ras signalling has previously been reported to protect cells from cell death, in particular in tumourigenic situations which arise from a loss of cell polarity [30]. We therefore wondered if blocking Ras signalling together with overexpression of Srp would lead to an increase in cell death comparable to what we see with overexpression of Sna alone. However, when we stained discs overexpressing Srp together with ras-RNAi in the anterior compartment for Death caspase-1 (Dcp1), we did not see any increase in the levels of cell death when compared to the wildtype posterior compartment (S3). This indicates that Ras signalling is not contributing to growth simply through preventing stress-induced cell death, and likely cooperates with Yki to induce uncontrolled tissue growth as recently reported in [31].
We next investigated how Srp, but not Sna, could elicit the activation of the Ras pathway. Srp is a key regulator of embryonic midgut morphogenesis [4, 32], thus it was intriguing to note that EGFR signaling through activation of its ligand vein (vn) is required during embryonic midgut development [33]. vn codes for one of the ligands of the EGFR [34], an RTK widely expressed in Drosophila, including the wing disc [35, 36], and ectopic activation of Vn in the wing triggers Ras activation through the EGFR [37]. We tested for vn activation by generating clones of Sna or Srp expressing cells in a background where wing disc cells have a lacZ enhancer trap insertion in the vn regulatory region (vn-lacZ). We found a massive upregulation of vn transcription within srp expressing clones (Fig 4K), but not with sna expressing clones (Fig 4I). In addition, because Vn is a secreted ligand it can thus bind the EGFR, and hence activate the Ras pathway, in the cells close to the ones in which it is transcribed; thus, activation of vn transcription can also account for the non-autonomous activation of the Ras pathway by Srp as revealed by dpERK staining in cells close to the srp expressing clones (Fig 4H, S4 Fig, arrowheads). Taken together, these data indicate that activation of Srp leads to tissue overgrowth through the combined activation of Yki and Ras signalling pathways (Fig 4M). In contrast, while Sna also activates the transcription of Yki targets, it does not activate the Ras signalling pathway, which is likely to be one of the causes of the difference in the effects of Sna and Srp on the final tissue size (Fig 4M).
To assess the role of Ras activation in the Srp overexpression phenotype we planned to reduce vn function in the Srp overexpression background by using the available UAS-vnRNAi construct. Unfortunately, this experiment turned not to be simple because of the chromosome location of all the constructs required. Thus, as an alternative, we resorted to an UAS-EGFRRNAi construct to impair Ras activation. It is well known that EGFR signalling is normally required for wing development and in particular that impairing EGFR signalling in an otherwise wild type wing impairs the normal rate of proliferation and the wing size [36, 38]. Accordingly, impairment of EGFR signalling in a srp overexpression background not only reverts the overgrowth phenotype but generates wings smaller than wild type indicating an absolute requirement of EGFR-mediated Ras activation for srp overgrowth (S3B Fig).
Discussion
EMT transcription factors are often found upregulated in human tumours (reviewed in [39]). Given their role in driving a transition from a polarised static epithelial cell to a migratory invasive cell state, much focus has been put on the pro-invasive and metastatic implications of their aberrant expression. However, tumour progression involves the progressive acquisition of many other biological capabilities including sustained proliferation, evasion of growth suppressors and resistance to cell death (reviewed in [40]), and there is increasing evidence suggesting that EMT transcription factors contribute to these earlier stages of tumour progression (reviewed in [10]). In this study we show that the Drosophila EMT-inducers Sna and Srp drive not only EMT, but also over-proliferation in a well-established epithelial tumour model. Sna-driven proliferation is accompanied by extensive cell death and a decrease in cell size, and thus the overall effects of aberrant Sna expression on tissue size are negligible. In contrast, Srp drives an increase in cell size as well as cell proliferation, but not cell death, leading to a profound overall increase in the size of the tissue, which is particularly evident upon transplantation, when the tissue has more time to grow. We find that both Sna and Srp repress crb transcription, which has previously been shown to induce a repression of the Hippo pathway and thus drive Yki-dependent overproliferation [17, 18]. Indeed, we show that both Sna and Srp activate Yki activity, which has previously been shown to drive excess proliferation in the wing disc. However, in addition to this, we find that Srp also activates the mitogenic Ras pathway, which has recently been shown to act synergictically with Yki to promote hyperproliferation and tumour development in the Drosophila wing disc [31]. Studies in breast cancer models and oesophageal epithelial cells have shown that the EMT transcription factors Twist and Zeb contribute to primary tumour growth through the activation of programs that prevent cells from undergoing oncogene-induced senescence and apoptosis [41–43]. Taken together with our results, this suggests that EMT transcription factors can contribute to the multistep process of tumour progression through the activation of different onco-promoting cell biological processes, and that this is both transcription factor and tumour dependent.
EMT transcription factors drive a loss of epithelial cell polarity, which has been shown to activate cell death pathways in a number of contexts. For example, scribble (scrib) mutant clones are completely eliminated from wild type discs through programmed cell death pathways [44]. Intriguingly, while overexpression of both Srp and Sna drives a loss of cell polarity, an increase in cell death is only seen with Sna, whereas Srp appears to correlate with an increase in cell survival. This is despite the fact that we find an increase in the transcription of the key apoptosis inhibitor Diap1 in both scenarios. Diap1 functions as an E3-ubiquitin ligase that protects cells from unwanted death by blocking the activity of the caspase DRONC and the Drosophila apoptotic protease-activating factor-1 (Apaf-1) homolog, Dark, and the relative levels of Diap1, Dronc and Dark are important in determining the outcome ie. cell survival vs cell death [45]. As we see a lot of cell death in the wing disc upon overexpression of Sna, despite a clear increase in Diap1 expression, this suggests that the levels of Diap1 induced are not sufficient to block the level of cell death induced by Sna. While we see a comparable increase in the levels of Diap1 upon Srp overexpression, Srp also activates Ras, which has been reported to protect cells with mutations in cell polarity genes from death [30]. However, reducing Ras signalling does not lead to an increase in cell death when Srp is overexpressed. We have previously see that ectopic Srp also induces expression of Forkhead (Fkh) [4], which has been reported to act as a survival factor in a number of Drosophila systems, including the midgut [46, 47]. These results therefore suggest that cells expressing ectopic Srp evade death through the upregulation of multiple cell survival factors.
Intriguingly, so-called EMT transcription factors such as Sna, Srp, Twist and Zeb proteins often activate many developmental pathways and processes of which a loss of cell polarity and EMT is only a part. They are all expressed in multiple tissues during development and play pleotrophic roles, depending on the context and time window in which they are activated. We want to emphasise the cell context dependence of the activity of these genes, which suggests that other genes may collaborate to the Srp and Sna induced transformations. For example, in Drosophila Sna activates an EMT in mesoderm cells during early stages of embryonic development [48], but later on it plays distinct roles during central nervous system [49] and peripheral eye development [50]. Srp is required for EMT in the Drosophila midgut, but also for midgut cell specification [48], and additionally plays multiple roles during specification and maturation of the haemocytes [51, 52]. Hence, it is not surprising that activation of such transcription factors outside the normal controls imposed during development can impinge on multiple cell features and signalling programs in addition to EMT, and thus play key roles in the initiation and development of primary tumours, rather than being limited to the steps of cancer cell invasion and metastatic spread. Additionally, it is worth noting that the effect of Srp activity on tissue overgrowth in the wing disc is due, at least in part, to the ectopic triggering of effector genes normally elicited by Srp in the midgut, one of its regular domains of expression.
We previously investigated the effects of triggering ectopic sna and srp in the Drosophila embryo, by driving their expression in ectodermal epithelial cells in which they are never normally expressed [53]. While we found that sna had no effect on ectoderm cell behaviour [53], a more recent study showed that when sna was expressed at high levels using a maternal driver, it triggers adherens junction disassembly in ectodermal cells, and in rare cases, the movement of some cells to inside the embryo [54]. Similarly, ectopic srp drives a loss of apicobasal polarity and junction disassembly, although with srp there is a profound migration of cells into the embryo. Remarkably, in the embryo we do not see any proliferation in these circumstances. Conversely, in wing discs we see overproliferation, but very little cell migration. Intriguingly, a "Go or Grow" hypothesis has been proposed which postulates that cell division and cell migration are temporally exclusive events and that tumor cells defer migration to divide and vice versa [55–59]. Our results suggest that EMT transcription factors can drive migration or proliferation, but tend to favour one over the other at any given time. Given the fact that EMT transcription factors are increasingly associated with cancer stem cells, it will be important to unravel when and how EMT promotes one over the other.
The transformation of a healthy cell into a cancerous one requires multiple mutations and cooperation between different oncogenic/tumor suppressor mutations. Not only can EMT transcription factors accelerate tumour progression by the activation of multiple biological processes, this can also be exacerbated through cooperative effects of the different pathways. For example, in breast cancer models cooperation between Twist and an active form of RAS is sufficient to trigger transformation of mammary epithelial cells into malignant cells exhibiting all the characteristic features of claudin-low tumors [60]. Similarly our results suggest that ectopic Sna in combination with situations where cells become resistant to cell death may have catastrophic effects. Remarkably, over-expression of Srp alone activates both the EGFR/Ras and Yki signalling pathways. Of note, over-activation of the Ras pathway in situations of compromised cell polarity often leads to dramatic tissue overgrowth, for example when oncogenic Ras is combined with a scribble mutation [44]. Furthermore, it has previously been shown that loss of Drosophila cell polarity regulators such as Scribble promotes epithelial tissue overgrowth and cooperation with the Ras pathway through impaired Hippo pathway signaling [61]. Thus the profound effects seen upon Srp activation are likely due to cooperation between these two pathways. GATA factors are increasingly found deregulated in human tumours, both at the invasive front and in primary lesions and are receiving increasing attention as onco-promoting genes (reviewed in [62]). Our work suggests that GATA factors could be activating multiple tumour promoting pathways, that act cooperatively both in early stages of primary tumour growth and later in driving invasion and metastasis.
Methods
Fly strains and genetics
Details for all genotypes and transgenes can be found in flybase (http://flybase.org) or in references listed here. Conditional activation of either RNAi or gene expression was achieved using the Gal4/Gal80ts system [63]. To misexpress in the anterior compartment Ci-Gal4; tubulinGal80ts was used, and in the wing disc proper nub-Gal4, UAS-srcGFP; tubulinGal80ts. Crosses were kept at 18 °C until late in L2 when larvae were shifted to 29 °C for 48 hours and dissected. Gal4 lines were crossed to the following UAS transgenes: UAS-srp (from D. Hoshizaki), UAS-sna (from J. Kumar); UAS-shotRNAi (#41858), UAS-cheerioRNAi (#35755), UAS-EGFRRNAi (#25781), UAS-rasRNAi (ras1—#29319, ras2—#34619, ras3—#35414), UAS-foxORNAi (#27656), UAS-merlinRNAi (#28007), UAS-fkhRNAi (#33760), UAS-moodyRNAi (#36821), UAS-psGEFRNAi (#33433) from Bloomington stock center; UAS-βvintegrinRNAi (#2503), UAS-ykiRNAi (#40497) from VDRC. Clones were generated by crossing ywhsflp; Tub>y+>gal4, UAS GFP to UAS-sna or UAS-srp, and larva were heat-shocked for 30 mins and fixed at 3rd instar after 48 hours. The following reporter lines were used: crbM11.M2 (crb-lacZ, from M.Alverof [16]), diap-lacZ [26] and vnrF264 (vn-lacZ).
Immunohistochemistry, fixed image acquisition and analysis
The following antibodies used were: rabbit anti-aPKC (1:500; Santa Cruz); rat anti-crb (1:500; gift from E.Knust); mouse anti-Dlg (1:500; Hybridoma Bank); rabbit anti-dpERK (1:200; Cell signalling); goat anti-GFP (1:500; Abcam); rabbit anti-RFP (1:300; Life Technologies); Phalloidin TRITC (1:200; Sigma Aldrich); mouse anti-Patched (1:100; Hybridoma Bank); rat anti-Srp (1:500 made in the Casanova lab). The TUNEL assay was performed using the In situ Cell Death Detection Kit (Roche). Cy2, Cy3 and Cy5-conjugated secondary antibodies were from Molecular Probes and were used at 1:200 dilutions, and discs were mounted in Vectashield containing DAPI. Confocal images were acquired with a Leica SP5. Images were analysed with Fiji software [National Institutes of Health (NIH) Bethesda, MD] and assembled into figures using both Fiji and the Adobe Photoshop software.
Allograft transplantations
Wing discs with nub-Gal4, UAS-srcGFP; tubulinGal80ts alone, or crossed to UAS-sna or UAS-srp were dissected from third instar larvae that had been kept at 18 °C until late in L2 and shifted to 29 °C for 48 hours, and transferred to cold PBS. Discs were fragmented into small pieces using Vannas scissors (World precision Instruments) and implanted into the abdomens of virgin females, as described in [22]. Fly hosts were kept at 25 °C and examined 21 d post-implantation.
Quantification of tissue growth
For measuring the effects on the wing disc proper, nub-Gal4, UAS-srcGFP; tubulinGal80ts was crossed to either UAS-sna or UAS-srp. Crosses were kept at 18 °C until late in L2 when larvae were shifted to 29 °C for 48 hours and dissected. The size of the GFP expressing area was then measured using Fiji software. For Anterior Posterior size ratios, the maximal width of the anterior compartment was divided by that of the posterior, again measured using Fiji software.
Supporting information
A-G Staining for polarity markers in control (A), nub-Gal4, UAS-Sna; tub-Gal80TS discs (B, D, E) and in nub-Gal4, UAS-Srp; tub-Gal80TS discs (C, F, G) 48 hours after shifting to 29°C, the permissive temperature. Staining for Crb (A-C) shows that Crb is lost from the nub expressing region upon ectopic Sna or Srp expression (B, C—dotted red lines denotes nub expressing region). D, E Staining for F-Act and Dlg show that cells towards the edges of the Sna expressing regions lose polarity (D, E, arrowheads). F, G Srp overexpression drives a dramatic loss of cell polarity throughout the nub expressing region, as seen by staining for F-Act and Dlg (F, G, arrowheads) Scale bars—100μm.
(TIF)
A, B, 3rd instar wing discs from either individuals expressing GFP to mark anterior the anterior compartment together with Sna (A) or together with both Sna and p35, which blocks cell death (B). Discs are stained for GFP (green) and F-Act (red). Transgenes are under the control of the ci-Gal4 driver and tub-Gal80TS and the discs are fixed 48 hours after shifting to 29°C. Scale bars—100μm. (C) Histogram plotting the A/P width ratio of wing primordia expressing either GFP alone (wt), Sna alone (sna), Sna together with GFP (sna, GFP) or Sna together with both GFP and p35 (sna, GFP, p35). Data are presented as mean ± SD. *P<0.005; paired t-test. There is no significant difference between wt and sna, GFP, p35.
(TIF)
A-C 3rd instar wing discs from either individuals expressing GFP to mark anterior the anterior compartment together with Srp (A, C) and RasRNAi (A); or just Srp (B). Transgenes are under the control of the ci-Gal4 driver and tub-Gal80TS and the discs are fixed 48 hours after shifting to 29°C. A Staining for Dcp1 to visualise cell death. D. Histogram plotting the A/P width ratio of wing primordia expressing either GFP alone (wt), Srp alone (srp) or Srp together with EGFRRNAi. Data are presented as mean ± SD. *P<0.005; paired t-test.
(TIF)
(A) Clones of UAS-srp were generated in an otherwise wild type background. Clones were induced by a 30 min heat shock and fixed after 24 hours. Staining for dpERK shows that Ras signalling is activated both inside (arrow) and outside (arrowhead) clones.
(TIF)
Acknowledgments
We are grateful to members of the Casanova, Franch-Marro, Casali, Arujo and Llimargas labs for helpful discussions and to Nicolás Martín, Yolanda Rivera and Esther Fuentes for technical assistance. Thanks to Panagiotis Giannios for critical reading of the manuscript and assistance with statistical analysis. We thank M. Alverof, D. Hoshizaki, E. Knust, J. Kumar, M. Milan, the Bloomington stock centre and DSHB for kindly sending us flies and reagents.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by grants from the Ministerio de Economía y competitividad (Grant Numbers BFU2015-66488-P to JC, BFU2015-73494-JIN to KC and CGL2014-55786-P to XF-M). This work was also supported by a grant from the Generalitat de Catalunya to the IRBB to JC, and by a Wellcome Trust/Royal Society Sir Henry Dale Award to KC (Grant number R/148777-11-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27(20):2192–206. Epub 2013/10/22. doi: 10.1101/gad.225334.113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye X, Weinberg RA. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25(11):675–86. doi: 10.1016/j.tcb.2015.07.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berx G, Raspe E, Christofori G, Thiery JP, Sleeman JP. Pre-EMTing metastasis? Recapitulation of morphogenetic processes in cancer. Clin Exp Metastasis. 2007;24(8):587–97. Epub 2007/11/06. doi: 10.1007/s10585-007-9114-6 . [DOI] [PubMed] [Google Scholar]
- 4.Campbell K, Whissell G, Franch-Marro X, Batlle E, Casanova J. Specific GATA factors act as conserved inducers of an endodermal-EMT. Dev Cell. 2011;21(6):1051–61. doi: 10.1016/j.devcel.2011.10.005 . [DOI] [PubMed] [Google Scholar]
- 5.Lim J, Thiery JP. Alternative path to EMT: regulation of apicobasal polarity in Drosophila. Dev Cell. 2011;21(6):983–4. doi: 10.1016/j.devcel.2011.11.017 . [DOI] [PubMed] [Google Scholar]
- 6.Shureiqi I, Zuo X, Broaddus R, Wu Y, Guan B, Morris JS, et al. The transcription factor GATA-6 is overexpressed in vivo and contributes to silencing 15-LOX-1 in vitro in human colon cancer. FASEB J. 2007;21(3):743–53. Epub 2006/12/15. doi: 10.1096/fj.06-6830com . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwei KA, Bashyam MD, Kao J, Ratheesh R, Reddy EC, Kim YH, et al. Genomic profiling identifies GATA6 as a candidate oncogene amplified in pancreatobiliary cancer. PLoS Genet. 2008;4(5):e1000081 doi: 10.1371/journal.pgen.1000081 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belaguli NS, Aftab M, Rigi M, Zhang M, Albo D, Berger DH. GATA6 promotes colon cancer cell invasion by regulating urokinase plasminogen activator gene expression. Neoplasia. 2010;12(11):856–65. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen F, Li J, Cai W, Zhu G, Gu W, Jia L, et al. GATA6 predicts prognosis and hepatic metastasis of colorectal cancer. Oncol Rep. 2013;30(3):1355–61. doi: 10.3892/or.2013.2544 . [DOI] [PubMed] [Google Scholar]
- 10.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–94. doi: 10.1038/ncb2976 . [DOI] [PubMed] [Google Scholar]
- 11.Olmeda D, Jorda M, Peinado H, Fabra A, Cano A. Snail silencing effectively suppresses tumour growth and invasiveness. Oncogene. 2007;26(13):1862–74. Epub 2006/10/18. doi: 10.1038/sj.onc.1209997 . [DOI] [PubMed] [Google Scholar]
- 12.Herranz H, Eichenlaub T, Cohen SM. Cancer in Drosophila: Imaginal Discs as a Model for Epithelial Tumor Formation. Curr Top Dev Biol. 2016;116:181–99. doi: 10.1016/bs.ctdb.2015.11.037 . [DOI] [PubMed] [Google Scholar]
- 13.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–15. . [DOI] [PubMed] [Google Scholar]
- 14.Tamori Y, Suzuki E, Deng WM. Epithelial Tumors Originate in Tumor Hotspots, a Tissue-Intrinsic Microenvironment. PLoS biology. 2016;14(9):e1002537 doi: 10.1371/journal.pbio.1002537 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EE. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21(4):436–49. doi: 10.1101/gad.1509007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herranz H, Stamataki E, Feiguin F, Milan M. Self-refinement of Notch activity through the transmembrane protein Crumbs: modulation of gamma-secretase activity. EMBO Rep. 2006;7(3):297–302. doi: 10.1038/sj.embor.7400617 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson EC, Pichaud F. Crumbs is required to achieve proper organ size control during Drosophila head development. Development. 2010;137(4):641–50. Epub 2010/01/30. doi: 10.1242/dev.041913 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107(36):15810–5. Epub 2010/08/28. doi: 10.1073/pnas.1004060107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidson FF, Steller H. Blocking apoptosis prevents blindness in Drosophila retinal degeneration mutants. Nature. 1998;391(6667):587–91. Epub 1998/02/19. doi: 10.1038/35385 . [DOI] [PubMed] [Google Scholar]
- 20.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120(8):2121–9. Epub 1994/08/01. . [DOI] [PubMed] [Google Scholar]
- 21.Schubiger G, Hadorn E. [Auto- and allotypic differentiation in vivo cultivated foreleg blastemas of Drosophila melanogaster]. Dev Biol. 1968;17(5):584–602. . [DOI] [PubMed] [Google Scholar]
- 22.Rossi F, Gonzalez C. Studying tumor growth in Drosophila using the tissue allograft method. Nat Protoc. 2015;10(10):1525–34. Epub 2015/09/12. doi: 10.1038/nprot.2015.096 . [DOI] [PubMed] [Google Scholar]
- 23.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet. 2005;37(10):1125–9. doi: 10.1038/ng1632 . [DOI] [PubMed] [Google Scholar]
- 24.Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330(6012):1824–7. doi: 10.1126/science.1195481 . [DOI] [PubMed] [Google Scholar]
- 25.Staley BK, Irvine KD. Hippo signaling in Drosophila: recent advances and insights. Dev Dyn. 2012;241(1):3–15. doi: 10.1002/dvdy.22723 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14(3):388–98. doi: 10.1016/j.devcel.2008.01.007 . [DOI] [PubMed] [Google Scholar]
- 27.Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19(4):507–20. Epub 2010/10/19. doi: 10.1016/j.devcel.2010.09.009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. The Biochemical journal. 2000;351 Pt 2:289–305. . [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth factors. 2006;24(1):21–44. doi: 10.1080/02699050500284218 . [DOI] [PubMed] [Google Scholar]
- 30.Igaki T, Pagliarini RA, Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol. 2006;16(11):1139–46. Epub 2006/06/07. doi: 10.1016/j.cub.2006.04.042 . [DOI] [PubMed] [Google Scholar]
- 31.Pascual J, Jacobs J, Sansores-Garcia L, Natarajan M, Zeitlinger J, Aerts S, et al. Hippo Reprograms the Transcriptional Response to Ras Signaling. Dev Cell. 2017;42(6):667–80 e4. Epub 2017/09/28. doi: 10.1016/j.devcel.2017.08.013 . [DOI] [PubMed] [Google Scholar]
- 32.Reuter R. The gene serpent has homeotic properties and specifies endoderm versus ectoderm within the Drosophila gut. Development. 1994;120(5):1123–35. Epub 1994/05/01. . [DOI] [PubMed] [Google Scholar]
- 33.Szuts D, Eresh S, Bienz M. Functional intertwining of Dpp and EGFR signaling during Drosophila endoderm induction. Genes Dev. 1998;12(13):2022–35. Epub 1998/07/03. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnepp B, Grumbling G, Donaldson T, Simcox A. Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes Dev. 1996;10(18):2302–13. . [DOI] [PubMed] [Google Scholar]
- 35.Clifford RJ, Schupbach T. Coordinately and differentially mutable activities of torpedo, the Drosophila melanogaster homolog of the vertebrate EGF receptor gene. Genetics. 1989;123(4):771–87. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diaz-Benjumea FJ, Garcia-Bellido A. Behaviour of cells mutant for an EGF receptor homologue of Drosophila in genetic mosaics. Proc Biol Sci. 1990;242(1303):36–44. Epub 1990/10/22. doi: 10.1098/rspb.1990.0100 . [DOI] [PubMed] [Google Scholar]
- 37.Wessells RJ, Grumbling G, Donaldson T, Wang SH, Simcox A. Tissue-specific regulation of vein/EGF receptor signaling in Drosophila. Dev Biol. 1999;216(1):243–59. doi: 10.1006/dbio.1999.9459 . [DOI] [PubMed] [Google Scholar]
- 38.Diaz-Benjumea FJ, Hafen E. The sevenless signalling cassette mediates Drosophila EGF receptor function during epidermal development. Development. 1994;120(3):569–78. Epub 1994/03/01. . [DOI] [PubMed] [Google Scholar]
- 39.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028 . [DOI] [PubMed] [Google Scholar]
- 40.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013 . [DOI] [PubMed] [Google Scholar]
- 41.Valsesia-Wittmann S, Magdeleine M, Dupasquier S, Garin E, Jallas AC, Combaret V, et al. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell. 2004;6(6):625–30. doi: 10.1016/j.ccr.2004.09.033 . [DOI] [PubMed] [Google Scholar]
- 42.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14(1):79–89. doi: 10.1016/j.ccr.2008.06.005 . [DOI] [PubMed] [Google Scholar]
- 43.Ohashi S, Natsuizaka M, Wong GS, Michaylira CZ, Grugan KD, Stairs DB, et al. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70(10):4174–84. doi: 10.1158/0008-5472.CAN-09-4614 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brumby AM, Richardson HE. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 2003;22(21):5769–79. Epub 2003/11/01. doi: 10.1093/emboj/cdg548 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Igaki T, Yamamoto-Goto Y, Tokushige N, Kanda H, Miura M. Down-regulation of DIAP1 triggers a novel Drosophila cell death pathway mediated by Dark and DRONC. J Biol Chem. 2002;277(26):23103–6. Epub 2002/05/16. doi: 10.1074/jbc.C200222200 . [DOI] [PubMed] [Google Scholar]
- 46.Tepass U, Fessler LI, Aziz A, Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development. 1994;120(7):1829–37. Epub 1994/07/01. . [DOI] [PubMed] [Google Scholar]
- 47.Myat MM, Andrew DJ. Fork head prevents apoptosis and promotes cell shape change during formation of the Drosophila salivary glands. Development. 2000;127(19):4217–26. Epub 2000/09/08. . [DOI] [PubMed] [Google Scholar]
- 48.Seifert JR, Lehmann R. Drosophila primordial germ cell migration requires epithelial remodeling of the endoderm. Development. 2012;139(12):2101–6. doi: 10.1242/dev.078949 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashraf SI, Hu X, Roote J, Ip YT. The mesoderm determinant snail collaborates with related zinc-finger proteins to control Drosophila neurogenesis. EMBO J. 1999;18(22):6426–38. doi: 10.1093/emboj/18.22.6426 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim HY, Tomlinson A. Organization of the peripheral fly eye: the roles of Snail family transcription factors in peripheral retinal apoptosis. Development. 2006;133(18):3529–37. doi: 10.1242/dev.02524 . [DOI] [PubMed] [Google Scholar]
- 51.Rehorn KP, Thelen H, Michelson AM, Reuter R. A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development. 1996;122(12):4023–31. . [DOI] [PubMed] [Google Scholar]
- 52.Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science. 2000;288(5463):146–9. . [DOI] [PubMed] [Google Scholar]
- 53.Campbell K, Casanova J. A role for E-cadherin in ensuring cohesive migration of a heterogeneous population of non-epithelial cells. Nat Commun. 2015;6:7998 doi: 10.1038/ncomms8998 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weng M, Wieschaus E. Myosin-dependent remodeling of adherens junctions protects junctions from Snail-dependent disassembly. J Cell Biol. 2016;212(2):219–29. doi: 10.1083/jcb.201508056 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giese A, Loo MA, Tran N, Haskett D, Coons SW, Berens ME. Dichotomy of astrocytoma migration and proliferation. International journal of cancer. 1996;67(2):275–82. doi: 10.1002/(SICI)1097-0215(19960717)67:2<275::AID-IJC20>3.0.CO;2-9 . [DOI] [PubMed] [Google Scholar]
- 56.Hatzikirou H, Basanta D, Simon M, Schaller K, Deutsch A. ‘Go or grow’: the key to the emergence of invasion in tumour progression? Mathematical medicine and biology: a journal of the IMA. 2012;29(1):49–65. doi: 10.1093/imammb/dqq011 . [DOI] [PubMed] [Google Scholar]
- 57.Garay T, Juhasz E, Molnar E, Eisenbauer M, Czirok A, Dekan B, et al. Cell migration or cytokinesis and proliferation?—revisiting the "go or grow" hypothesis in cancer cells in vitro. Experimental cell research. 2013;319(20):3094–103. doi: 10.1016/j.yexcr.2013.08.018 . [DOI] [PubMed] [Google Scholar]
- 58.Corcoran A, Del Maestro RF. Testing the "Go or Grow" hypothesis in human medulloblastoma cell lines in two and three dimensions. Neurosurgery. 2003;53(1):174–84; discussion 84–5. . [DOI] [PubMed] [Google Scholar]
- 59.Funk SE, Sage EH. The Ca2(+)-binding glycoprotein SPARC modulates cell cycle progression in bovine aortic endothelial cells. Proc Natl Acad Sci U S A. 1991;88(7):2648–52. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois-Cox S, Wierinckx A, et al. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS Genet. 2012;8(5):e1002723 doi: 10.1371/journal.pgen.1002723 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doggett K, Grusche FA, Richardson HE, Brumby AM. Loss of the Drosophila cell polarity regulator Scribbled promotes epithelial tissue overgrowth and cooperation with oncogenic Ras-Raf through impaired Hippo pathway signaling. BMC Dev Biol. 2011;11:57 doi: 10.1186/1471-213X-11-57 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lentjes MH, Niessen HE, Akiyama Y, de Bruine AP, Melotte V, van Engeland M. The emerging role of GATA transcription factors in development and disease. Expert reviews in molecular medicine. 2016;18:e3 doi: 10.1017/erm.2016.2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302(5651):1765–8. doi: 10.1126/science.1089035 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A-G Staining for polarity markers in control (A), nub-Gal4, UAS-Sna; tub-Gal80TS discs (B, D, E) and in nub-Gal4, UAS-Srp; tub-Gal80TS discs (C, F, G) 48 hours after shifting to 29°C, the permissive temperature. Staining for Crb (A-C) shows that Crb is lost from the nub expressing region upon ectopic Sna or Srp expression (B, C—dotted red lines denotes nub expressing region). D, E Staining for F-Act and Dlg show that cells towards the edges of the Sna expressing regions lose polarity (D, E, arrowheads). F, G Srp overexpression drives a dramatic loss of cell polarity throughout the nub expressing region, as seen by staining for F-Act and Dlg (F, G, arrowheads) Scale bars—100μm.
(TIF)
A, B, 3rd instar wing discs from either individuals expressing GFP to mark anterior the anterior compartment together with Sna (A) or together with both Sna and p35, which blocks cell death (B). Discs are stained for GFP (green) and F-Act (red). Transgenes are under the control of the ci-Gal4 driver and tub-Gal80TS and the discs are fixed 48 hours after shifting to 29°C. Scale bars—100μm. (C) Histogram plotting the A/P width ratio of wing primordia expressing either GFP alone (wt), Sna alone (sna), Sna together with GFP (sna, GFP) or Sna together with both GFP and p35 (sna, GFP, p35). Data are presented as mean ± SD. *P<0.005; paired t-test. There is no significant difference between wt and sna, GFP, p35.
(TIF)
A-C 3rd instar wing discs from either individuals expressing GFP to mark anterior the anterior compartment together with Srp (A, C) and RasRNAi (A); or just Srp (B). Transgenes are under the control of the ci-Gal4 driver and tub-Gal80TS and the discs are fixed 48 hours after shifting to 29°C. A Staining for Dcp1 to visualise cell death. D. Histogram plotting the A/P width ratio of wing primordia expressing either GFP alone (wt), Srp alone (srp) or Srp together with EGFRRNAi. Data are presented as mean ± SD. *P<0.005; paired t-test.
(TIF)
(A) Clones of UAS-srp were generated in an otherwise wild type background. Clones were induced by a 30 min heat shock and fixed after 24 hours. Staining for dpERK shows that Ras signalling is activated both inside (arrow) and outside (arrowhead) clones.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.