ABSTRACT
MHC-I exposes the intracellular contents to immune cells for surveillance of cellular health. Due to high genomic variation, individuals' immune systems differ in their ability to expose and eliminate cancer-causing mutations. These personalized immune blind spots create specific oncogenic mutation predispositions within patients and influence their prevalence across populations.
KEYWORDS: antigen presentation; cancer predisposition; cancer susceptibility prediction; cancer; human leukocyte antigen; immunology, immunoediting; immunotherapy; major histocompatibility complex; neoantigens
The Major Histocompatibility Complex Class I (MHC-I) is a molecule that displays intracellular peptides on the cell surface in all nucleated cells. It is now well established that MHC-I presents cancer-associated neoantigens to T-cells, allowing elimination of mature tumor cells through immune surveillance unless the tumor cells can evade this process.1-3 The concept of immunosurveillance dictates that tumor cells at early stages of tumor development should also be susceptible to elimination by the immune system.4 If the initiating cells successfully avoid elimination, they continue to evolve in an environment of selective pressure imposed by the immune system. Eventually the developing tumor will acquire mechanisms to suppress or evade immune-selection leading to escape and clinical presentation. In practice, it has not been possible to systematically study interactions between the immune system and developing tumors prior to clinical diagnosis.
Clinical successes of checkpoint blockade inhibitors have generated an enormous interest in understanding the characteristics of neoantigens found in tumors that have evaded the immune system. However the role of early interactions between the immune system and nascent tumors is also very relevant since it has the potential to determine patient susceptibilities to specific oncogenic mutations. Recurrent mutations that are enriched for early cancer driving events should be subject to the strongest immune selection due to the inability of early neoplastic cells to suppress immune action. Importantly, this initial immune-selection is completely dependent on MHC-I presentation of common oncogenic events, which varies across individuals based on the set of six MHC-I alleles carried. Thus, we hypothesized that patient-specific MHC-I variation would create individual differences in which cancer-causing mutations would be undetectable by the immune system and thus would drive tumorigenesis to clinically diagnosed disease.5
To systematically evaluate this hypothesis, we established a score representing a patient's ability to expose a mutation on the cell surface for recognition by the immune system. Existing tools rank peptides based on their binding affinity to a single MHC-I allele.6 We developed a Patient Harmonic-mean Best Rank (PHBR) score from ranked peptide binding affinities that accounts for the contribution of a patient's six MHC-I alleles and all possible peptides containing a specific residue, and validated it against mass spectrometry data for MHC-I alleles complexed with peptides.5 Higher PHBR scores are interpreted as poorer MHC-I presentation and lower PHBR scores are interpreted as better MHC-I presentation.
Next we collected a set of 1,018 mutations likely to represent early drivers of cancer by selecting recurrent mutations in known cancer genes,7 and procured genetic information from 9,176 cancer patients in The Cancer Genome Atlas. We typed the human leukocyte antigen locus of each patient to determine their MHC-I alleles.8-10 Finally, we calculated the PHBR scores for each patient-mutation combination, creating a matrix (9,176 patients x 1018 mutations) representing the ability of each patient to present each mutation in the driver space to the immune system. We found extensive patient heterogeneity of MHC-I presentation across the driver space.
Contrasting PHBR scores with the presence or absence of mutations, we found an enrichment for observed mutations at higher PHBR scores (poor MHC-I presentation) as compared to unobserved mutations. Upon observing a linear correlation between the logit mutation probability and the log-PHBR score, we modeled the relationship using an additive logistic regression model with non-linear effects to account for variation in patient mutation rates. Across 30 tumor types, each unit increase in log-PHBR lead to a 28% increase in odds of a patient acquiring the mutation. For more high frequency mutations, we observed the odds increased by 54.5%. Applying the model within specific tumor types found that over half of the types had odds ratios significantly larger than 1, with the largest effects observed in thyroid cancer where the odds of mutation increased by 151% per unit increase in log-PHBR. In contrast, MHC-I genotype did not increase the odds of observing passenger mutations or common germline polymorphisms.
After observing this early strong selection on oncogenic mutations within specific patients, we further evaluated the effect of MHC-I presentation on mutation frequencies across cancer patients using the same set of cancer driving mutations. We found a striking negative correlation, i.e. frequent oncogenic mutations were generally more poorly presented by MHC-I throughout the population. Relative to random mutations and germline polymorphisms, human MHC-I alleles as a whole presented a much smaller percentage of cancer driving mutations and a much higher percentage of viral and bacterial residues. Thus, oncogenic mutations are strongly biased to fall in regions of the genome poorly presented by the human MHC-I.
In conclusion, we report the first evidence that patient MHC-I genotype contains predictive information about which oncogenic mutations are more likely to occur in patient tumors. Our results suggest that to exist at high frequencies in human cancer, mutations must both provide a fitness advantage for tumor cells and be poorly presented by most human MHC-I alleles. Supporting this notion, passenger mutations, rather than drivers, have been the predominant source of confirmed neoantigens in human tumors. Immunogenic passengers are likely to have been acquired after tumor escape from immune surveillance. The relationship between MHC-I genotype and mutation probability was detected despite limitations in current state-of-the-art bioinformatic tools, suggesting that future improvements in MHC-I affinity prediction and genotyping are likely to boost the predictive power of MHC-I genotype. As more data accumulates, we expect to gain a more complete picture of the role of MHC-I in individual cancer susceptibility and predisposition.
Figure 1.
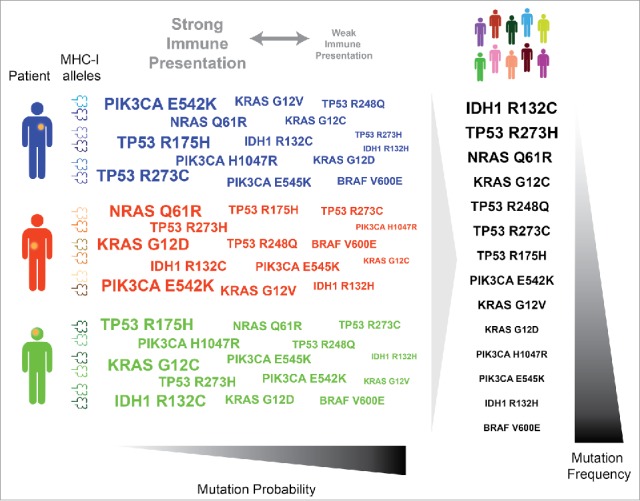
MHC-I genotype shapes the oncogenic landscape in tumors at the individual and population scale. Variation in MHC-I genotype amongst individuals results in individual differences in immune presentation and mutation probability (left). MHC-I variation of a population leads to a relationship between population immune presentation and population mutation frequency (right).
Funding Statement
This work was supported by the NSF under Grant # 2015205295 to R.M.; the NIH under Grant DP5-OD017937 to H.C.; and the NIH under Grant K99/R00CA191152 to J.F-B.
Disclosure statement
No potential conflicts of interest were disclosed.
Acknowledgments
The results published here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.”
References
- 1.Brahmer JR, Tykodi SS, Chow LQM, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al.. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al.. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al.. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6(10):715–727. doi: 10.1038/nri1936 [DOI] [PubMed] [Google Scholar]
- 5.Marty R, Kaabinejadian S, Rossell D, et al.. MHC-I Genotype Restricts the Oncogenic Mutational Landscape. Cell. 2017;171(6):1272–1283. doi: 10.1016/j.cell.2017.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8(1):33. doi: 10.1186/s13073-016-0288-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–962. doi: 10.1016/j.cell.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, Stevens J, Lane WJ, Dellagatta JL, Steelman S, et al.. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33(11):1152–1158. doi: 10.1038/nbt.3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, Raychaudhuri S, de Bakker PI. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8(6):e64683. doi: 10.1371/journal.pone.0064683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30(23):3310–3316. doi: 10.1093/bioinformatics/btu548 [DOI] [PMC free article] [PubMed] [Google Scholar]


