Abstract
Thalassotalic acids A–C and thalassotalamides A and B are new N-acyl dehydrotyrosine derivatives produced by Thalassotalea sp. PP2–459, a Gram-negative bacterium isolated from a marine bivalve aquaculture facility. The structures were elucidated via a combination of spectroscopic analyses emphasizing two-dimensional NMR and high-resolution mass spectral data. Thalassotalic acid A (1) displays in vitro inhibition of the enzyme tyrosinase with an IC50 value (130 μM) that compares favorably to the commercially-used control compounds kojic acid (46 μM) and arbutin (100 μM). These are the first natural products reported from a bacterium belonging to the genus Thalassotalea.
Graphical Abstract
Marine bacteria continue to be a highly productive source of new natural products with 408 new compounds reported from 2011–2013.1–3 While marine Gram-positive bacteria, especially actinomycetes, have garnered the most attention to date, genome mining and isolation-driven discovery efforts have raised awareness for the biosynthetic potential of marine Gram-negative bacteria (GNB).4, 5 Recognition that marine GNB produce such molecules as the didemnins,6, 7 and quite likely the bryostatins,8 highlights the potential for discovery of molecules with biomedical value. The marine γ-proteobacteria have been a particularly productive taxonomic group of GNB for natural product studies, yielding notable antibiotics such as andrimid and the moiramides, as well as the signaling molecule AI-2.9, 10 These studies and others portend the future success of discovery efforts aimed at marine GNB.
Thalassotalea is a new Gram-negative, aerobic, γ-proteobacteria genus.11 This genus was first characterized in 2014 following reclassification of members of the genus Thalassomonas.11 There have been only 10 species of Thalassotalea discussed in the literature and they have been primarily studied for their agarolytic properties, commensalism with marine animals, and pathogenic contribution to coral diseases.11–16 To date, there have been no published secondary metabolite studies on this genus.11–14
The enzyme tyrosinase is responsible for three transformations in the conversion of tyrosine to melanin, which is responsible for pigmentation of skin, hair, and eyes.17 It specifically catalyzes the conversion of L-tyrosine to 3,4-dihydroxyphenylalanine (L-DOPA), L-DOPA to dopaquinone, and 5,6-dihydroxyindole (DHI) to indole-quinone.17 Because of tyrosinase’s role in pigmentary diseases such as albinism and melasma, as well as cosmetic relevance to increase or decrease skin or hair color and pigmentation, compounds that can modulate tyrosinase have potential therapeutic and cosmetic applications.18 Additionally, tyrosinase is responsible for enzymatic browning of foods and inhibition of this process could lead to improved agricultural lifespans and nutrition.19
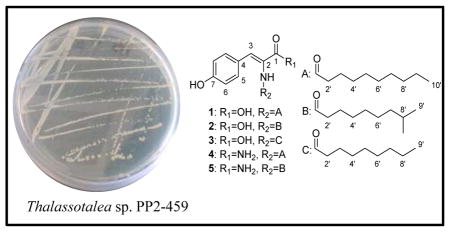
In this paper, we report new N-acyl dehydrotyrosine derivatives (1–5) produced during laboratory cultivation of Thalassotalea sp. PP2-459, formerly classified as Thalassomonas sp. PP2-459, a strain isolated from a carpet-shell clam (Ruditapes decussata) harvested in a bivalve hatchery located in Galicia (NW Spain).20 PP2-459 was previously determined to have quorum quenching ability against key bivalve aquaculture pathogens, including Vibrio anguillarum.20 Hence, PP2-459 has been proposed as a candidate for probiotic applications in aquaculture. Due to structural similarity with known tyrosinase inhibitors such as N-acylated tyrosines, 1–5 were evaluated for their inhibition of the tyrosinase enzyme.
Thalassotalea PP2-459 was cultured in YP seawater medium at 25 °C shaking and 200 rpm for two days. The cultures were centrifuged at 10,000 g for 10 min to substantially remove the cells, and the resulting supernatants were extracted with ethyl acetate. Purification efforts using reversed-phase (C18) chromatography methods yielded new compounds 1–5.
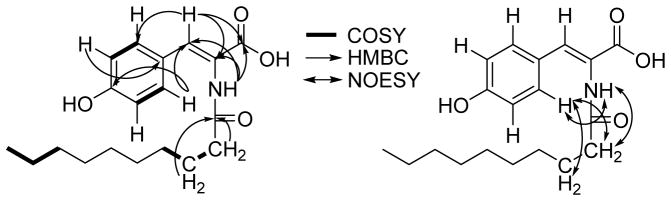
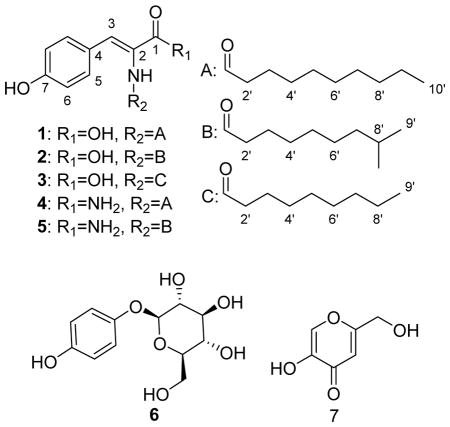
Thalassotalic acid A (1) was isolated as a light brown powder. The molecular formula was determined to be C19H27NO4 using HRESIMS (m/z 332.1866 [M-H]−, calcd for C19H26NO4, 332.1867), indicating 7 degrees of unsaturation. The presence of a decanoyl chain was supported by 1H-NMR resonances at δH 0.90 (t, 3H, H-10′), 1.29–1.31 (m, 12H, H-4′-H-9′), 1.59 (m, 2H, H-3′), and 2.28 (t, 2H, H-2′) that comprised a spin system in the 1H-1H COSY spectrum. 13C-NMR resonances at δC 172.0 (C-1′) and 166.7 (C-1) indicated the presence of two carbonyl carbons, the former belonging to the decanoyl chain based on HMBC correlations from H-2′ and H-3′. In the aromatic region, two doublets at δH 6.80 (H-6, J = 8.7 Hz) and 7.51 (H-5, J = 8.7 Hz) provided evidence for a para-substituted benzene ring. A HSQC correlation assigned a sharp singlet at δH 7.22 (H-3) as an olefinic methine attached to carbon at δC 132.2. HMBC correlations from H-5 to C-3 and H-3 to C-5 indicated that the C-3 methine was the benzylic position of the para-substituted aromatic ring. Consideration of the molecular formula and further HMBC correlations from the C-2-NH to C-1, C-2, and C-3, as well as correlations from H-3 to C-1 and C-2, established the α-β unsaturated tyrosine (Fig. 1). The (Z)-configuration of the double bond was determined based on NOE correlations shown in Fig. 1. TOF-MS/MS experiments further corroborated the proposed structure. Product ions were observed indicating the loss of CO2H (m/z 288.2, [M-45]), the decanoyl chain (m/z 178.0, [M-155]), and both the carboxylic acid and the acyl chain (m/z 134.0, [M-199]).
Figure 1.
Key COSY, HMBC, and NOESY correlations for thalassotalic acid A (1).
The structure of thalassotalic acid B (2) was determined by comparison to spectroscopic data of 1. A molecular ion at m/z 332.1872 [M-H]− indicated the same molecular formula as 1: C19H27NO4. A key difference in the 1H-NMR spectrum was the presence of a sharp doublet at δH 0.90 that integrated for 6 protons, suggesting a terminal branch in the aliphatic chain. Analysis of the HSQC spectrum revealed an aliphatic methine at δH 1.55 and δC 27.4, whose 1H-NMR resonance coupled to the C-8′ methyl groups in the COSY spectrum, thus confirming the terminal (CH3)2CH- group. The remaining structure was determined to be identical to 1 based on comparison of 13C and 2D NMR data and TOF-MS/MS evidence.
Thalassotalic acid C (3) displayed a HRESIMS ion (m/z 318.1699 [M-H]−, calcd for C18H24NO4, 318.1711) consistent with a molecular formula of C18H25NO4, which is one CH2 unit fewer than 1 and 2. Comparison of 1H- and 13C-NMR data with 1 and 2 revealed that 3 shared the same core tyrosine structure, but incorporated an unbranched chain containing one fewer methylene unit than 1. Thus, 3 was determined to be the nonanoyl analog of 1. TOF-MS/MS further corroborated this structure with the characteristic losses of CO2H (m/z 274.2, [M-45]), the nonanoyl chain (m/z 178.1, [M-141]), and both the carboxylic acid and the acyl chain (m/z 134.1 [M-185]).
Thalassotalamide A (4) was isolated as an amorphous brown powder. HRESIMS indicated that the molecular formula was C19H28N2O3 based on a molecular ion (m/z 331.2021 [M-H]−, calcd for C19H27N2O3, 331.2027). Thus, 4 had an additional NH and one less O than 1. Examination of the 1H-NMR spectrum of 4 indicated strong similarity to 1 based on resonances attributed to the p-substituted benzene ring (two doublets each integrating as 2H at δH 6.78 and 7.42) and unbranched aliphatic chain. The outstanding difference was two broad singlets at δH 7.05 and 7.28, indicating that 4 was likely the amide analog of 1. TOF-MS/MS confirmed this suspicion as fragment ions of m/z 288.2 [M-44] and m/z 134.1 [M-198] were observed, indicating that the α-β unsaturated carbonyl was an amide and not a carboxylic acid. Analysis of 13C- and 2D-NMR and further confirmed the structure of 4.
The structure of thalassotalamide B (5) was elucidated by comparison to spectral data of 2 and 4. HRESIMS indicated that the molecular formula of 5 was the same as 4 (C19H28N2O3) based on a molecular ion (m/z 331.2015 [M-H]−, calcd for C19H27N2O3, 331.2027). In the 1H-NMR spectrum, a doublet at δH 0.89 integrating 6H suggested that 5 had the same branched aliphatic chain as 2. Comparison of 13C-NMR data from the aliphatic chain of 2 to 5 confirmed they had the same 8-methyl nonanoyl aliphatic chain. Comparison of the remaining 13C-NMR data to those of 4, as well as the prominent loss of the amide in TOF-MS/MS (m/z 288.2), indicated that 5 was the amide derivative of 2.
N-acyl tyrosines have previously been studied for their effects on pigmentation of skin and hair.21–24 These compounds have been implicated in both increasing and decreasing pigmentation.21–24 Because of the similarity in structure to N-acyl tyrosines, we evaluated compounds 1–5 for their effects on tyrosinase enzyme activity compared to the positive controls arbutin (6) and kojic acid (7) that have been widely used in skin whitening products.25–27 Compounds 1, 2, and 3 exhibited tyrosinase enzyme inhibition with IC50 values of 130 ± 10 μM, 470 ± 10 μM, and 280 ± 10 μM, respectively (Table 2). The IC50 value for 1 compared favorably to the 6 (100 ± 10 μM), but was less effective than 7 (IC50 = 46 ± 2 μM). Compounds 4 and 5 were inactive at the maximum tested concentration (1 mM). These data support the requirement for a carboxylic acid and straight aliphatic chain for increasing enzyme inhibition within this structural class of tyrosinase inhibitors. While these N-acyl dehydrotyrosine compounds are not as potent as 6 or 7, 1 displays comparable tyrosinase inhibition and could be useful as a whitening agent or as an agricultural additive to prevent the browning of foods. 19
Table 2.
Tyrosinase inhibitory activity of compounds 1–5.
Positive controls
N-acyl derivatives of amino acids, including tyrosine, have been previously produced via heterologous expression of environmental DNA in Escherichia coli. These molecules are products of N-acyl amino acid synthases, which have been observed as single genes or as part of more complex biosynthetic sequences.28–30 While the compounds discussed here are structurally similar, the dehydrogenation across the α-β carbons of the tyrosine, the terminal amide functional group in 4 and 5, and the branched acyl chains in 2 and 5 represent biosynthetic variations not present in heterologously produced N-acyl tyrosines.29 Additionally, these are the first natural products reported from a bacterium belonging to the genus Thalassotalea.
EXPERIMENTAL SECTION
General Experimental Procedures
All chemical reagents and solvents were purchased from Sigma-Aldrich (St. Louis, MO) and acquired via Wilkem Scientific (Pawcatuck, RI). NMR experiments were conducted using an Agilent NMRS 500 MHz (Agilent Technologies, Santa Clara, CA) with (CD3)2SO as the solvent (referenced to residual DMSO at δH 2.54 and δC 39.5) at 25 °C. Electrospray ionization mass spectra (ESIMS) were acquired using an AB Sciex TripleTOF® 4600 spectrometer (Framingham, MA) in the negative ion mode. Flash chromatography was completed with a Combiflash Rf200 equipped with an 86 g C18 RediSep®Rf High Performance Gold column (Teledyne ISCO, Lincoln, NE). HPLC experiments were performed on a Hitachi Elite LaChrom system (Pleasanton, CA) equipped with a diode array detector (DAD) model L-2450, pump L-2130, and autosampler L-2200. Semi-preparative HPLC experiments were completed with a Waters XBridge™ Prep C18 5 μm, 10 × 250 mm column (Waters Corporation, Milford, MA).
Bacterial Strain and Culture Conditions
The bacteria strain (PP2-459) was isolated from a bivalve hatchery in Galicia, Spain and identified as a Thalassotalea sp. based on 16S rRNA sequence comparison (European Nucleotide Archive; accession number LN898116).20 It has been deposited in the Spanish Type Culture Collection (CECT) under restricted access. Cultures were maintained in YP seawater medium (1 g yeast extract, 5 g peptone, 22.5 g Instant Ocean (Spectrum Brands, Blacksburg, VA) per liter of deionized water) at 25 °C and shaking at 200 rpm.
Extraction and Isolation
A total of 17 L of bacterial cultures were cultivated and centrifuged at 10,000 g for 10 min to remove cells. The culture supernatants were extracted twice with equal volumes of ethyl acetate. The resultant organic layer was concentrated in vacuo to generate a total crude extract of 1.5 g.
The crude extract was adsorbed onto C18 resin (BAKERBOND™ Octadecyl (C18) 40 μm Prep LC Packing; J.T. Baker, Phillipsburg, NJ) and separated by reversed-phase flash chromatography. A linear gradient of 10 – 100% MeOH in H2O, each solvent modified with 0.1% formic acid (FA), afforded five fractions. Fraction 4 (135.6 mg) was further purified by reversed-phase C18 semi-preparative HPLC (25 min linear gradient from 40 – 78% MeOH in H2O (0.1% FA) to yield 5 pure compounds: 1 (28.3 mg, tR = 17.9 min), 2 (10.1 mg, tR = 16.9 min), 3 (7.8 mg, tR = 13.8 min), 4 (2.3 mg, tR = 16.0 min), and 5 (2.2 mg, tR = 15.1 min).
Thalassotalic acid A (1): off-white amorphous powder; UV (MeOH) λmax (log ε) 201 (4.16) 298 (3.99) nm; IR (ZnSe) υmax 3308, 3067 (br), 2917, 2850, 1688, 1656, 1601, 1584, 1271, 1167 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 332.1866 [M-H] −, (calcd for C19H26NO4, 332.1867).
Table 1.
NMR spectroscopic data (500 MHz, DMSO-d6) for thalassotalic acids A–C (1–3) and thalassotalamides A (4) and B (5).
| Thalassotalic acid A (1) | Thalassotalic acid B (2) | Thalassotalic acid C (3) | Thalassotalamide A (4) | Thalassotalamide B (5) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| position | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) |
| 1 | 166.7, C | 166.7, C | 166.8, C | 167.2, C | 167.2, C | |||||
| 1-OH/NH2 | 12.35, br s | 12.23, br s | 12.26, br s | 7.28/7.05, br s | 7.28/7.05, br s | |||||
| 2 | 124.2, C | 124.3, C | 124.5, C | 127.0, C | 127.0, C | |||||
| 2-NH | 9.23, s | 9.22, s | 9.20, s | 9.18, s | 9.19, s | |||||
| 3 | 132.2, CH | 7.22, s | 132.0, CH | 7.22, s | 131.7,a CH | 7.21, s | 128.6, CH | 7.06, s | 128.6, CH | 7.06, s |
| 4 | 124.7, C | 124.7, C | 124.8, C | 125.0, C | 125.0, C | |||||
| 5 | 131.7, CH | 7.51, d (8.7) | 131.7, CH | 7.51, d (8.6) | 131.7,a CH | 7.50, d (8.5) | 131.2, CH | 7.42, d (8.7) | 131.2, CH | 7.42, d (8.5) |
| 6 | 115.3, CH | 6.80, d (8.7) | 115.3, CH | 6.80, d (8.5) | 115.3, CH | 6.79, d (8.5) | 115.2, CH | 6.78, d (8.7) | 115.2, CH | 6.78, d (8.5) |
| 7 | 158.5, C | 158.5, C | 158.5, C | 158.0, C | 158.1, C | |||||
| 7-OH | 9.94, br s | 10.02, br s | 10.03, br s | 10.00, br s | 10.03, br s | |||||
| 1′ | 172.0, C | 171.9, C | 171.8, C | 172.0, C | 172.0, C | |||||
| 2′ | 35.1, CH2 | 2.28, t (7.2) | 35.2, CH2 | 2.28, t (7.4) | 35.2, CH2 | 2.28, t (7.2) | 35.2, CH2 | 2.32, t (7.5) | 35.2, CH2 | 2.32, t (7.5) |
| 3′ | 25.0, CH2 | 1.59, m | 25.0, CH2 | 1.59, m | 25.0, CH2 | 1.59, m | 24.7, CH2 | 1.58, m | 24.7, CH2 | 1.58, m |
| 4′ | 28.6–29.0, CH2 | 1.31, ma | 28.7, CH2 | 1.34, ma | 28.6–28.8, CH2 | 1.31, ma | 28.7–28.9, CH2 | 1.31, ma | 28.8, CH2 | 1.32, ma |
| 5′ | 28.6–29.0, CH2 | 1.31, ma | 29.1, CH2 | 1.30, ma | 28.6–28.8, CH2 | 1.31, ma | 28.7–28.9, CH2 | 1.31, ma | 29.1, CH2 | 1.28, ma |
| 6′ | 28.6–29.0, CH2 | 1.31, ma | 26.7, CH2 | 1.30, ma | 28.6–28.8, CH2 | 1.31, ma | 28.7–28.9, CH2 | 1.31, ma | 26.7, CH2 | 1.30, ma |
| 7′ | 28.6–29.0, CH2 | 1.31, ma | 38.4, CH2 | 1.19, m | 31.2, CH2 | 1.30, ma | 28.7–28.9, CH2 | 1.31, ma | 38.4, CH2 | 1.19, m |
| 8′ | 31.3, CH2 | 1.29, ma | 27.4, CH2 | 1.55, m | 22.1, CH2 | 1.32, ma | 31.3, CH2 | 1.29, ma | 27.4, CH2 | 1.55, m |
| 9′ | 22.1, CH2 | 1.30, ma | 22.5, CH3 | 0.90, d (6.6) | 13.9, CH3 | 0.90, t (7.0) | 22.1, CH2 | 1.30, ma | 22.5, CH3 | 0.89, d (6.5) |
| 10′ | 13.9, CH3 | 0.90, t (7.0) | 13.9, CH3 | 0.90, t (7.0) | ||||||
Overlapped signal
Thalassotalic acid B (2): brown amorphous powder; UV (MeOH) λmax (log ε) 201 (4.12) 299 (4.03) nm; IR (ZnSe) υmax 3326, 3118 (br), 2925, 2853, 1688, 1656, 1601, 1584, 1270, 1168 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 332.1872 [M-H] −, (calcd for C19H26NO4, 332.1867).
Thalassotalic acid C (3): brown amorphous powder; UV (MeOH) λmax (log ε) 202 (4.27) 299 (4.37) nm; IR (ZnSe) υmax 3317, 3067 (br), 2922, 2852, 1692, 1655, 1601, 1584, 1270, 1168 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 318.1699 [M-H] −, (calcd for C18H24NO4, 318.1711).
Thalassotalamide A (4): brown amorphous powder; 1H and 13C NMR data, see Table 1; HRESIMS m/z 331.2021 [M-H] −, (calcd for C19H27N2O3, 331.2027).
Thalassotalamide B (5): brown amorphous powder; UV (MeOH) λmax (log ε) 200 (4.26) 210 (4.19) 272 (3.59) nm; IR (ZnSe) υmax 3726, 3624, 3321, 2925, 2854, 1665, 1578, 1137 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 331.2015 [M-H] −, (calcd for C19H27N2O3, 331.2027).
Tyrosinase Inhibition Assay
Inhibitory effects of compounds 1–5 on mushroom tyrosinase were evaluated spectrophotometrically using L-tyrosine as a substrate according to our previously published method with minor modification.25 Tyrosinase inhibition assays were performed in 96-well microplate format using a SpectraMax M2 microplate reader (Molecular Devices, CA). Briefly, compounds were dissolved at various concentrations in 10% MeOH in phosphate buffer solution (PBS; 0.1 M, pH 6.8). Each well contained 40 μL of sample with 80 μL of PBS, 40 μL of tyrosinase (100 units/mL), and 40 μL L-tyrosine (2.5 mM). The mixture was incubated for 30 min at 37 °C, and absorbance was measured at 490 nm. Each sample was accompanied by a blank containing all components except L-tyrosine. Arbutin and kojic acid were used as positive controls. The results were compared with a control consisting of 10% MeOH in place of the test compounds. Experiments were completed in triplicate for each compound. The percentage of tyrosinase inhibition was calculated as: [(ΔAcontrol − ΔAsample)/ ΔAcontrol] × 100.
Supplementary Material
Acknowledgments
Instruments used for mass spectrometric and other analyses were supported through an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institute of Health (grant number 2 P20 GM103430). NMR data were acquired on instruments supported in part by the National Science Foundation EPSCoR Cooperative Agreement #EPS-1004057. J.C. was supported in part by the China Scholarship Council (File no. 201408330156).
Footnotes
Supporting Information Available: 1D- and 2D-NMR and HRESI mass spectra for compounds 1–5. The Supporting Information is available free of charge on the ACS Publications website at DOI:
References
- 1.Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. Nat Prod Rep. 2015;32:116–211. doi: 10.1039/c4np00144c. [DOI] [PubMed] [Google Scholar]
- 2.Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. Nat Prod Rep. 2014;31:160–258. doi: 10.1039/c3np70117d. [DOI] [PubMed] [Google Scholar]
- 3.Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. Nat Prod Rep. 2013;30:237–323. doi: 10.1039/c2np20112g. [DOI] [PubMed] [Google Scholar]
- 4.Machado H, Sonnenschein EC, Melchiorsen J, Gram L. BMC genomics. 2015;16:158. doi: 10.1186/s12864-015-1365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Still PC, Johnson TA, Theodore CM, Loveridge ST, Crews P. J Nat Prod. 2014;77:690–702. doi: 10.1021/np500041x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukimoto M, Nagaoka M, Shishido Y, Fujimoto J, Nishisaka F, Matsumoto S, Harunari E, Imada C, Matsuzaki T. J Nat Prod. 2011;74:2329–2331. doi: 10.1021/np200543z. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Kersten RD, Nam SJ, Lu L, Al-Suwailem AM, Zheng H, Fenical W, Dorrestein PC, Moore BS, Qian PY. J Am Chem Soc. 2012;134:8625–8632. doi: 10.1021/ja301735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu H, Patel A, Sherman DH, Haygood MG. J Nat Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 9.Mansson M, Gram L, Larsen TO. Mar Drugs. 2011;9:1440–68. doi: 10.3390/md9091440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isnansetyo A, Kamei Y. J Ind Microbiol Biotechnol. 2009;36:1239–48. doi: 10.1007/s10295-009-0611-2. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Tang K, Shi X, Zhang XH. Int J Syst Evol Micr. 2014;64:1223–1228. doi: 10.1099/ijs.0.055913-0. [DOI] [PubMed] [Google Scholar]
- 12.Stelling SC, Techtmann SM, Utturkar SM, Alshibli NK, Brown SD, Hazen TC. Genome Announc. 2014;2:e01231–14. doi: 10.1128/genomeA.01231-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S, Jung YT, Kang CH, Park JM, Yoon JH. Int J Syst Evol Micr. 2014;64:3676–3682. doi: 10.1099/ijs.0.067611-0. [DOI] [PubMed] [Google Scholar]
- 14.Hou T-T, Liu Y, Zhong Z-P, Liu H-C, Liu Z-P. Int J Syst Evol Micr. 2015 doi: 10.1099/ijsem.0.000637. [DOI] [PubMed] [Google Scholar]
- 15.Thompson F, Barash Y, Sawabe T, Sharon G, Swings J, Rosenberg E. Int J Syst Evol Micr. 2006;56:365–368. doi: 10.1099/ijs.0.63800-0. [DOI] [PubMed] [Google Scholar]
- 16.Ohta Y, Hatada Y, Miyazaki M, Nogi Y, Ito S, Horikoshi K. Curr Microbiol. 2005;50:212–216. doi: 10.1007/s00284-004-4435-z. [DOI] [PubMed] [Google Scholar]
- 17.Hearing VJ, Tsukamoto K. FASEB J. 1991;5:2902–2909. [PubMed] [Google Scholar]
- 18.Ando H, Kondoh H, Ichihashi M, Hearing VJ. J Invest Dermatol. 2007;127:751–761. doi: 10.1038/sj.jid.5700683. [DOI] [PubMed] [Google Scholar]
- 19.Parvez S, Kang M, Chung HS, Bae H. Phytother Res. 2007;21:805–816. doi: 10.1002/ptr.2184. [DOI] [PubMed] [Google Scholar]
- 20.Torres M, Romero M, Prado S, Dubert J, Tahrioui A, Otero A, Llamas I. Microbiol Res. 2013;168:547–554. doi: 10.1016/j.micres.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Guglielmini G. J Appl Cosmetol. 2006;24:55–61. [Google Scholar]
- 22.Guglielmini G. Cosmet Toiletries. 2004;119:61–65. [Google Scholar]
- 23.Kafara M, Arct J, Dzierzgowski S. J Appl Cosmetol. 2009;27:1–12. [Google Scholar]
- 24.Alvaro M, Cozzi R, Genovese A, Sedghi Zadeh H, Patent I. 2013/179098 A1. WO. 2013
- 25.Niesen DB, Ma H, Yuan T, Bach A, 2nd, Henry GE, Seeram NP. Nat Prod Commun. 2015;10:491–493. [PubMed] [Google Scholar]
- 26.Chang TS. Int J Mol Sci. 2009;10:2440–2475. doi: 10.3390/ijms10062440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YH, Avonto C, Avula B, Wang M, Rua D, Khan IA. J AOAC Int. 2015;98:5–12. doi: 10.5740/jaoacint.14-123. [DOI] [PubMed] [Google Scholar]
- 28.Brady SF, Clardy J. Org Lett. 2005;7:3613–3616. doi: 10.1021/ol0509585. [DOI] [PubMed] [Google Scholar]
- 29.Brady SF, Chao CJ, Clardy J. Appl Environ Microb. 2004;70:6865–6870. doi: 10.1128/AEM.70.11.6865-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Wagoner RM, Clardy J. Structure. 2006;14:1425–1435. doi: 10.1016/j.str.2006.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.