Abstract
RNA has the intrinsic propensity to form base pairs, leading to complex intramolecular and intermolecular helices. Direct measurement of base pairing interactions in living cells is critical to solving transcriptome structure and interactions, and investigating their functions (Lu and Chang, Curr Opin Struct Biol 36:142–148, 2016). Toward this goal, we developed an experimental method, PARIS (Psoralen Analysis of RNA Interactions and Structures), to directly determine transcriptome-wide base pairing interactions (Lu et al., Cell 165(5):1267–1279, 2016). PARIS combines four critical steps, in vivo cross-linking, 2D gel purification, proximity ligation, and high-throughput sequencing to achieve high-throughput and near-base pair resolution determination of the RNA structurome and interactome in living cells. In this chapter, we aim to provide a comprehensive discussion on the principles behind the experimental and computational strategies, and a step-by-step description of the experiment and analysis.
Keywords: Psoralen, Cross-linking, AMT (4′-aminomethyltrioxsalen hydrochloride), RNA structure, RNA–RNA interaction, 2D gel electrophoresis, Proximity ligation, High-throughput sequencing
1 Introduction
RNA structures and interactions are the building blocks of many macromolecular machines that drive basic cellular processes. Notable examples include the rRNAs and tRNAs in protein synthesis, spliceosomal snRNAs in intron removal, microRNAs in mRNA repression, small nucleolar and small cajal body RNAs (snoR-NAs/scaRNAs) in guiding RNA modifications, and ribozymes in catalyzing a variety of chemical reactions [1, 2]. The recent advent of high-throughput sequencing has led to the discovery of large numbers of long ncRNAs whose functions are under active investigations [3]. A picture is emerging for the lncRNAs where a multitude of sequence and structure motifs act as scaffolds in guiding the formation of functional protein–RNA complexes [4]. Even the previously well-studied smaller RNAs have been suggested to play more regulatory roles due to the variations in sequence, structure, expression pattern, RNA–protein complex compositions, and functions [5–8].
A detailed description of the physical basis of RNA molecules is an important step toward understanding their functions in physiology and pathology; however, structural analysis of RNA has been a daunting task for the entire RNA field. Since the first crystal structure was solved for the phenylalanine tRNA in the 1970s [9, 10], great progress has been made in the high-resolution analysis of many purified and “well-behaving” RNA molecules. Nevertheless, the vast majority of RNAs in living cells are large, highly dynamic and complex, and thus their structures are very challenging to solve by using most conventional methods, such as crystallography, nuclear magnetic resonance (NMR) and cryo-electron microscopy (cryo-EM). Recently developed chemical approaches, such as DMS-seq and icSHAPE, can measure RNA flexibility in cells [11–13], yet the measurements are one dimensional. In other words, interacting regions are not directly determined, but inferred based on the nucleotide reactivity. Modeling of structure with the flexibility measurement is often inaccurate, especially for long range, dynamic and complex structures [14].
On the molecular level, RNA structures and interactions are similar, both primarily made of stacking base pairs, or helices. To gain a global view of RNA helices in living cells, the key is to determine base pairing relationships, and the most commonly used chemical for such purpose is psoralen, a family of photo-cross-linkers that reversibly react with staggered pyrimidines on opposite strands [15]. Originally discovered in the early 1970s, psoralens were widely used for the analysis of the structures and interactions of abundant RNAs, such as rRNAs and snRNAs [16]. In conjunction with two-dimensional (2D) gel electrophoresis or northern blots using multiple probes, prominent psoralen cross-linking events can be used to determine base pairing partners, either within one RNA, or between two RNAs [17–20]. This classical method, however, is low throughput and laborious.
To directly determine double stranded RNA with high resolution on a global scale, we invented a new method, PARIS, which combines four critical techniques, psoralen cross-linking, 2D gel purification, proximity ligation, and high-throughput sequencing. The PARIS method employs a cell-permeable and reversible photo-cross-linker AMT (4′-aminomethyltrioxsalen) to covalently link RNA duplexes in living cells. The cross-linked RNA are partially digested with RNase and run through two-dimensional gels to selectively purify cross-linked RNA fragments, which make up a small fraction of all RNA fragments. Then proximity ligation is used to join the trimmed duplexes and the resulting chimeras can be sequenced and used to determine RNA structure and interaction. In this protocol, we provide a detailed step-by-step description, with explanations for the principle behind some critical steps. We also discussed some alternative solutions to tailor the analytic method for specific usage. Several of the library preparation steps have been optimized since the original protocol to make it more efficient [21].
2 Materials
2.1 Basic Reagents
All the materials used for RNA work should be RNase free. Common reagents and equipment are not listed here.
PBS.
S1 nuclease (e.g., ThermoFisher).
10 w/v % SDS (sodium dodecyl sulfate) in water.
Proteinase K (e.g., Ambion).
ShortCut RNase III (e.g., NEB).
Gel loading dye, orange, 6×.
TRIzol LS reagent (e.g., Life Technologies).
Chloroform.
100% ethanol.
80% ethanol.
Isopropanol.
100% DMSO.
3 M Na-Acetate pH 5.5.
Glycogen (e.g., GlycoBlue, 15 mg/mL).
RNase inhibitor (e.g., Thermo Scientific SuperaseIn and RiboLock).
T4 RNA ligase 1 (ssRNA ligase), high concentration (e.g., NEB).
RecJf exonuclease (e.g., NEB, cat. no. M0264).
5′ Deadenylase (e.g., NEB, cat. no. M0331).
Ultrapure TBE buffer, 10× (e.g., Life Technologies).
SequaGel UreaGel System, (National Diagnostics).
Ultrapure TEMED (e.g., Invitrogen).
Ammonium persulfate (APS).
Reverse transcriptase (e.g., Invitrogen SuperScript III).
10 mM Deoxynucleotide solution mix (dNTPs).
Magnetic streptavidin beads (e.g., Life Technologies Dyna-beads MyOne streptavidin C1).
Magnetic SPRI beads (e.g., Beckman-Coulter AMPure XP).
20% N-Lauroylsarcosine sodium salt solution (e.g., Sigma-Aldrich).
Deoxycholic acid sodium salt (e.g., Thermo Fisher Scientific).
D-Biotin.
RNase cocktail enzyme mix (Ambion, cat. no. AM2286).
RNase H (e.g., Enzymatics).
DNA ladder, 25 bp.
Denaturing PAGE loading buffer (e.g., Ambion Gel loading buffer II).
SYBR Gold nucleic acid gel stain, 10,000× (e.g., Life Technologies).
40 w/v % acrylamide–bis solution, 29:1.
CircLigase II ssDNA ligase (e.g., Epicentre, cat. no. CL9025K).
Phusion high-fidelity (HF) PCR master mix with HF buffer (e.g., NEB).
SYBR Green I nucleic acid gel stain, 10,000× (e.g., Life Technologies).
2.2 Oligos Used for Library Preparation
Preadenylated and 3′-biotin blocked RNA adapter (PAGE purified): /5rApp/AGATC GGAAG AGCGG TTCAG/3Biotin/. This designed adapter is important for efficient ligation on the 3′ end of RNA.
RT-primer-1 (DNA, standard desalting purification). /5phos/ WWW NNN ATCACG NNNNN TACCC TTCGC TTCACACACA AG/iSp18/GGATCC /iSp18/TACTG AACCGC, W = A/T and N = A/T/G/C are used to discriminate PCR duplicates, “ATCACG” is the specific experimental barcode, / iSp18/ is a spacer to prevent PCR from forming concatemers). The following is a list of 24 barcodes: 1. ATCACG, 2. CGATGT, 3. TTAGGC, 4. TGACCA, 5. ACAGTG, 6. GCCAAT, 7. CAGATC, 8. ACTTGA, 9. GATCAG, 10. TAGCTT, 11. GGCTAC, 12. CTTGTA, 13. AGTCAA, 14. AGTTCC, 15. ATGTCA, 16. CCGTCC, 17. GTCCGC, 18. GTGAAA, 19. GTGGCC, 20. GTTTCG, 21. CGTACG, 22. GAGTGG, 23. ACTGAT, 24. ATTCCT. The quality of the oligonucleotide synthesis should be verified by polyacrylamide gel electrophoresis; however, bulk PAGE purification before use in RT reactions is not necessary.
P3Tall v4 primer (PAGE purified): GGCAT TCCTG CTGAACCGCT CTTCC GATCT. P6Tall v4 primer (PAGE purified): CTCTT TCCCC TTGTG TGTGA AGCGA AGGGT.
P3-Solexa PCR primer (PAGE purified): 5′-CAAGC AGAAGACGGC ATACG AGATC GGTCT CGGCA TTCCT GCTGA ACCGC TCTTC CGATC T-3′.
P6-Solexa PCR primer (PAGE purified): 5′-AATGA TACGGCGACC ACCGA GATCT ACACT CTTTC CCTAC ACGAC GCTCT TCCGA TCT-3′.
P6_custom_SeqPrimer (PAGE purified): CACTC TTTCCCCTTG TGTGT GAAGC GAAGG GTA.
2.3 Regent Setup
AMT: dissolve 4′-aminomethyltrioxsalen in water at a final concentration of 1 mg/mL. Store it at −20 °C.
Urea/SDS solution containing 4 M urea and 0.1% SDS in water. Store it at room temperature.
10 w/v % APS: Dissolve 5 g of APS in a final volume of 50 mL of nuclease-free water. Store it at 4 °C for up to 6 months.
6 w/v % UreaGel denaturing PAGE solution. Prepare 1 L of UreaGel denaturing polyacrylamide gel stock solution by mixing 240 mL of UreaGel System concentrate, 660 mL of Urea-Gel System diluent, and 100 mL of UreaGel System buffer. Store it at 4 °C.
6 w/v % Native PAGE solution: Prepare gel stock solution by mixing 100 mL of 10× TBE solution and 150 mL of 40% acrylamide–bis solution in a final volume of 1 L of nuclease-free water. Store it at 4 °C.
Native PAGE solution for first dimension gel. To make 20 mL gel solution, use 6 mL from 40% stock (Bio-Rad 29:1), 2 mL 10× TBE, 12 mL water, 20 μL TEMED, and 200 μL 10% APS. Make it fresh for use.
Gel Crushing Buffer: 500 mM NaCl, 1 mM EDTA, pH 8.0, and 0.1% SDS.
Bead Binding Buffer. The final component concentrations are 100 mM Tris pH 7.0, 1 M NaCl, 10 mM EDTA, and 0.2 v/v % Tween 20. Prepare the buffer in nuclease-free water. Store it at 4 °C.
Streptavidin beads. Prepare 20 μL of magnetic streptavidin beads for each PARIS sample by washing the beads twice with 1 mL of bead binding buffer. After washing, resuspend the beads in 10 μL of bead binding buffer per PARIS sample, and store them on ice until needed.
Bead Wash Buffer. Final component concentrations are 100 mM Tris pH 7.0, 4 M NaCl, 10 mM EDTA, and 0.2 v/v % Tween 20. Prepare the buffer in nuclease-free water. Store it at 4 °C.
10 w/v % Sodium deoxycholic acid solution (NaDOC): Mix 5 g of deoxycholic acid sodium salt in 50 mL of nuclease-free water. Store it at room temperature.
RNase H buffer, 10×. Final component concentrations are 0.5 M HEPES pH 7.5, 0.75 M NaCl, 30 mM MgCl2, 1.25 w/v % N-lauroylsarcosine sodium salt, 0.25 w/v % NaDOC, and 50 mM DTT. Prepare the buffer in nuclease-free water. If a precipitate forms, briefly warm it at 37 °C so that it dissolves into solution. Freshly prepare the solution.
SYBR Green I, 25×. Make a 1:400 dilution from SYBR Green I nucleic acid gel stain (10,000×) in DMSO. Store it at −20 °C.
Proximity ligation mixture (10 μL/sample): 2 μL 10× T4 RNA ligase buffer, 5 μL T4 RNA ligase (high concentration), 1 μL SuperaseIn, 1 μL 10 mM ATP, and 1 μL water.
Adapter ligation mixture (10 μL/sample): 2 μL 10× T4 RNA ligase buffer, 1 μL 0.1 M DTT, 5 μL 50 v/v % PEG8000, 1 μL 10 μM preadenylated and 3′-biotinylated RNA adapter, and 1 μL High Concentration T4 RNA ligase 1.
Reverse transcriptase enzyme mix (8 μL/sample): 4 μL 5× First Strand Buffer, 0.75 μL 40 U/μL RiboLock, 1 μL 100 mM DTT, 1 μL dNTPs (10 mM each), and 1.25 μL SuperScript III reverse transcriptase.
RNaseA/T1/H elution mix (50 μL/sample): 5 μL 10× RNase H Buffer, 12.5 μL 50 mM D-biotin, 1 μL P3-Tallv2 primer, 1 μL RNase cocktail enzyme mix, 1 μL RNase H, and 30.5 μL RNase-free water.
Circularization reaction mix (4 μL/sample): 2 μL 10× CircLigase II buffer, 1 μL 50 mM MnCl2, and 1 μL CircLigase II. 19. qPCR reaction mixture (10.5 μL/sample): 10 μL 2× Phusion HF PCR master mix, 0.2 μL 25× SYBR Green I, and 0.25 μL P3/P6 TalI v4 PCR primer mix (20 μM forward and 20 μM reverse).
2.4 Equipment
10–15 well gel cassette (e.g., BioRad mini-Protean 3).
PAGE apparatus (e.g., CSB Scientific).
High-voltage electrophoresis power supply (e.g., Bio-Rad).
Freeze dryer/lyophilizer (e.g., Labconco).
Spectrophotometer (e.g., Thermo Scientific NanoDrop).
Agilent Bioanalyzer.
Thermomixer.
UV cross-linker (e.g., Stratagene Stratalinker 2400 model).
254 nm and 365 nm emission UV bulbs (e.g., Agilent Technologies).
Gel imager (e.g., Bio-Rad Gel Doc XR+).
300 nm blue light transilluminator (e.g., Clare Chemical Research Dark Reader).
qPCR machine capable of removal of individual samples amid reaction (e.g., Stratagene Mx3005p).
1.5 mL siliconized microcentrifuge tubes (e.g., MidSci).
0.5 mL 10 kDa cutoff Centrifugal Filter Unit (e.g., Millipore Amicon Ultra-0.5 10 k).
Centrifuge tube filters, 0.45 μm (e.g., Sigma-Aldrich Corning Costar Spin-X).
DNA purification columns (e.g., Zymo Research, DNA Clean & Concentrator-5 columns).
RNA purification columns (e.g., Zymo Research, RNA Clean & Concentrator-5 columns).
Magnetic Eppendorf-tube stand (e.g., Thermo Fisher DynaMag2).
2.5 Software
Java, Perl, and Python interpreters.
Trimmomatic [22].
icSHAPE pipeline (https://github.com/qczhang/icSHAPE).
fastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
STAR [23].
SAMtools [24].
samPairingCalling.pl (https://github.com/qczhang/paris).
mfold [25].
sam2ngmin.py (https://github.com/zhipenglu/).
IGV genome browser [26].
maf_extract_ranges_indexed.py (bxpython package, https://github.com/bxlab/bx-python).
RNAalifold [27].
SISSIz [28].
Alternativestructure.py (https://github.com/zhipenglu/duplex).
RNAcofold (from the Vienna RNA package).
3 Methods
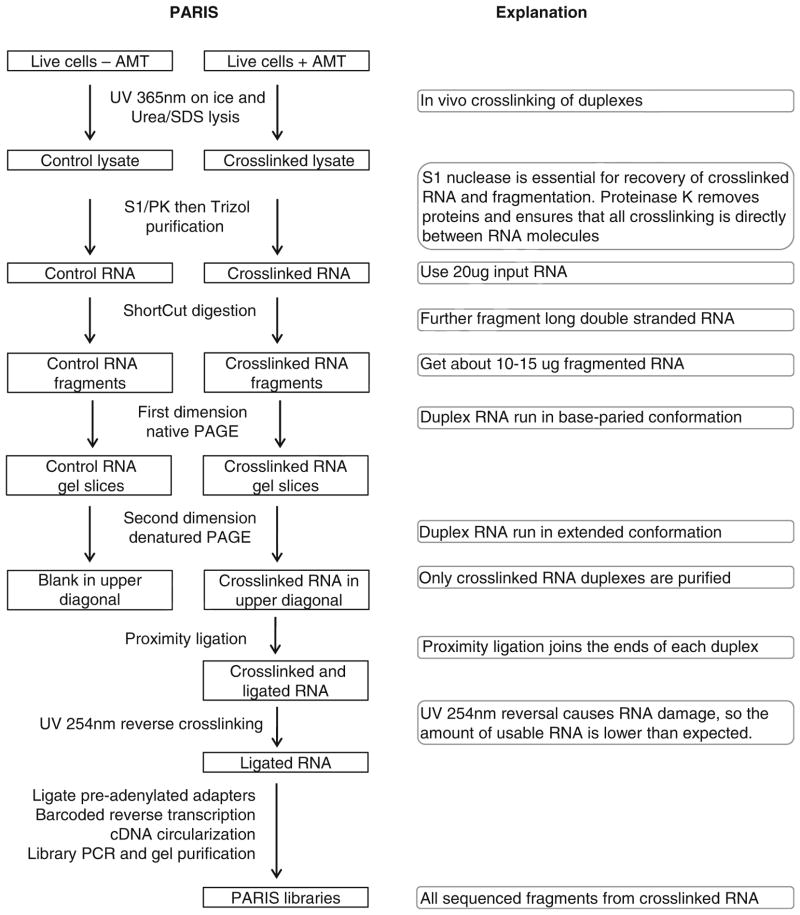
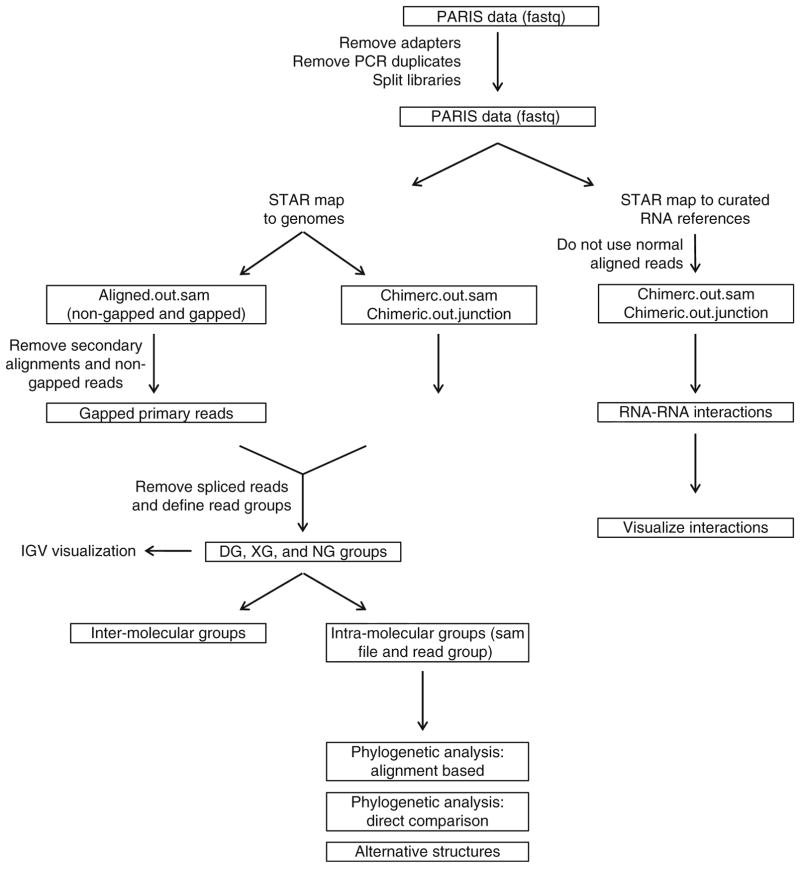
The following is a detailed description of the PARIS method (Fig. 1). Indicators of successful implementation are included for the critical steps. Standard experimental procedures like TRIzol extraction and ethanol precipitation of RNA are not described in details here and readers should refer to manufacturers’ manuals. The library preparation part (Subheadings 3.5 and 3.6) was adapted from a similar protocol for icSHAPE and modified [29]. The principles of data analysis have been described in the supplemental methods in the PARIS paper [21], and a step-by-step protocol is provided here (Fig. 4). Custom scripts for the analysis of PARIS data were published in Github: https://github.com/qczhang/paris, https://github.com/zhipenglu/duplex.
Fig. 1.
Outline of the PARIS experimental strategy. Major steps are explained on the right side
Fig. 4.
The PARIS analysis pipeline, modified from the PARIS paper [21]. Major analyses outlined here include DG and NG assembly, visualization of RNA structure data and models in IGV, two approaches of phylogenetic analysis, analysis of alternative structures, identification and visualization of RNA–RNA interactions
3.1 Psoralen Cross-Linking
Before performing psoralen cross-linking, design the experiment with the cross-linker capacity in mind. The Stratalinker 2400 model cross-linker can hold five 15 cm or ten 10 cm plates.
On the day of cross-linking, prepare the AMT cross-linking solution containing 0.5 mg/mL AMT and 1× PBS. Use PBS as control.
To cross-link RNA in vivo, adherent cells are cultured to ~70% confluent as usual. Remove media from tissue culture plates, wash with PBS and add 0.4 mL of PBS or AMT cross-linking solution to each 10 cm plate. Use 1 mL solution per 15 cm plate.
Incubate cells for 30 min at the normal cell culture conditions (e.g., 37 °C, 5% CO2 in incubator for mammalian cells). At the same time make sure the 365 nm light bulbs are installed in the cross-linker. Cross-linking in the presence of AMT loosens cells from the plate for certain cell lines, such as HEK 293T, and, therefore, care should be taken in adding and removing liquid from the plate.
Place ice trays in the cross-linker and put cell culture plates on ice. Irradiate cells with 365 nm UV, 15 cm away from the light bulbs. Swirl the plates every 10 min and make sure that they are horizontal and the liquid covers all the cells (see Note 1).
Remove cross-linking solution after cross-linking. Scrape off cells in 1 mL chilled PBS, transfer to 1.5 mL tubes and centrifuge at 4 °C, 400 × g for 5 min.
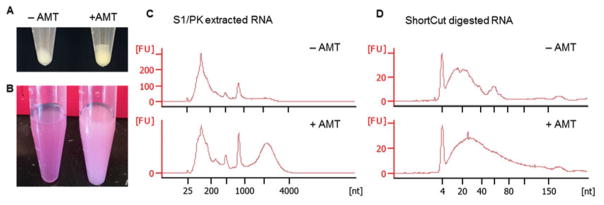
Remove PBS from the tubes, snap-freeze cell pellets on dry ice and store pellets at −80 °C. Control pellets are white and AMT cross-linked pellets are yellowish. The difference in color is an indicator for successful cross-linking (Fig. 2a, see Note 2) The pellet from a 10 cm plate should be around 30 μL, optimal for a single S1/PK reaction.
Fig. 2.
Example results of AMT cross-linking and RNA fragmentation. (a) AMT cross-linked cell pellets (right) have a darker color than the non-cross-linked ones (left). (b) After S1/PK digestion, adding TRIzol and chloroform, phase separation is faster for the non-cross-linked cells (left) than the cross-linked cells (right), so the cross-linked samples have a milky appearance. (c) S1/PK extraction produces a characteristic broad peak between 2000 and 4000 nt in the Bioanalyzer electrophoretic trace of cross-linked cells. The height of the broad peak is variable among cell types and different batches of experiments. (d) ShortCut RNase III digestion reduces RNA to a smaller size (usually below 150 nt) to facilitate 2D gel purification and library preparation
3.2 S1/PK Extraction and ShortCut Digestion
Resuspend cell pellet from a 10 cm plate in 200 μL urea/SDS solution. Control samples can be very viscous and hard to dissolve. Use a pipet to dislodge pellet and shake vigorously to suspend the pellet. AMT cross-linked cell pellets are easier to break up and resuspend in urea/SDS solution as the DNA is fragmented by UV 365 nm in the presence of AMT.
To the lysed cells add 60 μL 5× S1 nuclease buffer, 2 μL S1 nuclease and mix well. Expect ~300 μL final volume. Perform the S1 digestion at room temperature for 10 min with frequent pipetting or vortexing to break viscous material.
To the S1 digested lysate add 33 μL 10% SDS (final concentration 1%) and 2 μL proteinase K (PK, 20 mg/mL, final 0.1 μg/μL). Samples should become clear soon after adding SDS. Perform the PK digestion at 50 °C for 30 min in a thermomixer, at 1000 rpm to mix the suspension (see Note 3).
After PK digestion, add 0.9 mL TRIzol LS reagent and mix vigorously. Then add 180 μL chloroform and mix vigorously. Phase separation is faster in control samples than +AMT samples, another visible difference between control and cross-linked samples (Fig. 2b). Complete the RNA extraction following the standard TRIzol protocol.
Quantify the purified RNA using a spectrophotometer and analyze the quality using Bioanalyzer. An example Bioanalyzer trace file is shown in Fig. 2c. Also see the previous PARIS publication for another example [21]. The AMT cross-linked sample should have a broad peak between 2000 and 4000 nt for human, mouse, and Drosophila cells, an important indicator of successful cross-linking and RNA extraction (see Note 4).
Use 20 μg S1/PK purified RNA for ShortCut RNase III digestion, in order to reduce the RNA fragments to smaller size (see Note 5). Each ShortCut reaction in 50 μL volume includes 20 μg RNA, 4 μL ShortCut RNase III, 5 μL 200 mM MnCl2, and 5 μL ShortCut buffer. The reaction is incubated at 37 °C for 20 min.
After ShortCut digestion, purify RNA using the standard TRI-zol method and resuspend RNA in 15 μL water. Determine concentration of the samples by spectrophotometer and analyze size distribution using Bioanalyzer (Fig. 2d). Typically, the AMT cross-linked samples have a stronger tail above 100 nt than the control samples.
3.3 Purification of Cross-Linked RNA by 2D Gel
-
1
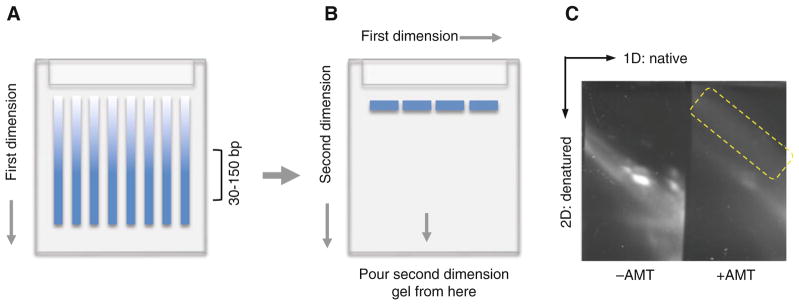
Prepare the 12% 1.5 mm thick native first dimension gel using the BioRad Mini-Protean 3 gel cassette (Fig. 3a). The 1.5 mm thick gel is used here to facilitate the removal of air bubbles from the second dimension. Use 15-well combs so that each lane is narrower and the second dimension has a higher resolution. After the gel solidifies, pull the comb very slowly to avoid the deforming well dividers.
-
2
To each 15 μL sample add 5 μL 6× Orange G loading dye. Load 3 μL dsRNA ladder as molecular weight marker. Run the first dimension gel at 100 V for 70 min in 0.5× TBE. Orange G should be 4/5 way to bottom. Usually we have a starting current of 15 mA and a starting power of 1.5 W.
-
3
After electrophoresis finishes, stain the gel with 2 μL SYBR Gold in 20 mL 0.5× TBE, incubate for 5 min. Image the gel using 300 nm transillumination (not the 254 nm epi-illumination, which reverses the psoralen cross-linking). Excise each lane between 30 and 150 bp from the first dimension gel (Fig. 3a). The second dimension gel can usually accommodate three gel splices.
-
4
Prepare the 20% 1.5 mm thick urea denatured second dimension gel using the UreaGel system. For 20 mL gel solution, use 16 mL UreaGel concentrate, 2 mL UreaGel diluent, 2 mL UreaGel buffer, 8 μL TEMED, and 160 μL 10% APS. Add TEMED and APS right before pouring the gel.
-
5
To make the second dimension gel, put the square plate horizontally and arrange gel slices in a “head-to-toe” manner with 2–5 mm gap between them (Fig. 3b). Leave 1 cm space at the top of the notched plate so that the second dimension gel would completely encapsulate the first dimension gel slices.
-
6
Apply 20–50 μL 0.5× TBE buffer on each gel slice to avoid air bubbles when placing the notched plate on top of the gel slices. Remove the excess TBE buffer after the cassette is assembled, and leave 2 mm space at the bottom of the notched plate to facilitate pouring the second dimension gel.
-
7
Pour and gel solution from the bottom of the plates, while slightly tilting the plates to one side to avoid air bubbles building up between the plates. If there are air bubbles, use the thin loading tips to draw them out.
-
8
Use ~60 °C prewarmed 0.5× TBE buffer to fill the electrophoresis chamber to facilitate denaturation of the cross-linked RNA. Run the second dimension at 30 W for 40 min to maintain high temperature and promote denaturation. Run the gel for 50 min. The voltage starts around 300 V and gradually increases to 500 V, while the current starts around 100 mA and gradually decreases to 60 mA.
-
9
After electrophoresis, stain the gel with SYBR Gold the same as the first dimension gel and image the gel using 300 nm transillumination (Fig. 3c).
-
8
Excise the gel containing the cross-linked RNA from the 2D gel and transfer it to a new 10 cm cell culture dish. Crush the gel by grinding with the cap of a 15 mL tube.
-
9
Add 300 μL crushing buffer to gel debris. Transfer the gel slurry to a 15 mL tube by shoveling with a cell scraper.
-
10
Add additional 1.2 mL crushing buffer and rotate at 4 °C overnight.
-
11
Transfer ~0.5 mL gel slurry to Spin-X 0.45 μm column. Spin at room temperature, 6000 rpm for 1 min. Continue until all gel slurry is filtered.
-
12
Aliquot 500 μL of the filtered RNA sample to an Amicon 10 k 0.5 mL column. Spin at 4 °C, 12,000 × g for 5 min. Repeat until all of the filtered RNA sample flowed through the column.
-
13
Wash the column with 300 μL water and spin the column at 4 °C, 12,000 × g for 5 min.
-
14
Invert and place the column in a new collection tube, and spin at 4 °C, 6000 × g for 5 min. Recover ~85 μL RNA from each column (~170 μL total from two columns).
-
15
Precipitate the RNA using the standard ethanol precipitation method, with glycogen as a carrier. Alternatively, the RNA can be purified using the Zymo RNA clean and concentrator-5 columns.
-
16
Reconstitute RNA in 11 μL water and dilute 1 μL RNA sample for Bioanalyzer analysis. The RNA sample should have a broad size distribution between 30 and 150 nt in the Bioanalyzer trace. The yield is typically 0.1–0.5% from 20 μg S1/PK extracted input RNA.
Fig. 3.
Diagram and example result for 2D gel purification of cross-linked and digested RNA. (a) First dimension native gel. Gel slices containing RNA around 30–150 bp are usually cut out from the first dimension for the second dimension. (b) Second dimension denatured gel. Gel slices are aligned between the glass plates before pouring the urea denatured gel solution at the bottom of the gel plates (as indicated by the arrow in the middle). (c) An example second dimension gel. The yellow-boxed area indicate the cross-linked RNA to be extracted for library preparation
3.4 Proximity Ligation and Photoreversal of Cross-Linking
Add 10 μL proximity ligation mixture to 10 μL of RNA, mix well and incubate at room temperature overnight.
Boil the ligation mixture at 95 °C for 2 min. After heat denaturation, the samples may turn turbid. Spin down the insoluble material at 6000 × g for 5 min and retain the supernatant.
To the 20 μL clarified ligation product, add 40 μL water, 1 μL GlycoBlue and 6 μL 3 M sodium acetate, and mix well.
Then add 150 μL pure ethanol and precipitate by centrifugation at 16,000 × g for 20 min.
Wash the pellet with 80% ethanol and resuspend in 12 μL water.
To reverse the AMT cross-linking, put the samples on a clean surface with ice beneath it. Irradiate with 254 nm UV for 15 min and recover about 10 μL RNA.
3.5 Adapter Ligation and Reverse Transcription
Add 10 μL adapter ligation mixture to 10 μL RNA and perform the adapter ligation reaction for 3 h at room temperature.
After adapter ligation add the following reagents to remove free adapters: 3 μL 10× RecJf buffer, 2 μL RecJf, 1 μL 5′ dead-enylase, 1 μL SuperaseIn, and 3 μL water. Incubate at 37 °C for 1 h.
Then purify RNA with Zymo RNA clean and Concentrator-5 or ethanol precipitation. Reconstitute RNA in 11 μL water.
To the purified RNA add 1 μL of custom RT primer 1 (with barcode).
Heat the samples to 70 °C for 5 min in a PCR block, cool the samples to 25 °C by stepping down 1 °C every 1 s (50 steps); hold at 25 °C.
Add 8 μL reverse transcriptase mix to the RNA and heat the samples at 25 °C for 3 min, 42 °C for 5 min, 52 °C for 30 min; hold at 4 °C.
Wash 10 μL of C-1 beads per sample with 100 μL Bead Binding Buffer three times.
Resuspend beads in 5 μL Bead Binding Buffer per sample. Add 5 μL bead suspension to the completed reverse transcription reaction.
Rotate at room temperature for 30 min.
Transfer bead suspension to new 1.5 mL tube. Let it settle on magnet and remove supernatant.
Add 500 μL Bead Wash Buffer to each sample and invert the tubes four times to mix.
Insert the samples into a magnetic stand for 1 min, and then remove the supernatant.
Wash 4 more times with 500 μL Bead Wash Buffer and twice with 500 μL 1× PBS.
Resuspend beads in 50 μL RNaseA/T1/H elution mix and incubate them at 37 °C for 30 min at 1000 rpm in a thermomixer.
Add 1 μL 100% DMSO and heat at 95 °C for 4 min. Briefly spin down samples and transfer eluted cDNA to a new tube.
Purify the cDNA using Zymo concentrator-5 columns.
Elute twice with 6 μL water to recover ~10 μL cDNA.
3.6 cDNA Circularization, Library PCR, and Sequencing
Add 4 μL circularization reaction mix to the cDNA sample and incubate them at 60 °C for 100 min, followed by 80 °C for 10 min. At the same time, warm up the lamps in the qPCR machine by starting the program (the warm-up takes about 20 min).
Add 10.5 μL qPCR reaction mix to each circularized cDNA sample. Transfer cDNA to optical PCR tubes (each tube should be separate so that individual tubes can be taken out of the qPCR machine when the fluorescence signal reaches a defined point).
Set up the following qPCR program. Choose SYBR, initial 95 °C, 45 s, 10 cycles of: 95 °C, 15 s; 65 °C, 30 s; 72 °C, 45 s, detect fluorescence at extension step (a set of nine cycles). Take sample out once amplification reaches exponential phase.
Transfer PCR product to 1.5 mL tube. Add 30 μL Ampure XP beads and 75 μL isopropanol.
Incubate for 5 min at RT.
Let the beads settle on the magnet for 5 min.
Remove the supernatant and wash the beads twice with 80% ethanol at RT.
Elute DNA with 10 μL water twice each tube (~20 μL eluate per sample). Pool elute and add 20 μL 2× Phusion HF mas-termix and 2 μL 20 μM P3/P5 Solexa PCR primer (final 1 μM).
Run PCR reaction (95 °C, 45 s; 3 cycles of 95 °C, 15 s; 65 °C, 20 s; 72 °C, 45 s; and 4 °C on hold).
Purify reaction by standard Zymo concentrator-5 column protocol.
Elute with 8 μL water and add 2 μL Orange G loading dye.
Run a 6% native TBE gel at 100 V for 75 min, until the dye just ran off the gel.
Stain gel in SYBR Gold for 3 min. Image gel at 0.5, 1, and 2 s exposure times. Cut out the DNA from 175 bp and above (corresponding to >40 bp insert).
Use a syringe needle to punch a hole in the bottom of a 0.65 mL tube.
Transfer the gel slice to 0.65 mL tube and insert into a 2 mL collection tube. Spin at room temperature, 16,000 × g for 5 min. The gel slice gets sheared into slurry by passing through the hole.
Remove the 0.65 mL tube and add 300 μL water to the slurry. Shake at 55 °C, 1000 rpm overnight in a thermomixer.
Pass the gel slurry through a Spin-X 0.45 μm column to recover the DNA library.
Add 5× volume Zymo DNA binding buffer and flow-through Zymo concentrator-5 column.
Wash with 200 μL Washing buffer once and elute twice with 8 μL water (recover ~15 μL library). Quantify library by a high-sensitivity Bioanalyzer assay.
Barcoded libraries can be pooled together for sequencing if necessary.
Sequence the libraries on an Illumina sequencer using standard conditions and the P6_Custom_seqPrimer. Usually, a 70 nt single end sequencing reaction is enough for PARIS. The multiplexing and random barcodes are sequenced together with the insert.
3.7 Basic Analysis: Trimming, Mapping, and Filtering
-
Sequencing data are usually provided in zipped fastq files. First, the reads are trimmed to remove the adapter sequences. If the data are multiplexed, split the library. See Fig. S1H of the PARIS paper for an overview of the analysis strategy [21]. The analysis outline is summarized in Fig. 4. Assume that the file name prefix is x. The following are a preferred set of file name conventions. Having standard file names makes the management of files easier.
x.fastq.gz: zipped raw data x_trim3.fastq: after removing the 3′ end adapter sequencesx_trim3_nodup.fastq: after removing PCR duplicatesx_trim3_nodup_bcnn.fastq: split libraries with barcode nn, such as 01, 02 . . .x_trim_nodup_bcnn.fastq: after removing 5′ end adapter sequences x_trim_nodup_bcnn_starSSAligned.out. sam: reads mapped to STAR index for species SS, normally aligned. x_trim_nodup_bcnn_starSSChimeric.out.sam: same as above, for the chimeric reads - Trim the 3′ end adapter sequences using Trimmomatic [22].
java -jar trimmomatic-0.32.jar SE -threads 16 -phred33 x.fastq.gz x_trim3.fastq ILLUMINACLIP: P6SolexaRC35.fa:3:20:10 SLIDINGWINDOW:10:30 MIN-LEN:28
- Remove the duplicates using the built-in random barcodes and readCollapse (from the icSHAPE pipeline).
readCollapse -U x_trim3.fastq -O x_trim3_nodup. fastq
- Split the trimmed and collapsed libraries using splitFastqLibrary (from the icSHAPE pipeline). L is a list of barcode and file name strings. D is the directory to store the split libraries
splitFastqLibrary -U x_trim3_nodup.fastq -l L -b 6:6 -s -d D
- Trim the 5′ adapter sequences. HEADCROP:17 option is used to trim off the designed random and multiplexing barcodes.
java -jar trimmomatic-0.32.jar SE -threads 16 -phred33 x_trim3_nodup_bc01.fastq x_trim_no-dup_bc01.fastq HEADCROP:17 MINLEN:20
Quality of the x_trim_nodup.fastq file was visualized using fastQC.
- Map reads to a genome index of choice using STAR [23] (see Notes 6 and 7). The parameters chosen here are to reduce the penalty for gapped reads and allow mapping of chiastic reads. Some of the options can be adjusted for specific purposes, such as –outFilterMultimapNmax and –alignSJoverhangMin.
STAR –runMode alignReads –genomeDir /seq/STAR/index/starSS/ –readFilesIn x_trim_nodup_bc01.fastq –outSAMtype SAM –outFileNamePrefix x_trim_no-dup_bc01_starSS –outReadsUnmapped Fastq –outSAMattributes All –alignIntronMin 1 –scoreGapNoncan -4 –scoreGapATAC -4 –chimSegmentMin 15 –chimJunctionO-verhangMin 15 –runThreadN 8
- Convert SAM to BAM format, sort the BAM file and extract primary mapped reads using SAMtools [24].
samtools view -b -F 0x900 -o x_trim_nodup_bc01_ starSSAligned_prim.out_sorted.bam x_trim_no-dup_bc01_starSSAligned.out_sorted.bam
- Extract gapped reads from the Aligned.out_sorted.bam file. The non-gapped reads do not contain the structure information and therefore discarded (see Note 8).
samtools view x_trim_nodup_bc01_starSSAligned_prim. out_sorted.bam | awk ‘$6 ~ /N/’ x_trim_nodup_bc01_starSSAligned_prim_N.out_sorted.sam
- To load SAM files into IGV for visualization, the SAMtools package is commonly used in the following three steps. These three steps can be used for any of the SAM files generated throughout the analysis.
samtools view -bS -o x.bam x.samsamtools sort x.bam x_sorted samtools index x_sorted.bam
3.8 Advanced Analysis and Visualization
-
Assemble duplex groups (DGs) using the samPairingCalling.pl script (https://github.com/qczhang/paris). Given the volume and complexity the RNA structures and RNA–RNA interactions. Here are two options for the assembly of DGs. First, the DGs can be assembled for all mapped reads. Second, DGs can be assembled for individual RNA transcripts. The second approach is preferred for detailed analysis since it is much faster than the first one. The processing first removed gapped reads that are gapped as a result of splicing and further removed PCR duplicates. Then the gapped reads are sorted by coordinates and then processed to obtain DGs with a two-step greedy algorithm. First, generate intermediate DGs. Each read is either added to an existing DG or used to establish a new DG based on the criteria: all reads in a DG must share at least 5 nt in both arms. Second, merge the intermediate DGs as long as the maximum gap of both arms is less than 10 nt and the maximum length of both arms of the final DG less than 30 nt. To guarantee the validity of the identified DG, we filter low quality DGs by two criteria: first, each DG must have two unique gapped reads that have different termini. Second, DG connection score(connection_A_B/sqrt(coverage_A*coverage_B), A and B representing the two arms) should be great than 0.01 (see Note 9).
perl samPairingCalling.pl -i x_Aligned_prim_N.sam -j x_Chimeric.out.junction -s x_Chimeric.out.sam -o x_geometric -g genome.fa -z chrom.sizes -a genome.gtf -t genome.fa -l 15 -p 2 -c geometric 1>x_geometric.stdout 2>x_geometric.log
Annotation of the command:x_Aligned_prim_N.sam: normal gapped reads x_Chimeric.out.sam and x_Chimeric.out.junction: chiastic reads and junctionsgenome.fa: genome referencechrom.sizes: two columns, chromosome name and sizegenome.gtf: genome annotation file from UCSC Genome Browsergeometric: using geometric mean of the coverage on the two arms for normalization.
This assembly step produces two main output files: *geometricsam and *geometric (while using the geometric option). The sam file can be used directly to assemble NGs in the next step, while the other file contains all connections that can be used for RNA–RNA interaction analysis (step 6).
- Assemble nonoverlapping groups (NGs). To efficiently pack the DGs in the IGV genome browser, the DGs are further assembled into NGs using sam2ngmin.py (from the “duplex” scripts, https://github.com/zhipenglu/). NG is a new custom-defined tag in the SAM file format. Then convert the NG-assembled SAM file to indexed BAM file for visualization on IGV as above (step 10). The *geometricsam file is produced from the DG assembly step.
python sam2ngmin.py x_trim_nodup_norm_starhg38_geometricsam x_trim_nodup_norm_starhg38_geometric_NG-min.sam
- The secondary structure model of an RNA can be prepared in a BED format, which is quite similar to the commonly used ‘connect’ format described in the mfold program [25]. This structure model can be uploaded to the IGV genome browser [26], in parallel with the PARIS DGs (Fig. 5). The file must include a track line specifying “track graphType=arc”. Each record line must contain the first three columns of a bed file: chrom, start and end, where the start and end represent the base pair. Note that the start position follows standard BED file convention and is zero-based (first base on a sequence is position 0). Note that one can import a known secondary RNA structure after converting to the arc type bed (e.g., https://github.com/zhipenglu/duplex/ct2bed.py to convert the connection format to the arc bed type file).The following small example represents a hypothetical stem loop (also described here: https://www.broadinstitute.org/igv/node/284).
track graphType=arc chr1 10 25 stemloop1 chr1 11 24 stemloop1 chr1 12 23 stemloop1 chr1 13 22 stemloop1 chr1 14 21 stemloop1 chr1 15 20 stemloop1
Visualization of RNA structure models and PARIS data on IGV genome browser. Upon loading files with DG, XG and NG tags to IGV (see Note 10), specify the following options by right clicking in the alignments panel. Color alignments by > tag > DG or XGGroup alignments by > tag > NG
For high-level visualization of RNA architecture, each DG can be represented by one arc (see Fig. 2c in the PARIS paper for an example [21]). To visualize all DGs in one transcript, extract the DGs from the *geometric file (from step 1). The start and end of the DG are used as the anchor points of the arc. The visualization is similar to the step 3 (see Note 11).
Analysis of RNA–RNA interactions. PARIS directly identifies all RNA–RNA interactions. The comprehensiveness of PARIS makes the analysis and visualization challenging. To identify RNA–RNA interactions from PARIS data with high confidence, reads are mapped to indices of specific subset of RNAs, each one as a small “chromosome”. For example, one can use all nonredundant human RNAs from Rfam, miRNAs from miRBase, and/or nonredundant RNAs from the human transcriptome annotations as references. After mapping reads to these references, chiastic reads were assembled into duplex groups and filtered to remove interactions within RNAs and ones with identical break points. Normal gapped reads are not used since these are all for RNA structures, not RNA–RNA interactions. The remaining chiastic reads are used to identify RNA–RNA interactions.
- To visualize the interaction between a pair of RNAs, use the following script. This analysis produces a pair of tracks for the interacting RNAs, highlighting the regions involved in interaction (see Fig. 6 for two examples). The SAM files from the previous step are used as the input. Parameters are defined within the following script.
python intrxn_specificity.py
- Phylogenetic analysis of RNA structure. The two arm intervals of each DG were used to extract multiple alignments from whole-genome alignments of 23 amniote vertebrate species (Ensembl, hg38 version) with the python script maf_extrac-t_ranges_indexed.py (bxpython package, https://github.com/bxlab/bx-python). RNAalifold was used to predict a consensus structure from the alignments for each DG with or without inter-arm base-pairing constraints [27]. The significance of each conserved structure was assessed using SISSIz shuffling with the RIBOSUM matrix [28].
./paris_covariation.sh
- For the direct comparison between human and mouse structures determined by PARIS, the mouse DGs were lifted from mm10 to hg38 coordinates using the liftOver utility and the mm10ToHg38.over.chain file (UCSC). The liftOver program was run with the following parameters. The minMatch was reduced from the default so that most regions can be properly aligned between species. In order to visualize the mouse PARIS reads on the human genome in IGV, the mouse PARIS reads were first converted to bed format using bedtools, lifted to hg38 coordinates, and then converted back to bam format using bedtools. It is noted that this strategy is limited by the quality of the available genome alignments, and improvement of these alignments is beyond the scope of the current study.
liftOver -minMatch=0.2 -minBlocks=0.2 -fudgeThick
- Analysis of alternative structures using the alternativestructure. py script (https://github.com/zhipenglu/duplex). Alternative structures are defined as helices that overlap on one arm by more than 50%. In practice, DGs were intersected with each other to identify pairs of DGs that have one pair of overlapped arms (left-left, left-right or right-right), but not two pairs at the same time. Inter-arm structures were predicted using RNAco-fold and significant overlapping of base pairs were used as another filter for alternative structures (at least 50% overlap). This script requires RNAcofold (from the Vienna RNA package) and python intervaltree module in proper paths. The x. bed file contains all the DGs in a BED format while the reference.fa contains the reference sequence. The x.alt is the output.
python alternativestructure.py x.bed reference.fa x.alt
Fig. 5.
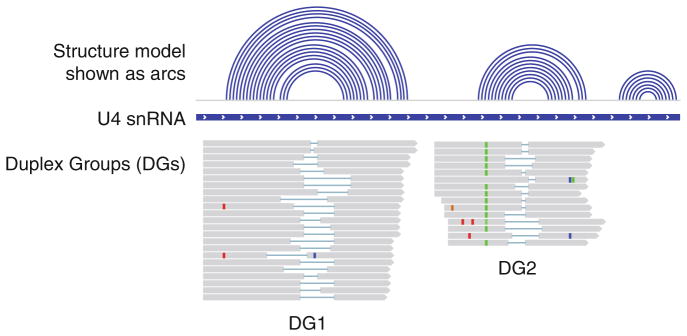
An example visualization of RNA structures in IGV. The structure model is in the linear format (quasi-concentric arcs for an RNA duplex). A subset of gapped reads is assembled into two DGs. The two DGs can be assembled into one NG, since they do not overlap with each other. For more examples, see the PARIS paper [21]
Fig. 6.
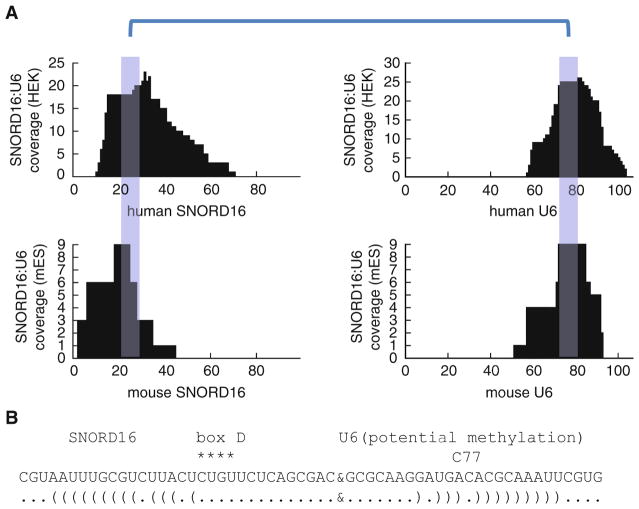
Analysis and visualization of a new interaction between SNORD16 (or U16) and U6 in human HEK 293T cells and mouse ES cells. (a) The blue highlighted regions are the sequences involved in base pairing. (b) The interaction model. Box D is a characteristic sequence motif in the C/D box snoRNAs. SNORD16 is known to guide modification of 18S rRNA (snoRNABase) [30]. Here, we show that it may also guide the modification of U6 snRNA at C77, the same site as the SNORD10 target (snoRNABase) [31], but the base pairing is less perfect compared to the SNORD10–U6 interaction. For more examples, see Fig. 5 and Fig. S6 the PARIS paper [21]
Acknowledgments
The authors wish to thank Dr. Howard Chang at Stanford University for his generous support and encouragement. Z.L. is a Layton Family Fellow of the Damon Runyon-Sohn Foundation Pediatric Cancer Fellowship Award (DRSG-14-15). This work was supported by the National Natural Science Foundation of China (31671355) and the National Thousand Young Talents Program of China to Q.C.Z., and by the Jump Start Award for Excellence in Research to Z.L. (Stanford University, award No. 106).
Footnotes
The 30 min irradiation time was chosen for strong cross-linking; however, shorter times may be useful for certain applications where rapid changes for RNA structures are expected. The cross-linking step performed on ice bed may cause cold shock and should be noted here. This condition, even though still likely to affect cell physiology, is very close to normal cell culture condition.
The quality and concentration of the solution can be measured by NanoDrop, which gives a characteristic spectrum with two major peaks at 250 and 300 nm [32]. The absorbance of AMT at 300 nm is 25,000/M cm [33].
Strong psoralen cross-linking turns RNA into extensive networks of interconnected molecules that are insoluble even under strong denatured and chaotropic conditions like TRIzol. Direct lysis of AMT cross-linked cells produces significant amount of insoluble material. S1 nuclease digestion is necessary to recover cross-linked RNA from the lysate. Psoralen cross-links RNA to protein to some extent and proteinase K digestion is also necessary to recover RNA from the lysate [34, 35]. S1 nuclease is resistant to certain denaturation conditions [36]. For example, S1 is active in 9 M urea. SDS can inactivate S1 activity at a concentration above 0.1% but proteins can complex with SDS to maintain S1 activity. When supplemented with manganese buffer, ShortCut converts double stranded RNA to 18–25 bp fragments with 5′-phosphate and 3′-hydroxyl groups that are can be directly used for proximity ligation.
The S1 and PK digestion of cell lysate is hard to control since the reaction is performed in suspension not solution. In general, the S1 digestion should greatly reduce the viscosity of the suspension. After S1/PK digestion, the addition of TRIzol should yield a clear solution. The RNA size distribution is variable especially regarding the height of the broad peak between 2000 and 4000 nt. Therefore, it is necessary to perform the cross-linking and digestion experiments at the same for a set of conditions that need to be compared.
Both S1 nuclease and ShortCut RNase III are chosen for the fragmentation because they produce 5′-P and 3′-OH that can be ligated directly in subsequent steps. In addition, the combination of S1 nuclease and ShortCut RNase III helps produce the maximal amount of cross-linked dsRNA fragments of appropriate size. RNase A and T1 do not produce clonable ends and therefore not used in these experiments.
The preprocessed PARIS sequencing reads can be mapped to the whole genome or a selected subset of gene, such as 45S rRNAs, mitochondrial rRNAs, snRNAs, and XIST. A STAR index is needed for each reference. To make these mini-reference STAR indices, it is important to note that a custom value is needed for the --genomeSAindexNbases option, calculated as min(14, log2(GenomeLength)/2 – 1) (see the STAR manual for details).
For a smaller subset of RNA interaction analysis, the STRA mapping parameter may need to be adjusted for better performance. For instance, while looking the interaction landscape of rRNAs, mapping parameter -outFilterMultimapNxax is set to 10 to reduce too ambiguous mapped reads.
The STAR mapping produces normally mapped reads (Aligned.out.sam) and chiastically mapped reads (Chimeric. out.junction and Chimeric.out.sam), all of which will be used for building RNA structures. Normal: LLLLLL-RRRRRR, chiastic: RRRRRR-LLLLLL, L for bases from the left arm, while R for the right arm. The Chimeric.out.sam is a naming convention from STAR, and it should not be confused with other definitions of “chimeric”; therefore, we use chiastic to refer to the RRRRRR-LLLLLL type alignments.
The coverage of the two arms in DG score calculation is different from the gapped reads connecting them, because each region could be covered by multiple DGs, and some of them are likely to be alternative structures, which are pervasive in the transcriptome.
Output files from the samPairingCalling.pl program contain both DG and XG (Chiastic Group) tags. The XG tag is used to differentiate normal gapped reads and chiastic reads, where the two arms are swapped in relative position. Chiastic reads also include additional reads that are mapped to different strands or different chromosomes. XG:i:0 for normal gapped, XG:i:1 for chiastic on the same strand of the same chromosome, and XG: i:2 for all others.
Visualization of entire bam files is not recommended since long-range and especially intermolecular interactions would extremely compress the DGs in arcs or read alignments. Instead, individual RNAs should be extracted from the bam files (aligned gapped reads) and bed files (arcs representing DGs) using the SAMtools and BEDtools programs. Examples and instructions can be downloaded from the following link https://www.dropbox.com/s/1oqkcfzlfafdahq/PARIS_visdata.tgz?dl=0.
Contributor Information
Zhipeng Lu, Department of Dermatology, Center for Personal Dynamic Regulomes, Stanford University, Stanford, CA 94305, USA.
Jing Gong, MOE Key Laboratory of Bioinformatics, Beijing Advanced Innovation Center for Structural Biology, Center for Synthetic and Systems Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Haidian, Beijing 100084, China.
Qiangfeng Cliff Zhang, MOE Key Laboratory of Bioinformatics, Beijing Advanced Innovation Center for Structural Biology, Center for Synthetic and Systems Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Haidian, Beijing 100084, China.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin IV. Non-coding RNA. Human molecular genetics. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 3.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Z, Matera AG. Developmental analysis of spliceosomal snRNA isoform expression. G3. 2014;5(1):103–110. doi: 10.1534/g3.114.015735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Reilly D, Dienstbier M, Cowley SA, Vazquez P, Drozdz M, Taylor S, James WS, Murphy S. Differentially expressed, variant U1 snRNAs regulate gene expression in human cells. Genome Res. 2013;23(2):281–291. doi: 10.1101/gr.142968.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia Y, JCM, Ackerman SL. Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell. 2012;148(1–2):296–308. doi: 10.1016/j.cell.2011.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Z, Guan X, Schmidt CA, Matera AG. RIP-seq analysis of eukaryotic Sm proteins identifies three major categories of Sm-containing ribonucleoproteins. Genome Biol. 2014;15(1):R7. doi: 10.1186/gb-2014-15-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SH, Quigley GJ, Suddath FL, McPherson A, Sneden D, Kim JJ, Weinzierl J, Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973;179(4070):285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- 10.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 11.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505(7485):701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilac-qua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505(7485):696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 13.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519(7544):486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Z, Chang HY. Decoding the RNA structurome. Curr Opin Struct Biol. 2016;36:142–148. doi: 10.1016/j.sbi.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JF, Hearst JE. Structure of E. coli 16S RNA elucidated by psoralen cross-linking. Cell. 1983;32(4):1355–1365. doi: 10.1016/0092-8674(83)90316-1. [DOI] [PubMed] [Google Scholar]
- 17.Ross A, Brimacombe R. Experimental determination of interacting sequences in ribosomal RNA. Nature. 1979;281(5729):271–276. doi: 10.1038/281271a0. [DOI] [PubMed] [Google Scholar]
- 18.Calvet JP, Pederson T. Heterogeneous nuclear RNA double-stranded regions probed in living HeLa cells by crosslinking with the psoralen derivative aminomethyltrioxsalen. Proc Natl Acad Sci U S A. 1979;76(2):755–759. doi: 10.1073/pnas.76.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvet JP, Pederson T. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell. 1981;26(3 Pt 1):363–370. doi: 10.1016/0092-8674(81)90205-1. [DOI] [PubMed] [Google Scholar]
- 20.Hausner TP, Giglio LM, Weiner AM. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 1990;4(12A):2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- 21.Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, Mesirov JP, Cech TR, Chang HY. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016;165(5):1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenz R, Bernhart SH, Honer Z, Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gesell T, von Haeseler A. In silico sequence evolution with site-specific interactions along phylogenetic trees. Bioinformatics. 2006;22(6):716–722. doi: 10.1093/bioinformatics/bti812. [DOI] [PubMed] [Google Scholar]
- 29.Flynn RA, Zhang QC, Spitale RC, Lee B, Mumbach MR, Chang HY. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat Protoc. 2016;11(2):273–290. doi: 10.1038/nprot.2016.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85(7):1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 31.Tycowski KT, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell. 1998;2(5):629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 32.Hearst JE. Psoralen photochemistry. Annu Rev Biophys Bioeng. 1981;10:69–86. doi: 10.1146/annurev.bb.10.060181.000441. [DOI] [PubMed] [Google Scholar]
- 33.Grass JA, Hei DJ, Metchette K, Cimino GD, Wiesehahn GP, Corash L, Lin L. Inactivation of leukocytes in platelet concentrates by photochemical treatment with psoralen plus UVA. Blood. 1998;91(6):2180–2188. [PubMed] [Google Scholar]
- 34.Sastry SS, Ross BM, P’Arraga A. Cross-linking of DNA-binding proteins to DNA with psoralen and psoralen furan-side monoadducts. Comparison of action spectra with DNA-DNA cross-linking. J Biol Chem. 1997;272(6):3715–3723. doi: 10.1074/jbc.272.6.3715. [DOI] [PubMed] [Google Scholar]
- 35.Sastry SS, Spielmann HP, Hoang QS, Phillips AM, Sancar A, Hearst JE. Laser-induced protein-DNA cross-links via psoralen furanside monoadducts. Biochemistry. 1993;32(21):5526–5538. doi: 10.1021/bi00072a006. [DOI] [PubMed] [Google Scholar]
- 36.Zechel K, Weber K. Degradation of nucleic acids in cell lysates by S1 nuclease in the presence of 9 M urea and sodium dodecyl-sulfate. Eur J Biochem. 1977;77(1):133–139. doi: 10.1111/j.1432-1033.1977.tb11651.x. [DOI] [PubMed] [Google Scholar]