Abstract
We provide evidence that the IFN-regulated member of the Schlafen (SLFN) family of proteins, SLFN5, promotes the malignant phenotype in glioblastoma multiforme (GBM). Our studies indicate that SLFN5 expression promotes motility and invasiveness of GBM cells, and that high levels of SLFN5 expression correlate with high grade gliomas and shorter overall survival in patients suffering from GBM. In efforts to uncover the mechanism by which SLFN5 promotes GBM tumorigenesis, we found that this protein is a transcriptional co-repressor of STAT1. Type-I IFN treatment triggers the interaction of STAT1 with SLFN5, and the resulting complex negatively controls STAT1-mediated gene transcription via interferon stimulated response elements (ISRE). Thus, SLFN5 is both an IFN-stimulated response gene and a repressor of IFN-gene transcription, suggesting the existence of a negative-feedback regulatory loop that may account for suppression of antitumor immune responses in glioblastoma.
Keywords: Interferon, Schlafen, SLFN5, Glioblastoma, Signaling
INTRODUCTION
Glioblastoma multiforme (GBM), an aggressive tumor with a highly heterogeneous genetic background, is the most common central nervous system (CNS) neoplasm in adults (1). Currently available therapies for GBM frequently fail due to the highly infiltrative GBM growth patterns and the presence of chemotherapy-resistant tumor initiating cancer stem cells (2, 3). Because of the high mortality and morbidity of these tumors and the limited treatment options, there is an urgent need for the development of novel therapeutic strategies to target and eliminate resistant cancer stem cells.
Type-I interferons (IFNs), specifically IFN-α and IFN-β, exhibit important antineoplastic properties in vitro and in vivo against a wide variety of malignancies (4, 5). There has been some evidence for Type-I IFN antitumor activity in GBM in vitro and in vivo, but this primarily involves studies of IFN combinations with chemotherapeutic agents (6). IFNβ treatment has been shown to sensitize human glioma cells to temozolomide (TMZ) treatment in vitro (7), and in some cases may have a beneficial therapeutic effect when incorporated in the therapeutic regimen of GBM patients (8). The efficacy of stand-alone IFN treatment is generally low, suggesting that some GBM cells may develop resistance to IFN-treatment in vivo (9).
The mechanisms of IFN-α/β signaling have been extensively defined. It is now well established that engagement of the Type-I IFN receptor, IFNAR, leads to STAT-dependent transcriptional activation of several interferon-stimulated genes (ISGs) that mediate the biological responses of Type-I IFNs (10, 11). Several mouse and human members of the Schlafen family of proteins are IFN inducible (reviewed in Mavrommatis et al. (12)). In previous studies we demonstrated that human Schlafen 5 (SLFN5) is a Type-I IFN regulated ISG in different cell types (13, 14). The protein is composed of an AAA domain, a unique SLFN box, and a predicted transcriptional regulatory area with a helix-turn-helix domain (COG2865) (12, 15). Other studies established that several SLFN genes are upregulated in melanoma and renal cell carcinoma cell lines following IFN treatment (13, 14). In the present study, we investigated the patterns of expression of different human SLFNs in GBM and examined the role of SLFN5 in GBM progression and the induction of IFN-induced biological responses. Our data establish that SLFN5 expression positively correlates with the GBM malignant phenotype and provide evidence for a novel mechanism by which this may occur, involving SLFN5-mediated repression of IFN-induced STAT1 transcriptional activity.
RESULTS
SLFN5 expression is associated with poor survival in GBM patients
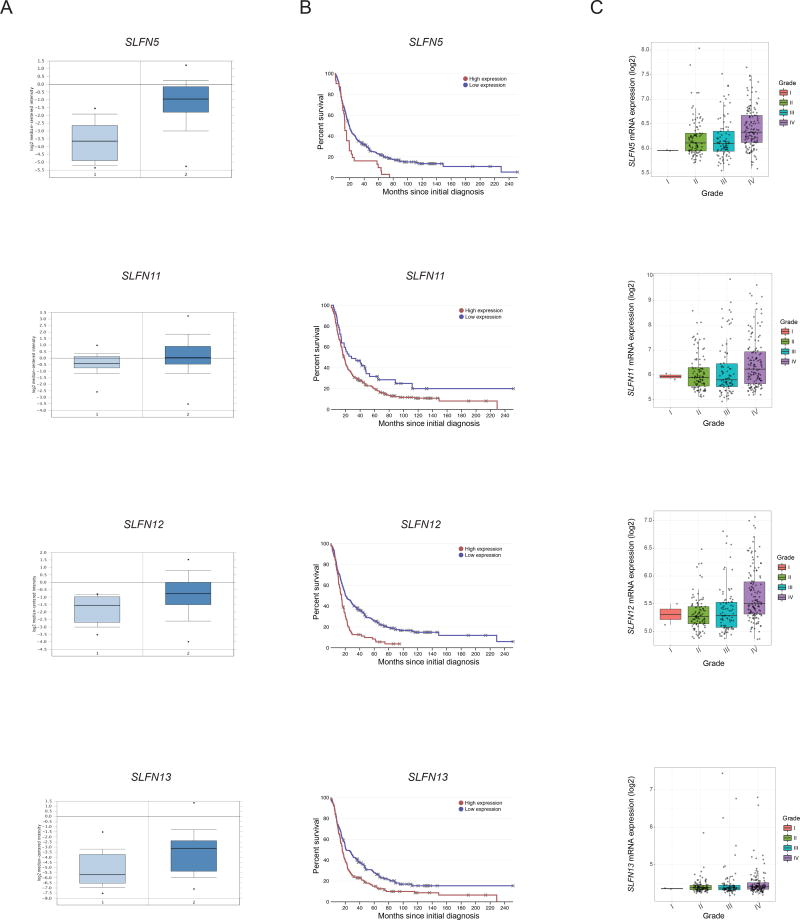
In initial studies we sought to define the patterns of expression of human SLFN genes in primary malignant cells from GBM patients, using publicly available microarray databases. We first assessed the relative expression levels of SLFN5, SLFN11, SLFN12 and SLFN13 genes in the Oncomine database (16), using data from the SUN (17) dataset. Differential expression analysis revealed a statistically significant increase in SLFN5 (5.6 fold difference, p =1.78e-10), and to a lesser extent SLFN11 (1.47 fold difference, p =0.004), SLFN12 (1.9 fold difference, p =1.19e-4), and SLFN13 (3.13 fold difference, p =4.81e-5) transcripts (Figure 1A). Next, we enquired whether high expression levels of SLFN genes correlate with poor survival in GBM patients using the REMBRANDT (REpository for Molecular BRAin Neoplasia DaTa) database (18). GBM patients expressing high levels of SLFN5 (p = 0.00528), SLFN11 (p = 0.0421), SLFN12 (p = 1.04e-5) and SLFN13 (p = 0.00249) had shorter overall survival compared with patients expressing low levels for the respective SLFN genes (Figure 1B). We further explored the relationship between SLFN5, SLFN11, SLFN12 and SLFN13 and glioma grade. We found that SLFN5, SLFN11 and SLFN12 expression levels increase with glioma grade and are highest in Grade IV (i.e., GBM), when compared to Grade I, Grade II or Grade III gliomas (Figure 1C).
Figure 1. Human SLFNs are overexpressed in primary cells from GBM patients and correlate with poor overall survival.
(A) SLFN5, SLFN11, SLFN12 and SLFN13 relative gene expression levels are shown in normal brain tissue (light blue, n = 23) versus GBM patient samples (dark blue, n = 81) using Sun et al. (17) dataset available through Oncomine database (16) (B) Kaplan-Meier survival plots showing percent of survival between GBM patients with high (n = 329) versus low (n = 523) expression levels of SLFN5, SLFN11, SLFN12 and SLFN13. Data were extracted from REMBRANDT database (18). (C) SLFN5, SLFN11, SLFN12 and SLFN13 expression data were analyzed using REMBRANDT-cohort of patients with Grade I, Grade II, Grade III, and Grade IV gliomas (GBM). Plots were generated using the GlioVis online tool (http://gliovis.bioinfo.cnio.es).
Type I IFN-dependent human SLFN expression in established and patient derived cell lines
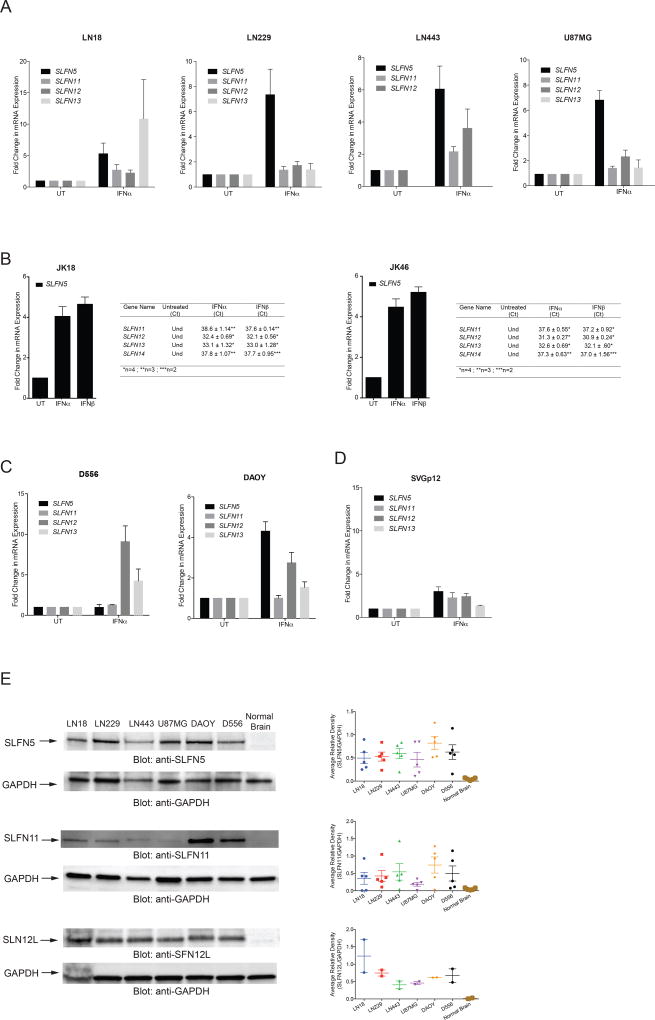
As previous studies from our group had demonstrated that SLFNs are ISGs in other tissues, we next evaluated the effects of Type-I IFN treatment on the expression of different SLFN genes in several malignant brain tumor cell lines. SLFN5 was the most prominent inducible gene in response to IFNα-treatment in most cases, while the inducible expression of SLFN11, SLFN12 and SLFN13 was more variable (Figures 2A–D). In patient-derived glioma stem cell (GSC) lines (19, 20), we found that SLFN5 was highly expressed, whereas SLFN11, SLFN12, SLFN13 and SLFN14 appeared to be expressed to a lesser extent (Figure 2B). Treatment with IFNα or IFNβ of GSCs markedly induced SLFN5 expression, confirming our observation in established GBM cell lines (Figure 2B). Interestingly, there was minimal induction of SLFN genes in normal astroglial cells (Figure 2D), consistent with selective IFN-dependent induction of SLFN5 expression in malignant brain tumor cells. Next, we analyzed the expression of different human SLFN proteins in different glioblastoma (LN18, LN229, LN443 and U87MG) and medulloblastoma (DAOY and D556) cell lines. SLFN5 expression was higher in all brain tumor cell lines compared to normal brain tissue (Figure 2E). Similarly, SLFN12L protein was expressed at higher levels in malignant cells compared to normal brain cells (Figure 2E), while there was less consistency for the levels of expression of SLFN11. Together, these results indicate that SLFN proteins, in particular SLFN5, are over-expressed in malignant brain tumor cells.
Figure 2. Type-I IFNs induce SLFN5 expression in vitro.
(A–D) Type-I IFN-dependent expression of human SLFN genes in GBM (A), patient-derived glioma stem cell lines (B), medulloblastoma cell lines (C), and human normal astrocytes SVGp12 (D). Indicated cells were left untreated (UT) or were treated with human IFNα or IFNβ for 6 hours. qRT-PCR analyses of the relative mRNA expression of SLFN5, SLFN11, SLFN12, SLFN13, and SLFN14 genes are shown. Data are expressed as fold change over untreated controls, and bar graphs represent means ± SEM of three independent experiments for LN18, LN443, SVGp12, and four independent experiments for JK18, JK46, DAOY, D556, LN229 and U87MG. (Und: undetected) (E) Left Panel, Expression of human SLFN proteins in GBM, medulloblastoma cell lines, and normal brain tissue lysates. The cells were lysed and equal amounts of whole cell lysates were resolved by SDS-PAGE. Immunoblots were probed with antibodies against SLFN5, SLFN11, SLFN12L and GAPDH, as indicated. Immunoblot images are representative of five independent experiments for SLFN5, SLFN11 and two independent experiments for SLFN12L. Right panels, bands from five SLFN5, SLFN11, or two SLFN12L independent experiments (including the blots shown) were quantified by densitometry using Image J software and normalized and reported relative to GAPDH.
Effects of SLFN5 on glioblastoma tumor formation, growth, and invasion
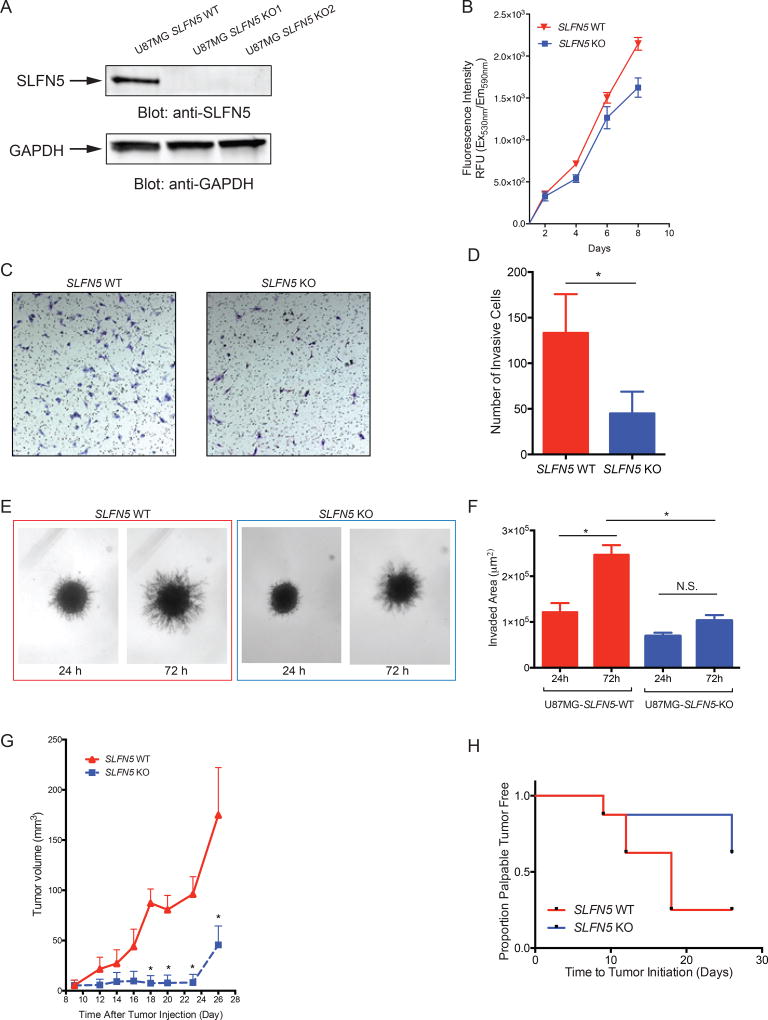
To define the effects of SLFN5 expression on GBM cellular proliferation, invasion and in vivo growth, we generated U87MG SLFN5 knockout (KO) cells. Plasmids encoding Cas9 and single guide RNAs (sgRNAs) targeting the SLFN5 gene (Figure S1A) were transfected into U87MG cells and clones were selected in puromycin. Western immunoblot analysis confirmed the lack of endogenous SLFN5 expression compared to U87MG SLFN5 wild type (WT) cells transfected with non-targeting sgRNA/Cas9-expressing plasmid (Figure 3A). To investigate the effects of targeted gene disruption of SLFN5 on cell proliferation and viability, we used an Alamar Blue-based cell viability assay. Decreased proliferation of SLFN5 KO cells compared to SLFN5 WT cells was observed in time course studies (Figure 3B). In addition, shRNA mediated SLFN5 knockdown in LN18 and U87MG cells reduced anchorage-independent growth in soft agar (Figure S1B). Next, we tested the invasive properties of SLFN5 WT and SLFN5 KO U87MG cells using matrigel-based (Figures 3C–D) and three-dimensional (3D) tumor spheroid invasion assays (21) (Figures 3E–F). Depletion of SLFN5 expression reduced the invasive properties of U87MG cells in both assays (Figures 3C–F), suggesting that SLFN5 plays an important role in GBM invasion. When U87MG SLFN5 WT and KO cells were injected subcutaneously into athymic nu/nu mice, we found that targeted disruption of SLFN5 gene expression delayed tumor formation and decreased tumor growth in vivo (Figures 3G–H).
Figure 3. SLFN5 is required for GBM tumorigenesis in vitro and in vivo.
(A) SLFN5 KO U87MG cells were established using CRISPR/Cas9 genome editing technology. U87MG SLFN5 WT, KO1 and KO2 cell lysates were resolved by SDS-PAGE and immunoblotted with anti-SLFN5 or anti-GAPDH antibodies, as indicated. (B) Alamar Blue based cell proliferation assay of U87MG SLFN5 WT and KO cells is shown. Data are expressed as means of relative fluorescent units (RFU) of 3 independent experiments ± SEM. (* p < 0.05). (C–D) U87MG SLFN5 WT and KO cells were subjected to invasion assays and cells that invaded through the matrigel-coated transwell inserts were stained and counted. Data are expressed as numbers of invading cells for each cell line and bar graphs represent means ± SEM of three independent experiments. All micrographs were taken at the same magnification. (E–F) Representative images and quantification of tumor cell invasion are shown. U87MG SLFN5 WT and KO spheres were subjected to 3-D matrigel invasion assay and the invaded area was measured after 24 and 72 hours. Data are expressed as invaded area by each cell line at the time points indicated and bar graphs represent means ± SEM of three independent experiments. One-way ANOVA was used to assess statistically significant differences between the groups (N.S.: not significant, * p < 0.05). All micrographs were taken at the same magnification. (G) U87MG SLFN5 WT and KO cells (1×106 cells in 100 µl of Matrigel) were injected into the right and left flank of nu/nu mice, respectively (n = 8). Tumor volume was measured every other day for 26 days. Graph represents means ± SEM of change in tumor volume (y axis) as a function of the time (x axis) after the injection. Multiple t-tests analysis was used to assess statistically significant differences between SLFN5 WT and KO tumors (* p < 0.05). (H) Time to tumor onset in mice injected with SLFN5 WT and KO cells. Time to tumor onset was assessed by physical palpitation. (See also Figure S1)
Effects of SLFN5 depletion on the glioblastoma transcriptome
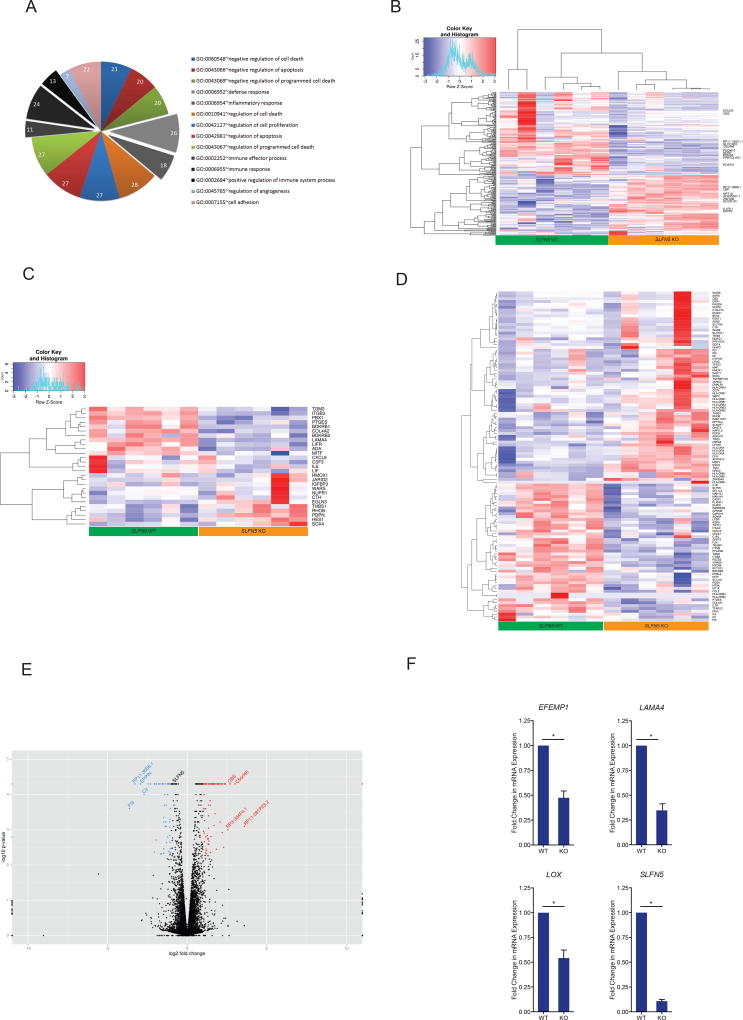
To gain further insight into the possible mechanism by which SLFN5 depletion mediates these observed biological changes, we performed a high-throughput paired-end RNA-sequencing (RNA-seq) on total RNA from SLFN5 depleted cells. To identify differentially regulated genes, we applied Cuffdiff analysis (see Supplemental Materials and Methods) to the entire data set. We identified 238 genes (Table S1) in total, 124 differentially expressed isoforms (Table S2) and 146 differentially expressed primary transcripts (Table S3) that met FDR criteria q value<10% of differential expression in KO cells compared to parental U87MG. To identify globally affected cellular processes and functions, we performed gene set enrichment analyses, using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 approach. Gene set enrichment analysis demonstrated over-representation of differentially expressed genes involved in the regulation of cell proliferation, cell activation, cell differentiation, cell death, and apoptosis processes, as well as the immune response and regulation of angiogenesis, cell adhesion and transcription factor activity (Figure 4A; Tables S4 and S5). To further characterize the differentially expressed genes in SLFN5 depleted U87MG cells, we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to determine biochemical pathway enrichment. There were several pathways significantly enriched in SLFN5 depleted cells compared to WT cells, including the TGF-beta signaling cascade, glycine, serine and threonine metabolism pathways, the complement and coagulation cascades, Jak-STAT signaling pathways, and ECM-receptor and cytokine-cytokine receptor interaction pathways (Figures S2, S3A and S3B and Table S6). Next, to gain functional insights into the gene interactions, we applied Ingenuity Pathway Analysis (IPA®) for any relationships that may exist among the differentially expressed genes. Cellular growth and proliferation (Figure S3C) along with cell cycle (Figure S3D) were identified as the top-scoring IPA gene interaction networks.
Figure 4. Effects of SLFN5 on the GBM transcriptome.
(A) GO term analysis using DAVID identifies enriched biological processes among SLFN5 regulated genes involved in the regulation of cell proliferation, cell activation, cell differentiation, cell death, and apoptosis processes, as well as the immune response and regulation of angiogenesis, cell adhesion and transcription factor activity. Cluster names are based on enriched GO annotations. Hierarchical clustering of global differentially expressed genes (B), cell cycle and proliferation related genes (C) and IFN-responsive genes (D) in both SLFN5 WT and KO U87MG cells that was performed with the R 3.2.2 software by using heatmap.2 function. Colored rectangles represent mRNA abundance levels of the transcript. The intensity of the color is proportional to the standardized values from each RNA-seq measurement, as indicated on the bar below the heat map image (q < 0.1). (E) A Volcano plot of differentially expressed genes for KO versus WT cells (F) qRT-PCR validation of genes identified by RNA-Seq. Total RNA was isolated, and changes in EFEMP1, LAMA4, LOX and SLFN5 mRNA gene expression were measured by qRT-PCR. Data are expressed as fold change over WT controls and represent means ± SEM of three independent experiments for LOX, four independent experiments for SLFN5 and LAMA4, and six independent experiments for EFEMP1 (* p < 0.05). (See also Figures S2–3 and Tables S1–7)
It is well established that many mammalian genes have multiple transcript variants and protein isoforms, mediated by alternative splicing (22). Studying these dynamic events requires an efficient way of detecting these changes at the transcript level (23, 24). Here we applied a complementary analytical approach, Cuffdiff, an algorithm that robustly tracks the dynamics of individual transcript expression (25). The gene set enrichment analysis at the isoform level revealed regulation of inflammation-associated processes and negative regulation of cell proliferation or angiogenesis (Figure 4A, Table S7). Consistent with these data, differential transcript expression of SLFN5 target genes, as well as cell cycle and proliferation related genes, revealed distinct clustering of SLFN5 KO and WT cells (Figure 4B and 4C), suggesting a regulatory role for SLFN5 at the transcriptional level. Because SLFN5 is associated with IFN signaling, we analyzed the Interferome v2.01 database (26) to interrogate the effects of depletion of SLFN5 on IFN signaling-associated genes. We identified a large subset of genes primarily involved with the IFN pathway (115/238, 48.3%), that differentially cluster in WT and KO cells (Figure 4D). A Volcano plot of differentially expressed genes for KO versus WT cells revealed significant downregulation of SLFN5 in edited U87MG cells (Figure 4E). We validated downregulation of EFEMP1 (27), LOX (28) and LAMA4 (29), previously identified as genes that promote malignant phenotypes, along with SLFN5 (Figure 4F). Collectively, these results underscore the involvement of SLFN5 in coordinating proliferation and inflammation signals through regulation of critical nodes in GBM.
SLFN5 interacts with STAT1 and exhibits ISRE repressor activity
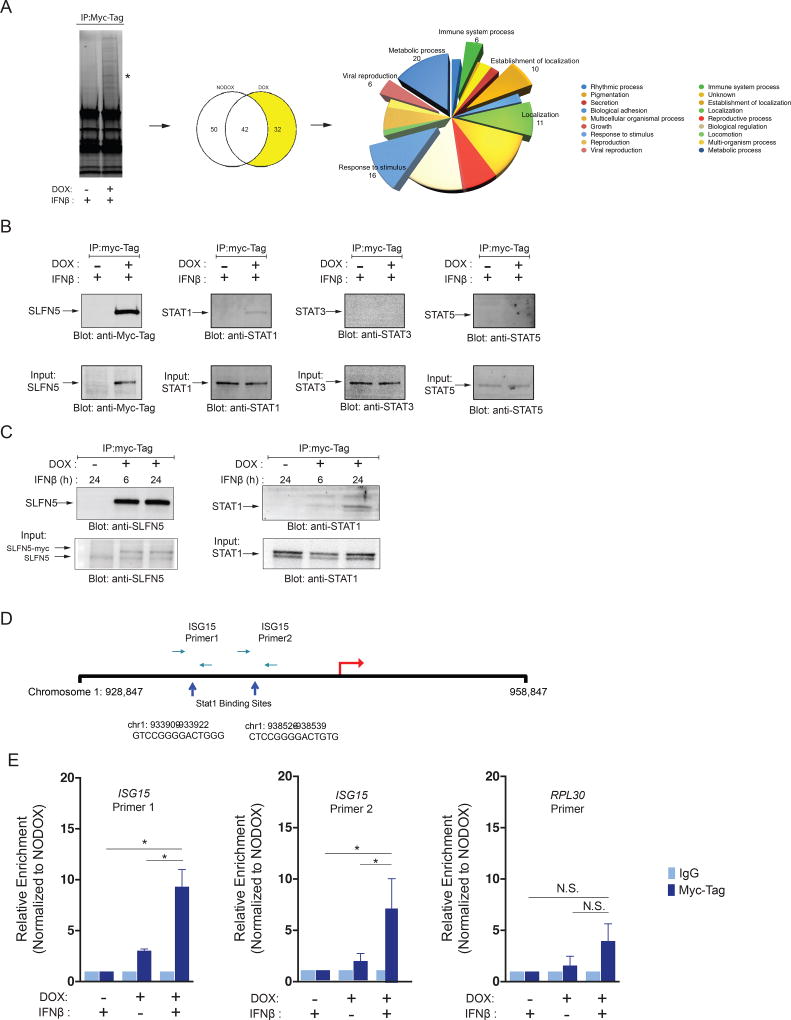
To identify the potential mechanism(s) by which SLFN5 exerts its repressor effects on ISGs, we undertook studies to determine SLFN5-protein interactions using mass spectrometry analysis of associated proteins in cells overexpressing SLFN5. SLFN5-protein complexes immunoprecipitated from 293T cells carrying inducible Myc-tagged SLFN5 were subjected to SDS-PAGE and visualized with BioRad Stain-Free technology (Figure 5A, left panel). Next, the protein complexes were extracted and subjected to mass spectrometry analysis using the Velos Orbitrap platform that generates sequence information on individual peptides, later converted into 32 unique proteins (Figure 5A, middle panel and Table S8). Classification according to functional relationships between the putative SLFN5 interactors revealed enrichment of proteins involved in the immune response, response to stimulus, viral reproduction, metabolic process and establishment of localization GO terms (Figure 5A, right panel). Among the proteins detected in association with SLFN5 by the mass spectrometry analysis, was STAT1, a key element of Type-I IFN-mediated signaling and the IFN-response (4, 10). To determine if SLFN5 interacts directly with IFN-activated STAT1, co-immunoprecipitation analyses were performed in the same 293T cells carrying inducible Myc-tagged SLFN5. The association of SLFN5 with protein members of the STAT family was established by immunoblot analysis, using anti-STAT1, anti-STAT3 and anti-STAT5 antibodies in immunoprecipitated protein lysates (Figure 5B). SLFN5 was found to be associated with STAT1, but not STAT3 or STAT5 (Figure 5B) and the signal was more intense after 24 hours of IFNβ treatment (Figure 5C). Collectively, our findings suggested that SLFN5 protein exhibits specificity for STAT1, suggesting a potential role for SLFN5 in the regulation of STAT1-mediated Type-I IFN-induced transcriptional activation of ISGs.
Figure 5. Identification of proteins and complexes associated with SLFN5.
(A) Purification of SLFN5 associated protein complexes. Total extracts, prepared from 293T cells expressing doxycycline inducible SLFN5-Myc-Tag, were subjected to immunoprecipitation (IP) using anti-Myc-Tag magnetic beads. Immunoprecipitated proteins were resolved by SDS-PAGE and visualized using Bio-Rad stain-free gels (left panel). Asterisk indicates SLFN5-Myc complexes. Lanes were excised and subjected to mass spectrometry analysis (middle panel). Venn diagram showing the number of proteins identified by MS analysis that overlap and differ between the doxycycline treated and untreated samples (right panel). Protein set enrichment analysis demonstrated overrepresentation of proteins involved in metabolic processes, immune system, biological regulation, localization and response to stimulus processes. (B) Cells carrying doxycycline inducible SLFN5-Myc-Tag fusion protein were grown in the presence or absence of doxycycline for 72 hours, followed by IFNβ treatment for 3 hours. Equal amounts of lysates from (No-DOX) and (DOX) treated HEK-293T cells were subjected to Immunoprecipitation using anti-c-Myc magnetic beads, and immune complexes were resolved by SDS-PAGE and immunoblotted with anti-Myc, anti-STAT1, anti-STAT3, or anti-STAT5 antibodies, as indicated (top panel). Total lysates from the corresponding experiments shown in the upper panels were resolved by SDS-PAGE and immunoblotted with the same corresponding antibodies, to assess for protein input (bottom panel). (C) Cells carrying doxycycline inducible SLFN5-Myc-Tag fusion protein were grown in the presence or absence of doxycycline for 72 hours, followed by different time points of IFNβ treatment, as indicated. Immunoprecipitated complexes were analyzed by SDS-PAGE and probed with the indicated primary antibodies. (D) ISG15 locus and ChIP primer set locations. Blue arrowheads indicate the Stat1-binding site. The ISG15 transcription start site is indicated by red arrow. Paired arrows indicate the positions of ChIP primer sets. (E) Cells carrying doxycycline inducible SLFN5-Myc fusion protein were grown in the presence or absence of doxycycline for 72 hours and left untreated or treated with IFNβ for 3 hours, as indicated. Chromatin-protein complexes were immunoprecipitated with anti-myc-Tag or appropriate IgG isotype antibodies used as controls. Immunoprecipitated DNA samples were analyzed by qPCR using ISG15 primer set 1, ISG15 primer set 2 and, RPL30 primer set. Data are expressed as relative enrichment over untreated (No-DOX) and are represented as means ± SD of two independent experiments. Two-way Anova analysis followed by Tukey’s multiple comparison test was used to assess statistically significant differences between experimental conditions. (N.S.: not significant; * p < 0.05; See also Table S8)
To further understand and define the functional outcome of this SLFN5-STAT1 interaction, we examined whether there is enrichment of SLFN5 in the promoters of STAT1 target genes, using chromatin immunoprecipitation (ChIP) assays. For this, we employed the ISG15 promoter that has multiple STAT1 binding sites (30, 31) (Figure 5D). We used a lentiviral-based doxycycline inducible gene expression system to overexpress Myc-tagged SLFN5 fusion protein in HEK-293T cells. Cells were either mock-treated or treated with doxycycline for 72 hours prior to the start of IFNβ treatment. Lysates of IFNβ-treated Myc-tagged SLFN5 overexpressing cells and control HEK-293T cells were immunoprecipitated with ChIP grade antibodies for Myc-Tag or control mouse immunoglobulin G (IgG). After column purification, the precipitated chromatin DNA fragments were used as templates for quantitative real time RT-PCR, with two specific ChIP grade primers for the indicated binding sites (Figure 5D). The specificity of the assay was monitored by including a primer set that targets a non-STAT1 binding site (Figure 5E, right panel). There was no detectable enrichment for non-specific regions with the mouse IgG antibody. In the absence of doxycycline treatment there was no significant enrichment of SLFN5-containing complexes to the ISG15 promoter sites 1 and 2 (Figure 5E, left and middle panels). However, there was significant enrichment of SLFN5-containing complexes to both ISG15 promoter sites in HEK-293T overexpressing SLFN5 cells treated with IFNβ (Figure 5E left and middle panel). Taken altogether, these data strongly suggest that SLFN5 is present together with STAT1 in a complex that binds to ISRE elements in the promoters of ISGs.
Crispr/Cas9-mediated SLFN5 gene disruption enhances IFNβ-dependent gene transcription
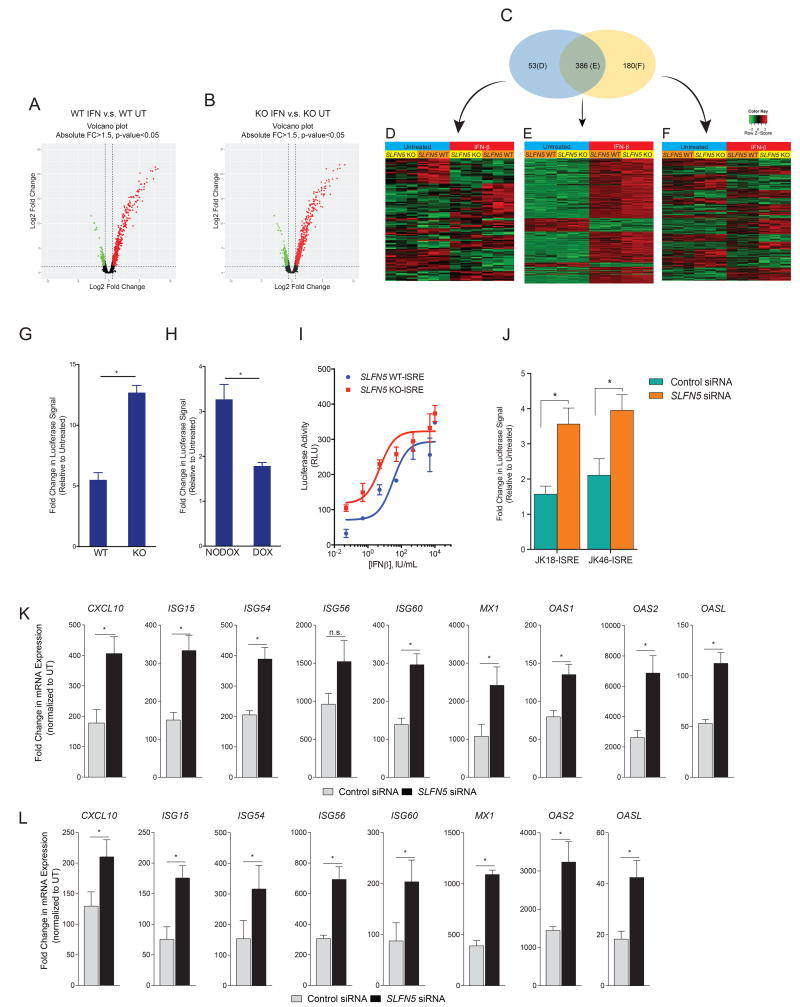
Having demonstrated that SLFN5 interacts with STAT1 upon IFN treatment, we further examined the role of SLFN5 in IFN-induced ISG transcription. We employed HumanHT-12 v4 Expression BeadChip microarrays to identify genes differentially induced by IFNs in SLFN5 WT and SLFN5 KO U87MG cells. Putative probes with fold change threshold > 1.5, significance level of Benjamini and Hochberg false discovery rate < 0.05 were deemed to be significantly differentially expressed. Analysis of gene expression profiles from three independent experiments revealed that 439 genes were upregulated in IFN-treated SLFN5 WT cells (Figure 6A), whereas 566 genes were upregulated in IFN-treated SLFN5 KO cells (Figure 6B). 53 induced genes were detected in SLFN5 WT, but not in SLFN5 KO cells (Figures 6C and 6D, Table S9). 180 uniquely induced genes were identified only in the SLFN5 KO cells, but not in SLFN5 WT cells (Figures 6C and 6F, Table S9). Importantly, among the 386 genes induced in both SLFN5 WT and SLFN5 KO cells (Figures 6C and 6E), the expression of 295 of these genes was significantly higher in the SLFN5 KO compared to the SLFN5 WT U87MG cells (Table S9), suggesting potential repressive effects of SLFN5 on Type-I IFN signaling for the majority of IFN-stimulated genes. In contrast to genes whose transcription was upregulated by SLFN5 depletion, a smaller set of genes was downregulated and there was very little overlap between these genes in SLFN5 WT and SLFN5 KO cells. To further assess the canonical signaling pathways the up-regulated genes might be involved in, we performed pathway enrichment analysis using the Ingenuity Pathway Analysis (IPA) tool (32) (Table S10). Overall, depletion of SLFN5 led to the transcriptional enhancement of nearly 3/4 of the up-regulated ISGs (295 out of 386), establishing a key regulatory role for SLFN5 in IFN-dependent transcriptional responses, which play key roles in antitumor cellular responses (33).
Figure 6. Crispr/Cas9-mediated deletion of SLFN5 enhances IFNβ-induced ISG mRNA expression.
Cells were incubated in the presence or absence of human IFNβ for 6 hours. The gene expression profiles of untreated and IFNβ-treated cells were compared in three independent experiments, using Human HT-12 v4 Expression BeadChip and Illumina iScan. Volcano plots of differentially expressed genes after IFNβ treatment are shown for (A) SLFN5 WT and (B) SLFN5 KO U87MG cells. 439 genes were differentially induced between untreated and IFNβ-treated SLFN5 WT cells, whereas 566 genes were differentially induced between untreated and IFN-β-treated SLFN5 KO U87MG cells. (C) Venn diagram of microarray data shows common genes differentially expressed in SLFN5 WTs (blue ellipse) and in SLFN5 KO U87MG cells (yellow ellipse) after treatment with IFNβ. (D) Hierarchical clustering of differentially expressed genes only in SLFN5 WT U87MG cells. (E) Hierarchical clustering of differentially expressed genes in both SLFN5 WT and SLFN5 KO cells upon IFNβ treatment. Differences in the effects of IFNβ treatment between SLFN5 WT and SLFN5 KO cells are seen for 386 genes, 295 of these genes are characterized by a less efficient IFNβ-driven transcription in SLFN5 WT U87MG cells. (F) Hierarchical clustering of differentially expressed genes only in SLFN5 KO U87MG cells. All annotations presented here are based on statistical analyses and are presented with p values after FDR. Only annotations with p values < 0.05 are shown. (G) U87MG SLFN5 WT and SLFN5 KO cells were transduced with an ISRE-luciferase promoter construct carrying lentivirus. Stably transduced cells were seeded in 96-well plates in triplicated technical replicates and the next day cells were incubated for 6 hours in the presence or absence of human IFNβ, and luciferase activity was measured. Data are expressed as fold increase of luciferase activity in response to IFNβ, treatment over control untreated samples for each condition. Bar graphs show means ± SEM of three independent experiments. (H) U87MG cells carrying doxycycline-inducible SLFN5-myc-Tag fusion protein were transduced with an ISRE-luciferase promoter construct carrying lentivirus. Stably transduced cells were grown in the presence and absence of DOX for 48 hours, and then seeded in 96-well plates in triplicate technical replicates and the next day cells were incubated for 6 hours in the presence or absence of human IFNβ, and luciferase activity was measured. Data are expressed as fold increase of luciferase activity in response to IFNβ treatment over control untreated samples for each condition. Bar graphs show means ± SEM of three independent experiments. (* p < 0.05) (I) SLFN5 WT and KO U87MG were stably transduced with the ISRE-luciferase reporter carrying viral particles, as described in methods. Equal numbers of stably transduced reporter cells were seeded in 96-well plates in triplicates and the next day cells were incubated with escalating concentrations of human IFNβ as indicated, and luciferase activity was measured. Data are expressed as fold increase of luciferase activity in response to IFNβ, treatment over control untreated samples for each condition. (J) ISRE-luciferase reporter carrying GSC lines, JK18 (JK18-ISRE) and JK46 (JK46-ISRE), were transfected with SLFN5 siRNA or control siRNA. 24 hours after transfection equal number of cells were seeded in 96-well plate. Cells were incubated for 6 hours in the presence or absence of human IFNβ, and luciferase activity was measured. Data are expressed as fold increase of luciferase activity in response to IFNβ treatment over control untreated samples for each condition per cell line. Bar graphs show means ± SEM of three independent experiments. (* p < 0.05) (K–L) qRT-PCR analyses of the relative mRNA expression of ISGs after IFNβ stimulation in JK18 (K) and JK46 (L) GSC lines after siRNA transfection are shown. Expression levels of the indicated genes were determined using RPL19 for normalization. Data are expressed as fold change over untreated samples and represent means ± SEM of three independent experiments for ISG56 and four for CXCL10, ISG15, ISG54, ISG60, MX1, OAS1, OAS2, and OASL. (n.s.: not significant, * p < 0.05) (See also Figure S4 and Tables S9–10)
We also examined whether SLFN5 elicits differential transcriptional regulation via IFN-stimulated response elements (ISRE) in luciferase reporter assays, by establishing stable ISRE-luciferase reporter expression in SLFN5 WT and KO U87MG cells and JK18, JK46 GSC lines. Treatment with IFNβ for 6 hours led to the induction of luciferase in SLFN5 WT cells, but this induction was significantly enhanced in the absence of SLFN5, suggesting that SLFN5 can inhibit IFNβ-induced STAT1 activity (Figure 6G and J). Conversely, ectopic expression of SLFN5 in a different set of ISRE reporter cells led to decreased luciferase activity (Figure 6H). When we examined the effects of escalating doses of IFN for a prolonged duration of treatment, we found that in reporter cells with SLFN5 gene disruption, there was enhanced responsiveness, as measured by ISRE driven luciferase activity (Figure 6I). As our data demonstrated regulatory effects of SLFN5 on ISRE reporter activity, we determined the effects of SLFN5 depletion on protein expression of the ISG15 gene product, a post-translational protein modifier and important player in the control of IFN responses (34). Cells carrying doxycycline-inducible SLFN5 shRNA were grown in the presence or absence of doxycycline, and then treated with IFNβ, after which ISG15 protein expression was determined by immunoblotting. As shown in Figure S4A, protein expression of ISG15 was induced in response to IFNβ in cells with endogenous SLFN5, but this expression increased further in cells in which SLFN5 was knocked down using doxycycline-inducible SLFN5 shRNA. To further establish the role that SLFN5 protein plays in regulation of ISG15 expression, we ectopically expressed SLFN5 by using a doxycycline-inducible promoter (Figure S4B) in U87MG cells and examined the effects on ISG15 protein expression. Consistent with our knockdown studies, ectopic expression of SLFN5 substantially decreased the levels of IFN-dependent expression of ISG15 protein (Figure S4B).
To directly examine the function of SLFN5 in Type-I IFN induced gene activation and to validate the microarray results, SLFN5 WT and SLFN5 KO U87MG, JK18 and JK46 cells were treated with IFNβ for the indicated time, or left untreated, and the induction of known ISGs (35, 36) was analyzed by quantitative real-time RT-PCR. As expected, IFNβ treatment led to a robust induction of several key ISGs. We found the induction of ISG54, ISG56, ISG60 and OASL by IFNβ was significantly enhanced in SLFN5 KO cells compared with WT cells (Figure S4C). Similar to GBM cell lines, combination of SLFN5 knockdown and IFNβ treatment resulted in enhanced induction of CXCL10, ISG15, ISG54, ISG56, ISG60, MX1, OAS1, OAS2 and OASL in JK18 and JK46 GSCs (Figures 6K and 6L), establishing a critical role for SLFN5 in Type-I IFN-signaling in GBM, including their GSC population.
SLFN5 disruption enhances Type-I IFN mediated antineoplastic responses
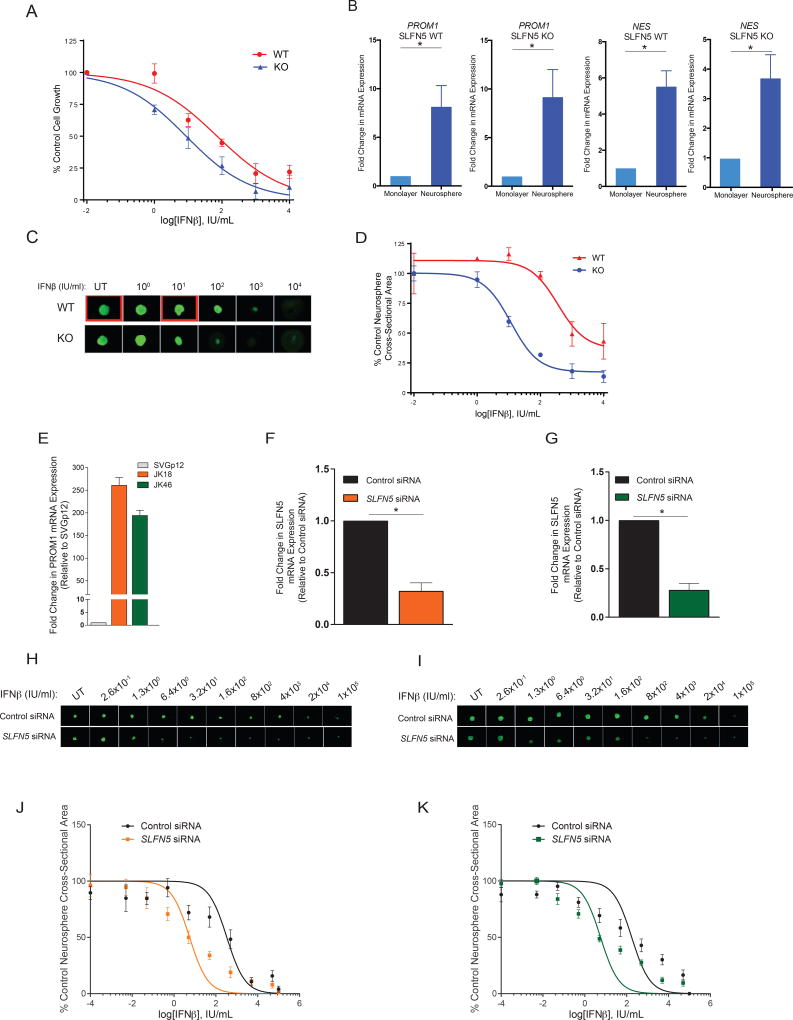
To determine the potential regulatory effects of SLFN5 on proliferation and survival of GBM, we assessed IFN sensitivity in SLFN5 WT and SLFN5 KO U87MG cells using the Alamar Blue viability assay. Depletion of SLFN5 in U87MG cells resulted in enhanced cellular sensitivity to Type-I IFN-induced anti-proliferative responses (Figure 7A). Next, we asked whether SLFN5 depletion sensitizes GSCs to the anti-proliferative effects of Type-I IFNs. For this purpose, glioma stem-like cells (GSLC) were cultured in sphere forming conditions, as previously described (37, 38). Culturing cells under these conditions resulted in enrichment for stem cell markers, such as CD133-encoding PROM1 and NESTIN-encoding NES as compared with monolayer-cultured cells (Figure 7B) (38–40). Upon exposure to increasing doses of IFNβ, there was reduced GSLC-derived sphere formation, which was clearly more pronounced in SLFN5 depleted spheres (Figures 7C and 7D). We next studied the effects of SLFN5 knockdown in the patient derived GSC lines, JK18 and JK46, which express high levels of the stem cell marker CD133-encoding PROM1 gene (Figure 7E). For this purpose, we used SLFN5 siRNA in JK18 and JK46 GSC lines (Figure 7F–G). While only a minimal inhibitory effect on neurosphere size was observed after treatment with IFNβ alone, the combination of IFNβ treatment and SLFN5 knockdown resulted in greatly impaired neurosphere growth, as indicated by a substantial decrease in neurosphere size in patient-derived GSC lines, confirming our observations in U87MG cells (Figure 7H–K). These results strongly suggest that down-regulation of SLFN5 leads to enhanced cellular sensitivity to Type-I IFN-induced anti-proliferative responses in glioma stem-like cancer cells, suggesting that SLFN5 is an important negative regulator of the IFN response in glioma stem cells.
Figure 7. Deletion of SLFN5 enhances IFN-dependent antineoplastic responses.
(A) Equal numbers of SLFN5 WT and SLFN5 KO U87MG cells were incubated with the indicated concentrations of human IFNβ, and cellular proliferation was assessed after 6 days using Alamar Blue assay. Data shown represent means ± SEM of three independent experiments. (B) SLFN5 WT and SLFN5 KO U87MG cells were grown for 7–8 days as monolayer or in cancer stem cell media as neurosphere cultures. Total RNA was isolated, and CD133-encoding PROM1 and NESTIN-encoding NES mRNA gene expression was measured by qRT-PCR. Data are expressed as fold increase over monolayer-grown cells and represent means ± SEM of three independent experiments for both PROM1 and NES in SLFN5 WT cells, and three independent experiments for NES, and two for PROM1 in SLFN5 KO cells. (* p < 0.05). (C) SLFN5 WT and SLFN5 KO U87MG neurosphere cells were seeded into round-bottom 96-well plates (1500 cells per well) and treated with the indicated concentrations of IFNβ or left untreated (UT). After 6 days, neurospheres were stained with 0.1 µg/mL acridine orange for 1 h. Neurosphere images were acquired using a BioTek Cytation3 multimode microplate reader (GFP channel). (D) Neurosphere cross-sectional areas of SLFN5 WT and SLFN5 KO U87MG were analyzed using the Gen5 Imager Software. Data are expressed as percentages of control untreated sample values and represent means ± SEM of three independent experiments, including the one shown in panel C. (E) Total RNA was isolated from SVGp12, JK18 and JK46 cells, and CD133-encoding PROM1 mRNA gene expression was measured by qRT-PCR. Data are expressed as fold increase over normal astrocyte SVGp12 cells and represent means ± SD of two independent experiments. (F–G) siRNA mediated knockdown of SLFN5, using specific siRNAs in JK18 (F) or JK46 (G) GSC lines. Knockdown was assessed by qRT-PCR, using RPL19 for normalization. Results represent means ± SEM of three independent experiments. (* p < 0.05) (H–I) Control siRNA- and SLFN5 siRNA-transfected JK18 (H) or JK46 (I) GSC lines were seeded into round-bottom 96-well plates and treated with the indicated concentrations of IFNβ or left untreated (UT). After two weeks, neurospheres were stained with 0.1 µg/mL acridine orange for 1 h. Neurosphere images were acquired using a BioTek Cytation3 multimode microplate reader (GFP channel). (J–K) Neurosphere cross-sectional areas of control siRNA- or SLFN5 siRNA-transfected JK18 (J) and JK46 (K) GSC lines were analyzed using the Gen5 Imager Software. Data are expressed as percentages of control untreated sample values and represent means ± SEM of four independent experiments, including the ones shown in panels H and I.
DISCUSSION
In recent years there has been increasing attention directed to the Schlafen family of genes and proteins. This reflects the accumulating evidence that these proteins play key roles in several cellular functions and accumulating data suggest their involvement in the pathophysiology of both neoplastic and non-neoplastic disorders. The original description of the Schlafen gene family arose in the mouse system (12). Although the initial mouse work described four mouse members, there are now over 10 mouse Slfns that have been identified and at least 5 human SLFNs (12, 13, 41, 42). All Schlafens are characterized by the presence of a Slfn box/domain that is unique for this family, but whose precise function still remains to be defined (12, 42). The 3 different groups of Schlafens are defined by the size of the proteins and by the presence of unique domains, such as the SWADL domain (Ser-Trp-Ala-Asp-Leu) in groups II and III, a C-terminal extension helicase motif in group III, and a nuclear localization signal in group III (12, 15).
There is evidence that the function of human members of the Schlafen family mediate important biological responses, including antiviral responses. Specifically, SLFN11 blocks HIV-1 production, via its ability to inhibit expression of viral proteins (43). Other studies have implicated SLFNs in the regulation of malignant cell proliferation. It has been shown that SLFN5 inhibits anchorage-independent growth and invasion of malignant melanoma cells (13). SLFN5 has also been implicated in the control of motility and invasiveness of renal cell carcinoma cells, a function that reflects its suppressive effects on expression of the matrix metalloproteinase genes MMP-1 and MMP-13, and several other genes involved in the control of malignant cell motility (14). Such genes include PCDHGB2, PCDHB11, PPL, JUP, KIF1C, and TPM4 (14). Notably, these effects of SLFN5 appeared to be specific to SLFN5, since SLFN11, SLFN12 and SLFN13 did not exert any such suppressive effects (14). Other studies have shown that SLFN11 exhibits a novel function as a sensitizer of malignant cells to different types of chemotherapy (44, 45).
In the present study we provide the first evidence implicating SLFN5 in glioblastoma tumorigenesis and demonstrate a novel function of SLFN5 as a negative regulator of STAT1-dependent transcriptional activation of ISGs. In initial studies, we found that SLFNs are inducible by Type-I IFNs in malignant brain tumor cells. Remarkably, and to our surprise, we found that Crispr/Cas9-driven deletion of SLFN5 enhances Type-I IFN induced antitumor activity in glioblastoma and leads to suppression of sphere formation in sphere-cultured glioma and patient derived cell lines, consistent with inhibitory effects on the malignant glioma stem cell population. In efforts to define the functional mechanisms mediating these responses, we found that knockout or knockdown of SLFN5 leads to enhanced expression of ISGs, consistent with a suppressive role of SLFN5 on ISG transcription in glioblastoma cells. Using mass spectrometry analysis, we found that STAT1 associates with SLFN5 in a Type-I IFN-dependent manner and is present in complexes that bind ISRE elements in the promoters of ISGs. Thus, SLFN5 appears to act as a repressor of STAT1-driven gene transcription, probably via its direct interaction with the protein. Consistent with this, SLFN5 shows enrichment at the sites that STAT1 interacts within the promoters of the Type-I IFN inducible ISGs, and targeted disruption of the SLFN5 gene results in an enhanced global transcriptional response after IFNβ stimulation. These findings for the first time define a member of the Schlafen family as a negative regulator of the Type-I IFN response in a given malignant phenotype. This raises the question of whether targeting of SLFN5 may provide a unique approach to treat resistant glioblastoma and/or to eliminate glioma cancer stem cells.
Beyond our findings, there appear to be other examples where human SLFNs may promote tumorigenesis in other systems. For instance, high SLFN5 expression was found to positively correlate with progression to gastric cancer in patients with intestinal metaplasia (46). On the other hand, it appears that in other tumors SLFN5 may have opposing effects and this variability in elicited biological response may reflect the transcription factors and genes targeted by the repressor activity of SLFN5. In the case of renal cell carcinoma patients, high levels of SLFN5 are associated with better overall prognosis, possibly a reflection of the repressive effects of SLFN5 on the genes for MMP1 and MMP3 and other genes that promote cell motility. Thus, selective targeting of SLFN5 for therapeutic purposes in malignancies in the future may require precise analysis of other associated factors and design of therapeutic targeting in the context of the specific role of SLFN5 in a particular tumor. Our data suggest that in the case of glioblastoma, a highly fatal malignancy, SLFN5 targeting can be exploited for the development of desperately needed therapeutic approaches. However, for this to be possible, further biochemical characterization of the SLFN5 protein and the function of its various motifs, as well as the SLFN5-containing protein complexes, are necessary. An important issue that, if addressed, may provide the basis for the development of specific SLFN-targeting agents, would be the characterization of the function of the Slfn box motifs that are highly specific for SLFNs, but whose function(s) remain unknown. Also, more detailed biochemical analysis and identification of other transcription factors, beyond STAT1, that may interact directly with SLFN5 and other SLFNs, may provide unique opportunities and new targets for the treatment of glioblastoma and other malignancies.
MATERIALS AND METHODS
Additional materials and methods can be found in the Supplemental Information.
Materials
Recombinant human IFNα was obtained from Hoffmann-La Roche (USA). Recombinant human IFNβ was obtained from Biogen Idec (USA). Adult human normal brain lysate (ab30078) was obtained from Abcam (USA). The antibodies against STAT1, STAT3, STAT5 and Myc-Tag, were obtained from Cell Signaling Technology (USA). SLFN5, SLFN11 and SLFN12L antibodies were obtained from Sigma Aldrich (USA). GAPDH antibody was obtained from Millipore (USA). For RNAi mediated gene silencing, cells were transfected with double stranded control non-targeting or SLFN5 targeting RNA oligonucleotides (Santa Cruz Biotechnology, Santa Cruz, CA.). For transfection, Amaxa™ nucleofector kits from Lonza were used according to the manufacturer's instructions.
Cell lines
Glioblastoma cell lines, LN18, LN229, LN443 and U87MG; medulloblastoma cell lines, DAOY and D556, human embryonic kidney 293T cells were all maintained in DMEM and SVGp12 cells were maintained in EMEM, all supplemented with 10% fetal bovine serum and antibiotics. All cell lines were regularly tested for mycoplasma contamination. LN18, LN229, LN443, U87MG, DAOY and D556 cell lines were subjected to short tandem repeat (STR) analysis to ensure genetic stability and authenticated where published reference STR profiles were available (Genetica DNA Laboratories).
Patient-derived GSC lines JK18 and JK46 were obtained from Dr. John A. Kessler (Northwestern University) (19, 20) and were cultured in DMEM/F12 (Thermo Fisher, Carlsbad, CA), supplemented with B27 (2%, Thermo Fisher), penicillin and streptomycin (1%, Sigma Aldrich, St. Louis, MO), Heparin (5 µg/ml, Sigma-Aldrich, St. Louis, MO), EGF (20 ng/ml), and basic FGF (20 ng/ml, Peprotech) and grown as suspension in non-cell culture treated flasks with filter caps.
CRISPR-Cas9 approach to generate SLFN5 knockout U87MG cells
The guide RNA (gRNAs) sequences and Cas9 carrying plasmids were purchased from Santa Cruz Biotechnologies (USA). The gRNA/Cas9 expression constructs were transfected into U87MG cells using Lipofectamine 3000 transfection reagent according to manufacturer’s instructions (Thermo Fisher, USA) 48 hours after transfection, U87MG cells were cultured in the presence of puromycin (1µg/ml). Approximately 2 weeks later, puromycin-resistant cells were expanded. Lack of SLFN5 protein expression in U87MG knockout cells was confirmed by Western blotting with SLFN5-specific antibody.
Lentiviral production and titration
Second generation, integration-deficient viral particles pseudotyped with the VSV-G envelope glycoprotein were produced by transient co-transfection of the relevant transfer plasmid and pMD2.VSV-G, and psPAX2 packaging plasmids into Lenti-X 293T cell line (Clontech). Supernatant containing virus was harvested 48 hours after transfection, centrifuged at 500 × g for 10 minutes at room temperature and then filtered with a 0.45 µm sterile filter to remove the cellular debris. The filtered medium was then concentrated using Lenti-X concentrator (Clontech), and the titer was estimated using the Lenti-X GoStix (Clontech). The particles were then aliquoted and stored at −80°C.
Statistical Analysis
Unless otherwise specified, GraphPad PRISM v6.0 software for Mac (GraphPad Software, Inc) and Student’s t-test were used to perform statistical analyses of the data. Error bars are SEM of averaged values for each experimental condition obtained from indicated number of experiments. Sample size was chosen based on similar experiments in previous publications. Similar variance is reflected by small SEM with equal n between the different conditions of a same experiment. p values < 0.05 were considered statistically significant between means.
Supplementary Material
Acknowledgments
The work was supported in part by NIH grants CA161196, CA77816, and CA155566 and by grant 5I01CX000916 from the Department of Veterans Affairs. A.D.A was supported in part by NIH training grant T32CA070085, D.S was supported in part by NIH training grant T32CA080621, and P.L. was supported in part by National Science Centre, Poland Grant 2016/22/M/NZ2/00548. We thank Dr. John A. Kessler for providing the JK18 and JK46 GSC lines.
Footnotes
Competing interests:
The authors declare no competing financial interests.
Data and materials availability:
The microarray data was deposited to the Gene Expression Omnibus (GEO) repository under accession number GSE88771. The RNA-Seq data was deposited to the NCBI's Sequence Read Archive (SRA) repository under the registered BioProject PRJNA341338. The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information files are available from the corresponding author upon request.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 3.Desjardins A, Rich JN, Quinn JA, Vredenburgh J, Gururangan S, Sathornsumetee S, et al. Chemotherapy and novel therapeutic approaches in malignant glioma. Front Biosci. 2005;10:2645–68. doi: 10.2741/1727. [DOI] [PubMed] [Google Scholar]
- 4.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 5.Platanias LC. Interferons and their antitumor properties. J Interferon Cytokine Res. 2013;33(4):143–4. doi: 10.1089/jir.2013.0019. [DOI] [PubMed] [Google Scholar]
- 6.Wakabayashi T, Hatano N, Kajita Y, Yoshida T, Mizuno M, Taniguchi K, et al. Initial and maintenance combination treatment with interferon-beta, MCNU (Ranimustine), and radiotherapy for patients with previously untreated malignant glioma. J Neurooncol. 2000;49(1):57–62. doi: 10.1023/a:1006405512579. [DOI] [PubMed] [Google Scholar]
- 7.Natsume A, Ishii D, Wakabayashi T, Tsuno T, Hatano H, Mizuno M, et al. IFN-beta down-regulates the expression of DNA repair gene MGMT and sensitizes resistant glioma cells to temozolomide. Cancer Res. 2005;65(17):7573–9. doi: 10.1158/0008-5472.CAN-05-0036. [DOI] [PubMed] [Google Scholar]
- 8.Motomura K, Natsume A, Kishida Y, Higashi H, Kondo Y, Nakasu Y, et al. Benefits of interferon-beta and temozolomide combination therapy for newly diagnosed primary glioblastoma with the unmethylated MGMT promoter: A multicenter study. Cancer. 2011;117(8):1721–30. doi: 10.1002/cncr.25637. [DOI] [PubMed] [Google Scholar]
- 9.Bradley NJ, Darling JL, Oktar N, Bloom HJ, Thomas DG, Davies AJ. The failure of human leukocyte interferon to influence the growth of human glioma cell populations: in vitro and in vivo studies. Br J Cancer. 1983;48(6):819–25. doi: 10.1038/bjc.1983.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fish EN, Platanias LC. Interferon receptor signaling in malignancy: a network of cellular pathways defining biological outcomes. Mol Cancer Res. 2014;12(12):1691–703. doi: 10.1158/1541-7786.MCR-14-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porritt RA, Hertzog PJ. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 2015;36(3):150–60. doi: 10.1016/j.it.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Mavrommatis E, Fish EN, Platanias LC. The schlafen family of proteins and their regulation by interferons. J Interferon Cytokine Res. 2013;33(4):206–10. doi: 10.1089/jir.2012.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katsoulidis E, Mavrommatis E, Woodard J, Shields MA, Sassano A, Carayol N, et al. Role of interferon {alpha} (IFN{alpha})-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J Biol Chem. 2010;285(51):40333–41. doi: 10.1074/jbc.M110.151076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassano A, Mavrommatis E, Arslan AD, Kroczynska B, Beauchamp EM, Khuon S, et al. Human Schlafen 5 (SLFN5) Is a Regulator of Motility and Invasiveness of Renal Cell Carcinoma Cells. Mol Cell Biol. 2015;35(15):2684–98. doi: 10.1128/MCB.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int Immunol. 2004;16(10):1535–48. doi: 10.1093/intimm/dxh155. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9(4):287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–67. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srikanth M, Das S, Berns EJ, Kim J, Stupp SI, Kessler JA. Nanofiber-mediated inhibition of focal adhesion kinase sensitizes glioma stemlike cells to epidermal growth factor receptor inhibition. Neuro Oncol. 2013;15(3):319–29. doi: 10.1093/neuonc/nos316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng H, Lopez GY, Kim CK, Alvarez A, Duncan CG, Nishikawa R, et al. EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis. J Clin Invest. 2014;124(9):3741–56. doi: 10.1172/JCI73093. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Court W, et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321(5891):956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 23.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Pal S, Bi Y, Tchou J, Davuluri RV. Isoform level expression profiles provide better cancer signatures than gene level expression profiles. Genome Med. 2013;5(4):33. doi: 10.1186/gm437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41(Database issue):D1040–6. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Cao CJ, Huang LL, Ke ZF, Luo CJ, Lin ZW, et al. EFEMP1 promotes the migration and invasion of osteosarcoma via MMP-2 with induction by AEG-1 via NF-kappaB signaling pathway. Oncotarget. 2015;6(16):14191–208. doi: 10.18632/oncotarget.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji F, Wang Y, Qiu L, Li S, Zhu J, Liang Z, et al. Hypoxia inducible factor 1alpha-mediated LOX expression correlates with migration and invasion in epithelial ovarian cancer. Int J Oncol. 2013;42(5):1578–88. doi: 10.3892/ijo.2013.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagato S, Nakagawa K, Harada H, Kohno S, Fujiwara H, Sekiguchi K, et al. Downregulation of laminin alpha4 chain expression inhibits glioma invasion in vitro and in vivo. Int J Cancer. 2005;117(1):41–50. doi: 10.1002/ijc.21102. [DOI] [PubMed] [Google Scholar]
- 30.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2(4):383–93. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 31.Reich N, Evans B, Levy D, Fahey D, Knight E, Jr, Darnell JE., Jr Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci U S A. 1987;84(18):6394–8. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–30. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16(3):131–44. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 34.Ritchie KJ, Zhang DE. ISG15: the immunological kin of ubiquitin. Semin Cell Dev Biol. 2004;15(2):237–46. doi: 10.1016/j.semcdb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Gil MP, Bohn E, O'Guin AK, Ramana CV, Levine B, Stark GR, et al. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci U S A. 2001;98(12):6680–5. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3(11):900–11. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 37.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 38.Eckerdt F, Alvarez A, Bell J, Arvanitis C, Iqbal A, Arslan AD, et al. A simple, low-cost staining method for rapid-throughput analysis of tumor spheroids. Biotechniques. 2016;60(1):43–6. doi: 10.2144/000114372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon CH, Kim MJ, Kim RK, Lim EJ, Choi KS, An S, et al. c-Jun N-terminal kinase has a pivotal role in the maintenance of self-renewal and tumorigenicity in glioma stem-like cells. Oncogene. 2012;31(44):4655–66. doi: 10.1038/onc.2011.634. [DOI] [PubMed] [Google Scholar]
- 40.Zhu T, Li X, Luo L, Wang X, Li Z, Xie P, et al. Reversion of malignant phenotypes of human glioblastoma cells by beta-elemene through beta-catenin-mediated regulation of stemness-, differentiation- and epithelial-to-mesenchymal transition-related molecules. J Transl Med. 2015;13:356. doi: 10.1186/s12967-015-0727-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bustos O, Naik S, Ayers G, Casola C, Perez-Lamigueiro MA, Chippindale PT, et al. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene. 2009;447(1):1–11. doi: 10.1016/j.gene.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neumann B, Zhao L, Murphy K, Gonda TJ. Subcellular localization of the Schlafen protein family. Biochem Biophys Res Commun. 2008;370(1):62–6. doi: 10.1016/j.bbrc.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 43.Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491(7422):125–8. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A. 2012;109(37):15030–5. doi: 10.1073/pnas.1205943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Companioni Napoles O, Tsao AC, Sanz-Anquela JM, Sala N, Bonet C, Pardo ML, et al. SCHLAFEN 5 expression correlates with intestinal metaplasia that progresses to gastric cancer. J Gastroenterol. 2016 doi: 10.1007/s00535-016-1202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.