Abstract
Oxidant stress modifies T lymphocyte activation and function. Previous work demonstrated that murine T cell specific Keap1 deletion enhances antioxidant capacity and protects from experimental acute kidney injury. Here, we used CRISPR technology to develop clinically translatable human T cell-specific KEAP1 deletion. Delivery of KEAP1 exon 2 specific Cas9:gRNA in Jurkat T cells led to significant (~70%) editing and upregulation of NRF2 regulated antioxidant genes NQO1 (upto 11 fold), HO1 (upto 11 fold) and GCLM (upto 2 fold). In primary human T cells, delivery of KEAP1 exon 2 target site 2-specific ATTO 550-labeled Cas9:gRNA edited KEAP1 in ~40% cells and significantly (p≤0.04) increased NQO1 (16 fold), HO1 (9 fold) and GCLM (2 fold) expression. To further enrich KEAP1 edited cells, ATTO 550 positive cells were sorted 24h after electroporation. Assessment of ATTO 550 positive cells showed KEAP1 editing in ~55% cells. There was no detectable off-target cleavage in top 3 predicted genes in the ATTO 550 positive cells. Gene expression analysis found significantly (p≤0.01) higher expression of NQO1 mRNA in ATTO 550 positive cells compared to control cells. Flow cytometric assessment showed increased (p≤0.01) frequency of CD4, CD25 and CD69 expressing KEAP1 edited cells whereas frequency of CD8 (p≤0.01) and IL-17 (p≤0.05) expressing cells was reduced compared to control cells. Similar experimental conditions resulted in significant KEAP1 editing, increased antioxidant gene expression and frequency of CD69 and IL-10 positive cells in highly enriched KEAP1 edited Treg cells. KEAP1 edited T cells could potentially be used for treating multiple human diseases.
Introduction
T lymphocytes in concert with other immune mediators elicit adaptive immune responses following an antigen exposure. In addition to mounting antigen-specific immune response, T lymphocytes sense and respond to varying oxygen concentrations (1, 2). Significant experimental and clinical data indicates T lymphocyte involvement during ischemia reperfusion (IR)-induced tissue injury and repair, where oxidative stress dependent mechanisms appear to modulate T cell responses (3, 4).
Previous research demonstrated that T lymphocyte specific genetic deletion of Keap1 (kelch like-ECH-associated protein 1), used to upregulate nuclear Nrf2 (nuclear factor erythroid-derived 2 like 2) activation, significantly enhanced antioxidant responses, while adoptive transfer of Keap1-deficient T lymphocytes protected wild type (WT) mice from experimental IR-induced acute kidney injury (AKI) (5). KEAP1 is an adapter protein for the E3 ubiquitin ligase complex that tags NRF2 for ubiquitination and proteasomal degradation (6). NRF2 is a b-ZIP transcription factor that regulates multiple pro-survival genes including NQO1 (NADPH dehydrogenase quinone 1) and HO1 (heme oxygenase 1) and thus an attractive therapeutic target for various oxidative stress-related diseases (7-9). Although, genetic deletion of Keap1 using Cre/lox system effectively increases T lymphocyte specific Nrf2 activity, which renders protection from IR injury in mice, this method is not clinically viable. Therefore, we harnessed CRISPR (clustered regularly interspaced short palindromic repeats) technology as a novel tool for ex vivo KEAP1 editing in primary human T cells to develop T lymphocyte based antioxidant therapy with potential for clinical translation.
Genome editing using CRISPR technology, comprising of a Cas9 (Streptococcus pyogenes derived RNA guided endonuclease) protein and a gene specific guide RNA (gRNA), allows effective knock-out and knock-in of virtually any gene (10-12). In spite of its immense success to edit genome in large number of cell types and initial approval to use in human clinical trial to treat certain cancers, the delivery of Cas9:gRNA or the ribonucleoprotein (RNP) complex in some cell types such as primary human T lymphocytes has been challenging (13). Moreover, targeting genes that encode for intracellular proteins poses additional difficulty in term of identification and enrichment of the edited cells. Nonetheless, some research groups have reported successful use of the CRISPR technology to knock-out CXC chemokine receptor type 4 (CXCR4) and programmed cell death receptor 1 (PD1) as well as targeted nucleotide replacement (all expressed on cell surface) in human CD4+ T cells (14-18).
Here, we present data to demonstrate successful targeting of KEAP1 gene in primary and immortalized human T cells that significantly enhances their antioxidant potential. Our data show that CRISPR based KEAP1 editing results in significant upregulation of NRF2 dependent antioxidant genes. KEAP1 editing was also found to induce immunological changes in T lymphocytes in addition to an increased antioxidant gene expression. Additionally, this study presents a strategy to enrich edited cells while targeting genes that encode intracellular proteins. This KEAP1 editing and enrichment strategy in purified regulatory T (Treg) cells resulted in significant KEAP1 gene editing, upregulated NRF2 regulated antioxidant genes and induced immunological changes compared to control Treg cells. Successful expansion of KEAP1 edited cells can lead to the development of novel, ready to use, immune cell based antioxidant therapy for a broad range of human diseases.
Materials and Methods
Jurkat T cell culture
Jurkat E6-1 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA) and cultured in RPMI 1640 containing 10% FBS, 10 mM HEPES and 100 U/ml penicillin and streptomycin. A total of 2×105 cells were used per electroporation for each experimental condition.
Human T cell isolation and culture
Primary T cells were isolated from blood collected from healthy individuals by Ficoll gradient centrifugation and negative selection using EasySep human T cell isolation kit (STEMCELL Technologies, Cambridge, MA). Treg cells were isolated using CD4+CD25+CD127dim/− Treg cell isolation kit (Miltenyi Biotech, Auburn, CA). T cells and Treg cells were cultured in CTS OpTmizer T cell expansion media (ThermoFisher, Waltham, MA) containing 2% OpTmizer T-Cell expansion supplement, 10 mM HEPES and 100 U/ml penicillin and streptomycin and stimulated with plate bound anti-CD3 (10μg/ml) and anti-CD28 (10μg/ml) in the presence of interleukin 2 (IL-2) (50–1000 U/ml) for 48h prior to electroporation as described elsewhere (15). A total of 5×106 cells were used for each electroporation. These studies were approved by Johns Hopkins institutional review board.
Cas9:guide RNA delivery and editing analysis
All CRIPSR related reagents were purchased from IDT (San Jose, CA). Cas9:gRNA complex was prepared immediately before each experiment. Briefly, KEAP1, exon 2 specific CRISPR RNAs (crRNAs) were mixed in equimolar concentrations (200 μM) with trans-activating crRNA (tracrRNA) and allowed to form a gRNA (Table 1). A complex of Cas9 (1.5 or 3 μM) and gRNA (1.8 or 3.6μM) along with Alt-R Cas9 electroporation enhancer oligo (1.8 or 3.6μM) was electroporated with a Neon transfection kit and device (Invitrogen). Control cells were electroporated in the absence of Cas9:gRNA complex. Electroporation efficiency was assess by eGFP mRNA (TriLink Biotechnologies, San Diego, CA) or ATTO 550 labeled tracrRNA. Editing of KEAP1 gene as well as top three off-target genes was estimated by Surveyor mutation detection assay at different time points after electroporation using target specific primers (Table 2 and 3).
Table 1.
List of crRNA sequences used for targeting KEAP1, exon 2.
| Target site 1 (T1) | AGCCGCCCGCGGTGTAGATC |
| Target site 2 (T2) | CTACCTGGTCAAGATCTTCG |
| Target site 3 (T3) | GGAAGTTCGGCGTCAACGAG |
Table 2.
List of PCR primers used for detecting KEAP1 specific editing.
| Sense | AGCCGCCCGCGGTGTAGATC |
| Anti-sense | CTACCTGGTCAAGATCTTCG |
Table 3.
List of PCR primers used for detecting off target editing in top three predicted genes.
| PIGS (NM_033198) | |
| Sense | GGTAGATGGAAGGCACAGTAAG |
| Anti-sense | CCTGACAGACAAAGCCAACTA |
| HELZ2 (NM_001037335) | |
| Sense | GAGACGCAGTGAAGGAAGAC |
| Anti-sense | GTCCACAGTGAAGGTCAAGAA |
| TNIP1 (NM_001258456) | |
| Sense | TCAGCGGAGTGAAAGGATTG |
| Anti-sense | AGAGAAGAAGGGAGGGAGAAA |
Enrichment of KEAP1 edited primary T cells
ATTO 550 positive cells were flow sorted using MoFlo XDP (Beckman Coulter, Indianapolis, IN) cell sorter 24h after Cas9:gRNA electroporation. Propidium iodide (PI) was added before sorting to exclude dead cells. Sorted cells were assessed visually with Leica fluorescent microscope for purity and Surveyor mutation detection assay for KEAP1 editing.
Phenotypic and intracellular cytokine analysis of edited cells
Fluorochrome conjugated antibodies to following human antigens were used for flow cytometric analysis of KEAP1 edited cells: TCR-BV421(BioLegend, San Diego, CA), CD4-PerCP-Cy5.5 (BD Biosciences, Franklin Lakes, NJ), CD8-APC (BioLegend, San Diego, CA), CD25-BV605 (eBioscience, San Diego, CA), FoxP3-APC (eBioscience, San Diego, CA), or Alexa488 (BioLegend, San Diego, CA) CD69-APC-Cy7 (BD Biosciences, Franklin Lakes, NJ), IFNγ-PE (BD Biosciences, Franklin Lakes, NJ), TNFα-FITC (BD Biosciences, Franklin Lakes, NJ), IL4-AlexaFluor 488 (BioLegend, San Diego, CA) IL-10-PE or APC (eBioscience, San Diego, CA), and IL17-BV421 or PE (BioLegend, San Diego, CA). T lymphocytes (~5×105) were stimulated with leukocyte activation cocktail (BD Pharmigen, San Jose, CA) containing PMA (Phorbol 12-Myristate 13-Acetate), ionomycin and brefeldin A before staining for surface markers and intracellular cytokines. Labelled samples were analyzed with LSRII flow cytometer (BD Biosciences, Franklin Lakes, NJ). Unstained and unstimulated samples were used to correctly identify and gate cell populations during analysis using FlowJo software (Tree Star Inc., Ashland, OR).
Antioxidant gene expression analysis
Total RNA from purified T cells was isolated with RNeasy mini kit (Qiagen, Valencia, CA) and reverse transcribed using RevertAid first strand cDNA synthesis kit (ThermoFisher, Waltham, MA). Gene specific Taqman primer and probe sets (Applied Biosystems) were used to assess transcriptional status of NQO1, HO1, GCLM and GCLC in CFX96 real time PCR (BioRad, Hercules, California). The expression value for each gene was normalized to β-actin and the relative fold expression values calculated using δδCT method.
Statistical analysis
Data are presented as mean ± standard error of mean (SEM) or standard deviation (SD), and are compared by a paired, two-tailed student t test for a single comparison between two groups. Statistical significance of difference was defined as a p value ≤ 0.05.
Results
Delivery of KEAP1 specific CRISPR/Cas9 results in NRF2 activity in Jurkat T cell
We used Jurkat cells (clone E6-1), a lymphoblastic human T cell line, to optimize KEAP1 editing using CRISPR technology. We targeted 3 different target sites against KEAP1 exon 2 using two different electroporation conditions (1600V/10ms/3pulse and 1700V/10ms/3pulse) based on the previous published literature to determine most effective KEAP1 editing (Table 1 and (16, 17)). There was no effect on cell viability 72h after electroporation under these electroporation conditions, though we observed fewer cells after 1700V indicating an initial deleterious effect of high voltage electroporation on cell viability (Supplemental figure 1). Flow cytometric analysis showed comparable electroporation efficiencies (≥95% GFP positive cells) under these electroporation conditions (Figure 1A). Genomic cleavage analysis indicated KEAP1 editing in ~70% cells under 1600V and ~65% editing in 1700V (Figure 1B). Analysis of NRF2 target genes 72h after electroporation of Cas9:gRNA complex resulted in significant (p≤0.05) increases in NQO1 (upto 11 fold) and HO1 (upto 11 fold) for all three target sites and under both electroporation conditions where as GCLM (upto 2 fold) levels increased mildly for all three target sites under 1700V electroporation condition (Figure 1C). Similarly, delivery of all 3 KEAP1 specific Cas9:gRNA complexes under these conditions resulted in a significant (p≤0.05) increase in NQO1 (upto 21 fold), HO1 (upto 40 fold), GCLM (upto 6 fold) and GCLC (upto 3 fold) expression in primary human T cells, though electroporation efficiency was less (<70% compared to > 95% GFP positive cells) than that of Jurkat cells (Supplemental figure 2). In general, primary T cells were more vulnerable at high voltage electroporation as we observed low cell number and reduced NRF2 activation at 1700V compared to 1600V.
Figure 1.
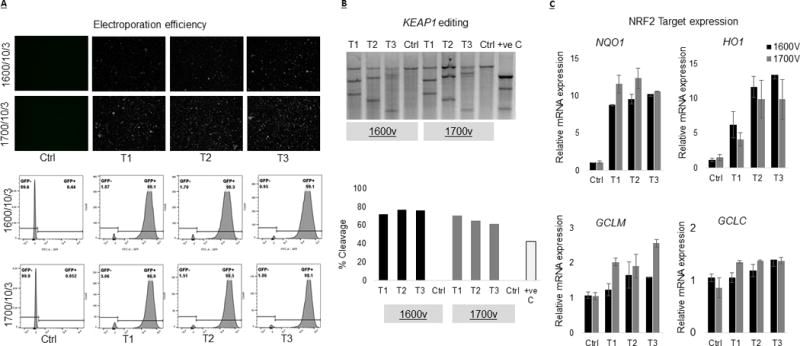
Delivery of KEAP1 specific RNP complex resulted in gene editing and NRF2 activation in Jurkat T cells. We tested guide RNA against three different target sites against KEAP1, exon 2. The RNP complexes were delivered using Neon Transfection system at two different electroporation conditions (1600V/10ms/3pulse and 1700V/10ms/3pulse). (A) Flow cytometric analysis of GFP positive cells 72h after electroporation showed ≥95% electroporation efficiencies under both electroporation conditions. (B) Genomic cleavage analysis indicated KEAP1 editing in ~70% cells under 1600V and ~65% editing in 1700V. (C) Real time PCR based assessment of NRF2 target genes 72h after RNP complex delivery showed significant (p≤0.05) increases in NQO1 (upto 11 fold), HO1 (upto 11 fold) and GCLM (upto 2 fold) under these electroporation condition.
KEAP1 editing using CRISPR/Cas9 upregulates NRF2 activity in primary human T cell
Since we observed significant editing with all three KEAP1 sfpecific gRNA tested in Jurkat T cells and primary T cell in our optimization studies, we decided to use target site 2 (T2) specific gRNA in primary T cells. Furthermore, we used ATTO 550-labeled tracrRNA to directly measure electroporation of RNP complex in primary T cells instead of eGFP mRNA that does not directly represent Cas9:gRNA complex electroporation properties. ATTO 550-labeled tracrRNA is an effective tool for monitoring transfection efficiency using microscopy and flow cytometry as it forms an integral part of the RNP complex thus providing more reliable electroporation measurement of RNP complex. Labeling of tracrRNA with ATTO 550 does not affect specificity of gRNA and activity of Cas9 protein. Flow cytometric analysis of primary T cells 24h after electroporation showed upto ~80% ATTO 550 positive cell confirming RNP complex delivery. KEAP1 editing analysis showed cleavage in about 40% of RNP treated cells in comparison to control cells. Moreover, we observed a significant increase in the expression of NRF2 target genes NQO1 (p≤0.01; 16 fold), HO1 (p≤0.01, 9 fold) and GCLM (p≤0.04; 2 fold) in primary T cell, 72h after RNP complex electroporation, compared to control cells.
Enrichment of KEAP1 edited primary T cells
Since there is no established method to enrich edited cells in situations where the target gene encodes for an intracellular protein, we decided to use ATTO 550 labeled tracrRNA for enrichment of KEAP1 edited primary T cell. We sorted ATTO 550 positive cells 24h after electroporation of RNP complex. Analysis of KEAP1 editing, 90h after sorting, using Surveyor mutation enzyme showed cleavage in ~55% ATTO 550 positive T cells (n=4) with no detectable editing in the ATTO 550 negative cells, indicating that labeled tracrRNA could be useful to enrich edited cells under these conditions. In addition to the on-target editing effects of RNP complex on KEAP1 gene we also examined for potential off-target effects of CRISPR mediated KEAP1 editing. We selected top 3 off-target genes, identified using CRISPR design tool (crispr.mit.edu). Mutation detection analysis showed no detectable off-target effect for the selected genes in this study (Figure 3C). Real-time PCR based NRF2 target gene expression analysis of ATTO 550 positive cells showed significantly (p≤0.01) higher expression of NQO1 (6.5 fold) in comparison to control cells, 120h after RNP delivery. There was no significant difference in the expression of other NRF2 target genes between ATTO 550 positive and control cells 120h after RNP delivery.
Figure 3.
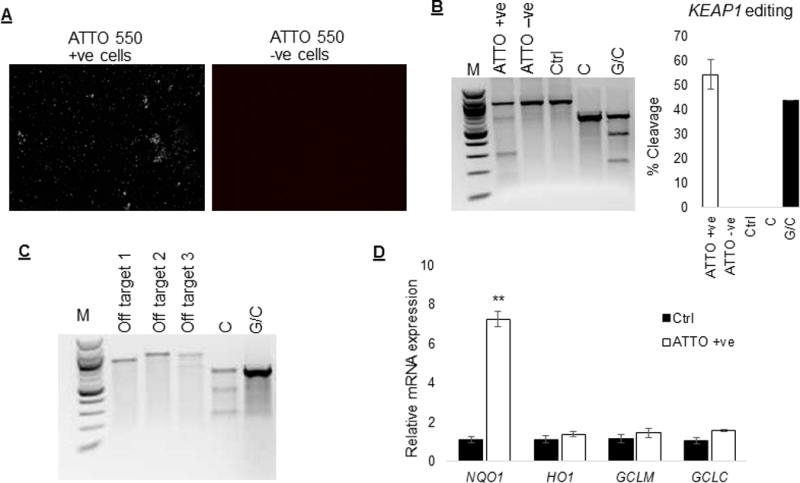
Enrichment of KEAP1 edited primary human T cells. Since ATTO 550 tracrRNA forms an integral part of the RNP complex we sorted ATTO550 positive cells (n=4) 24h after electroporation and assessed KEAP1 editing and Nrf2 activity. (A) Fluorescent microscopic images of ATTO 550 positive and negative cells 90h after sorting. (B) Assessment of KEAP1 editing, 90h after sorting (120h after electroporation), showed cleavage in ~ 55% ATTO 550 positive T cells with no detectable editing in the ATTO 550 negative cells or control cells. (C) Surveyor enzyme based genomic mutation detection analysis showed no detectable off-target effect in the ATTO 550 positive cells. (D) Real-time PCR based gene expression analysis showed significantly (p≤0.01) higher expression of NQO1 mRNA in ATTO 550 positive cells compared to control cells. There was no difference in the mRNA level of other NRF2 target genes between ATTO 550 positive and control cells at this time point.
KEAP1 editing induces immunological changes in primary T cells
To further understand the functional effects of KEAP1 editing on primary T cells we investigated the expression of CD4, CD8, CD25, CD69, and T cell specific intracellular cytokines in KEAP1 edited cell (ATTO 550 positive) and control cells. KEAP1 editing significantly increased the frequency of CD4+ (47.5%±3.1% vs 24.1%±3%; p≤0.01) T cells whereas frequency of CD8+ (47.8%±3.4% vs 68.5%±2.5%; p≤0.01) T cells was reduced compared to control cells, 120h post RNP delivery. Furthermore, KEAP1 edited T cells expressed significantly higher level of CD25 (14.9%±2% vs 6.3%±0.4%; p≤0.01) and CD69 (9.1%±1.2% vs 1%±0.3%; p≤0.01) in comparison to control cells (Figure 4A). We found no significant difference in frequencies of cells producing TNF-α (13.5%±3.4% vs 9%±2.4%; p=0.36), IFN-γ (9.3%±1.7% vs 10.8%±1.3%; p=0.54), IL-4 (13.7%±4.4% vs 6.5%±1.6%; p=0.20) or IL-10 (14.4%±4.5% vs 4.7%±1.3%; p= 0.09) between KEAP1 edited and control cells. However, IL-17 production was significantly reduced (6.1%±1.1% vs 21.3%±5.5%; p=0.04) in KEAP1 edited cells (Figure 4B).
Figure 4.
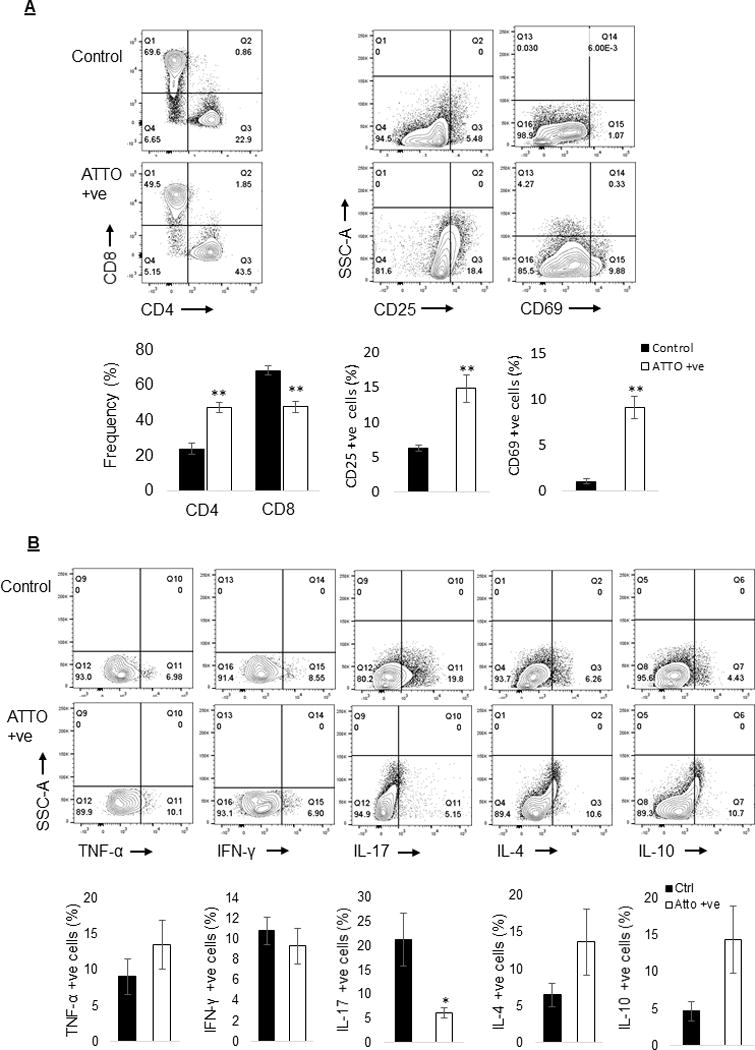
KEAP1 editing induced immunological changes in primary human T cells. (A) KEAP1 editing significantly increased the frequency of CD4 cells whereas frequency of CD8 cells was reduced compared to control cell, 5 days post RNP delivery. Furthermore, KEAP1 edited T cells expressed significantly higher level of CD25 and CD69 in comparison to control cells. (B) The frequency of IL-17 producing cells was significantly reduced in ATTO 550 positive KEAP1 edited cells in comparison to control cells. There was no difference in frequency of cell producing TNF-α, IFN-γ, IL-4, IL-10 between KEAP1 edited and control cells.
Treg specific KEAP1 editing upregulates antioxidant genes, CD69 and IL-10
We next tested this KEAP1 editing and enrichment strategy in purified human Treg cells. We used magnetic beads to enrich CD4+CD25+CD127dim/− Treg cells that contained over 90% cells positive for CD25 and Foxp3 (Figure 5A). Delivery of KEAP1 specific (T2) Cas9:gRNA complex resulted in KEAP1 editing in ~35% cells (Figure 5B) and significant increase in NRF2 target genes NQO1 (p≤0.001), HO1(p≤0.05) and GCLM (p≤0.01) mRNA levels (Figure 5C). Furthermore, flow sorting based enrichment of ATTO 550 positive cells improved cleavage to ~63% cells (Figure 5D & E, n=3). Flow cytometric analysis of enriched KEAP1 edited Treg cells showed increased frequency of CD69 (52.0%±2.7% vs 41.3%±2.8%, p≤0.05) and IL-10 (5.4%±1.3% vs 1.8%±0.4%, p≤0.05) expressing cells compared to control Treg cells (Figure 5F). Frequency of TNF-α (32.5%±2.7% vs 31.1%±3.1%; p=0.75), IFN-γ (8.9%±1.1% vs 8.9%±0.9%; p=0.97), or IL-17 (2.2%±0.4% vs 1.0%±0.4%; p= 0.10) producing cells was not significantly different between KEAP1 edited and control Treg cells.
Figure 5.
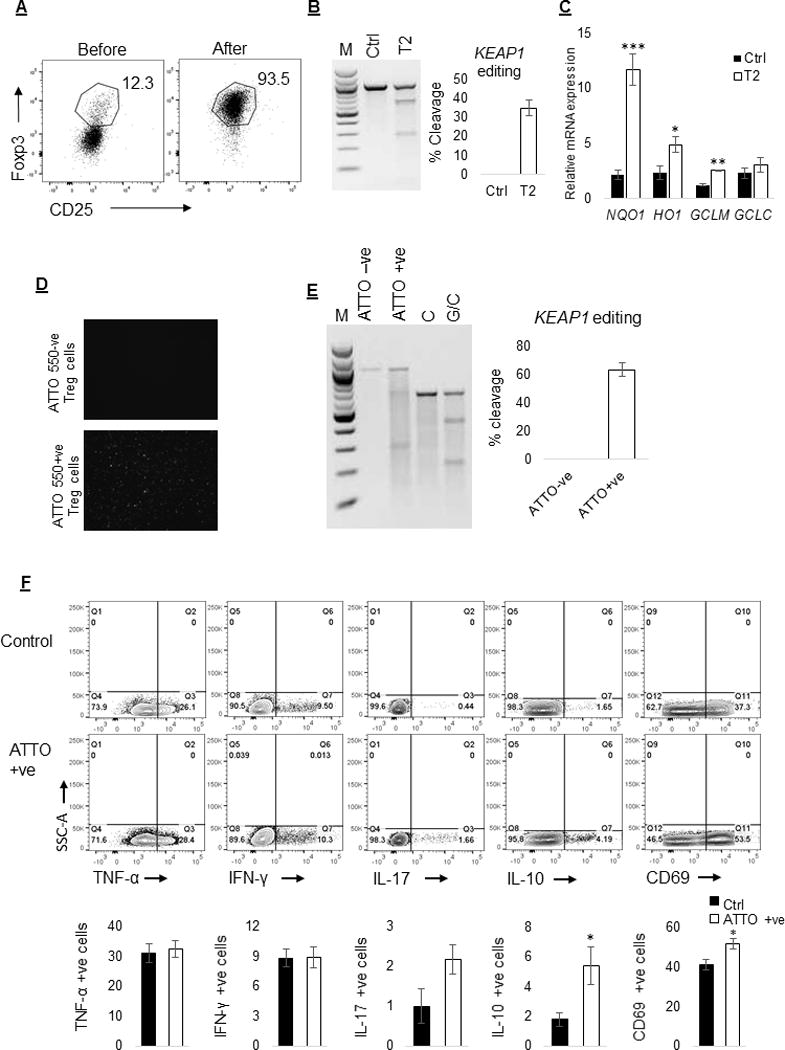
Treg specific KEAP1 editing increases NRF2 regulated antioxidant gene expression and frequency of CD69 and IL-10 positive cells. (A) Isolation of CD4+CD25+CD127dim/− cells using magnetic beads resulted in the enrichment of highly purified (>90%) Treg cells that were positive for CD25 and Foxp3. (B) Delivery of KEAP1 specific Cas9:gRNA complex resulted in KEAP1 editing in ~35% cells. (C) KEAP1 edited cells had significantly increased NQO1 (p≤0.001; 5 fold), HO1 (p≤0.05; 2 fold) and GCLM (p≤0.01; 2 fold) mRNA levels compared to control cells. (D) ATTO 550 positive, KEAP1 edited cells were enriched using flow sorting and assessed using fluorescent microscope. (E) Enrichment of ATTO 550 positive KEAP1 edited cells improved percent cleavage to ~63% cells. (F) Flow cytometric analysis of ATTO 550 positive KEAP1 edited Treg cells showed increased frequency of CD69 (52.0±2.7 vs 41.3%±2.8, p≤0.05) and IL-10 (5.4±1.3 vs 1.8±0.4, p≤0.05) positive cells compared to control Treg cells. The frequency of TNF-α (32.5%±2.7% vs 31.1%±3.1%; p=0.75), IFN-γ (8.9%±1.1% vs 8.9%±0.9%; p=0.97), and IL-17 (2.17%±0.4% vs 1.0%±0.4%; p= 0.10) positive cells was not different between KEAP1 edited and control Treg cells.
Discussion
CRISPR based genome editing allows specific gene targeting and is revolutionizing the field of experimental medicine (19). Our present study demonstrates that CRISPR technology can be used to successfully engineer both immortalized and primary human T cells for therapeutic enhancement of NRF2 regulated antioxidant capacity. The rationale to edit T lymphocyte KEAP1 was based on our previous data in T cell specific Keap1 deficient mice that demonstrated significant upregulation of Nrf2 regulated antioxidant gene expression and protection from IR induced AKI (5) and myocardial injury (unpublished observation). Adoptive transfer of T cells with augmented antioxidant activity protected kidneys from AKI and improved survival in WT mice, indicating that transfer of engineered T cells with enhanced antioxidant activity could potentially be used as immune cell based therapy for various oxidative stress driven diseases.
In this study, we first optimized KEAP1 editing conditions in Jurkat T cells and subsequently targeted KEAP1 in primary human T cells as well as purified Treg cells. Present experimental conditions and enrichment strategy using fluorescently labeled tracrRNA resulted in KEAP1 editing in about 55% primary T cells and 63% Treg cells. Despite modest editing efficiency, we observed significant augmentation of NRF2 target gene expression in total T cell as well as purified Treg cell population. This increase in basal antioxidant gene expression was expected and comparable to that of Keap1 deficient T cells from our conditional knock out mice and other Nrf2 activation studies using pharmacologic activators (6, 20-22). We found multiple antioxidant genes (NQO1, HO1 and GCLM) upregulated 72h after Cas9:gRNA delivery but only NQO1 remained elevated at 120h time point indicating that the edited cells attain a more stable transcriptional status following an initial surge under basal conditions. In addition to an increased antioxidant gene expression, we observed distinct phenotypic (increased CD4 and reduced CD8 cell frequency in total T cell population) and functional (high CD25 and CD69 and low IL-17 positive cell frequency in total T cells and high CD69 and IL-10 positive cell frequency in purified Treg cells) differences between the KEAP1 edited primary T cell and control cells. The exact reason for increased CD4 and reduced CD8 frequency is not clear but could be due to better electroporation of ATTO 550 labeled RNP complex in CD4 cells than CD8 cells. Although, some studies suggest attenuated pro-inflammatory response upon NRF2 activation, Morzadec et al. found no effect of NRF2 activation on cytokine secretion by human T cells (20, 23, 24). Thus, it is not entirely clear how KEAP1/NRF2 modulates cytokine expression in T cells, however KEAP1 editing in primary T cells may induce regulatory features that increase anti-inflammatory and suppressive functions.
CRISPR based KEAP1 editing may induce additional changes that were not investigated in this study. For example, reactive oxygen species (ROS) production by activated T cells triggers glutathione (GSH) response to scavenge increasing ROS and prevent cellular damage (25) Therefore, KEAP1 editing could modulate metabolic integration and reprogramming during inflammatory T cell responses. Another possible affect could be on PD-1 that inhibits T cell activation (26). Furthermore, KEAP1 editing may affect epigenetic elements such as histone deacetylases (HDACs) and histone acetyltransferases that modulate suppressive function, mainly of Treg cells. In addition to these, KEAP1 editing may modulate T cell differentiation via Notch signaling. Notch signaling has been shown to be a critical regulator of T cell differentiation that was found to improve hematopoietic progenitor stem cell (HSPC) function and myelosuppression following radiation exposure in an Nrf2 dependent manner (27).
Apart from T cell specific KEAP1 editing, CRISPR/Cas9 based KEAP1 editing can be carried out in additional cell types. KEAP1 has been shown to modulate metabolic shift from oxidative to glycolytic energy production during iPSC reprogramming (28). Therefore, editing of KEAP1 using CRISPR technology may be a useful tool for controlling iPSC reprogramming. Furthermore, CRISPR based gene editing appears to produce more robust functional effects (29). KEAP1 ablation using CRISPR technology has also been found to modulate sensitivity of lung cancer for kinase targeted therapy (30). In a recent study Zagorski et al. employed CRISPR/Cas9 to delete NRF2 in Jurkat T cell line and found significant reduction in NQO1 mRNA but no immunological effect at steady state (24).
Even though we did not observe any detectable off-target effects of CRISPR based KEAP1 editing, this study lacks a comprehensive sequence based analysis of non-specific effects in KEAP1 edited T cells. Lack of suitable animal models further limits functional characterization of these KEAP1 edited human T cells in in vivo disease models. Moreover, this study lacks investigation into NRF2 independent effects of KEAP1 editing. In spite of its limitations CRISPR based KEAP1 editing appears to be promising for permanent and specific NRF2 activation compared to reversible and non-specific pharmacologic or small interfering RNA (siRNA) based approaches (31). Successful expansion of KEAP1 edited T cells is expected to result in the development of a novel T cell based antioxidant therapy.
Supplementary Material
Figure 2.
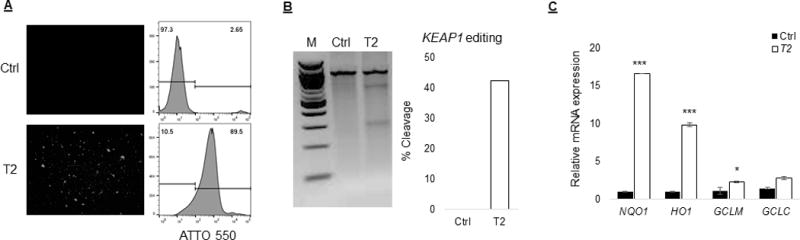
KEAP1 editing resulted in significant NRF2 activation in primary T cells. We used target site 2 (T2) specific RNP complex containing ATTO 550 labeled tracrRNA in primary T cells (n=3). ATTO 550 labeled tracrRNA forms an integral part of the RNP complex and provided better assessment of transfection efficiency. (A) Microscopic and flow cytometric analysis of primary T cell 24h after electroporation showed upto ~80% cells received RNP complex. (B) KEAP1 editing analysis using Surveyor mutation assay showed cleavage in about 40% cells that received RNP complex in comparison to control cells. (C) There was a significant increase in NQO1 (p≤0.01; 16 fold), HO1 (p≤0.01; 9 fold) and GCLM (p≤0.04; 2 fold) mRNA expression in primary T cell, 72h after RNP complex electroporation, compared to control cells.
Acknowledgments
The authors are thankful to all the volunteers for donating blood samples for this study and a research gift from Mr. Rogelio Miro of Panama.
This work was funded by grants from National Institutes of Health (RO1 DK111209) and Living Legacy Foundation to HR
References
- 1.Clever D, Roychoudhuri R, Constantinides MG, Askenase MH, Sukumar M, Klebanoff CA, Eil RL, Hickman HD, Yu Z, Pan JH, Palmer DC, Phan AT, Goulding J, Gattinoni L, Goldrath AW, Belkaid Y, Restifo NP. Oxygen Sensing by T Cells Establishes an Immunologically Tolerant Metastatic Niche. Cell. 2016;166:1117–1131.e14. doi: 10.1016/j.cell.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Ciuceis C, Agabiti-Rosei C, Rossini C, Airo P, Scarsi M, Tincani A, Tiberio GA, Piantoni S, Porteri E, Solaini L, Duse S, Semeraro F, Petroboni B, Mori L, Castellano M, Gavazzi A, Agabiti-Rosei E, Rizzoni D. Relationship between different subpopulations of circulating CD4+ T lymphocytes and microvascular or systemic oxidative stress in humans Relationship between different subpopulations 344 of circulating CD4+ T lymphocytes and microvascular or systemic oxidative stress in humans. Blood Press. 2017:1–9. doi: 10.1080/08037051.2017.1292395. [DOI] [PubMed] [Google Scholar]
- 3.Burne MJ, Daniels F, El Ghandour A, Mauiyyedi S, Colvin RB, O’Donnell MP, Rabb H. Identification of the CD4(+) T cell as a major pathogenic factor in ischemic acute renal failure. J Clin Invest. 2001;108:1283–1290. doi: 10.1172/JCI12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandolfo MT, Jang HR, Bagnasco SM, Ko GJ, Agreda P, Satpute SR, Crow MT, King LS, Rabb H. Foxp3+ regulatory T cells participate in repair of ischemic acute kidney injury. Kidney Int. 2009;76:717–729. doi: 10.1038/ki.2009.259. [DOI] [PubMed] [Google Scholar]
- 5.Noel S, Martina MN, Bandapalle S, Racusen LC, Potteti HR, Hamad AR, Reddy SP, Rabb H. T Lymphocyte-Specific Activation of Nrf2 Protects from AKI. J Am Soc Nephrol. 2015;26:2989–3000. doi: 10.1681/ASN.2014100978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noel S, Hamad AR, Rabb H. Reviving the promise of transcription factor Nrf2-based therapeutics for kidney diseases. Kidney Int. 2015;88:1217–1218. doi: 10.1038/ki.2015.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu QQ, Wang Y, Senitko M, Meyer C, Wigley WC, Ferguson DA, Grossman E, Chen J, Zhou XJ, Hartono J, Winterberg P, Chen B, Agarwal A, Lu CY. Bardoxolone methyl (BARD) ameliorates ischemic AKI and increases expression of protective genes Nrf2, PPARgamma, and HO-1. Am J Physiol Renal Physiol. 2011;300:F1180–92. doi: 10.1152/ajprenal.00353.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright MM, Kim J, Hock TD, Leitinger N, Freeman BA, Agarwal A. Human haem oxygenase-1 induction by nitro-linoleic acid is mediated by cAMP, AP-1 and E-box response element interactions. Biochem J. 2009;422:353–361. doi: 10.1042/BJ20090339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ano S, Panariti A, Allard B, O’Sullivan M, McGovern TK, Hamamoto Y, Ishii Y, Yamamoto M, Powell WS, Martin JG. Inflammation and airway hyperresponsiveness after chlorine exposure are prolonged by Nrf2 deficiency in mice. Free Radic Biol Med. 2017;102:1–15. doi: 10.1016/j.freeradbiomed.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han X, Liu Z, Jo MC, Zhang K, Li Y, Zeng Z, Li N, Zu Y, Qin L. CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci Adv. 2015;1:e1500454. doi: 10.1126/sciadv.1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hultquist JF, Schumann K, Woo JM, Manganaro L, McGregor MJ, Doudna J, Simon V, Krogan NJ, Marson A. A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Rep. 2016;17:1438–1452. doi: 10.1016/j.celrep.2016.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 17.Liang X, Potter J, Kumar S, Ravinder N, Chesnut JD. Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J Biotechnol. 2017;241:136–146. doi: 10.1016/j.jbiotec.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Su S, Zou Z, Chen F, Ding N, Du J, Shao J, Li L, Fu Y, Hu B, Yang Y, Sha H, Meng F, Wei J, Huang X, Liu B. CRISPR-Cas9-mediated disruption of PD-1 on human T cells for adoptive cellular therapies of EBV positive gastric cancer. Oncoimmunology. 2016;6:e1249558. doi: 10.1080/2162402X.2016.1249558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyagi A, Lu A, Humphreys BD. Gene Editing: Powerful New Tools for Nephrology Research and Therapy. J Am Soc Nephrol. 2016;27:2940–2947. doi: 10.1681/ASN.2016020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rockwell CE, Zhang M, Fields PE, Klaassen CD. Th2 skewing by activation of Nrf2 in CD4(+) T cells. J Immunol. 2012;188:1630–1637. doi: 10.4049/jimmunol.1101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turley AE, Zagorski JW, Rockwell CE. The Nrf2 activator tBHQ inhibits T cell activation of primary human CD4 T cells. Cytokine. 2015;71:289–295. doi: 10.1016/j.cyto.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M, Reddy NM, Higbee EM, Potteti HR, Noel S, Racusen L, Kensler TW, Sporn MB, Reddy SP, Rabb H. The Nrf2 triterpenoid activator, CDDO-imidazolide, protects kidneys from ischemia-reperfusion injury in mice. Kidney Int. 2014;85:134–141. doi: 10.1038/ki.2013.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morzadec C, Macoch M, Sparfel L, Kerdine-Romer S, Fardel O, Vernhet L. Nrf2 expression and activity in human T lymphocytes: stimulation by T cell receptor activation and priming by inorganic arsenic and tert-butylhydroquinone. Free Radic Biol Med. 2014;71:133–145. doi: 10.1016/j.freeradbiomed.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Zagorski JW, Maser TP, LIby KT, Rockwell CE. Nrf2-dependent and -independent effects of tBHQ, CDDO-Im, and H2O2 in human Jurkat T cells as determined by CRISPR/Cas9 gene editing. J Pharmacol Exp Ther. 2017 doi: 10.1124/jpet.116.238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mak TW, Grusdat M, Duncan GS, Dostert C, Nonnenmacher Y, Cox M, Binsfeld C, Hao Z, Brustle A, Itsumi M, Jager C, Chen Y, Pinkenburg O, Camara B, Ollert M, Bindslev-Jensen C, Vasiliou V, Gorrini C, Lang PA, Lohoff M, Harris IS, Hiller K, Brenner D. Glutathione Primes T Cell Metabolism for Inflammation. Immunity. 2017;46:675–689. doi: 10.1016/j.immuni.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 26.Pan F, Fan H, Liu Z, Jiang S. T cell signaling targets for enhancing regulatory or effector function. Sci Signal. 2012;5:pe32. doi: 10.1126/scisignal.2003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JH, Thimmulappa RK, Kumar V, Cui W, Kumar S, Kombairaju P, Zhang H, Margolick J, Matsui W, Macvittie T, Malhotra SV, Biswal S. Nrf2-mediated Notch pathway activation enhances hematopoietic reconstitution following myelosuppressive radiation. J Clin Invest. 2014;124:730–741. doi: 10.1172/JCI70812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawkins KE, Joy S, Delhove JM, Kotiadis VN, Fernandez E, Fitzpatrick LM, Whiteford JR, King PJ, Bolanos JP, Duchen MR, Waddington SN, McKay TR. Nrf2 Orchestrates the Metabolic Shift during Induced Pluripotent Stem Cell Reprogramming. Cell Rep. 2016;14:1883–1891. doi: 10.1016/j.celrep.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gonen M, Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krall EB, Wang B, Munoz DM, Ilic N, Raghavan S, Niederst MJ, Yu K, Ruddy DA, Aguirre AJ, Kim JW, Redig AJ, Gainor JF, Williams JA, Asara JM, Doench JG, Janne PA, Shaw AT, McDonald RE, Iii, Engelman JA, Stegmeier F, Schlabach MR, Hahn WC. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife. 2017;6 doi: 10.7554/eLife.18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt HH, Stocker R, Vollbracht C, Paulsen G, Riley D, Daiber A, Cuadrado A. Antioxidants in Translational Medicine. Antioxid Redox Signal. 2015;23:1130–1143. doi: 10.1089/ars.2015.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


