Abstract
Host-bacterial interactions at mucosal surfaces require recognition of the bacteria by host cells enabling targeted responses to maintain tissue homeostasis. It is now well recognized that an array of host-derived pattern recognition receptors (PRRs), both cell-bound and soluble, are critical to innate immune engagement of microbes via microbial-associated molecular patterns (MAMP). This report describes the use of a nonhuman primate model to evaluate changes in the expression of these sensing molecules related to aging in healthy gingival tissues. Macaca mulatta aged 3-24 years were evaluated clinically and gingival tissues obtained, RNA isolated and microarray analysis conducted for gene expression of the sensing pattern recognition receptors (PRRs). The results demonstrated increased expression of various PRRs in healthy aging gingiva including extracellular (CD14, CD209, CLEC4E, TLR4), intracellular (NAIP, IFIH1, DAI) and soluble (PTX4, SAA1) PRRs. Selected PRRs were also correlated with both bleeding on probing (BOP) and pocket depth (PD) in the animals. These findings suggest that aged animals express altered levels of various PRRs that could affect the ability of the tissues to interact effectively with the juxtaposed microbial ecology, presumably contributing to an enhanced risk of periodontitis even in clinically healthy oral mucosal tissues with aging.
Keywords: Aging, oral mucosa, microbial sensing, pattern recognition receptors, nonhuman primates, periodontitis
INTRODUCTION
Mucosal tissues are colonized by an extremely dense and diverse microbiota of commensal bacteria, as well as occasionally having to interact with pathogenic microorganisms. These sites continuously sample foreign material via various cells types, including macrophages (mФ) and dendritic cells (DCs). These are innate immune cells within the skin and mucosa, including oral and gingival epithelium, that respond rapidly to infection, carrying crucial information about the infection to lymph nodes to trigger an immune response (Kopitar, et al., 2006, Nestle, et al., 1994, Makino, et al., 2001, Jotwani, et al., 2001, Cutler and Jotwani, 2006). Historically, both of these APC types were identified to effectively engage microbes using a repertoire of pattern recognition receptors (PRRs)(Hemmi and Akira, 2005, Benko, et al., 2008), which recognize distinct classes of microorganism-associated molecular patterns (MAMPs), including a range of bacterial, viral, and fungal pathogens, through engagement of LPS, LTA, and nucleic acid (e.g., CpG, DNA, dsRNA) ligands(Blach-Olszewska, 2005, Wollenberg, et al., 2002, Kawai and Akira, 2006, Kumar, et al., 2009). However, it is now recognized that these PRRs exist on virtually all types of host cells with variation in the quality and quantity that are expressed on particular cell-types (Hajishengallis, et al., 2012). Moreover, the literature demonstrates that the PRRs exist as cell associated [e.g. Toll-like receptors (TLRs), nucleotide-binding oligomerization domains (NODs)] and soluble PRRs (e.g. C-reactive protein, serum amyloid A) that are critical to innate immunity (Mogensen, 2009) and help link innate and adaptive immune responses (Werling and Jungi, 2003, Kumar, et al., 2011).
While there are numerous reports regarding PRR expression and functions with aging, generally the existing data remains somewhat unclear regarding aging effects on these critical members of the innate immune system (Shaw, et al., 2013, Shaw, et al., 2011). Studies in mice and humans have suggested that the expression and functions of various TLR and other PRRs such as RIG-I and NLRP3 are altered with aging, usually decreasing in level and/or functional capabilities. Beyond adaptive immune senescence that has been uniformly described in aging, these findings contribute to the concept of “inflamm-aging” that emphasizes a dysregulation and exacerbated destructive inflammatory responses with aging (Shaw, et al., 2013, Franceschi, et al., 2000, Fulop, et al., 2013). Dunston and Griffiths (Dunston and Griffiths, 2010) suggested that a poor inflammatory response via TLR activation still allowed dysregulated inflammation in aging related to ineffective clearance of pathogens and less well controlled activation of macrophages. Interestingly, Sun et al. (Sun, et al., 2012) demonstrated a less LPS tolerigenic macrophage in aging with challenge by an oral pathogen, P. gingivalis, apparently related to altered TLR2 and TLR expression/function.
The periodontium of the oral cavity is a multi-component tissue comprised of epithelium, connective tissue, a capillary vascular bed, and alveolar bone. Disease of this structure, ie. periodontitis, results in chronic inflammation (measured by redness and bleeding), soft tissue destruction (described as clinical attachment loss) and bone resorption, that is generally reflected in increased probing pocket depth. The current paradigm of periodontitis is a chronic destructive inflammatory response with oral bacterial biofilms triggering an influx of host immune cells and release of cytokines/chemokines and inflammatory lipids that have direct effects on the integrity of the tissues that comprise the periodontium. A consistent clinical finding in periodontitis is substantial increases in incidence, extent, and severity of disease with aging (Baelum and Lopez, 2013, Eke, et al., 2012). Moreover, a vast literature exists describing the range of molecular inflammatory, innate immune and adaptive immune responses in human health and disease (Ebersole, et al., 2013, Bartold and Van Dyke, 2013, Hajishengallis, 2015, Garlet, et al., 2014, Souza and Lerner, 2013), as well as a number of animal models that have been used to assess causality (de Molon, et al., 2013, Graves, et al., 2012, Oz and Puleo, 2011, Madden and Caton, 1994, Holt, et al., 1988).
This report uses a nonhuman primate model of naturally-occurring periodontitis to document alterations in expression of these microbial sensing molecules. The hypothesis to be tested was that aging effects on clinically healthy gingival tissues are detected within the pattern of PRR expression and may be related to the aging susceptibility for this mucosal tissue disease.
METHODS
Nonhuman primate model and Oral Clinical Evaluation
Rhesus monkeys (Macaca mulatta) (n=41; 21 females and 20 males) housed at the Caribbean Primate Research Center (CPRC) at Sabana Seca, Puerto Rico, were used in these studies. Healthy animals (5–14/group) were distributed by age into four groups: ≤3 years (young), 3–7 years (adolescent), 12–16 years (adult) and 18–23 years (aged). The nonhuman primates are typically fed a 20% protein, 5% fat, and 10% fiber commercial monkey diet (diet 8773, Teklad NIB primate diet modified: Harlan Teklad). The diet is supplemented with fruits and vegetables, and water is provided ad libitum in an enclosed corral setting.
A protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Puerto Rico, enabled anesthetized animals to be examined for clinical measures of periodontal including probing pocket depth (PD), and bleeding on probing (BOP) as we have described previously (Ebersole, et al., 2008).
Tissue sampling and gene expression microarray analysis
A buccal gingival sample from healthy tissue from the premolar/molar maxillary region of each animal was taken using a standard gingivectomy technique, and maintained frozen in RNAlater solution. Total RNA was isolated from each gingival tissue using a standard procedure as we have described and tissue RNA samples submitted to the microarray core to assess RNA quality and analyze the transcriptome using the GeneChip® Rhesus Macaque Genome Array (Affymetrix) or GeneChip® Rhesus Gene 1.0 ST Array (Affymetrix) (Meka, et al., Gonzalez, et al., 2011). Individual samples were used for gene expression analyses. Table 1 lists the microbial sensing gene set examined in this report.
Table 1.
Gene expression targets for microbial sensing molecules
| Gene ID | Gene Title |
|---|---|
| Surface Cell Associated PRRs | |
| CD14 | Binds to monomeric lipopolysaccharide and delivers it to the MD-2/TLR4 complex; also soluble molecule |
| CD209 (DC-SIGN) | Surface of immature dendritic cells (DCs) and involved in initiation of primary immune response |
| CLEC7A | C-Type Lectin Domain Family 7, Member A, beta-1,3-linked and beta-1,6-linked glucans from fungi |
| CLEC4E | C-Type Lectin domain family 4, member E, mycobacteria is via trehalose 6,6′-dimycolate; SAP130, a nuclear protein that is released by dead or dying cells |
| CLEC4M (L-SIGN) | C-Type Lectin Domain Family 4, Member M, parasites to viruses |
| CLEC6A (Dectin-2) | C-Type Lectin Domain Family 6, Member A, alpha-mannans on C.albicans hypheas |
| COLEC12 | Collectin Sub-Family Member 12, Scavenger receptor for phagocytosis of Gram-positive, Gram-negative bacteria and yeast |
| FCN2 | Ficolin (Collagen/Fibrinogen Domain Containing Lectin) 2 (Hucolin), Complement-activating lectin, phagocytosis of S.typhimurium by neutrophils |
| MARCO | Macrophage Receptor With Collagenous Structure, class A scavenger receptor for both Gram-negative and Gram-positive bacteria |
| MRC1 | Mannose Receptor, C Type 1, bind high-mannose structures on the surface of potentially pathogenic viruses, bacteria, and fungi for phagocytosis |
| TLR1 | Toll-like receptor 1, diacylated and triacylated lipopeptides |
| TLR2 | Toll-like receptor 2, bacterial lipoproteins and other microbial cell wall components |
| TLR4 | Toll-like receptor 4, bacterial lipopolysaccharide (LPS) |
| TLR5 | Toll-like receptor 5, bacterial flagellins |
| TLR6 | Toll-like receptor 6, Gram-positive bacteria and fungi |
| Intracellular Associated PRRs | |
| AIM2 | Cytosolic double-stranded DNA |
| ARHGEF2 | Rho/Rac Guanine Nucleotide Exchange Factor (GEF) 2, intracellular sensing system along with NOD1 for the detection of microbial effectors during cell invasion |
| IFIH1 (MDA5) | Interferon induced with helicase C domain 1, RIG-1-like receptor family, viral sensor |
| LGP2 (DEXH58) | DEXH (Asp-Glu-X-His) Box Polypep tide 58, RIG-1-like receptor family, viral sensor |
| NAIP (BIRC1) | Baculoviral IAP repeat-containing protein 1, effects apoptosis |
| NLRC4 (IPAF) | NLR family CARD domain-containing protein 4, inflammasome |
| NOD1 | Nucleotide-binding oligomerization domain 1, intracellular bacterial lipopolysaccharides (LPS), senses peptidoglycan (PGN)-derived muropeptides |
| NOD2 | Nucleotide-binding oligomerization domain 2, intracellular bacterial lipopolysaccharides (LPS) by recognizing the muramyl dipeptide |
| RIG-1 (DDX58) | Retinoic acid-inducible gene 1, RIG-1-like receptor family, viral sensor |
| TLR3 | Toll-like receptor 3, nucleotide-sensing for double-stranded RNA |
| TLR7 | Toll-like receptor 7, nucleotide-sensing for single-stranded RNA |
| TLR8 | Toll-like receptor 8, G-rich oligonucleotides |
| TLR9 | Toll-like receptor 9, nucleotide-sensing for unmethylated cytidine-phosphate-guanosine (CpG) dinucleotides |
| ZBP1/DAI | Z-DNA Binding Protein 1, cytoplasmic sensor binds to foreign DNA and induces type-I interferon |
| Soluble PRRs | |
| CRP (PTX1) | C-reactive protein, promotes agglutination, bacterial capsular swelling, phagocytosis and complement fixation |
| FCN1 | Ficolin (Collagen/Fibrinogen Domain Containing) 1, Complement-activating lectin, 9-O-acetylated 2–6-linked sialic acid derivatives and to various glycans |
| MBL2 | Mannose-Binding Lectin (Protein C) 2, Soluble, soluble mannose-binding lectin or mannose-binding protein found in serum |
| PTX3 | Pentraxin 3, Long, mediating agglutination, complement activation, and opsonization |
| PTX4 | Pentraxin 4, Long, mediating agglutination, complement activation, and opsonization |
| SAA1 | Serum Amyloid A1 |
Based upon the microarray outcomes we selected 5 genes and performed a qPCR analysis using a standard technique in our laboratory employing a Roche 480 LightCycler (Gonzalez, et al., 2014). Primers were prepared for SAA1 (forward - CAGCGATGCCAGAGAGAATATC; reverse – CAGCGATGCCAGAGAGAATATC; amplicon 121 bp), CD14 (forward – GCCCTAAACTCCCTCAATCTG; reverse – CAGTCTGTTGCAGCTGAGAT; amplicon 99 bp), NAIP (forward - GGCTCTTGGATGCAGATGATA; reverse – ATGATTGGAGAGAACGGCAATA; amplicon 112 bp), TLR2 (forward - GATGCTGCCATTCTTGTTCTTC; reverse – CAGGTAGGTCTTGGTGTTCATT; amplicon 99 bp) and ZBP1/DAI (forward – TCAGCCCATCACTCGAAAC; reverse – ATGAGGCTTCATCCACATAGTC; amplicon 113 bp) genes, designed using software PrimerQuest at Integrated DNA Technologies website (www.idtdna.com) and were synthesized by Integrated DNA Technologies, Inc (Coralville, IA). The level of message was determined (Gonzalez, et al., 2013) and those levels compared across the RNA samples prepared from each of the age groups.
Data analysis
Normalization of values across the chips was accomplished through signal intensity standardization across each chip using Affymetrix PLIER algorithm. The arrays contained matched and mismatched pairs allowing the MAS 5 algorithm to be used. For each gene we first determined differences in expression across the groups using ANOVA (version 9.3, SAS Inc., Cary, NC). The healthy aged tissues were then compared to the other age groups using a t-test and accepting a p-value <0.05 for significance. Because of the cost of these types of nonhuman primate experiments and availability of primates of the various ages, we did not have sufficient samples to identify if the relationship between age and gene expression could be treated using a linear model, thus the subjects were classified and ANOVA was used for analysis. The choice of Least Significant Difference for multiple comparisons (ANOVA followed by t-tests) provided maximum power given our necessarily small sample sizes. We determined a correlation with aging in healthy tissues using a Spearman Rank correlation analysis that was fit to the gene expression by age. A p-value ≤ 0.05 was used to evaluate the significance of the correlation. The data has been uploaded into the ArrayExpress data base (www.ebi.ac.uk) under accession number: E-MTAB-1977.
RESULTS
While all of the animals were categorized as periodontally healthy based upon mouth mean bleeding index and pocket depths, across the age range variations in these clinical parameters were noted. Figure 1 summarizes the relationship of these clinical parameters to age of the animals. As is shown, there was a negligible relationship between age and bleeding; however, the pocket depth measures were significantly positively correlated with age.
Figure 1.
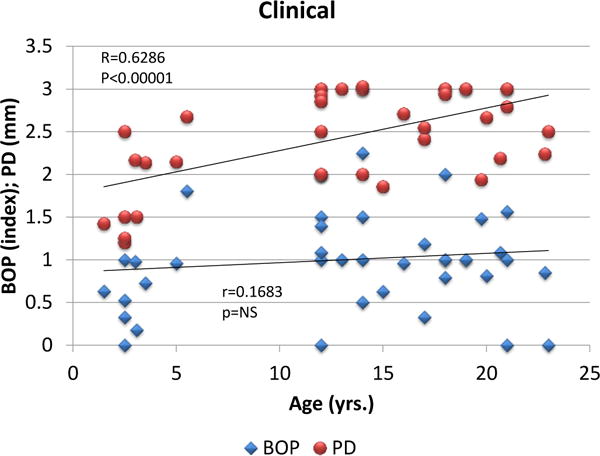
Correlation of mouth mean bleeding on probing (BOP) and pocket depth (PD) measures related to age of the animals. Each point represents one animal.
We then evaluated the expression of the array of PRRs particularly related to changes that could be detected in healthy gingival tissues from aged animals versus the expression patterns in all other age groups. Figure 2 shows that surface PRRs, including CD14, CD209, CLEC4E and TLR4 were significantly increased in the healthy aging gingival tissues. Intracellular receptors IFIH1, NAIP and ZBP1/DAI were also significantly increased in aging. An array of soluble PRRs was also examined in health with aging and the changes observed included a significant up-regulation and high signal for SAA1 in the healthy aged tissues and a decrease in expression of PTX4. Table 2 provides a summary of the qPCR analysis of selected genes as a validation of the microarray results. The findings demonstrate an agreement in level differences for 4/5 genes using these 2 techniques.
Figure 2.
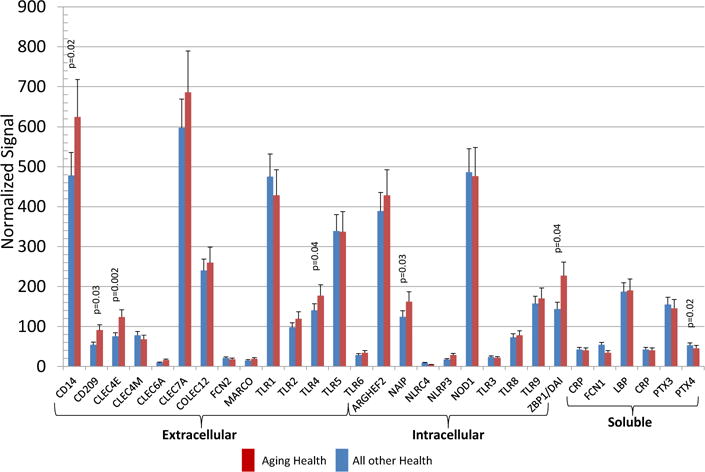
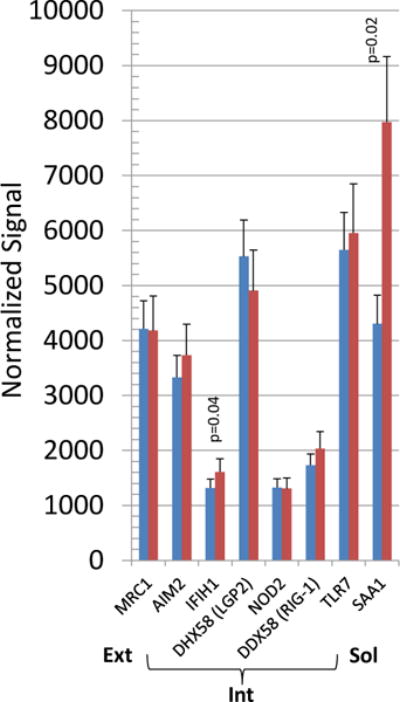
Distribution of gene expression of PRRs in healthy aging gingival tissues compared to other age groups. The bars denote the mean levels in healthy aged (n=6), and all other healthy (n-17). Significant differences are denoted by p-values.
Table 2.
Comparison of gene expression profiles using qPCR and microarray analyses. Values represent fold-difference compared to Adult Healthy tissue message levels assigned a value of 1.0. GAPDH was employed as a housekeeping gene for normalization of the results.
| GENE ID | AGED HEALTH |
|---|---|
|
| |
| SAA1 | |
| qPCR | 6.74 ± 1.14 |
| GeneChip | 2.71 ± 1.11 |
|
| |
| CD14 | |
| qPCR | 1.73 ± 0.46 |
| GeneChip | 1.28 ± 0.30 |
|
| |
| NAIP | |
| qPCR | 5.67 ± 2.34 |
| GeneChip | 2.16 ± 0.28 |
|
| |
| TLR2 | |
| qPCR | 2.44 ± 1.75 |
| GeneChip | 1.14 ± 0.22 |
|
| |
| ZBP1/DAI | |
| qPCR | 2.38 ± 0.83 |
| GeneChip | 2.59 ± 1.04 |
Figure 3 depicts the distribution of PRR sensing gene expression levels plotted for significance and relative change comparing healthy aged tissues with healthy gingival tissues from all other age groups. The results identify a select group of PRRs that are up-regulated in gingival tissues from healthy aging animals, with a more limited number that appeared to be down-regulated.
Figure 3.
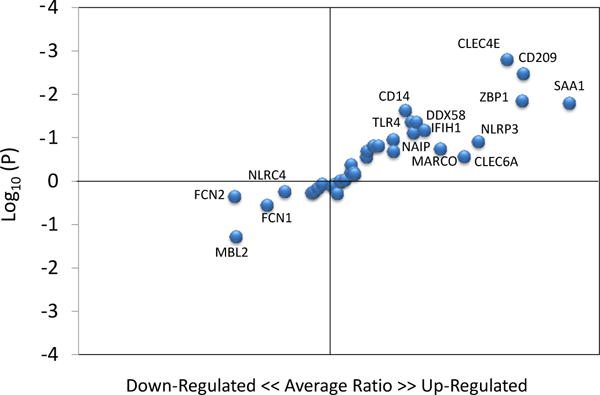
Identification of gene expression profiles in healthy gingival tissues comparing aged tissues to all other age groups. Points denote individual genes plotted as fold-difference with aging and p-value.
Figure 4 provides a depiction of the PRR gene expression profiles that demonstrated a significant correlation with age in healthy gingival tissues. Of the 38 genes, 4 genes (CD14, CLEC6A, CLEC7A, ZPB1/DAI) were all significantly positively correlated with aging in healthy tissues. Additionally, FCN1, CLEC4M, and PTX4 were negatively correlated demonstrating lower levels of expression in the aged gingival tissues.
Figure 4.
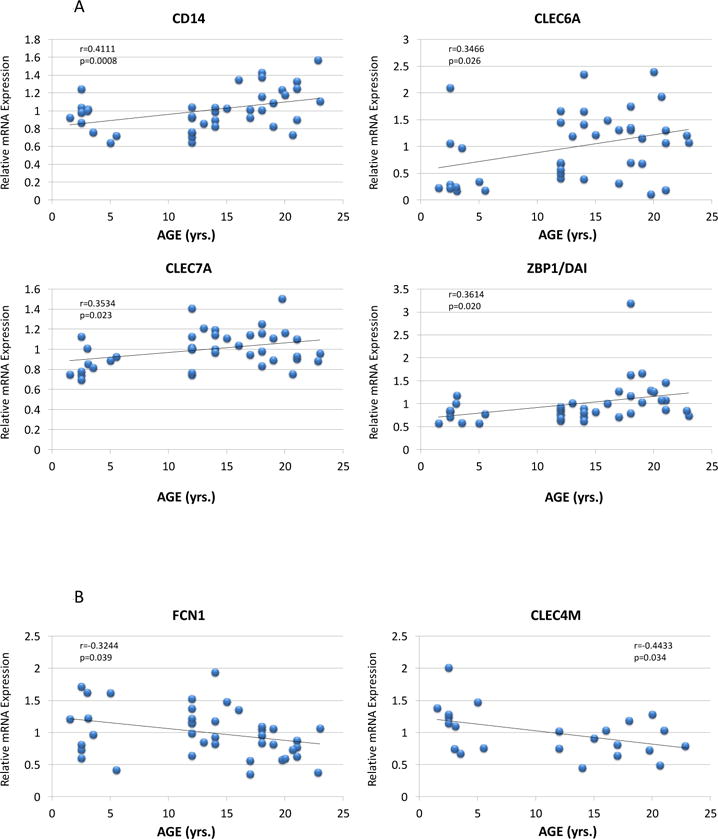
Correlation of PRR expression in healthy gingival tissues for genes that significantly correlated with age. Each point denotes the values from 1 animal (n=41). Genes CLEC4M (Rhesus Macaque Genome Array; n=23) and PTX4 (Rhesus Gene 1.x ST Exon Array; n=18) are only currently annotated for one of the microarray chips that were used in the experiments.
The relationship of the gene expression profiles of these microbial sensing PRRs to clinical presentation of the animals over the lifespan was examined. Generally very limited correlations were observed between the expression of any of the PRRs and the clinical parameters of bleeding on probing (BOP) and pocket depth (PD) within the healthy gingiva from young to aged animals. Only CLEC7A and ZBP1/DAI demonstrated a significant relationship with pocket depth, and NAIP and NOD2 with bleeding on probing indices in the healthy gingival tissues (Figure 5).
Figure 5.
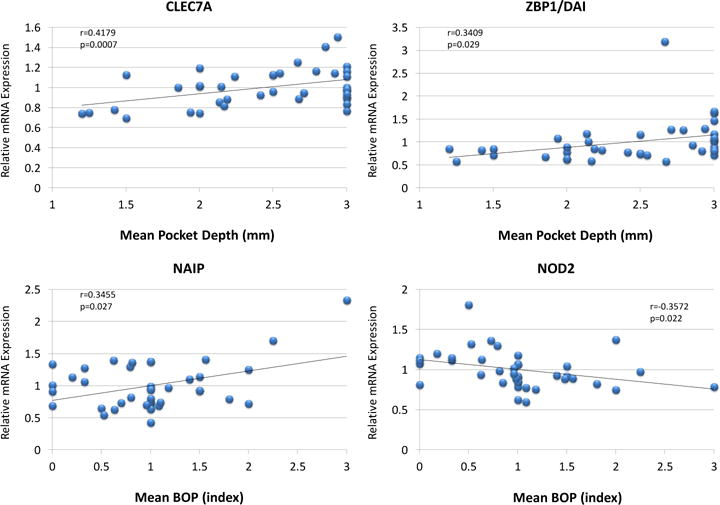
Correlation of PRR gene expression in healthy gingival tissues related to mouth mean PD or BOP. Each point denotes the values for 1 animal (n=41).
Recognizing that the response to a complex stimulus in the gingival tissues is a multifaceted and interconnected tuning of the systems biology to manage the noxious bacterial challenge, we interfaced the various findings to create a schematic network of relationships among the PRRs in healthy gingival tissues related to the aging process. For this comparison we calculated correlations of gene expression across all PRRs as they related to aging of the animals. The results in Figure 6 demonstrate that 5 of the PRRs (ZBP1, TLR1, TLR7, AIM2, COLEC12) are highly networked with numerous other PRR gene expression profiles (5 or more) in the healthy gingival tissues. An additional, 11 genes were also significantly correlated with expression of multiple (3 or 4) other PRRs as noted by the network connecting lines in the figure. Interestingly, TLR7 was somewhat unique in that it was exclusively negatively correlated with other PRRs in the healthy tissues. As such, the expression of 21 of the PRR genes appeared rather independent of this larger family of microbial recognition molecules in the gingival tissues.
Figure 6.
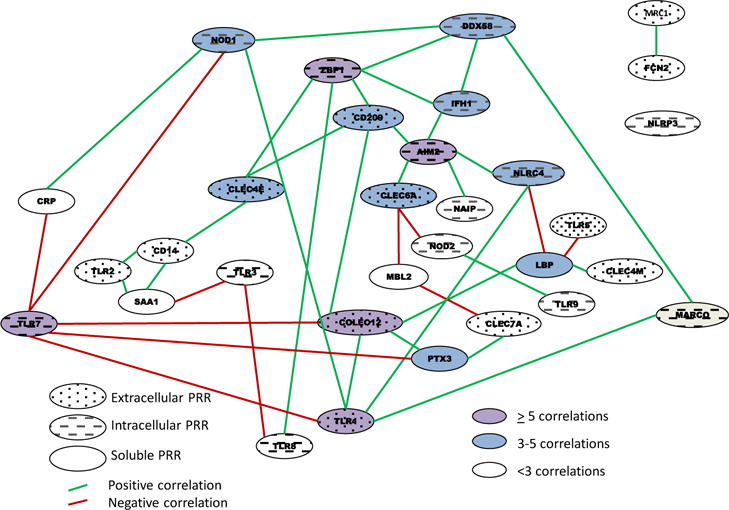
Schematic of network of gene expression correlations among the array of PRRs in healthy gingival tissues. The characteristics of the PRRs are identified by speckled (surface PRRs), slanted lines (intracellular PRRs), and solid (soluble PRRs) ovals. The green lines denote a significantly positive (at least p<0.05) correlation and red line denotes a significant negative correlation in expression of 2 genes. Pink highlights are genes with 5 or more correlations, blue highlights signify genes with 3–4 correlations, and white highlights denote 1–2 correlations.
DISCUSSION
Effective innate immune responses, both locally and systemically, depend upon the availability of an array of microbial pattern recognition receptors (PRR) that interact with bacteria, viruses, and fungi through specific ligands (PAMPs, MAMPs) that are structural motifs of their cell membranes, cell walls, or genomes (Kumar, et al., 2011). The engagement of these PRRs, both cell-associated and soluble are critical for management of microbial challenge of the host and are directly involved in downstream signaling of an array of host innate immune, inflammatory, and even adaptive immune cells and molecules (Mogensen, 2009). While the PRRs are nonspecific, in that they engage the structural motifs from a range of microbes, there is some specificity in that individual PRRs tend to recognize specific motifs that are unique to bacteria, viruses or fungi (Kolli, et al., 2013, Kumar, et al., 2013, Min, et al., 2013).
A major family of the cell-associated PRRs are the Toll-like receptors (TLRs), with 10 TLR genes identified in humans (TLR1-10) (Qian and Cao, 2013). Each of the TLRs has a somewhat different microbial molecular pattern with which it engages TLR1’s ligand is multiple triacyl lipopeptides on bacteria. TLR2 forms heterodimers with TLR1, TLR6 or non-TLR receptors [e.g. CD36, CD14, CLEC7A (Dectin-1)] to engage specific molecular ligand structures that include, mycobacterial and mycoplasma diacylated lipopeptides, lipoteichoic acid, triacylated lipopeptides, β-glucan of fungi, and heat shock protein 70 (van Bergenhenegouwen, et al., 2013). TLR3, TLR7, and TLR8 all appear to primarily engage nucleic acids related to detection of viral infections (Qian and Cao, 2013, Frazao, et al., 2013). The primary ligand for TLR4 has been identified as the lipopolysaccharides (LPS) from most Gram-negative bacteria, although TLR4 can bind to heat shock proteins, fibrinogen, and hyaluronic acid fragments of the host (Park and Lee, 2013). TLR5 has specificity for flagellin of bacteria (Qian and Cao, 2013, Frazao, et al., 2013). Finally, TLR9 also recognizes unique patterns of nucleic acids, specifically binding to unmethylated CpG sequences that are found in bacteria and viruses (Qian and Cao, 2013, Frazao, et al., 2013). Generally, TLRs were not altered in healthy aging gingival tissues, except, TLR4 levels were up-regulated in the aging tissues. In contrast, the TLRs were significantly correlated with an array of PRRs in the tissues of the animals, with an interesting finding of TLR7 in particular being negatively correlated with multiple PRRs (CRP, NOD1, COLEC12, TLR1, PTX3). It is not clear why this seemingly unique feature exists with the TLR7, albeit there are reports of decreased TLR7 levels in irritable bowel syndrome patients (Sainathan, et al., 2012, Brint, et al., 2011), elevated stimulation of TLR7 leading to a proinflammatory response in the gut (Dillon, et al., 2010), and that the normal intestinal microbiota can regulate TLR7 pathways and help trigger a proper activation of inflammasomes related to respiratory responses (Pang and Iwasaki, 2011, Wu, et al., 2013). Finally, in the absence of TLR7-mediated responses, the immune milieu of the host is modified with effects on both respiratory epithelium functions (Kaiko, et al., 2013). Thus, these networks in the gingival tissues support that TLR7 may have an important role in regulating the homeostatic responses in these tissues and contributing to maintaining gingival health with aging.
The CD14 molecule exists anchored to the membrane of many immune system cells and in a soluble form generally after shedding or direct secretion. This molecule acts as co-receptor with TLR4 and MD2 for bacterial LPSs through recognition of diacylated lipopeptides (Park and Lee, 2013, Laine, et al., 2005, Jin and Lee, 2008). The soluble form of CD14 confers LPS-responsiveness to cells not expressing CD14 (Litvack and Palaniyar, 2010). The expression of CD14 in the gingival tissues was significantly elevated in healthy aging tissues and correlated significantly with aging, but was generally unrelated to the level of bleeding or pocket depth in the healthy animals.
The C-type lectin domain family members share a common structure with roles in inflammation and immune responses. The C-type lectin receptors are often endocytic receptors, and may have direct signaling through NF-κB activation or may act indirectly by modulating TLRs. CLEC4E functions as cell-surface receptor for a wide variety of ligands such as nuclear proteins from damaged cells, fungi (mannose residues) and mycobacteria (trehalose 6,6′-dimycolate). CLEC4M encodes L-SIGN that is 77% identical to CD209 (DC-SIGN), as well as being involved in the innate immune system and recognizes divergent pathogens from parasites to viruses (Yan, et al., 2013, Wevers, et al., 2013, Sukhithasri, et al., 2013, Hoving, et al., 2013). This receptor also interfaces with the coagulation system through binding of von Willebrand Factor (Rydz, et al., 2013). CLEC7A (Dectin-1), which is primarily expressed on immune cells, functions as a PRR for a variety of beta-1,3-linked and beta-1,6-linked glucans from fungal cell walls and some bacteria as part of innate immune responses, and collaborates with TLR2 to elicit inflammatory responses. Engagement of Dectin-1 leads to a cellular respiratory burst, production of arachidonate metabolites, maturation of dendritic cells, phagocytosis and an array of cytokine/chemokine production, particularly related to development of Th17 cells (Lyakh, et al., 2008, Reid, et al., 2009). CLEC6A (Dectin-2) is part of the dendritic cell immunoreceptor family that binds high-mannose carbohydrates and alpha-mannans on C. albicans hyphae, thus, playing an important role in host defense by inducing Th17 cell differentiation (Kerscher, et al., 2013). CLEC6A and CLEC7A were significantly positively correlated with age, while CLEC4M was decreased with aging. Also, CLEC7A was positively with clinical pocket depth. Finally, all of the C-lectin type PRRs were correlated with gene expression of multiple other PRRs in the tissue, suggesting a potential role in maintaining tissue homeostasis even during aging.
Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin (DC-SIGN), or CD209 is a C-type lectin receptor primarily expressed by macrophages and dendritic cells that plays an important role in innate and adaptive immunity (Zhou, et al., 2006, Khoo, et al., 2008). The receptor engages mannose type glycoproteins, commonly found on viruses, bacteria and fungi to initiate phagocytosis. Binding of ligand to DC-SIGN also causes DC interactions with endothelial cells and ac(Gonzalez, et al., 2011, Gonzalez, et al., 2013)tivation of CD4+ T cells. Numerous reports have emphasized the role of DCs in periodontitis, often focusing on their role in innate immunity. However, the changes in aging healthy gingival tissues may support a contribution to the adaptive immune system that helps to maintain a healthy homeostasis or related to a delayed apoptosis of these cells with aging in these tissues, as we have identified previously (Gonzalez, et al., 2011, Gonzalez, et al., 2013).
Retinoic acid-inducible gene I-like receptors (RLRs) are cytoplasmic RNA helicases that recognize RNA species and are critical for host antiviral responses through interferon induction (Kumar, et al., 2013, Eisenacher and Krug, 2012). These cytosolic PRRs are represented by three members: RIG-I, melanoma differentiation associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). RIG-I (DDX58) generally recognizes various ssRNA viruses, and binds to small dsRNA species (Onoguchi, et al., 2011). LGP2 (DHX58) appears to have a function more relevant to innate antiviral immunity. LGP2 has been found to be essential for producing effective antiviral responses against many viruses that are recognized by RIG-I and MDA5. Examination of type I IFN production related to LGP2 suggests that it can negatively or positively regulate RIG-I and MDA5 responses, depending on the type of RNA virus infection (Kato, et al., 2011). Generally, these types of receptors have had limited exposure in studies of periodontitis. However, we identified a significant decrease in expression of LGP2 in healthy aged gingival tissues potentially suggesting an increased risk of viral challenge to these tissues that could alter local immune reactivity and risk of bacterial triggering of periodontitis (Slots, 2010).
The nucleotide-binding oligomerization domain-containing protein receptors (NLRs) are intracellular PRRs, and include NOD1, NOD2, NLRC4 (NLR family CARD domain-containing protein 4), NLRP3 (NLR family, pyrin containing domain 3; NALP3) and NAIP (NLR family, apoptosis inhibitory protein; BIRC1). NOD1 and NOD2 recognize intracellular bacterial cell products of peptidoglycan (PGN), i.e. D-γ-glutamyl-meso-DAP (mDAP) and muramyl dipeptide (MDP) leading to activation of NF-κB. In addition, the cytokines TNF and IL-32 synergize with the NOD agonists, mDAP and MDP, to enhance IL-1β production (Wen, et al., 2013). NALP3 responds to multiple stimuli to form a multi-protein complex termed the inflammasome, which promotes the release of the IL-1 family of cytokines (Martinon, et al., 2009). NLRC4 (IPAF) indirectly senses specific proteins from pathogenic bacteria and fungi and responds by assembling an inflammasome complex that promotes caspase-1 activation, cytokine production, and macrophage pyroptosis. NAIP is a sensor component of the NLRC4 inflammasome that recognizes intracellular bacteria pathogens and is anti-apoptotic by inhibiting the activities of CASP3, CASP7 and CASP9 (Gong and Shao, 2012, Kofoed and Vance, 2012). These intracellular PRRs demonstrated a range of variation in the gingival tissues. NAIP was significantly elevated in healthy aged tissues. This is consistent with our previous findings of decreased apoptotic gene profiles in aging gingival tissues and periodontitis (Gonzalez, et al., 2011, Gonzalez, et al., 2013).
Soluble PRRs are produced by macrophages and hepatocytes and generally are considered as antimicrobial by binding to the cell wall of various microorganisms, enhancing phagocytosis and complement activation, leading to both destruction and processing for antigen presentation (Kumar, et al., 2013, Litvack and Palaniyar, 2010). Complement receptors, collectins, ficolins and the family of pentraxins [e.g. Serum Amyloid A (SAA), C-Reactive Protein (CRP)] are all secreted by cells as soluble PRRs. Pentraxins are divided into short proteins, such as CRP and serum amyloid protein, and long proteins, such as PTX3 and PTX4 (Deban, et al., 2009, Agrawal, et al., 2009). The serum levels of these soluble PRRs increase in response to inflammatory cytokines; therefore these PRRs are also referred to as acute-phase proteins (Ebersole and Cappelli, 2000). The pentraxin family members are considered as part of innate immunity and function as primitive antibodies by mediating agglutination, complement activation, and opsonization of pathogens. No differences were observed in CRP gene expression. The SAA1 protein may help repair damaged tissues, act as an antibacterial agent, and signal the migration of immune cells to sites of infection. It was significantly upregulated in healthy aged gingival tissues. PTX4 was significantly under-expressed in healthy aging tissues and demonstrated a significant negative correlation with aging. The PTX3 soluble PRR is a prototypical member of this group that is an acute phase reactant and is produced and released by mononuclear phagocytes, dendritic cells, fibroblasts, and endothelial cells in response to TLR engagement, and the action of various host factors (TNFα, IL-1ß), and has been suggested to contribute to controlling autoimmune responses and fixing complement (Deban, et al., 2009). A few reports have described elevated levels of PTX3 in gingival tissues (Lakshmanan, et al., 2013) and gingival crevicular fluid and plasma (Pradeep, et al., 2011, Fujita, et al., 2012), leading to a suggestion that these types of soluble PRRs may provide some diagnostic potential for periodontal disease. However, no other reports are available that have examined PTX4 related to oral health or disease.
Z-DNA-binding protein 1 (ZBP1 or DAI/DLM-1) recognizes DNA in the cytoplasm as an antiviral mechanism. Once ZBP1 is activated, it increases the production of antiviral cytokines, particularly the type I interferons (Vilaysane and Muruve, 2009, Yanai, et al., 2009). Expression of this PRR was significantly elevated in healthy aging tissues and it was positively correlated with aging, and positively correlated with the mean pocket depth across the periodontally healthy population of animals. While this PRR has generally been considered as a molecule with a role in antiviral innate immunity, its ability to increase type I interferons that may be related to a more general immunomodulatory response, could suggest a more direct role in bacterial responses, or may indicate some involvement of altered local viral reactivation/challenge to the gingival tissues during disease, as has been suggested in human periodontitis (Slots, 2010, Contreras, et al., 2014).
The findings of this study provide some evidence that variation in the expression of PRRs in gingival tissues of healthy aged individuals may be reflective of a biological pattern of tissue change with aging that could increase the risk for developing disease, as has been reported in humans and nonhuman primates (Kye, et al., 2012, Gonzalez, et al., 2012, Chung, et al., 2011, Hajishengallis, 2010, Huttner, et al., 2009, Miller, et al., 1995). The relationship of some of these microbial detection molecules related to expression of some “physiologic” inflammatory changes even in healthy tissues suggested a more complex role for this family of “detectors” than may have been previously considered in models targeting a single PRR with in vitro systems and a mono-microbial challenge.
Acknowledgments
This project was supported by National Institute of Health grants P20GM103538 and UL1TR000117, and P20GM103436. We express our gratitude to the Caribbean Primate Research Center (CPRC) supported by grant P40RR03640, and the Microarray Core of University Kentucky for their invaluable technical assistance. The authors declare that there are no conflicts of interest related to this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The Authors declare that there is no conflict of interest.
References
- Kopitar AN, Ihan Hren N, Ihan A. Commensal oral bacteria antigens prime human dendritic cells to induce Th1, Th2 or Treg differentiation. Oral Microbiol Immunol. 2006;21:1. doi: 10.1111/j.1399-302X.2005.00237.x. [DOI] [PubMed] [Google Scholar]
- Nestle FO, Thompson C, Shimizu Y, Turka LA, Nickoloff BJ. Costimulation of superantigen-activated T lymphocytes by autologous dendritic cells is dependent on B7. Cell Immunol. 1994;156:220. doi: 10.1006/cimm.1994.1166. [DOI] [PubMed] [Google Scholar]
- Makino M, Utsunomiya A, Maeda Y, Shimokubo S, Izumo S, Baba M. Association of CD40 ligand expression on HTLV-I-infected T cells and maturation of dendritic cells. Scand J Immunol. 2001;54:574. doi: 10.1046/j.1365-3083.2001.01003.x. [DOI] [PubMed] [Google Scholar]
- Jotwani R, Palucka AK, Al-Quotub M, Nouri-Shirazi M, Kim J, Bell D, Banchereau J, Cutler CW. Mature dendritic cells infiltrate the T cell-rich region of oral mucosa in chronic periodontitis: in situ, in vivo, and in vitro studies. J Immunol. 2001;167:4693. doi: 10.4049/jimmunol.167.8.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler CW, Jotwani R. Dendritic cells at the oral mucosal interface. J Dent Res. 2006;85:678. doi: 10.1177/154405910608500801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Akira S. TLR signalling and the function of dendritic cells. Chem Immunol Allergy. 2005;86:120. doi: 10.1159/000086657. [DOI] [PubMed] [Google Scholar]
- Benko S, Magyarics Z, Szabo A, Rajnavolgyi E. Dendritic cell subtypes as primary targets of vaccines: the emerging role and cross-talk of pattern recognition receptors. Biol Chem. 2008;389:469. doi: 10.1515/bc.2008.054. [DOI] [PubMed] [Google Scholar]
- Blach-Olszewska Z. Innate immunity: cells, receptors, and signaling pathways. Arch Immunol Ther Exp (Warsz) 2005;53:245. [PubMed] [Google Scholar]
- Wollenberg A, Mommaas M, Oppel T, Schottdorf EM, Gunther S, Moderer M. Expression and function of the mannose receptor CD206 on epidermal dendritic cells in inflammatory skin diseases. J Invest Dermatol. 2002;118:327. doi: 10.1046/j.0022-202x.2001.01665.x. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Krauss JL, Liang S, McIntosh ML, Lambris JD. Pathogenic microbes and community service through manipulation of innate immunity. Adv Exp Med Biol. 2012;946:69. doi: 10.1007/978-1-4614-0106-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling D, Jungi TW. TOLL-like receptors linking innate and adaptive immune response. Vet Immunol Immunopathol. 2003;91:1. doi: 10.1016/s0165-2427(02)00228-3. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- Shaw AC, Goldstein DR, Montgomery RR. Age-dependent dysregulation of innate immunity. Nat Rev Immunol. 2013;13:875. doi: 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AC, Panda A, Joshi SR, Qian F, Allore HG, Montgomery RR. Dysregulation of human Toll-like receptor function in aging. Ageing Res Rev. 2011;10:346. doi: 10.1016/j.arr.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Fulop T, Larbi A, Witkowski JM, Kotb R, Hirokawa K, Pawelec G. Immunosenescence and cancer. Crit Rev Oncog. 2013;18:489. doi: 10.1615/critrevoncog.2013010597. [DOI] [PubMed] [Google Scholar]
- Dunston CR, Griffiths HR. The effect of ageing on macrophage Toll-like receptor-mediated responses in the fight against pathogens. Clin Exp Immunol. 2010;161:407. doi: 10.1111/j.1365-2249.2010.04213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li H, Yang MF, Shu W, Sun MJ, Xu Y. Effects of aging on endotoxin tolerance induced by lipopolysaccharides derived from Porphyromonas gingivalis and Escherichia coli. PLoS One. 2012;7:e39224. doi: 10.1371/journal.pone.0039224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baelum V, Lopez R. Periodontal disease epidemiology - learned and unlearned? Periodontol. 2013;62:37. doi: 10.1111/j.1600-0757.2012.00449.x. 2000. [DOI] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ, Cdc Periodontal Disease Surveillance workgroup: James Beck, G.D.R.P. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- Ebersole JL, Dawson DR, 3rd, Morford LA, Peyyala R, Miller CS, Gonzalez OA. Periodontal disease immunology: ‘double indemnity’ in protecting the host. Periodontol. 2013;62:163. doi: 10.1111/prd.12005. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartold PM, Van Dyke TE. Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontol. 2013;62:203. doi: 10.1111/j.1600-0757.2012.00450.x. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlet GP, Sfeir CS, Little SR. Restoring host-microbe homeostasis via selective chemoattraction of Tregs. J Dent Res. 2014;93:834. doi: 10.1177/0022034514544300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza PP, Lerner UH. The role of cytokines in inflammatory bone loss. Immunol Invest. 2013;42:555. doi: 10.3109/08820139.2013.822766. [DOI] [PubMed] [Google Scholar]
- de Molon RS, de Avila ED, Cirelli JA. Host responses induced by different animal models of periodontal disease: a literature review. J Investig Clin Dent. 2013;4:211. doi: 10.1111/jicd.12018. [DOI] [PubMed] [Google Scholar]
- Graves DT, Kang J, Andriankaja O, Wada K, Rossa C., Jr Animal models to study host-bacteria interactions involved in periodontitis. Front Oral Biol. 2012;15:117. doi: 10.1159/000329675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oz HS, Puleo DA. Animal models for periodontal disease. J Biomed Biotechnol. 2011;2011:754857. doi: 10.1155/2011/754857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden TE, Caton JG. Animal models for periodontal disease. Methods Enzymol. 1994;235:106. doi: 10.1016/0076-6879(94)35135-x. [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole J, Felton J, Brunsvold M, Kornman KS. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. 1988;239:55. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- Ebersole JL, Steffen MJ, Gonzalez-Martinez J, Novak MJ. Effects of age and oral disease on systemic inflammatory and immune parameters in nonhuman primates. Clin Vaccine Immunol. 2008;15:1067. doi: 10.1128/CVI.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meka A, Bakthavatchalu V, Sathishkumar S, Lopez MC, Verma RK, Wallet SM, Bhattacharyya I, Boyce BF, Handfield M, Lamont RJ, Baker HV, Ebersole JL, Kesavalu L. Porphyromonas gingivalis infection-induced tissue and bone transcriptional profiles. Mol Oral Microbiol. 25:61. doi: 10.1111/j.2041-1014.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, Stromberg AJ, Huggins PM, Gonzalez-Martinez J, Novak MJ, Ebersole JL. Apoptotic genes are differentially expressed in aged gingival tissue. J Dent Res. 2011;90:880. doi: 10.1177/0022034511403744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, Novak MJ, Kirakodu S, Orraca L, Chen KC, Stromberg A, Gonzalez-Martinez J, Ebersole JL. Comparative analysis of gingival tissue antigen presentation pathways in ageing and periodontitis. J Clin Periodontol. 2014;41:327. doi: 10.1111/jcpe.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, Novak MJ, Kirakodu S, Orraca L, Chen KC, Stromberg A, Gonzalez-Martinez J, Ebersole JL. Comparative analysis of gingival tissue antigen presentation pathways in ageing and periodontitis. J Clin Periodontol. 2013 doi: 10.1111/jcpe.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolli D, Velayutham TS, Casola A. Host-Viral Interactions: Role of Pattern Recognition Receptors (PRRs) in Human Pneumovirus Infections. Pathogens. 2013:2. doi: 10.3390/pathogens2020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Ingle H, Prasad DV, Kumar H. Recognition of bacterial infection by innate immune sensors. Crit Rev Microbiol. 2013;39:229. doi: 10.3109/1040841X.2012.706249. [DOI] [PubMed] [Google Scholar]
- Min W, Kim WH, Lillehoj EP, Lillehoj HS. Recent progress in host immunity to avian coccidiosis: IL-17 family cytokines as sentinels of the intestinal mucosa. Dev Comp Immunol. 2013;41:418. doi: 10.1016/j.dci.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Qian C, Cao X. Regulation of Toll-like receptor signaling pathways in innate immune responses. Ann N Y Acad Sci. 2013;1283:67. doi: 10.1111/j.1749-6632.2012.06786.x. [DOI] [PubMed] [Google Scholar]
- van Bergenhenegouwen J, Plantinga TS, Joosten LA, Netea MG, Folkerts G, Kraneveld AD, Garssen J, Vos AP. TLR2 & Co: a critical analysis of the complex interactions between TLR2 and coreceptors. J Leukoc Biol. 2013;94:885. doi: 10.1189/jlb.0113003. [DOI] [PubMed] [Google Scholar]
- Frazao JB, Errante PR, Condino-Neto A. Toll-like receptors’ pathway disturbances are associated with increased susceptibility to infections in humans. Arch Immunol Ther Exp (Warsz) 2013;61:427. doi: 10.1007/s00005-013-0243-0. [DOI] [PubMed] [Google Scholar]
- Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med. 2013;45:e66. doi: 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainathan SK, Bishnupuri KS, Aden K, Luo Q, Houchen CW, Anant S, Dieckgraefe BK. Toll-like receptor-7 ligand Imiquimod induces type I interferon and antimicrobial peptides to ameliorate dextran sodium sulfate-induced acute colitis. Inflamm Bowel Dis. 2012;18:955. doi: 10.1002/ibd.21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brint EK, MacSharry J, Fanning A, Shanahan F, Quigley EM. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol. 2011;106:329. doi: 10.1038/ajg.2010.438. [DOI] [PubMed] [Google Scholar]
- Dillon SM, Rogers LM, Howe R, Hostetler LA, Buhrman J, McCarter MD, Wilson CC. Human intestinal lamina propria CD1c+ dendritic cells display an activated phenotype at steady state and produce IL-23 in response to TLR7/8 stimulation. J Immunol. 2010;184:6612. doi: 10.4049/jimmunol.1000041. [DOI] [PubMed] [Google Scholar]
- Pang IK, Iwasaki A. Inflammasomes as mediators of immunity against influenza virus. Trends Immunol. 2011;32:34. doi: 10.1016/j.it.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Jiang ZY, Sun YF, Yu B, Chen J, Dai CQ, Wu XL, Tang XL, Chen XY. Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza A virus infection. Curr Microbiol. 2013;67:414. doi: 10.1007/s00284-013-0380-z. [DOI] [PubMed] [Google Scholar]
- Kaiko GE, Loh Z, Spann K, Lynch JP, Lalwani A, Zheng Z, Davidson S, Uematsu S, Akira S, Hayball J, Diener KR, Baines KJ, Simpson JL, Foster PS, Phipps S. Toll-like receptor 7 gene deficiency and early-life Pneumovirus infection interact to predispose toward the development of asthma-like pathology in mice. J Allergy Clin Immunol. 2013;131:1331. doi: 10.1016/j.jaci.2013.02.041. [DOI] [PubMed] [Google Scholar]
- Laine ML, Morre SA, Murillo LS, van Winkelhoff AJ, Pena AS. CD14 and TLR4 gene polymorphisms in adult periodontitis. J Dent Res. 2005;84:1042. doi: 10.1177/154405910508401114. [DOI] [PubMed] [Google Scholar]
- Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29:182. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Litvack ML, Palaniyar N. Review: Soluble innate immune pattern-recognition proteins for clearing dying cells and cellular components: implications on exacerbating or resolving inflammation. Innate Immun. 2010;16:191. doi: 10.1177/1753425910369271. [DOI] [PubMed] [Google Scholar]
- Yan H, Ohno N, Tsuji NM. The role of C-type lectin receptors in immune homeostasis. Int Immunopharmacol. 2013;16:353. doi: 10.1016/j.intimp.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Wevers BA, Geijtenbeek TB, Gringhuis SI. C-type lectin receptors orchestrate antifungal immunity. Future Microbiol. 2013;8:839. doi: 10.2217/fmb.13.56. [DOI] [PubMed] [Google Scholar]
- Sukhithasri V, Nisha N, Biswas L, Anil Kumar V, Biswas R. Innate immune recognition of microbial cell wall components and microbial strategies to evade such recognitions. Microbiol Res. 2013;168:396. doi: 10.1016/j.micres.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Hoving JC, Wilson GJ, Brown GD. Signalling C-Type lectin receptors, microbial recognition and immunity. Cell Microbiol. 2013 doi: 10.1111/cmi.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydz N, Swystun LL, Notley C, Paterson AD, Riches JJ, Sponagle K, Boonyawat B, Montgomery RR, James PD, Lillicrap D. The C-type lectin receptor CLEC4M binds, internalizes, and clears von Willebrand factor and contributes to the variation in plasma von Willebrand factor levels. Blood. 2013;121:5228. doi: 10.1182/blood-2012-10-457507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyakh L, Trinchieri G, Provezza L, Carra G, Gerosa F. Regulation of interleukin-12/interleukin-23 production and the T-helper 17 response in humans. Immunol Rev. 2008;226:112. doi: 10.1111/j.1600-065X.2008.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DM, Gow NA, Brown GD. Pattern recognition: recent insights from Dectin-1. Curr Opin Immunol. 2009;21:30. doi: 10.1016/j.coi.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher B, Willment JA, Brown GD. The Dectin-2 family of C-type lectin-like receptors: an update. Int Immunol. 2013;25:271. doi: 10.1093/intimm/dxt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Chen Y, Hao L, Zhang Y. DC-SIGN and immunoregulation. Cell Mol Immunol. 2006;3:279. [PubMed] [Google Scholar]
- Khoo US, Chan KY, Chan VS, Lin CL. DC-SIGN and L-SIGN: the SIGNs for infection. J Mol Med (Berl) 2008;86:861. doi: 10.1007/s00109-008-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez OA, John Novak M, Kirakodu S, Stromberg AJ, Shen S, Orraca L, Gonzalez-Martinez J, Ebersole JL. Effects of aging on apoptosis gene expression in oral mucosal tissues. Apoptosis. 2013;18:249. doi: 10.1007/s10495-013-0806-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenacher K, Krug A. Regulation of RLR-mediated innate immune signaling–it is all about keeping the balance. Eur J Cell Biol. 2012;91:36. doi: 10.1016/j.ejcb.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Onoguchi K, Yoneyama M, Fujita T. Retinoic acid-inducible gene-I-like receptors. J Interferon Cytokine Res. 2011;31:27. doi: 10.1089/jir.2010.0057. [DOI] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Slots J. Human viruses in periodontitis. Periodontol. 2010;53:89. doi: 10.1111/j.1600-0757.2009.00325.x. 2000. [DOI] [PubMed] [Google Scholar]
- Wen H, Miao EA, Ting JP. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity. 2013;39:432. doi: 10.1016/j.immuni.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- Gong YN, Shao F. Sensing bacterial infections by NAIP receptors in NLRC4 inflammasome activation. Protein Cell. 2012;3:98. doi: 10.1007/s13238-012-2028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed EM, Vance RE. NAIPs: building an innate immune barrier against bacterial pathogens. NAIPs function as sensors that initiate innate immunity by detection of bacterial proteins in the host cell cytosol. Bioessays. 2012;34:589. doi: 10.1002/bies.201200013. [DOI] [PubMed] [Google Scholar]
- Gonzalez O, Novak MJ, Orraca L, Martinez-Gonzalez J, Stromberg AJ, Ebersole JL. Apopotosis gene expression in healthy and oral mucosal tissues with aging. Apoptosis. 2013 doi: 10.1007/s10495-013-0806-x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deban L, Bottazzi B, Garlanda C, de la Torre YM, Mantovani A. Pentraxins: multifunctional proteins at the interface of innate immunity and inflammation. Biofactors. 2009;35:138. doi: 10.1002/biof.21. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Singh PP, Bottazzi B, Garlanda C, Mantovani A. Pattern recognition by pentraxins. Adv Exp Med Biol. 2009;653:98. doi: 10.1007/978-1-4419-0901-5_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole JL, Cappelli D. Acute-phase reactants in infections and inflammatory diseases. Periodontol. 2000;23:19. doi: 10.1034/j.1600-0757.2000.2230103.x. 2000. [DOI] [PubMed] [Google Scholar]
- Lakshmanan R, Jayakumar ND, Sankari M, Padmalatha O, Varghese S. Estimation of Pentraxin-3 Levels in the Gingival Tissues of Chronic And Aggressive Periodontitis Participants-An in Vivo Study. J Periodontol. 2013 doi: 10.1902/jop.2013.120718. [DOI] [PubMed] [Google Scholar]
- Pradeep AR, Kathariya R, Raghavendra NM, Sharma A. Levels of pentraxin-3 in gingival crevicular fluid and plasma in periodontal health and disease. J Periodontol. 2011;82:734. doi: 10.1902/jop.2010.100526. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Ito H, Sekino S, Numabe Y. Correlations between pentraxin 3 or cytokine levels in gingival crevicular fluid and clinical parameters of chronic periodontitis. Odontology. 2012;100:215. doi: 10.1007/s10266-011-0042-1. [DOI] [PubMed] [Google Scholar]
- Vilaysane A, Muruve DA. The innate immune response to DNA. Semin Immunol. 2009;21:208. doi: 10.1016/j.smim.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Yanai H, Savitsky D, Tamura T, Taniguchi T. Regulation of the cytosolic DNA-sensing system in innate immunity: a current view. Curr Opin Immunol. 2009;21:17. doi: 10.1016/j.coi.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Contreras A, Botero JE, Slots J. Biology and pathogenesis of cytomegalovirus in periodontal disease. Periodontol. 2014;64:40. doi: 10.1111/j.1600-0757.2012.00448.x. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye W, Davidson R, Martin J, Engebretson S. Current status of periodontal risk assessment. J Evid Based Dent Pract. 2012;12:2. doi: 10.1016/S1532-3382(12)70002-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez O, Tobia C, Ebersole J, Novak MJ. Caloric restriction and chronic inflammatory diseases. Oral Dis. 2012;18:16. doi: 10.1111/j.1601-0825.2011.01830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HY, Lee EK, Choi YJ, Kim JM, Kim DH, Zou Y, Kim CH, Lee J, Kim HS, Kim ND, Jung JH, Yu BP. Molecular inflammation as an underlying mechanism of the aging process and age-related diseases. J Dent Res. 2011;90:830. doi: 10.1177/0022034510387794. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. Too old to fight? Aging and its toll on innate immunity. Mol Oral Microbiol. 2010;25:25. doi: 10.1111/j.2041-1014.2009.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner EA, Machado DC, de Oliveira RB, Antunes AG, Hebling E. Effects of human aging on periodontal tissues. Spec Care Dentist. 2009;29:149. doi: 10.1111/j.1754-4505.2009.00082.x. [DOI] [PubMed] [Google Scholar]
- Miller DR, Aufdemorte TB, Fox WC, Waldrop TC, Mealey BL, Brunsvold MA. Periodontitis in the baboon: a potential model for human disease. J Periodontal Res. 1995;30:404. doi: 10.1111/j.1600-0765.1995.tb01294.x. [DOI] [PubMed] [Google Scholar]


