Abstract
B-1 cells are a unique subset of B cells that are positively selected for expressing autoreactive BCRs. We isolated RNA from peritoneal (B-1a, B-1b, B-2) and splenic (B-1a, marginal zone, follicular) B cells from C57BL/6 mice and used 5′-RACE to amplify the IgH variable region (IGHV) using massively parallel sequencing. By analyzing 379,000 functional transcripts, we demonstrate that B-1a cells use a distinct and restricted repertoire. All B-1 cell subsets, especially peritoneal B-1a cells, had a high proportion of sequences without N additions, suggesting predominantly prenatal development. Their transcripts differed markedly and uniquely contained VH11 and VH12 genes, which were rearranged only with a restricted selection of D and J genes, unlike other V genes. Compared to peritoneal B-1a, the peritoneal B-1b repertoire was larger, had little overlap with B-1a, and the majority of sequences contained N additions. Similarly, the splenic B-1a repertoire differed from peritoneal B-1a sequences, having more unique sequences and more frequent N additions, suggesting influx of B-1a cells into the spleen from non-peritoneal sites. Two CDR3s, previously described as antibodies to bromelain-treated RBCs, comprised 43% of peritoneal B-1a sequences. We show that a single chain variable fragment (scFv) designed after the most prevalent B-1a sequence, binds oxidation-specific epitopes (OSEs) such as the phosphocholine (PC) of oxidized phospholipids. In summary, we provide the IGHV library of six murine B cell subsets, including for the first time a comparison between B-1a and B-1b cells, and highlight qualities of B-1 cell antibodies that indicate unique selection processes.
Introduction
Ly-1+ (CD5+) B cells, later named B-1 cells for their early appearance in ontogeny, have many unique characteristics (1, 2). In contrast to conventional B-2 cells, B-1 cells develop in the fetal liver, produce so-called “natural” antibodies (NAbs2) even in a germ-free environment, react to antigen independent of cognate T-cell help, and their antibody production can be stimulated by non-antigen-specific signals (e.g. TLR agonists) (3–5). A phenotypically similar subset, termed B-1b cells, has been described, which shares similar surface markers with B-1a cells, but does not express CD5 (6). In contrast to B-1a cells, B-1b cells are able to expand clonally in response to antigen and can be reconstituted from a single hematopoietic stem cell from adult bone marrow, suggesting that B-1b cells develop from different stem cells than B-1a cells (7–9). B-1 cells also are the predominant B cell subset in the peritoneal cavity and B-1a cells can migrate to the spleen in response to LPS, where they differentiate and secrete antibody (10, 11). Their antibodies form a first-line response against infections (e.g. S. pneumoniae), are involved in clearing apoptotic debris, and can protect from atherosclerosis by binding to oxidation-specific epitopes (OSEs) of oxidized LDL (OxLDL) and apoptotic cells (12, 13). We have postulated that these innate NAbs can be thought of as innate pattern recognition receptors (PRRs) that have been selected to provide homeostasis against endogenous danger associated molecular patterns (DAMPs) such as OSEs, as well as against pathogen associated molecular patterns (PAMPs) as found on exogenous pathogens. A combination of these functions can be mediated by only a single antibody, e.g. by the phosphocholine-binding T15/E06 IgM NAb (14, 15).
Each B cell develops its BCR during development in the fetal liver or adult bone marrow by rearrangement of V, D, and J gene segments (only V and J for the light chain). During rearrangement, the junctions are modified by terminal deoxynucleotidyl transferase (TdT) activity, which adds random non-templated nucleotides (N additions) to the ends of gene segments, and exonucleases, which can shorten each segment (16). Notably, TdT is active only after birth and neonatal mouse Ig lack N additions (17, 18). The regions constituting the antigen-binding site of the antibody are the highly variable complementary-determining regions (CDRs). Of these, the most diverse is the CDR3 region of the Ig heavy chain, which spans both the V-D and the D-J junction. It is the major site determining the antibody’s specificity and a substitution in only one amino acid in the CDR3 of the heavy chain can alter reactivity with its antigen (19, 20).
Originally, B-1a cell antibody sequences were studied by sequencing of hybridomas derived from CD5+ B cells (21, 22). They were described as antibodies with high analogy to germline sequence and a low number of N additions (18). A high number of these cells bound to bromelain-treated red blood cells (BrRBCs), binding a hidden “auto-antigen” exposed by proteolysis (21). Most of these anti-BrRBC sequences contained the gene segments VH11 and JH1 (23). Phosphatidylcholine competed antibody binding to BrRBCs, indicating that this might be the exposed epitope or a molecular mimic (13, 24).
Later, Kantor et al. analyzed the repertoire of single-cell-sorted peritoneal B-1a, B-1b, and B-2 cells by sequencing the Ig heavy chain variable region (IGHV) of 55–70 cells per subset, giving further insight into the properties of the B-1 cell repertoire (25). Vale et al. showed that transcripts of peritoneal VH5 (VH 7183) sequences had properties and CDR3 sequences that were uncommonly found in splenic or bone marrow sequences (26). In a study describing functional differences of B-1a cells with different levels of surface expression of plasma cell alloantigen 1 (PC-1), Wang et al. recently showed some disparities in VH usage and N additions of PC-1hi as opposed to PC-1lo B-1a cells (27). All of these studies used traditional sequencing techniques, which only allowed the identification of a few hundred sequences at a time. With the advance of massively parallel next-generation sequencing and new amplification methods for the IGHV, it is now possible to analyze a much higher number of sequences (28, 29). In fact, during preparation of this manuscript, an article by Yang et al. was published, where next-generation sequencing was used to show, among other findings, that the repertoire of B-1a cells becomes more restricted as mice age (30).
In this report, we analyze the repertoire of peritoneal (B-1a, B-1b, B-2) and splenic (B-1a, marginal zone (MZ), and follicular (FO)) B cell subsets by using a non-biased 5′-RACE reverse transcription step to generate cDNAs including all potential IGHV sequences, which were then determined by massively parallel sequencing. We used RNA as starting material as a representation of the functional repertoire (i.e. actually expressed sequences). Our studies demonstrate that B-1a cells have a unique repertoire with higher usage of VH11 and VH12 and that these sequences have distinct properties compared to other antibody sequences. All B-1 subsets were shown to have significantly more N-addition-free transcripts compared to B-2 cells. Additionally, we compare the repertoires between all the 6 subsets analyzed, finding notable overlaps, but also surprising differences in the antibodies they express.
Materials and Methods
Mice
10-week old female C57BL/6 mice were purchased from Charles River Laboratories and maintained in our vivarium for 2 weeks with a 12-hour light cycle and ad libitum access to water and food. Mice were sacrificed for analysis at the age of 12 weeks. All animal experiments were performed according to the NIH guidelines and were approved by the UC San Diego Animal Subjects Committee.
Peritoneal and splenic cell isolation
Peritoneal lavage was performed on sacrificed mice using 10 ml isolation buffer (PBS with 1% FCS, 10 mM EDTA). Spleens were homogenized with a syringe and cells were passed through a 70 μm cell strainer using isolation buffer to obtain a single cell suspension. All further steps were done at 4°C or on ice. Red blood cells were lysed using RBC lysis buffer (BioLegend, San Diego, CA) for 3 minutes. Both peritoneal and splenic cells were subsequently centrifuged at 400 g for 5 minutes, resuspended in sorting buffer (PBS with 5 mM EDTA, 2 mM HEPES, 1% FCS) and counted.
2–3 mice were used for each sort and their cells pooled in order to attain sufficient amounts of RNA for 5′ RACE and sequencing. To avoid overrepresentation of one single mouse in that pool, an equal ratio of peritoneal to splenic cells from each mouse was used. Cell isolations were done on 3 separate days and sequencing libraries were prepared independently for each of the experiments and later combined, culminating in one database with sequences from 8 mice.
FACS isolation of B cell subsets
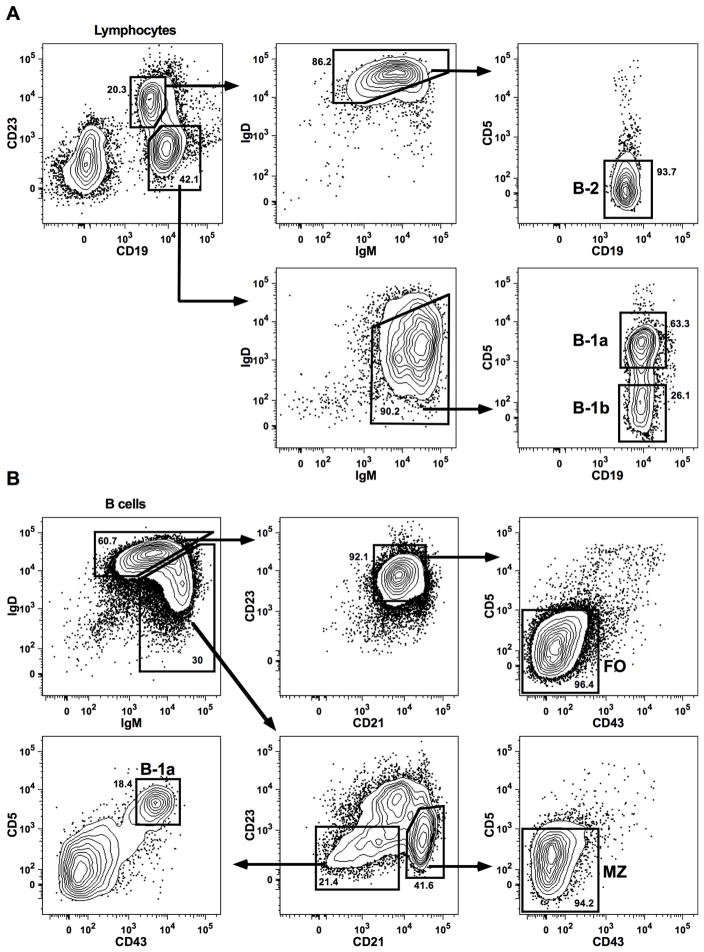
Peritoneal and splenic cells were first incubated with anti-CD16/32 antibody to block Fc receptors and then incubated for 30 minutes with antibodies specific for: CD19 (clone: 1D3, fluorochrome: APC-Cy7), CD23 (B3B4, FITC), IgM (II/41, APC), IgD (11-26c.2a, BV421), and CD5 (53–7.3, PE-Cy7) for 30 minutes. For splenic cells, antibodies to CD43 (S7, PE) and CD21 (7G6, BV605) were used additionally. 7-AAD was added to determine cell viability. All reagents were purchased from BD Biosciences (San Jose, CA) or BioLegend (San Diego, CA). FACS was done at the flow cytometry core of the La Jolla Institute for Allergy & Immunology (LIAI), and cells were sorted into fetal calf serum. Single, live lymphocytes were gated based on scatter properties and 7-AAD staining. The gating strategy for peritoneal and splenic B cell subsets was modified from published methods (Fig. 1) (31, 32) and criteria are indicated below:
Figure 1. Sorting of peritoneal and splenic B cell subsets.
(A) In the peritoneal cavity, B-1a cells were defined as CD19hi, CD23−, IgMhi, IgDlo, CD5+, B-1b cells as CD19hi, CD23−, IgMhi, IgDlo, CD5−, and B-2 cells as CD19mid, CD23+, IgMlo, IgDhi, CD5−. (B) From splenic CD19+ lymphocytes, B-1a cells were selected based on their surface markers IgMhi, IgDlo, CD21−/lo, CD23−, CD43+, CD5+, marginal zone cells as IgMhi, IgDlo, CD21hi, CD23−/lo, CD43−, CD5−, and follicular cells as IgMlo, IgDhi, CD21mid, CD23hi, CD43−, CD5−.
Peritoneal cell subsets (Fig. 1A):
B-1a cells: CD19hi CD23− IgMhi IgDlo CD5+
B-1b cells: CD19hi CD23− IgMhi IgDlo CD5−
B-2 cells: CD19mid CD23+ IgMlo IgDhi CD5−
Splenic cell subsets (Fig. 1B):
B-1a cells: CD19+ IgMhi IgDlo CD23− CD21−/lo CD43+ CD5+
Marginal zone (MZ) cells: CD19+ IgMhi IgDlo CD23−/lo CD21hi CD43− CD5−
Follicular (FO) cells: CD19+ IgMlo IgDhi CD23hi CD21mid CD43− CD5−
RNA isolation, 5′ RACE, and sequencing
Immediately after sorting, cells were centrifuged and resuspended in TRIzol and RNA was isolated by spin column centrifugation (Direct-zol™ RNA MiniPrep Kit, Zymo Research, Irvine, CA). Reverse transcription with template switching was performed using the 5′ RACE SMARTer kit (Clontech, Mountain View, CA) and a specific primer encoding the μ chain (5′-ATGGCCACCAGATTCTTATCAGAC-3′) (33). The resulting cDNA product of the IGHV with the Clontech oligonucleotide sequence at the 5′ end was amplified by two-step PCR with HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA). The first PCR reaction was designed with a primer containing both a region complementary to the constant region of the μ chain in proximity to the JH region (underlined) and to the Illumina adaptor sequence (bold) (5′-GACGTGTGCTCTTCCGATCTGGGAAGACATTTGGGAAGGACTG-3′) and a primer mix complementary to the oligonucleotide used for template switching (underlined) and to the Illumina adaptor sequence (bold) (5′-ACACGACGCTCTTCCGATCTAAGCAGTGGTATCAACGCAGAGT-3′ and 5′-ACACGACGCTCTTCCGATCT-3′). PCR products were loaded onto a 1% agarose gel. After electrophoresis, the IGHV product at 450–600 bp was extracted using MinElute Gel Extraction Kit (Qiagen, Hilden, Germany). A second PCR reaction was then used to add the remaining Illumina adaptor sequence and unique sample indexes (5′-AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ and 5′-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC-3′ where NNNNNNNN is the unique index sequence).
Sequencing was performed at the Institute for Genomic Medicine at UC San Diego on the Illumina MiSeq using paired-end read sequencing with a length of 2x300 bp.
Massively parallel sequencing data analysis
Raw sequencing data was groomed and quality trimmed at the 3′ ends of each of the paired-end reads for a Phred score >30 with Galaxy (34). Quality trimmed reads were joined in their overlapping regions using FLASH (35). The dataset of joined sequences was uploaded to IMGT/HighV-QUEST (36, 37). Data from the obtained spreadsheets were then analyzed using the Ig analysis tool (IgAT) (38). Specific sequence subsets were extracted based on certain characteristics (N addition, VH usage) by custom Python scripts (available at https://github.com/maxchang/IMGT-subsets). Data from the IMGT files or IgAT output was plotted and analyzed with Prism (Graphpad, San Diego, CA). Only functional sequences (i.e. without a stop codon) were included in the analysis. For each of the B cell subsets, 3 separate pools were prepared, each of which contained B cells from 2–3 mice, and each pool was separately sequenced. For the main part of the analysis, we combined the data generated from these 3 individual experiments into one dataset for each B cell subtype. In an additional analysis, joined sequences were analyzed with the Antibody Mining Toolbox to obtain a list of expressed CDR3 amino acid sequences along with their frequency in each subset (39). All raw sequence data were uploaded to the NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/ - BioProject accession number PRJNA418221).
Anti-BrRBC scFv generation
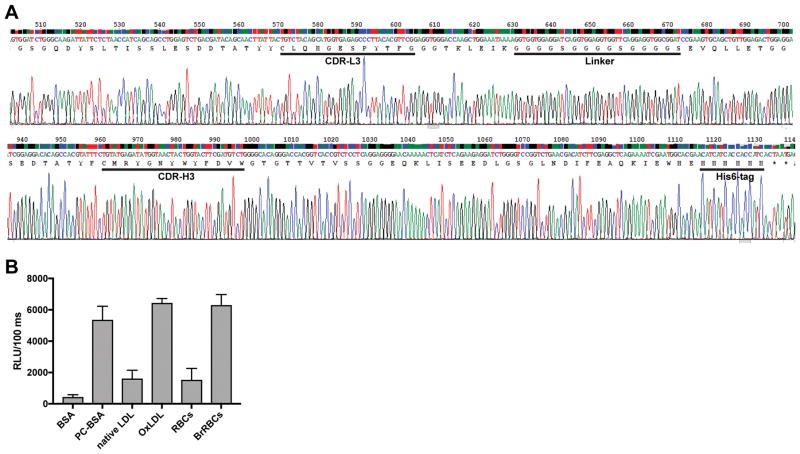
To characterize the binding specificity of the most common CDR3 sequence observed in peritoneal and splenic B-1a cells (CMRYGNYWYFDVW, V11-D2-J1), we generated a cDNA for a single-chain variable fragment (scFv) using the original IGHV bearing this CDR3, paired with the light chain variable region from the hybridoma sequences from the study originally describing this sequence (21). This was constructed by gene fragment synthesis of heavy and light chain variable regions and Gibson assembly (Integrated DNA Technologies, San Diego, CA) in a construct with a flexible 15-amino acid long linker connecting the heavy and light chain domains and designated as XQ11-scFv, which is shown in Fig. 10. A His6-tag was inserted into the C-terminus of the scFv, and the fusion construct cloned into a pFUSE mammalian expression vector (Invivogen, San Diego, CA) under the control of an hEF1-HTLV promoter. This was transiently expressed in HEK293T cells cultured first in DMEM containing 4.5 g/L glucose, 10% FBS, 100 μg/ml Zeocin, and 15 μg/ml blasticidin, and then serum-free media. The culture supernatant was concentrated using an Amicon Ultra Centrifugal Filter Device (Millipore, Burlington, MA) and then used in an ELISA to evaluate binding to antigens.
Figure 10. XQ11-scFv binds BrRBCs, phosphocholine (PC), and OxLDL.
(A) The scFv XQ11 was designed based on the reported IGHV and light chain variable region of the most prevalent B-1a antibody and a His6-tag was added. It was expressed in HEK293 cells as described in Methods. (B) Chemiluminescent ELISA of culture supernatant. XQ11-scFv bound, in addition to BrRBCs as previously reported, OSEs on PC-BSA and OxLDL. This suggests that the previously reported binding to phosphatidylcholine is likely mediated through the PC head group that is also prominently displayed on OxLDL. Because different amounts of antigen of a given type are plated, direct comparisons of binding should only be done between related antigens, e.g. BSA vs. PC-BSA, etc. Shown are mean + SD in triplicates, representative of two independent experiments.
Enzyme-linked immunosorbent assay
Chemiluminescent ELISA of XQ11 binding to indicated antigens was performed in 96 well microtiter plates as previously described (15). Red blood cells (RBCs) were obtained from C57BL/6 mice and either used for the experiment as native RBCs or incubated with 0.5% bromelain (Acros, Geel, Belgium) solution for 10 minutes at 37°C. Cells were then washed and stored in PBS at 4°C. For ELISA, 2x105 cells per well were plated. Native LDL and OxLDL were prepared as described (15). BSA was from Sigma-Aldrich (St. Louis, MO), and phosphocholine-BSA (PC-BSA) from Biosearch Technologies (Petaluma, CA). XQ11 binding to various antigens was detected using anti-His alkaline phosphatase conjugated antibody from Sigma-Aldrich.
Results
VDJ usage differs substantially between B cell subsets
Peritoneal B-1a, B-1b, and splenic B-1a cells, collectively referred to as B-1 cells, and peritoneal B-2, splenic MZ, and FO cells, collectively referred to as conventional B cells, were isolated from female C57BL/6 mice at 12 weeks of age. B cell subsets were isolated by flow cytometry using established criteria as defined in Methods (Fig. 1) and the purity of the isolated pools averaged >99% in post-sort analysis. Utilizing an unbiased 5′-RACE amplification strategy, we generated cDNAs of the IGHV and used these to perform massively parallel sequencing, with a yield of 711,760 total reads after quality trimming and joining of reads, as described in Methods. From these, we obtained 378,944 functional IGHV sequences, an average of 63,157 sequences per B cell subset (range 48,579 to 81,579, Fig 2). We analyzed the repertoire of these subsets on the level of total RNA transcripts as an estimate of the functional repertoire. The terminology used in this report is according to the IMGT nomenclature (40, 41).
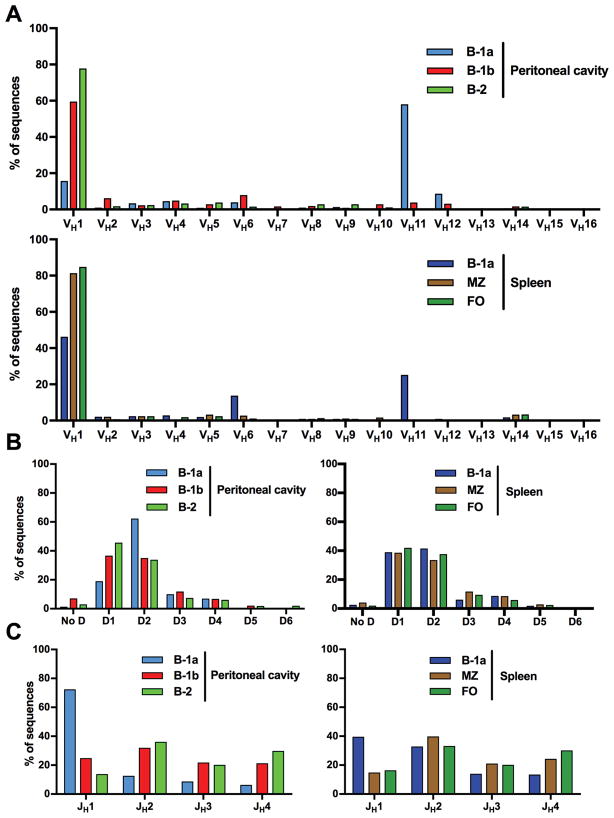
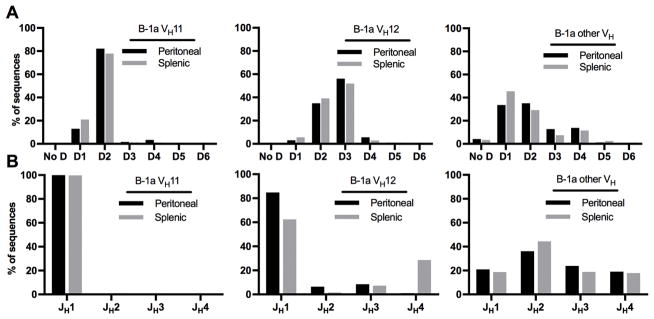
Figure 2. B-1a cells preferentially express VH11, D2, and JH1 gene segments.
(A) Shown here is the expression of each VH family as percent of total functional transcripts in each subset. Remarkably, peritoneal and splenic B-1a cells expressed a substantial proportion of VH11 transcripts and also some VH12 sequences. On the other hand, conventional B cells (peritoneal B-2, splenic MZ, FO) did not use either of these V genes more than 0.2% in any subset. Concomitantly with the increased expression of VH11, there was relatively decreased expression of segments from the VH1 family in B-1a cells compared to other B cell subsets. (B) All populations used mainly D1 and D2 segments, but peritoneal B-1a cells preferentially expressed segments of the D2 family compared to all other subsets. (C) Both peritoneal and, less pronounced, splenic B-1a cells expressed sequences with the JH1 gene segment more frequently than conventional B cells that preferentially expressed JH2.
Number of reads analyzed for each cell type: Peritoneal Cavity: B-1a: n=59,703, B-1b: n=66,030, B-2: n=48,579; Spleen: B-1a: n=81,571, MZ: n=70,304, FO: n=52,749.
VH gene segments (Fig. 2A)
Major V-D-J gene expression for IGHV sequences of each B cell subset is shown in Figure 2 and more detailed analyses of VJ rearrangements can be found in Supplemental Figures 1–3. Genes of the VH1 family (the biggest V family, consisting of 53 functional genes) were the most commonly used VH genes in all B cell subsets besides B-1a cells. They ranged from 59.6% in B-1b cells to 84.8% in follicular cells, whereas they only comprised 15.8% and 46.2% of peritoneal and splenic B-1a sequences, respectively. B-1a cells expressed VH11 genes substantially more frequently than other B cell subsets (58.1% and 25.2% of peritoneal and splenic B-1a sequences, respectively) (Fig. 2A). All of these were of the VH11-2 gene segment (Suppl. Figs. 1 and 2). B-1b cells also expressed VH11 transcripts, but to a relatively limited extent (3.9% of B-1b sequences). Additionally, 8.7% of peritoneal B-1a sequences contained VH12, compared to 3.2% of B-1b and 1.0% of splenic B-1a sequences, respectively. Of note, conventional B cells showed little to no expression of VH11 or VH12 segments (<0.2% in all subsets). This confirms prior studies showing preferential expression of VH11 and VH12 in B-1 cells (23, 25, 42). In our analysis however, we show that expression of both these prototypic B-1 cell VH genes was significantly higher in B-1a cells than in B-1b cells.
D gene segments (Fig. 2B)
In most B cell populations, D1 and the D2 family were the most common D genes and were expressed to a similar extent (33.5–45.7% of transcripts). However, peritoneal B-1a cells exhibited preferential expression of genes from the D2 family (62.4%) with an accompanying decreased use of D1 (19.0%). In contrast, D3 and D4 genes were present only to a limited extent (5.9–11.8%) in all subsets. The increase in D2 family genes in peritoneal B-1a cells was mostly driven by higher expression of D2-1 and D2-5 with decreased expression of other D2 genes compared to other sequences. Less prominently, this pattern was also present in splenic B-1a cells (Suppl. Fig. 4).
JH gene segments (Fig. 2C)
Conventional B cells and B-1b cells also expressed J genes in similar proportions, using mainly JH2 (32.0–39.8%) and comparable proportions of JH1 (13.9–24.9%), JH3 (20.2–21.9%), and JH4 (21.3–30.1%). However, peritoneal and, to a lesser extent, splenic B-1a cells expressed JH1 more frequently than other subsets (72.4 and 39.6% respectively), with an accompanying decrease in expression of other JH genes. Interestingly, JH1 was previously noted to be the most commonly expressed JH gene among neonatal cells, further indicating a possible fetal origin of sequences with this gene (43).
CDR3s of B-1 cells are less hydrophobic, shorter and have fewer N additions than those of conventional B cells
Hydrophobicity
We used the Kyte-Doolittle scale to compare average CDR3 hydrophobicity. All B-1 subsets expressed CDR3s with a lower average hydrophobicity (−0.18 ± 0.001 to −0.21 ± 0.001) compared to conventional B cell sequences, whose means also were in a relatively narrow range for all subsets, but at a higher hydrophobicity (−0.09 ± 0.002 to −0.12 ± 0.001) (Fig. 3A). Of note, Vale et al. reported similar findings when analyzing VH5 sequences in BALB/c mice. (26) Different studies by the same group found that forcibly expressing a highly charged CDR3 led to decreases in B-1 cell populations with an increase in peritoneal B-2 and splenic MZ cells (44). In contrast to this increase in conventional B cells with expression of a highly charged CDR3, we find that in our studies these subsets actually express sequences with higher CDR3 hydrophobicity compared to B-1 cells.
Figure 3. B-1 sequences are less hydrophobic, have a shorter CDR3 and D segment, and less frequently contain N additions.
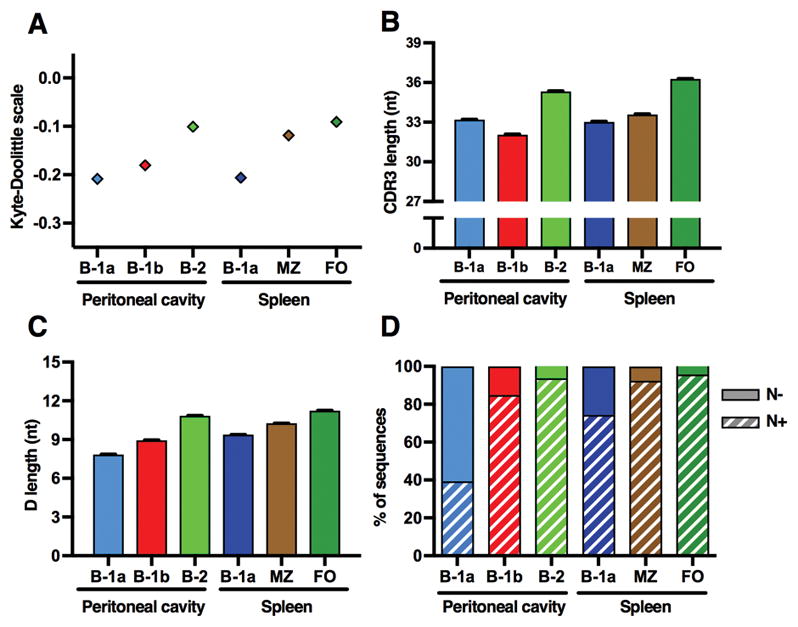
(A) Average CDR3 hydrophobicity of B-1 cell CDR3s was in a narrow range and notably lower compared to conventional B cells. (B) CDR3s of peritoneal B-2 cells and splenic follicular cells were longer compared to all other subsets. (C) A similar trend was seen with D length, which increases from B-1a to B-1b to B-2 cells in the peritoneal cavity, and also from B-1a to MZ to FO cells in the spleen. Each value is displayed as mean ± SEM (error bars were too small to be accurately displayed for hydrophobicity). (D) Sequences were analyzed in regard to containing N additions at either the V-D or D-J junction in the presence of an identifiable D gene. Peritoneal B-1a cells had by far the largest fraction of N− sequences, compared even to splenic B-1a cells, consistent with fetal origin of peritoneal B-1a cells. Splenic B-1a cells had far fewer N− sequences, consistent with postnatal development as might occur with enrichment of B-1a cells from the bone marrow.
One-way ANOVA: hydrophobicity: peritoneal vs. splenic B-1a cells: p > 0.05. Comparisons between all other subsets: p < 0.0001. CDR3 length: all comparisons: p < 0.0001. D length: all comparisons: p < 0.0001. The number of reads for each cell type is provided in legend of Fig 2.
CDR3 and D length
CDR3s were shorter in B-1 and MZ cells (32.05 ± 0.03 to 33.59 ± 0.02 nt) compared to peritoneal B-2 and splenic FO cells (35.33 ± 0.04 and 36.27 ± 0.04 nt, respectively) (Fig. 3B). A similar trend was seen in regard to D length. Peritoneal B-1a CDR3s contained the shortest (7.85 ± 0.01 nt) and splenic FO cells the longest D segments (10.85 ± 0.02 and 11.24 ± 0.02 nt). B-1b (8.95 ± 0.02 nt), splenic B-1a (9.38 ± 0.01 nt), MZ (10.26 ± 0.02 nt), and peritoneal B-2 (10.85 ± 0.02 nt) D lengths fell in between these subsets (Fig. 3C).
N additions
We categorized all transcripts with an identifiable D segment as either N+ or N−, based on the presence of N additions at either the V-D or D-J junction. In the peritoneal cavity, 60.6% of peritoneal B-1a transcripts were N−, much more than in any other subset, suggesting a much greater contribution of fetal development to this compartment. In contrast, only 15.1% and 25.6% of B-1b and splenic B-1a transcripts respectively, were without N additions. N− sequences were prevalent only to a very limited extent in all conventional B cell subsets (4.3 to 7.6%) (Fig. 3D).
Comparison of N+ vs. N− sequences reveals similarities between B-1 cell subsets
We extracted N− and N+ sequences from the database and analyzed them individually for each subset. Most notably, the fraction of VH1 family genes was relatively low in both peritoneal and splenic B-1a N− sequences, while showing a higher proportion of VH11 sequences (Fig. 4A). Furthermore, all B-1 cell N− transcripts showed substantially lower average CDR3 hydrophobicity in a tight range (−0.268 ± 0.005 to −0.292 ± 0.001) compared to N− sequences of conventional B cells and N+ sequences of all cell subsets (−0.086 ± 0.003 to −0.170 ± 0.001) (Fig. 4B). Average CDR3 lengths were similar in N− and N+ B-1a transcripts (32.50 ± 0.01 to 34.13 ± 0.04) and were also longer than those of N− sequences of other subsets (27.86 ± 0.11 to 29.36 ± 0.14). Notably, B-2 and follicular N+ sequences had the longest average CDR3 length (36.27 ± 0.03 and 36.91 ± 0.04, respectively) (Fig. 4C).
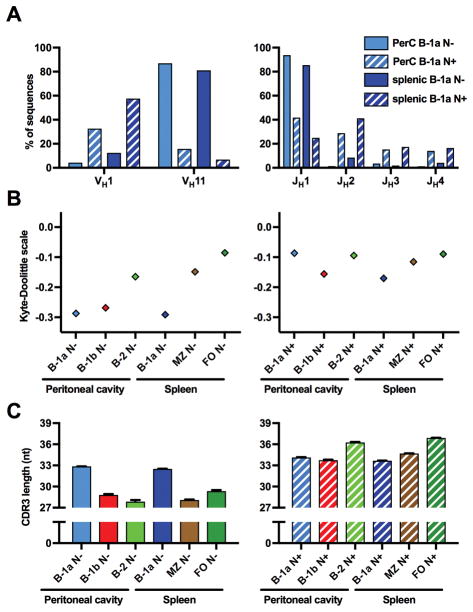
Figure 4. B-1a N+ sequences differ substantially from N− sequences.
(A) Both peritoneal and splenic B-1a N− sequences contained VH11 to a much higher amount than the respective N+ sequences. JH1 was expressed more frequently and was the predominant J segment in B-1a N− sequences. In other cell subsets, VDJ usage was not substantially different between N− and N+ sequences (not shown). (B) B-1 N− CDR3s were on average substantially less hydrophobic than the CDR3s of all other subsets, including B-1 N+ sequences. (C) CDR3s of N− sequences were markedly shorter on average, except in B-1a sequences, for which N+ and N− CDR3s were roughly the same length.
Each value in A is displayed as a proportion of total sequences. In B and C, values are displayed as mean ± SEM (error bars were too small to be accurately displayed for hydrophobicity).
One-way ANOVA: Hydrophobicity: p > 0.05: PerC B-1a N− vs. splenic B-1a N−, PerC B-1a N+ vs. B-2 N+, PerC B-1a N+ vs. FO N−, PerC B-1a N+ vs. FO N−, B-1b N+ vs. B-2 N−, B-2 N− vs. splenic B-1a N+, B-2 N− vs. MZ N−, B-2 N+ vs. FO N−, B-2 N+ vs. FO N−, B-2 N+ vs. FO N+, FO N− vs. N+. p < 0.01: all other comparisons. CDR3 length: p > 0.05: B-1b N− vs. FO N−, B-1b N+ vs. splenic B-1a N+, B-2 N− vs. MZ N−). p < 0.001: all other comparisons.
B-1a cells have distinct preferential V-D-J rearrangements
VH and JH rearrangements
We examined the frequency of the VH and JH rearrangements in the analyzed B cell subsets. Both peritoneal and splenic B-1a cells showed distinct rearrangements compared to conventional B cells, expressing a high number of VH11/JH1 recombinations, with peritoneal B-1a cells also using a considerable number of VH12/JH1 sequences. These findings highlight the restricted repertoire in both peritoneal and splenic B-1a cells. In contrast, peritoneal B-1b and B-2 transcripts, as well as splenic MZ and FO sequences displayed a much more diverse set of VJ rearrangements (Supplemental Figures 1 and 2). Because VH11 and VH12 sequences account for more than 50% of the B-1a transcripts, we also present the data for peritoneal and splenic B-1a cells in a form that excludes VH11 and VH12 transcripts to better visualize all other clones (Suppl. Fig. 3).
DJ usage of VH11 and VH12 B-1a sequences
VH11 and VH12 have been previously described as prototypical V genes of B-1a cells (45–47). Transgenic mice for either of these V genes have a marked increase of their B-1 cell population and of phosphatidylcholine-binding antibodies (48). In contrast, conventional B cells showed very little to no expression of these V genes in our studies. We further investigated the properties these specific V genes have in B-1a cells, which appeared to be the most unique of all subsets. Of note, all of the VH11/VH12 sequences in our database contained the genes VH11-2 and VH12-3, respectively (Supplemental Figs. 1 and 2).
Overall, D and J expression were very similar between peritoneal and splenic B-1a subsets, indicating that a substantial number of sequences expressed might be the same in both subsets. Whereas B-1a sequences used different DJ genes than other B cell subsets, sequences that contained neither VH11 nor VH12 showed comparable D gene rearrangement to conventional B cells or B-1b cells (Fig. 5A). The expression of D1 and D2 in non-VH11/VH12 sequences in B-1a cells was similar to those in conventional B cells. However, rearrangements with VH11 and VH12 appeared to utilize a very restricted set of both D and J segments. In VH11 sequences, there was a striking predominance of rearrangement with D2 genes, and to a smaller extent also with D1, but almost never rearranged with D3 or D4 genes. Furthermore, we found that VH12 sequences preferentially contained D2 or D3. In fact, D3 was the most common D gene in VH12 sequences, but was found to rearrange only rarely with other VH genes (Fig. 5A).
Figure 5. VH11 and VH12 segments rearrange with a restricted set of D and J genes.
This analysis examines the usage of D and J genes in VH11, VH12, and all other VH gene segment sequences. (A) VH11 most frequently rearranged with D2 genes, and to a smaller extent D1, while VH12 sequences contained mainly D2 and D3. B-1a sequences with other V genes use D genes in similar proportions as other B cell subsets (compare to Figure 2B). (B) JH usage was even more restricted within these sequences. While VH11 sequences exclusively contained JH1, VH12 sequences also rearranged with other JH segments to a noteworthy amount. As with D usage, B-1a sequences with other V genes showed a similar rearrangement with J genes as conventional B cells (compare to Figure 2C).
The number of reads for each gene usage: Peritoneal cavity: VH11: n=34,672, VH12: n= 5,206, other: n=19,825; spleen: VH11: n=20,508, VH12: n=790, other: n=60,077.
There was a similar restricted pattern in J usage of VH11 or VH12 sequences (Fig. 5B). Whereas non-VH11/VH12 sequences showed a slight predominance of JH2 and almost equal usage of the remaining 3 J genes (again similar to conventional B cells and B-1b cells), the rearrangement of VH11 and VH12 with J genes was very restricted: VH11 almost exclusively and VH12 predominately rearranged with JH1. Interestingly, 28.7% of VH12 sequences rearranged with JH4 in splenic B-1a cells, whereas peritoneal B-1a VH12 only rarely recombined with JH4 (0.4%), indicating different selection processes or developmental origin (Fig. 5B).
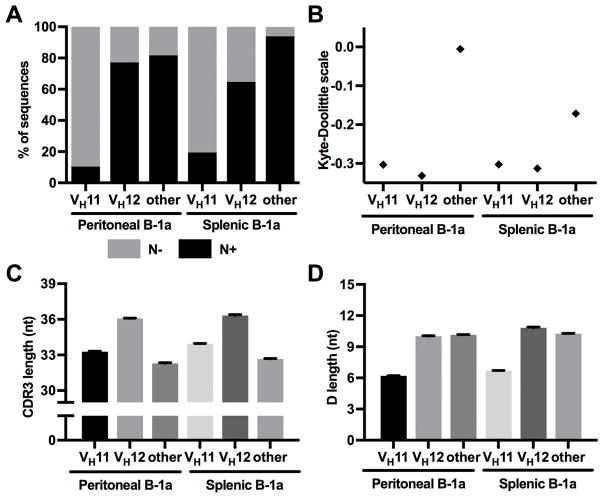
CDR3s of VH11 and VH12 sequences differ from other sequences
While we showed above that a substantial proportion of B-1a sequences have no N additions, we next investigated if there is a difference in N additions between sequences that have different V genes. In fact, we found that most VH11 sequences did not contain N additions and this was true for both peritoneal and splenic B-1a cells, whereas for VH12 sequences, the proportion of N− sequences was lower and similar to non-VH11/VH12 sequences (Fig. 6A). VH11 and VH12 sequences were also much less hydrophobic than other sequences (Fig. 6B). Interestingly, peritoneal B-1a CDR3s that did not contain VH11 or VH12 were the most hydrophobic subset in our analysis, even more hydrophobic than all N+/N− subsets in Fig. 4B. This increased hydrophobicity in non-VH11/VH12 sequences was not seen in splenic B-1a CDR3s whose hydrophobicity was in a range comparable to most conventional B cell (both N+ and N−) and B-1 N+ sequences, as shown in Fig. 4B. Furthermore, CDR3s of VH12 sequences were longer than the CDR3s of VH11 or non-V11/V12 sequences and in a similar range as B-2/FO cells (Fig. 6C). Interestingly, D length was the shortest in VH11 sequences, whereas the D segment of VH12 and non-V11/V12 sequences was approximately one amino acid longer (Fig. 6D).
Figure 6. CDR3s of VH11 and VH12 B-1a sequences have distinct properties.
(A) VH11 sequences had a substantially higher proportion of N− sequences in both peritoneum and spleen compared to sequences with other VH genes. (B) VH11 and VH12 sequences were markedly less hydrophobic, whereas other B-1a sequences fell into a similar range as sequences from other (conventional) B cell subsets. Non-V11/V12 subtype peritoneal B-1a sequences were also more hydrophobic than average conventional B cell sequences (compare to Fig. 4B). Interestingly, this was not the case for splenic B-1a cell sequences. (C, D) Both the CDR3 region and D segment were longer in VH12 sequences (similar to peritoneal B-2 and splenic FO cells in Figure 3B), and shorter in the remaining sequences. VH11 sequences also had a substantially shorter average D segment.
All values in sections C+D are displayed as mean ± SEM. Error bars were too small to be accurately displayed in B (hydrophobicity).
Hydrophobicity: peritoneal VH11 vs. splenic VH11, peritoneal VH11 vs. splenic VH12, peritoneal VH12 vs. splenic VH12, splenic VH11 vs. splenic VH12: p>0.05. All other comparisons: p<0.0001
CDR3 length: peritoneal VH12 vs. splenic VH12: p>0.05; all other comparisons: p<0.0001
D length: peritoneal other vs. peritoneal VH12: p>0.05; all other comparisons: p<0.01
Clonotype analysis reveals similar patterns of VH11 and VH12 sequences
To determine if the rather unique properties of VH11 and VH12 sequences might be caused by the high expression of very few antibody sequences, we also analyzed sequences based on clonotypes instead of total sequences. One “IMGT clonotype (AA)” is defined as using the same VDJ genes and having the same CDR3 amino acid sequence. As opposed to total sequence numbers, in this analysis, each unique clonotype was regarded as occurring only once in the analysis. We found similar rearrangements of VH11 and VH12 clonotypes with D and J genes, and similar CDR3 properties to those found on the analysis of total sequences, confirming that the preferential recombinations and qualities described above are unlikely to be skewed by the high expression of only a few clones of antibodies (Suppl. Fig. 5).
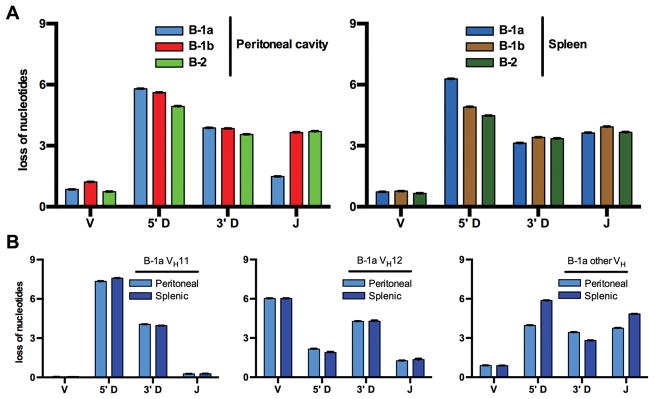
B-1a sequences are differently modified by exonucleases
During rearrangement, not only are nucleotides inserted, but they can also be removed from the end of each gene segment by exonucleases. We found that peritoneal B-1a sequences had less loss of JH nucleotides than other B cell subsets, similar to a previous report by Kantor et al. in peritoneal B-1a sequences from BALB/c mice (25). This could be explained by the high fraction of VH11 and VH12 sequences that on average lost only 0.3 and 1.3 nucleotides of their J gene, respectively. These numbers were very similar in both peritoneal and splenic B-1a cells (Fig. 7). On the other hand, peritoneal and splenic B-1a sequences had on average slightly more nucleotides excised from the 5′ D end. This is an attribute of their VH11 sequences, which had a higher 5′ D loss than any other sequence subset analyzed. Remarkably however, almost all VH11 gene segments were completely preserved with an average loss of only 0.04 and 0.07 (peritoneal and splenic, respectively) nucleotides. On the other hand, VH12 sequences lost an average of 6 nucleotides, considerably more than any other group analyzed (Fig. 7).
Figure 7. Loss of nucleotides at sites of VDJ rearrangement.
(A) Peritoneal B-1a sequences had a substantially lower loss of J nucleotides, and excision of more nucleotides at the 5′ D location was noted in splenic B-1a sequences. (B) Within B-1a transcripts, there were marked differences depending on the V gene used. Whereas virtually all VH11 sequences contained the complete VH gene in the CDR3, VH12 sequences had on average 6 nucleotides removed from their VH segment.
A: V loss: B-2 vs. splenic B-1a: p>0.05; B-2 vs. MZ: p<0.05; all other comparisons: p<0.0001; 5′D loss: B-2 vs. MZ: p>0.05; all other: p<0.0001; 3′D loss: peritoneal B-1a vs. B-1b: p>0.05; all other: p<0.0001; J loss: B-1b vs. B-2, B-1b vs. splenic B-1a, B-1b vs. FO, B-2 vs. FO, splenic B-1a vs. FO: p>0.05; B-2 vs. splenic B-1a: <0.01; all other: p<0.0001.
CDR3 amino acid sequences overlap between different B cell subsets
B-1 cells have been thought to have a unique repertoire of IgM antibodies. We thus investigated whether B-1 cells, and B-1a in particular, would contain distinct CDR3 amino acid sequences and in general, to what extent the expressed sequences overlap between B cell populations. The Antibody Mining Toolbox was used to extract CDR3 sequences from the database. For this analysis, we included only CDR3 amino acid sequences that occurred at least 5 times in the whole dataset across all subsets to reduce the probability that a given sequence might be a result of amplification or sequencing error. By this definition, there were 8,820 unique CDR3s that accounted for 71% of the total functional reads (267,840 of 378,944 total reads).
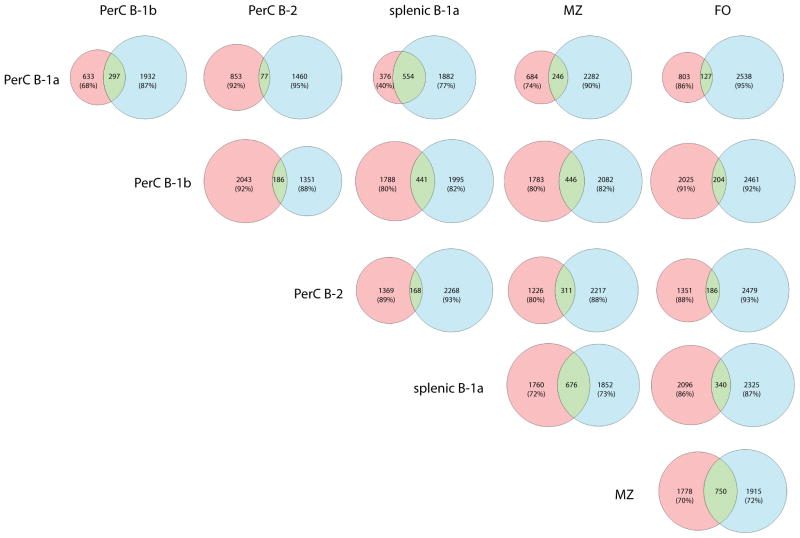
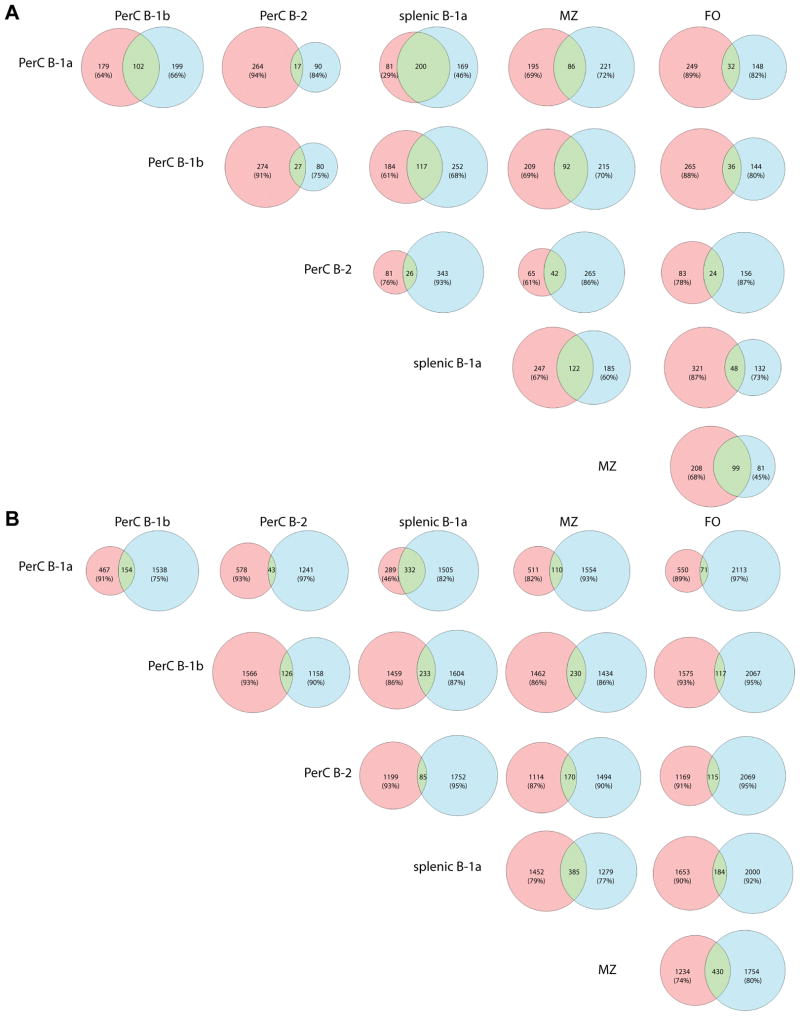
Figure 8 presents a series of Venn diagrams that examine the shared repertoire of unique CDR3 sequences in the different B cell subsets. In this diagram, the unique and shared repertoire of CDR3 sequences for each B cell subset in both the peritoneum and spleen are compared. In each horizontal line, the pink color identifies the unique CDR3 sequences of the indicated cell type on the left margin of that line, while the blue color identifies unique sequences of the cell type in the header of that column. The green field indicates the shared CDR3s of these two subpopulations. A shared sequence was defined as one expressed at least once in each of the subsets being compared.
Figure 8. CDR3 amino acid sequence analysis reveals insights into the relationship of B cell populations.
CDR3 amino acid sequences were extracted and compared.as described in the Results section. In each horizontal line, the pink color identifies the unique CDR3 sequences of the indicated cell type on the left margin of that line, while the blue color identifies unique sequences of the cell type in the header of that column. The green field indicates the shared CDR3s of these two subpopulations. Only sequences that occur more than 5 times across all cell subsets were used in these analyses, which includes 8,820 unique CDR3 sequences. A shared sequence was defined as one expressed at least once in each of the subsets being compared. We indicated the percentage of sequences the non-shared sequences make up of each cell subset in every comparison.
Most remarkably, 59.5% of unique peritoneal B-1a CDR3 amino acid sequences were also present in the splenic B-1a repertoire. However, splenic B-1a cells seemed to have a much more diverse CDR3 repertoire and also a considerable number of sequences that did not occur in the peritoneal B-1a pool (consistent with our other analyses above). Interestingly, the B-1b CDR3 repertoire had a similar overlap with B-1a cells in the peritoneal cavity and spleen, as well as with MZ cells. Relatively few shared sequences were found between B-1b cells and B-2/FO cells. Furthermore, peritoneal B-2 cells seemed to have a very unique repertoire with only little overlap to all other subsets. They actually shared the highest proportion of sequences with MZ cells, while surprisingly their repertoire barely overlapped with that of splenic follicular cells.
It has been assumed that the B-1a population of the peritoneum represents the reservoir of unique B-1a cells, which then migrate to the spleen to transform to plasma cells and secrete IgM as needed. Indeed, 59.6% of the peritoneal B-1a sequences were found in splenic B-1a cells as well. However, our analysis shows that the repertoire of the splenic B-1a compartment exceeded that of peritoneal B-1a cells, such that splenic B-1a cells had a 5-fold larger repertoire of unique CDR3 transcripts compared to peritoneal B-1a cells (1882 vs. 376 CDR3s, respectively), with 554 additional shared CDR3s between the two subsets. In a separate analysis not shown, even if one looked at the number of unique CDR3s that occurred only 1–3 times, the splenic B-1a transcripts were still substantially more numerous. This might suggest that the spleen could be a reservoir of unique B-1a cells, perhaps being seeded not only from the various tissue compartments that B-1a cells are thought to reside in, such as peritoneal and pleural cavities and the omentum, but also the bone marrow (2, 49, 50). In addition, peritoneal B-1a cells also shared 31.9% of their CDR3s with B-1b cells and 26.5% with MZ cells, but only small proportions with B-2 and FO cells (8.3 and 13.7%, respectively).
The repertoire of splenic B-1a cells, on the other hand, overlapped to a modest degree with MZ cells (27.8%) and to a smaller amount with B-1b (18.1%) and FO cells (14.0%). However, again the overlap with B-2 cells was much lower (8.4%), similar to the small overlap between peritoneal B-1a and B-2 cells.
The B-1b CDR3 repertoire, on the other hand, shared sequences with B-1a cells (both peritoneal and splenic) and MZ cells (13.3%, 19.8%, 20.0%, respectively). However, there was little overlap of B-1b cells with B-2 (8.3%) or follicular cells (9.1%). Another notable finding was that the CDR3 repertoire of peritoneal B-2 cells was the most unique compared to the other B cell subsets studied. The B-2 cell repertoire shared 20.2% of its sequences with MZ, but only 5–11% with B-1 cell subsets and, interestingly, only 12.1% with those of FO cells.
The analyses above were performed using all sequences. To better visualize clonotypes that are developmentally related among the B cell subsets, we performed a similar type analysis using N− and N+ transcripts separately (Figure 9). In these analyses, the majority of B-1a N− sequences were shared between peritoneal and splenic B-1a cells, but the splenic B-1a subset contained many more N+ sequences than peritoneal B-1a cells, consistent with more postnatal development of splenic B-1a cells. Interestingly, there is relatively high overlap of N− sequences between B-1 and MZ cells (30.6% of peritoneal B-1a, 30.6% of B-1b, and 33.1% of splenic B-1a, respectively). However, there was little overlap between B-1 cells and conventional B cells – only 6–13% of the B-1 N− sequences were found in the B-2 or FO repertoires. This could indicate prenatal selection processes that are similar for B-1 and MZ cells, but do not include B-2 and FO cells. Complicating this picture, a substantial number of conventional B cell N− sequences were shared with MZ cells (39.2% of B-2 and 55.0% of FO, respectively), thereby making the N− repertoire of MZ cells a hybrid of B-1 and conventional B cell repertoires (Fig. 9A). On the N+ level, besides B-1a cells in the peritoneal cavity and spleen, there was also notable overlap in the repertoires of MZ cells with splenic B-1a cells and FO cells (23.1% and 25.8% of MZ sequences, respectively). Here, MZ cells shared fewer sequences with the peritoneal subsets (7–14% of MZ sequences), suggesting a postnatal separation of the peritoneal and splenic B cell repertoires (Fig. 9B).
Figure 9. Overlap of N− and N+ CDR3 repertoires.
Applying the same rules as in Figure 8, we separately compared sequences with and without N additions. (A) First, only N− sequences were analyzed. Most notably there was marked overlap between the N− repertoires of peritoneal and splenic B-1a cells. These cell subsets shared more N− sequences than there are unique sequences in each of the populations. The MZ subset shared a significant number of N− sequences with all other subsets (14–40% of its N− sequences). B-2 cells have a low absolute number of N− sequences – however, 39.2% of these B-2 N− sequences also occur in MZ cells, which is the closest relationship the B-2 repertoire has with any other cell type. FO cells shared a substantial amount of their N− sequences with MZ cells, but notably only few with B-2 cells. (B) Analyzing only N+ sequences, there again was a large number of sequences shared between peritoneal and splenic B-1a cells, but the splenic B-1a subset included many more N+ sequences than the peritoneal. There was also marked overlap between splenic B-1a and MZ cells – in absolute numbers, they shared more N+ sequences than peritoneal and splenic B-1a cells. MZ cells also shared a significant portion of their transcripts with FO cells, which was the biggest absolute number of shared N+ sequences between any subset. Peritoneal B-2 cells proved to be an isolated subset, having the most overlap with MZ cells, but still at a low quantity of only 13.2% of their N+ repertoire.
Two peptide CDR3 sequences account for a large number of B-1a sequences
In Table 1, we show a list of the most common peptide CDR3s in each subset. Remarkably, we found that two specific peptide CDR3 sequences, which differ only in one amino acid (CMRYGNYWYFVW, CMRYSNYWYFDVW), made up a large proportion of total B-1a CDR3 sequences. Together, these sequences accounted for 43.4% of the total number of peritoneal B-1a and 15.9% of splenic B-1a peptide sequences, and thus, are a major contributor of the VH11 pool in B-1a cells. They were the most common B-1a transcripts in each of the 3 independent B cell sorts, as well as in a pilot experiment on peritoneal B-1a cells performed on 10 female C57BL/6 mice bred in our vivarium, as opposed to the purchased mice used in the studies reported in this paper. Both sequences are known V11-D2-J1 sequences that have been previously described and studied as prototypic B-1 cell antibodies binding to BrRBCs and phosphatidylcholine (21, 42). These same sequences were also recently reported by Yang et al. as consistently among the top 10 recurring CDR3 sequences in splenic B-1 samples in both specific-pathogen-free and germ-free mice (30).
Table 1. Most frequently expressed CDR3 peptides by B-cell subset.
Remarkably, 2 CDR3s (both V11-D2-J1) that differ only by one amino acid are by far the most frequently expressed peptide sequences in both peritoneal and splenic B-1a cells. The low numbers in MZ and FO cells can be explained by their diverse repertoire with less predominance of a few antibodies, as seen in the repertoires of B-1 cell subsets.
| Peritoneal B-1a cells | Peritoneal B-1b cells | Peritoneal B-2 cells | |||
|---|---|---|---|---|---|
| CMRYGNYWYFDVW | 17336 | CARNSGSYYGSSDWYFDVW | 3301 | CARSDITTVVADYW | 2089 |
| CMRYSNYWYFDVW | 8825 | CARWGELGLWFAYW | 2491 | CARGADYEGGYYAMDYW | 1925 |
| CMRYGSYWYFDVW | 1832 | CARGTTVPDFDYW | 1840 | CARDGATVVAKRDYAMDYW | 1448 |
| CMRYGSSYWYFDVW | 1188 | CACNYDAMDYW | 1835 | CASYYGSSHFDYW | 974 |
| CARGQFITTVVAFDYW | 979 | CARDYDFDYW | 1422 | CARGGVANYYFDYW | 956 |
| CAGDSWGYWYFDVW | 973 | CARHGSSYEDWYFDVW | 1128 | CARVRGHAMDYW | 909 |
| CASSNYAWFAYW | 863 | CTRDYYGSDYW | 1047 | CARDRNYGMRNPFDYW | 884 |
| CMRYGPYWYFDVW | 784 | CAGDRDGYWYFDVW | 1006 | CASDGNLDDYAMDYW | 834 |
| CARSWHYGSSYVIAYW | 677 | CMRSRGDYW | 982 | CARDYYADYW | 784 |
| CMRYSSYWYFDVW | 564 | CASGGELPFAYW | 956 | CAILGHPYW | 767 |
| Splenic B-1a cells | Marginal Zone cells | Follicular cells | |||
| CMRYSNYWYFDVW | 7367 | CARGGHYYGGFDYW | 122 | CARRRNYDYAMDYW | 187 |
| CMRYGNYWYFDVW | 6314 | CARMELGRDYAMDYW | 68 | CARPVVANYAMDYW | 151 |
| CTAPAGSSDYW | 3529 | CARDGYDYFDYW | 66 | CARDGSSYVGYAMDYW | 123 |
| CTGPGSSLGDW | 3090 | CARGVFDVW | 65 | CARGDYYAMDYW | 107 |
| CAKGSNYVDW | 2458 | CARDGDYGSRRAMDYW | 60 | CAVDSSGPYYYAMDYW | 106 |
| CMRYGSSYWYFDVW | 2369 | CARRAGTFFDYW | 60 | CARWGYGSSGYW | 99 |
| CTADPNAYW | 1756 | CTKDSSGSYYFDYW | 57 | CAREGGFPW | 94 |
| CARGGHYYGGFDYW | 1525 | CTGGALYAMDYW | 54 | CARSITTVVARDYFDYW | 82 |
| CTRSNYYGRSPSYFDVW | 1012 | CTRYPTGPESYFDYW | 53 | CTPYTNAFAYW | 81 |
| CMRYDSNYWYFDVW | 911 | CARSGYYGFDYW | 50 | CTGLAWFAYW | 80 |
A single-chain variable fragment (scFv) of a common B-1a antibody binds BrRBCs and OSEs
To characterize the binding specificity of the most common CDR3 sequence, CMRYGNYWYFVW, as described in Methods, we generated a cDNA for an scFv antibody using the original IGHV bearing this CDR3, paired with the light chain variable region from the original hybridoma as described by Reininger et al. (21), which we termed XQ11-scFv (Fig. 10A). Its cDNA was transfected into HEK293 cells and the binding characteristics of XQ11-scFv secreted into the culture medium was characterized by chemiluminescent assay. In earlier studies, the IgM antibody had been shown to bind to BrRBCs and it was postulated that this was to a cryptic epitope that might be represented by binding to phosphatidylcholine present on liposomes (21, 24). As shown in Fig. 10B, XQ11-scFv binds to murine BrRBCs as originally reported, but we now show that it binds specifically to the phosphocholine (PC) epitope present on BSA, i.e. to the headgroup of phosphatidylcholine. In contrast, XQ11 does not bind to unmodified BSA. Importantly, XQ11 also binds prominently to OxLDL, a rich source of oxidized phospholipid (OxPL), which prominently displays such PC. The binding of XQ11 to OxLDL and OxPL is analogous to the binding of the prototypic IgM E06 (or IgA T15) natural antibody, which binds the PC of OxLDL or the PC present on the cell wall of S. pneumoniae (14, 15). We have previously described PC in this context as an OSE and shown that IgM natural antibodies to PC attenuate atherosclerosis development (15, 51). Of interest, XQ11-scFv also appears to bind to a limited extent to the starting preparation of murine RBCs not treated with bromelain, perhaps consistent with the concept that RBCs steadily accumulate OSEs with aging (52).
Discussion
In this study, we utilized massively parallel sequencing to define the complete IGHV repertoire of peritoneal (B-1a, B-1b, B-2) and splenic (B-1a, MZ and FO) B cell subsets from female C57BL/6 mice 3 months of age. B-1 cells in particular are a unique subset of lymphocytes whose repertoire is believed to have developed through natural selection and whose antibodies have important homeostatic and housekeeping functions. We have suggested that in particular a substantial subset of these IgM NAbs are directed to OSEs and not only provide homeostasis to OSEs found on OxLDL but also on apoptotic cells and microvesicles, which otherwise would be both immunogenic and pro-inflammatory (reviewed in (13)). We have also suggested that because such innate IgM represent soluble PRRs, their selection has been additionally influenced in order to provide homeostasis against PAMPs of pathogens. A prototypic example of such an IgM NAb is the B-1 cell derived T15/E06 idiotype antibody that was first identified for its binding to phosphocholine (PC) on the cell wall of S. pneumoniae, and which provides optimal protection to mice against lethal infection with S. pneumoniae infection (20, 53). Additionally, we have shown that E06 provides homeostasis by neutralizing inflammatory properties of microvesicles and apoptotic cells bearing PC containing oxidized phospholipids (OxPL) (12, 54), and restricts atherosclerosis by both inhibiting uptake of OxLDL by macrophages and by preventing inflammatory properties of OxPL (14, 15, 55). In a similar manner, we have shown that an even greater number of both murine and human cord blood IgM NAb bind to other OSEs, and in particular malondialdehyde type adducts (12, 13, 51). Of course, it has been long known that B-1 cell antibodies provide the first line of protection against many bacterial and viral pathogens (7, 56, 57). Furthermore, it has been reported that the titers of such innate IgM NAbs decline with age, and could thus contribute to a general weakening of innate immune responses with aging (58, 59). Thus, knowing the baseline repertoire and understanding how the B-1 cell antibody repertoire changes with aging and with disease can give insight into beneficial functions of these antibodies that could eventually be used in humans by passive or active immunization strategies.
In a recent elegant publication, Yang et al. traced the early fate of B-1a cells and demonstrated that B-1a represent a B cell lineage whose IGHV recombinations represent fetal cells, as their IGHV rearrangements are primarily without N additions typical of post-natal TdT activity (30). They found a paucity of N additions in splenic B-1a cells up to 6 days of life, which slowly increases until weaning at ~ 3 weeks. Thereafter, the proportion of splenic B-1a cells with N additions appears to stabilize with ~ 80% showing multiple insertions. Our data are consistent with these observations, as 74% of the splenic B-1a sequences contained N additions. Interestingly however, in contrast to the splenic B-1a population, we found that about 60% of all peritoneal B-1a rearrangements still lacked N additions at 3 months of age, and it was particularly striking that among the VH11 sequences that dominated the B-1a peritoneal repertoire at that age, about 90% of the sequences lack N additions, consistent with a fetal origin of these cells. It might thus be surmised that the peritoneal B-1a population is more isolated developmentally than the splenic B-1a pool. Furthermore, Vale et al. previously demonstrated that very few VH5 (VH7183) sequences from the neonatal liver have N additions, while almost all sequences obtained from adult bone marrow contained N insertions (26). Additionally, Holodick et al. showed recently that B-1a cells derived from adult bone marrow progenitors in fact had a higher number of N additions compared to fetal liver progenitor cells (60). It should also be noted that the total unique expressed repertoire of the peritoneal B-1a cells was only 930 peptide sequences in this analysis, which is the smallest number of unique sequences among all the B cell subsets studied. This is even more striking when it is realized that two sequences alone accounted for 43% of all expressed sequences, as noted below. This restricted repertoire, which is nearly 5-fold smaller than that of the splenic B-1a population, plus the largely prenatal origin of these cells, is consistent with a very protected and isolated set of “innate” soluble PRRs that are the products of natural selection mediated by both innate DAMPs and exogenous PAMPs and are tasked with maintenance of homeostatic functions against “conserved” antigens.
As noted above, it is also of considerable interest that B-1a cells in the spleen have a larger repertoire than peritoneal B-1a sequences, and about 75% of these sequences contain N additions, quite different than seen with peritoneal B-1a cells. This suggests that at least by 3 months of age, the majority of the splenic B-1a cell population is postnatal in origin, consistent with findings of Yang et al (30). One might thus speculate that the spleen could act as a “melting pot” of prenatal B-1a cells residing in the peritoneal cavity and postnatal B-1a cells, the latter of which might be greatly enriched from other sites such as the bone marrow (50, 60). B-1a cells from the peritoneum migrate to the spleen when activated (11), whereas the vascular architecture of the spleen could enhance the possibility of bone marrow-derived B-1a cells to take up residence in the splenic pulp. Further studies tracking B-1a cells from the fetal liver and bone marrow are needed to confirm these hypotheses.
Yang et al. (30) utilized a deep sequencing strategy to report on the IGHV repertoire of various B cell subsets of varying ages in C57BL/6 mice including B-1a cells, but their studies did not include B-1b cells. Thus, the current results are the first to include a full IGHV characterization of all peritoneal and splenic B cell subsets including the B-1b subset. It has long been discussed how closely B-1a and B-1b cells are related as their surface markers only differ in expression of CD5. In striking contrast to the peritoneal B-1a population, we found in the current study that about 85% of the IGHV sequences from peritoneal B-1b cells from 3-month-old mice had N additions, and thus are likely to have developed postnatally. In addition, the VH11 sequences that were expressed so prominently in peritoneal B-1a cells were very rarely found in B-1b sequences, further distinguishing B-1a from B-1b populations. Indeed, a recent study by Ghosn et al. showed that a single adult hematopoietic stem cell could replenish the B-1b, but not the B-1a cell population (9). Further evidence of a distinct origin of B-1a and B-1b may also be seen in the Venn diagrams of Fig. 8, which compare unique sequences in different B cell subsets. Note that the shared repertoire of peritoneal B-1a and B-1b CDR3 peptide sequences is very small: From among 2,229 unique B-1b sequences, there are only 297 (13.3%) shared sequences, further distinguishing the unique origins of these two seemingly closely related B-1 cell subsets. Despite this difference in the overall functional repertoire, it appears that both B-1a and B-1b cell compartments secrete IgM that are anti-inflammatory and prevent atherosclerosis development and progression. Indeed we and others have shown that B-1a cells provide atheroprotective IgM (13) and more recently, that B-1b antibodies also decrease atherosclerosis and protect against obesity-associated inflammation (61, 62). Interestingly, the overlap of the B-1b repertoire with MZ sequences was larger than with peritoneal B-1a and at about the same level as with splenic B-1a sequences. Splenic B-1a cells, on the other hand, also shared a significant number of sequences with MZ cells. However, while MZ cells also share a significant portion of their repertoire with splenic B-1a cells, interestingly, this was less so with peritoneal B-1a cells. On a more detailed analysis, MZ cells share substantial parts of their N− repertoire with all other subsets while there was little overlap in N− sequences between B-1 and B-2/FO cells. Therefore, MZ cells could be regarded as the cell type that “bridges the gap” between B-1 and conventional cells on a repertoire level. However, this interaction did not appear as strong on the N+ level, suggesting that these cells have more similarities in repertoire development prenatally, while the repertoires diverge postnatally.
Other B cell subsets thought to be closely related are peritoneal B-2 cells and splenic FO cells. Interestingly, as shown in the Venn diagrams of Fig. 8, the shared repertoire between peritoneal B-2 and splenic FO cells is very small and B-2 cells seem to be very isolated in their repertoire. In a prior study, B-2 cells transplanted into peritoneal cavities of Rag−/− mice led them to acquire a B-1b like phenotype, suggesting there might be some form of relationship or interchange between them (63). Notably, this could not be confirmed on a repertoire base in our mice, as we found very little overlap between peritoneal B-1b and B-2 cells. Of note, the highest numbers of sequences B-2 cells share with any other subset are MZ cells, both on the level of N− and N+ sequences. Thus, one might speculate that B-2 cells represent MZ cells in a different environment, rather than FO cells. However, even this amount was low (~20% of B-2 sequences) and B-2 cells notably lack the overlap of their repertoire with B-1 cells that MZ cells show, as noted above.
As suggested by the literature and confirmed by our studies, VH11 and VH12 are V genes commonly expressed by B-1a cells, but not conventional B cells. They showed a remarkable specificity for rearrangement with certain D and J genes that were used to a lesser extent in all other sequences, indicating that these are conserved sequences that need a very specific rearrangement to lead to the formation of a mature B cell. The high number of transcripts without N additions in VH11 sequences (both on the level of total sequences and clonotypic analysis) further suggests that prenatal development of VH11 is predominant in B-1a cells. Interestingly, about 75% of VH12 transcripts contained N additions, indicating that this V gene is also selected postnatally to a much greater extent. In accordance to these findings, Gu et al. showed that JH1 (which is the predominant J gene of VH11 sequences) is the most common J gene in neonatal pre-B cells, but not later in life (43). In line with these observations, we found that the majority of N− sequences in B-1a cells (~85–95%) contained this J gene, while B-1a N+ sequences contained this V gene at a much lower frequency (~25–40%).
In our analysis, B-1 cell CDR3s had a lower hydrophobicity compared to conventional B cells, mainly driven by their N− sequences, consistent with a previous report by Vale et al. of peritoneal VH5 sequences in BALB/c mice (26). Interestingly, the same group showed that forced expression of a highly charged CDR3 led to significantly decreased B-1a cell numbers and a slight increase in peritoneal B-2 and splenic MZ cells (44). This is in apparent contrast to our study in which we found lower hydrophobicity of B-1 cell CDR3s compared to conventional B cells. However, it has been shown that C57BL/6 mice (used in our study) and BALB/c mice (used in the cited reports) regulate highly hydrophobic or highly charged CDR3s differently, which might at least partially account for these results (64). Of the B-1a sequences, VH11/VH12 sequences had a substantially lower average CDR3 hydrophobicity compared to other sequences. Non-VH11/VH12 sequences of B-1a cells in fact had an average CDR3 hydrophobicity in a range much closer to N+ sequences or the N− sequences of conventional cells, indicating that the VH11 and VH12 sequences contribute strongly in giving B-1a cells the unique repertoire properties found in this study.
Notably, many of the most highly expressed sequences in the report of Yang et al. contained short CDR3s of 5 or less amino acids, almost exclusively using JH2 genes (30). However, we did not make the same observation in our data where only 581 (0.15%) of all transcripts had a CDR3 length of 5 amino acids or less (11 JH1, 215 JH2, 319 JH3, and 36 JH4 sequences, respectively). We used a 5′-RACE amplification method of RNA as opposed to multiplex PCR used by Yang et al., but in theory this should not have any influence on these findings. With C57BL/6, we used the same strain of mice, but only studied a narrow age range. Also, as noted below, unlike the findings of Yang et al., there was only infrequent expression of the E06/T15 idiotype in our mice. Thus, one might speculate that differences in antibody expression between our data and that of others is in part due to differences in strains and ages of mice studied. Additionally, different antigen exposures of the mouse colonies could have caused further disparities in the antibody repertoire.
One of the most striking and unique qualities of B-1a cells in both the peritoneal cavity and the spleen is the expression of substantial numbers of VH11 sequences, which were very rarely found in other B cell subsets, including B-1b. This is consistent with previous reports that have found high numbers of these sequences in hybridomas generated from B-1a cells (23). The proportion of VH11 sequences was higher than in studies using BALB/c mice for analysis. However, this is in line with Seidl et al. (42), who found much higher proportions of VH11 in phosphatidylcholine-binding B-1 cells of C57BL/6 mice compared to BALB/c mice. This V gene has also been reported to primarily rearrange with JH1, a finding that we can also confirm, and is commonly found in antibodies that bind BrRBCs and phosphatidylcholine (24, 65). Yang et al. in fact reported that the expression of VH11 in splenic B-1a cell sequences increases substantially at 2–6 months of age compared to younger mice (30). Strikingly, in our 3-month-old mice, we identified two CDR3 sequences that were the most dominant sequences in B-1a cells both in the peritoneal cavity and in the spleen, accounting for 43.4% and 15.9% of all B-1a CDR3s, respectively. These exact CDR3 amino acid sequences have also been previously reported to bind BrRBCs and phosphatidylcholine present on liposomes, making them prototypical B-1a sequences (21). They were also highly expressed in all of the B-1a populations reported by Yang et al. as well, including in mice reared in a germ-free environment (30). These sequences were the most common B-1a transcripts not only in the mice used in this study, but also in a pilot experiment performed only on peritoneal cells, pooled from 10 female C57BL/6 mice that were bred in our vivarium. Their overwhelming dominance in B-1a cells warrants further studies about their target epitopes and their possible important physiological functions in vivo.
Our laboratory is especially interested in the role of B-1 cells in generating IgM antibodies that target what we have termed OSEs. We discovered that up to 20–30% of all IgM in non-infected mice, and in human umbilical cord blood, bind to various OSEs, such as PC, oxidized phospholipids (OxPL), malondialdehyde (MDA), malondialdehyde acetaldehyde adducts (MAA), 4-hydroxynonenal (4-HNE) and others (51). We have observed that all of these bind not only to a variety of oxidatively modified structures, but to apoptotic cells as well. We have thus suggested that they are part of the innate immune response providing homeostasis against oxidative stress. A major purpose in undertaking this study was to provide a basal library of the B-1 cell IGHV sequences at steady state. In further studies, we are exploring the impact on these sequences to perturbations of oxidative stress, such as high-fat high-cholesterol feeding, to determine which clones expand, and to annotate the epitopes to which such clones bind.
Surprisingly, the classic E06/T15 idiotype occurred only infrequently in our analysis, most commonly in MZ cells (45 transcripts) and rarely in splenic B-1a and peritoneal B-1b cells (1 and 4 transcripts, respectively). However, we show that the most frequently observed CDR3 amino acid sequence CMRYGNYWYFVW, which together with a sequence that differs by only one amino acid accounted for an astounding 43% of all unique B-1 peritoneal sequences, encodes for an OSE binding antibody. As noted above, this antibody previously was noted to bind to a “cryptic site” on BrRBCs and to also bind phosphatidylcholine present on liposomes (21, 24). We have prepared XQ11-scFV containing the heavy and light chain segments described in the original hybridomas, and while confirming that it does bind BrRBCs, we found it is an example of an antibody targeting an OSE – the PC of OxLDL and PC-BSA, which is also bound by T15/E06 (15). We also have preliminary data that it binds to lipoprotein(a), a lipoprotein that is known to be greatly enriched in OxPL (data not shown) (66). It is of interest that it also bound modestly to “native” RBCs. RBCs are known to be under increased oxidative stress as they age and it is likely that they accumulate OSEs as they age (52). The binding of IgM targeting OSEs, such as OxPL, may in time mediate clearance of the aged RBCs. Furthermore, this antibody, like other antibodies that bind OSEs, may play generalized roles in maintaining homeostasis against oxidative stress and in providing protection against inflammation and atherosclerosis (15, 51, 67). Further studies will be needed to test these suggestions.
In summary, we utilized an unbiased 5′ RACE amplification strategy with massively parallel sequencing to define the IGHV functional repertoire of all murine peritoneal and splenic B cells. We could analyze a total of about 379,000 productive transcripts from 6 B cell subsets including peritoneal B-1a, B-1b and B-2 cells, and splenic B-1a, MZ and FO cells. An early analysis has revealed that B-1a cell sequences differed remarkably from other subsets, adding further evidence for a distinct selection process. Furthermore, we found that the peritoneal B-1a compartment also appeared to be a unique compartment derived predominantly prenatally, and quite distinct from B-1b cells, which appear to be separate and derived postnatally. Interestingly, peritoneal B-2 cells appear to be the most isolated subset based on their CDR3 repertoire. Massively parallel sequencing is a powerful tool to analyze the B cell repertoire and can be used in further studies to investigate how the repertoire changes during development of environmental influences or disease states, and with aging, giving further insight into these exceptional cells and their roles in innate and acquired immunity.
Supplementary Material
Acknowledgments
Grant Support
We gratefully acknowledge the following grant support: NIH HL P01-088093 (TP, XQ, SH, CKG, CB, JLW) HL 119828 (JLW), HL R35-135737 and HL P01 136275 (JLW), U19 Al106754 (CB, MC), DRC P30DK063491 (KJ) and the Leducq Transatlantic Network Grant (CKG).
Footnotes
Abbreviations:
IGHV: IgH variable region
NAb: natural antibody
PRR: pattern-recognition receptor
DAMP: danger-associated molecular pattern
PAMP: pathogen-associated molecular pattern
OSE: oxidation-specific epitope
BrRBCs: bromelain-treated red blood cells
PC-1: plasma cell alloantigen 1
MZ: marginal zone
FO: follicular
OxLDL: oxidized LDL
PC: phosphocholine
OxPL: oxidized phospholipids
Disclosures: XQ and JLW are co-inventors and receive royalties from patents owned by the University of California San Diego on oxidation-specific antibodies. JLW is a consultant for Ionis Pharmaceuticals. The other co-authors have no conflicts of interest.
References
- 1.Hayakawa K, Hardy RR, Parks DR, Herzenberg LA. The “Ly-1 B” cell subpopulation in normal immunodefective, and autoimmune mice. The Journal of experimental medicine. 1983;157:202–218. doi: 10.1084/jem.157.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nature reviews Immunology. 2011;11:34–46. doi: 10.1038/nri2901. [DOI] [PubMed] [Google Scholar]
- 3.Hardy RR. B-1 B cell development. Journal of immunology. 2006;177:2749–2754. doi: 10.4049/jimmunol.177.5.2749. [DOI] [PubMed] [Google Scholar]
- 4.Bekeredjian-Ding I, Jego G. Toll-like receptors--sentries in the B-cell response. Immunology. 2009;128:311–323. doi: 10.1111/j.1365-2567.2009.03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin F, Kearney JF. B-cell subsets and the mature preimmune repertoire. Marginal zone and B1 B cells as part of a “natural immune memory”. Immunological reviews. 2000;175:70–79. [PubMed] [Google Scholar]
- 6.Kantor AB, Stall AM, Adams S, Herzenberg LA, Herzenberg LA. Differential development of progenitor activity for three B-cell lineages. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:3320–3324. doi: 10.1073/pnas.89.8.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alugupalli KR, Leong JM, Woodland RT, Muramatsu M, Honjo T, Gerstein RM. B1b lymphocytes confer T cell-independent long-lasting immunity. Immunity. 2004;21:379–390. doi: 10.1016/j.immuni.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 8.Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 2005;23:7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Ghosn EE, Yamamoto R, Hamanaka S, Yang Y, Herzenberg LA, Nakauchi H, Herzenberg LA. Distinct B-cell lineage commitment distinguishes adult bone marrow hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5394–5398. doi: 10.1073/pnas.1121632109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha SA, Tsuji M, Suzuki K, Meek B, Yasuda N, Kaisho T, Fagarasan S. Regulation of B1 cell migration by signals through Toll-like receptors. The Journal of experimental medicine. 2006;203:2541–2550. doi: 10.1084/jem.20061041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Tung JW, Ghosn EE, Herzenberg LA, Herzenberg LA. Division and differentiation of natural antibody-producing cells in mouse spleen. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4542–4546. doi: 10.1073/pnas.0700001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang MK, Binder CJ, Miller YI, Subbanagounder G, Silverman GJ, Berliner JA, Witztum JL. Apoptotic cells with oxidation-specific epitopes are immunogenic and proinflammatory. The Journal of experimental medicine. 2004;200:1359–1370. doi: 10.1084/jem.20031763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binder CJ, Papac-Milicevic N, Witztum JL. Innate sensing of oxidation-specific epitopes in health and disease. Nature reviews Immunology. 2016;16:485–497. doi: 10.1038/nri.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw PX, Horkko S, Chang MK, Curtiss LK, Palinski W, Silverman GJ, Witztum JL. Natural antibodies with the T15 idiotype may act in atherosclerosis, apoptotic clearance, and protective immunity. The Journal of clinical investigation. 2000;105:1731–1740. doi: 10.1172/JCI8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binder CJ, Horkko S, Dewan A, Chang MK, Kieu EP, Goodyear CS, Shaw PX, Palinski W, Witztum JL, Silverman GJ. Pneumococcal vaccination decreases atherosclerotic lesion formation: molecular mimicry between Streptococcus pneumoniae and oxidized LDL. Nature medicine. 2003;9:736–743. doi: 10.1038/nm876. [DOI] [PubMed] [Google Scholar]
- 16.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annual review of biochemistry. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 17.Gregoire KE, Goldschneider I, Barton RW, Bollum FJ. Ontogeny of terminal deoxynucleotidyl transferase-positive cells in lymphohemopoietic tissues of rat and mouse. Journal of immunology. 1979;123:1347–1352. [PubMed] [Google Scholar]
- 18.Feeney AJ. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. The Journal of experimental medicine. 1990;172:1377–1390. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 20.Vale AM, Kapoor P, Skibinski GA, Elgavish A, Mahmoud TI, Zemlin C, Zemlin M, Burrows PD, Nobrega A, Kearney JF, Briles DE, Schroeder HW., Jr The link between antibodies to OxLDL and natural protection against pneumococci depends on D(H) gene conservation. The Journal of experimental medicine. 2013;210:875–890. doi: 10.1084/jem.20121861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reininger L, Ollier P, Poncet P, Kaushik A, Jaton JC. Novel V genes encode virtually identical variable regions of six murine monoclonal anti-bromelain-treated red blood cell autoantibodies. Journal of immunology. 1987;138:316–323. [PubMed] [Google Scholar]
- 22.Pennell CA, Arnold LW, Haughton G, Clarke SH. Restricted Ig variable region gene expression among Ly-1+ B cell lymphomas. Journal of immunology. 1988;141:2788–2796. [PubMed] [Google Scholar]
- 23.Hardy RR, Carmack CE, Shinton SA, Riblet RJ, Hayakawa K. A single VH gene is utilized predominantly in anti-BrMRBC hybridomas derived from purified Ly-1 B cells. Definition of the VH11 family. Journal of immunology. 1989;142:3643–3651. [PubMed] [Google Scholar]
- 24.Mercolino TJ, Arnold LW, Haughton G. Phosphatidyl choline is recognized by a series of Ly-1+ murine B cell lymphomas specific for erythrocyte membranes. The Journal of experimental medicine. 1986;163:155–165. doi: 10.1084/jem.163.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kantor AB, Merrill CE, Herzenberg LA, Hillson JL. An unbiased analysis of V(H)-D-J(H) sequences from B-1a, B-1b, and conventional B cells. Journal of immunology. 1997;158:1175–1186. [PubMed] [Google Scholar]
- 26.Vale AM, Tanner JM, Schelonka RL, Zhuang Y, Zemlin M, Gartland GL, Schroeder HW., Jr The peritoneal cavity B-2 antibody repertoire appears to reflect many of the same selective pressures that shape the B-1a and B-1b repertoires. Journal of immunology. 2010;185:6085–6095. doi: 10.4049/jimmunol.1001423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Shin DM, Abbasi S, Jain S, Kovalchuk AL, Beaty N, Chen S, Gonzalez-Garcia I, Morse HC., 3rd Expression of plasma cell alloantigen 1 defines layered development of B-1a B-cell subsets with distinct innate-like functions. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20077–20082. doi: 10.1073/pnas.1212428109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nature biotechnology. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Wang C, Yang Q, Kantor AB, Chu H, Ghosn EE, Qin G, Mazmanian SK, Han J, Herzenberg LA. Distinct mechanisms define murine B cell lineage immunoglobulin heavy chain (IgH) repertoires. eLife. 2015:4. doi: 10.7554/eLife.09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tung JW, Parks DR, Moore WA, Herzenberg LA, Herzenberg LA. Identification of B-cell subsets: an exposition of 11-color (Hi-D) FACS methods. Methods in molecular biology. 2004;271:37–58. doi: 10.1385/1-59259-796-3:037. [DOI] [PubMed] [Google Scholar]
- 32.Yenson V, Baumgarth N. Purification and immune phenotyping of B-1 cells from body cavities of mice. Methods in molecular biology. 2014;1190:17–34. doi: 10.1007/978-1-4939-1161-5_2. [DOI] [PubMed] [Google Scholar]
- 33.Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 34.Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A, Galaxy T. Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010;26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giudicelli V, Lefranc MP. IMGT/junctionanalysis: IMGT standardized analysis of the V-J and V-D-J junctions of the rearranged immunoglobulins (IG) and T cell receptors (TR) Cold Spring Harbor protocols. 2011;2011:716–725. doi: 10.1101/pdb.prot5634. [DOI] [PubMed] [Google Scholar]
- 37.Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods in molecular biology. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- 38.Rogosch T, Kerzel S, Hoi KH, Zhang Z, Maier RF, Ippolito GC, Zemlin M. Immunoglobulin analysis tool: a novel tool for the analysis of human and mouse heavy and light chain transcripts. Frontiers in immunology. 2012;3:176. doi: 10.3389/fimmu.2012.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Angelo S, Glanville J, Ferrara F, Naranjo L, Gleasner CD, Shen X, Bradbury AR, Kiss C. The antibody mining toolbox: an open source tool for the rapid analysis of antibody repertoires. mAbs. 2014;6:160–172. doi: 10.4161/mabs.27105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giudicelli V, Lefranc MP. Ontology for immunogenetics: the IMGT-ONTOLOGY. Bioinformatics. 1999;15:1047–1054. doi: 10.1093/bioinformatics/15.12.1047. [DOI] [PubMed] [Google Scholar]
- 41.Lefranc MP. Antibody nomenclature: from IMGT-ONTOLOGY to INN definition. mAbs. 2011;3:1–2. doi: 10.4161/mabs.3.1.14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seidl KJ, Wilshire JA, MacKenzie JD, Kantor AB, Herzenberg LA, Herzenberg LA. Predominant VH genes expressed in innate antibodies are associated with distinctive antigen-binding sites. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2262–2267. doi: 10.1073/pnas.96.5.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu H, Forster I, Rajewsky K. Sequence homologies, N sequence insertion and JH gene utilization in VHDJH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. The EMBO journal. 1990;9:2133–2140. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ippolito GC, Schelonka RL, Zemlin M, Ivanov, Kobayashi R, Zemlin C, Gartland GL, Nitschke L, Pelkonen J, Fujihashi K, Rajewsky K, Schroeder HW., Jr Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production. The Journal of experimental medicine. 2006;203:1567–1578. doi: 10.1084/jem.20052217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carmack CE, Shinton SA, Hayakawa K, Hardy RR. Rearrangement and selection of VH11 in the Ly-1 B cell lineage. The Journal of experimental medicine. 1990;172:371–374. doi: 10.1084/jem.172.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardy RR, Wei CJ, Hayakawa K. Selection during development of VH11+ B cells: a model for natural autoantibody-producing CD5+ B cells. Immunological reviews. 2004;197:60–74. doi: 10.1111/j.0105-2896.2004.0100.x. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Clarke SH. Positive selection focuses the VH12 B-cell repertoire towards a single B1 specificity with survival function. Immunological reviews. 2004;197:51–59. doi: 10.1111/j.0105-2896.2004.0098.x. [DOI] [PubMed] [Google Scholar]
- 48.Arnold LW, Pennell CA, McCray SK, Clarke SH. Development of B-1 cells: segregation of phosphatidyl choline-specific B cells to the B-1 population occurs after immunoglobulin gene expression. The Journal of experimental medicine. 1994;179:1585–1595. doi: 10.1084/jem.179.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baldan A, Gonen A, Choung C, Que X, Marquart TJ, Hernandez I, Bjorkhem I, Ford DA, Witztum JL, Tarling EJ. ABCG1 is required for pulmonary B-1 B cell and natural antibody homeostasis. Journal of immunology. 2014;193:5637–5648. doi: 10.4049/jimmunol.1400606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi YS, Dieter JA, Rothaeusler K, Luo Z, Baumgarth N. B-1 cells in the bone marrow are a significant source of natural IgM. European journal of immunology. 2012;42:120–129. doi: 10.1002/eji.201141890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chou MY, Fogelstrand L, Hartvigsen K, Hansen LF, Woelkers D, Shaw PX, Choi J, Perkmann T, Backhed F, Miller YI, Horkko S, Corr M, Witztum JL, Binder CJ. Oxidation-specific epitopes are dominant targets of innate natural antibodies in mice and humans. The Journal of clinical investigation. 2009;119:1335–1349. doi: 10.1172/JCI36800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar D, Rizvi SI. Markers of oxidative stress in senescent erythrocytes obtained from young and old age rats. Rejuvenation Res. 2014;17:446–452. doi: 10.1089/rej.2014.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Briles DE, Forman C, Hudak S, Claflin JL. Anti-phosphorylcholine antibodies of the T15 idiotype are optimally protective against Streptococcus pneumoniae. The Journal of experimental medicine. 1982;156:1177–1185. doi: 10.1084/jem.156.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsiantoulas D, Perkmann T, Afonyushkin T, Mangold A, Prohaska TA, Papac-Milicevic N, Millischer V, Bartel C, Horkko S, Boulanger CM, Tsimikas S, Fischer MB, Witztum JL, Lang IM, Binder CJ. Circulating microparticles carry oxidation-specific epitopes and are recognized by natural IgM antibodies. Journal of lipid research. 2015;56:440–448. doi: 10.1194/jlr.P054569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horkko S, Bird DA, Miller E, Itabe H, Leitinger N, Subbanagounder G, Berliner JA, Friedman P, Dennis EA, Curtiss LK, Palinski W, Witztum JL. Monoclonal autoantibodies specific for oxidized phospholipids or oxidized phospholipid-protein adducts inhibit macrophage uptake of oxidized low-density lipoproteins. The Journal of clinical investigation. 1999;103:117–128. doi: 10.1172/JCI4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi YS, Baumgarth N. Dual role for B-1a cells in immunity to influenza virus infection. The Journal of experimental medicine. 2008;205:3053–3064. doi: 10.1084/jem.20080979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bachmann MF, Zinkernagel RM. Neutralizing antiviral B cell responses. Annual review of immunology. 1997;15:235–270. doi: 10.1146/annurev.immunol.15.1.235. [DOI] [PubMed] [Google Scholar]
- 58.Holodick NE, Vizconde T, Hopkins TJ, Rothstein TL. Age-Related Decline in Natural IgM Function: Diversification and Selection of the B-1a Cell Pool with Age. Journal of immunology. 2016;196:4348–4357. doi: 10.4049/jimmunol.1600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothstein TL. Natural Antibodies as Rheostats for Susceptibility to Chronic Diseases in the Aged. Frontiers in immunology. 2016;7:127. doi: 10.3389/fimmu.2016.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holodick NE, Vizconde T, Rothstein TL. B-1a cell diversity: nontemplated addition in B-1a cell Ig is determined by progenitor population and developmental location. Journal of immunology. 2014;192:2432–2441. doi: 10.4049/jimmunol.1300247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosenfeld SM, Perry HM, Gonen A, Prohaska TA, Srikakulapu P, Grewal S, Das D, McSkimming C, Taylor AM, Tsimikas S, Bender TP, Witztum JL, McNamara CA. B-1b Cells Secrete Atheroprotective IgM and Attenuate Atherosclerosis. Circulation research. 2015;117:e28–39. doi: 10.1161/CIRCRESAHA.117.306044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harmon DB, Srikakulapu P, Kaplan JL, Oldham SN, McSkimming C, Garmey JC, Perry HM, Kirby JL, Prohaska TA, Gonen A, Hallowell P, Schirmer B, Tsimikas S, Taylor AM, Witztum JL, McNamara CA. Protective Role for B-1b B Cells and IgM in Obesity-Associated Inflammation, Glucose Intolerance, and Insulin Resistance. Arteriosclerosis, thrombosis, and vascular biology. 2016;36:682–691. doi: 10.1161/ATVBAHA.116.307166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hastings WD, Tumang JR, Behrens TW, Rothstein TL. Peritoneal B-2 cells comprise a distinct B-2 cell population with B-1b-like characteristics. European journal of immunology. 2006;36:1114–1123. doi: 10.1002/eji.200535142. [DOI] [PubMed] [Google Scholar]
- 64.Khass M, Buckley K, Kapoor P, Schelonka RL, Watkins LS, Zhuang Y, Schroeder HW., Jr Recirculating bone marrow B cells in C57BL/6 mice are more tolerant of highly hydrophobic and highly charged CDR-H3s than those in BALB/c mice. European journal of immunology. 2013;43:629–640. doi: 10.1002/eji.201242936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cox KO, Hardy SJ. Autoantibodies against mouse bromelain-modified RBC are specifically inhibited by a common membrane phospholipid, phosphatidylcholine. Immunology. 1985;55:263–269. [PMC free article] [PubMed] [Google Scholar]
- 66.Bergmark C, Dewan A, Orsoni A, Merki E, Miller ER, Shin MJ, Binder CJ, Horkko S, Krauss RM, Chapman MJ, Witztum JL, Tsimikas S. A novel function of lipoprotein [a] as a preferential carrier of oxidized phospholipids in human plasma. Journal of lipid research. 2008;49:2230–2239. doi: 10.1194/jlr.M800174-JLR200. [DOI] [PubMed] [Google Scholar]
- 67.Miller YI, Choi SH, Wiesner P, Fang L, Harkewicz R, Hartvigsen K, Boullier A, Gonen A, Diehl CJ, Que X, Montano E, Shaw PX, Tsimikas S, Binder CJ, Witztum JL. Oxidation-specific epitopes are danger-associated molecular patterns recognized by pattern recognition receptors of innate immunity. Circulation research. 2011;108:235–248. doi: 10.1161/CIRCRESAHA.110.223875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.