Abstract
Purpose
Conventional sequences for metabolite transverse relaxation quantification all generally measure signal changes at different echo times (TEs). However, quantification results obtained via these conventional methods can be very different and are highly dependent on the type of sequence being applied. TE-dependent effects such as diffusion, macromolecule baseline, and J-coupling modulation contribute significantly to these differences. Here, we propose a novel technique—MARZSS (Multiple flip Angle pulse driven Ratio of longitudinal Steady States)—for preparing magnetization with T2/T1 weighting. Using premeasured T1 values, T2 values for metabolites can thereby be determined. The measurement procedure does not require varying TE and is TE independent; T2, diffusion, and J-coupling effects induced by the readout sequence are cancelled.
Method
Longitudinal steady states (Mzss) at different flip angles were prepared with trains of radio frequency (RF) pulses interspersed with field gradients. The resulting spatially modulated longitudinal magnetization was acquired with a PRESS readout module. A new linear equation for quantification of MARZSS was derived from Bloch equations.
Results
By implementing this readout independent method, T2 measurement of brain metabolites at 7T was demonstrated through Bloch simulations, phantom and in-vivo experiments.
Conclusions
The proposed MARZSS technique can be used to largely avoid multi-TE associated interference, including diffusion, macromolecule s, and J modulation. This MARZSS technology, which is uniquely insensitive to readout sequence type and TE, is a promising technique for more accurately probing in vivo metabolite relaxation.
Keywords: MR spectroscopy, brain metabolite T2 relaxation, J-coupling, glutamate relaxation, spatially modulated magnetization, DANTE
Introduction
Nuclear magnetic resonance spectroscopy (MRS) allows noninvasive detection of endogenous metabolites in the human brain and can provide valuable metabolic and physiological information. MRS is also a promising tool for diagnosing metabolic disorders and other diverse pathological conditions in human brain, including epilepsy, multiple sclerosis, stroke, cancer, and psychiatric disease [1–4]. The concentrations and relaxation time of metabolites can be used to detect abnormalities in brain regions that appear normal in magnetic resonance imaging (MRI), as well as to characterize pathology underlying MRI-visible abnormalities [5]. For example, many neurological and psychiatric diseases alter the cellular environment, which may be reflected in changes of metabolite T2 relaxation times [6–7].
In general, two types of techniques are conventionally adopted for T2 quantification of metabolites in brain. Sequences with simple spin or stimulated echoes [8–10] can be used to measure signals at step-incremented echo times (multi-TEs) for T2 quantification. Carr-Purcell-Meiboom-Gill (CPMG) sequences [11, 12] are used in a similar manner by repeating more (Carr-Purcell) CP blocks. However, the quantification results obtained via these two types of conventional methods are very different and depend significantly on the sequence type being used. In some circumstances, quantification results may differ even when the same sequence is used, due to selection of parameters such as the TE steps during the spin echo technique or the inter-pulse delay used during the CPMG sequence.
Thus, given that MR exponential decay signal from the multi-TE approaches can be complicated under many scenarios (e.g., confounding spin evolution due to scalar couplings, significant diffusion weighting during readout, or very fast T2 relaxation of many 31P MRS signals), a technique capable of generating variable T2 weighting without changing TE would be highly valuable.
Tissue signals at steady states driven by rapid pulse trains are frequently used in MRI techniques to quantify different components of tissue water [13]. Balanced steady-state free precession (b-SSFP) serves as a high signal to noise ratio (SNR) imaging tool to generate images with T2/T1 weighted contrasts. Quantitative maps can be extracted from those T2/T1 weighted images acquired with two or multiple flip angles. However, b-SSFP cannot be directly used to quantify MRS spectra because b-SSFP may have undesirable frequency selective effects; these were previously attributed to the periodic nature of DANTE pulses [14]. Specifically, DANTE pulses interspersed with gradients were previously used as a preparation module for spatial tagging of MR images [15] as well as black blood contrast imaging [16]. When DANTE preparation is used for black blood imaging applications, spatial modulation of longitudinal magnetization (Mz) could give rise to periodic intensity variations in images (dark bandings). To avoid visualizing the dark bandings in the black blood images, the gradient moment in DANTE pulses could be adjusted so that banding size was smaller than voxel size. Under those circumstances, voxel signal amplitude could be quantified [17]. This DANTE-prepared imaging application suggests that the spatial modulation of metabolites at steady state Mzss may also be similarly quantifiable.
This study proposes a novel method— MARZSS (Multiple flip Angle pulse driven Ratio of longitudinal Steady States)—for quantifying spatially modulated magnetization of metabolites. The prepared longitudinal magnetization of longitudinal steady states (Mzss) weighted by T2/T1 can be acquired by any readout sequence. When T1s of metabolites are pre-determined from other techniques, such as inversion recovery or saturation recovery, T2s and spin densities of metabolites can be correspondingly calculated. Because it is a quantifiable contrast preparation technique, MARZSS does not require varying TE for metabolite relaxation quantification and is independent of the readout module. We hypothesized that using the proposed MARZSS technique instead of conventional multi-TE or CPMG methods would allow us to avoid the readout-sequence-dependent variations associated with diffusion weighting, macromolecule baseline, and J-coupling modulation.
Theory
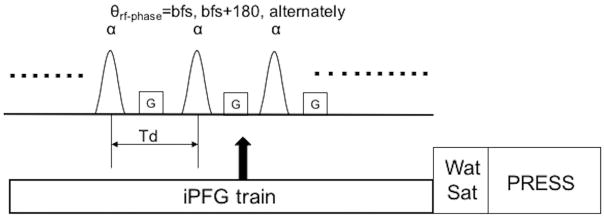
In order to quantify the longitudinal Mzss of metabolites at steady state by different flip angles, we derived a new linear equation from Bloch equations. Considering a train of radio frequency (RF) pulses interspersed with field gradient (iPFG; shown in Fig. 1), the resulting steady state magnetization (Mzss) is given by [14, 17],
Figure 1.
Diagram of the proposed Multiple flip Angle pulse driven Ratio of longitudinal Steady States (MARZSS) module. θ is the phase of the radio frequency (RF) pulse where bfs is the base frequency shift decided by frequency offset of the pulse. α is the flip angle of the pulse train. G is gradient along the z direction. Td is the interpulse delay of the pulse train. iPFG is the interleaved pulse field gradient. WatSat is the water saturation module. PRESS is the readout module.
| Eq. 1 |
where E1=exp(−Td/T1), E2 = exp(−Td/T2), n is an integer, α is the flip angle of a single RF pulse, and M0 is the thermal equilibrium signal intensity. Td is the inter-pulse delay time, which is much shorter than the metabolite T1 and T2; within these limitations, E1=1−Td/T1 and E2=1−Td/T2, and θ is the position-dependent linear phase angle induced inside the voxel by the applied field gradient and local field inhomogeneity.
For Eq. 1 to be valid, the minimal gradient moment GdG in Fig. 1 must be much greater than π/(γΔr), where γ is the gyromagnetic ratio, dG is the duration of the applied gradient, and Δr is the voxel size. The above integral yields a closed-form expression for Mzss:
| Eq. 2 |
where and .
Eq. 2 can be rearranged into a linear equation:
| Eq. 3 |
where , which is the ratio of signal amplitudes from readout sequence. Eq. 3 suggests that T1/T2 of metabolites can be calculated by linear fitting. More importantly, Eq. 3 is independent of readout sequence and local inhomogeneity. Because TE in readout is a fixed parameter, the real or complex exponential terms (or effects) induced by readout sequence (for instance, exp(−TE/T2), exp(−bD), or J evolution f(J,σ)) are common factors that are cancelled in ratio of longitudinal steady states (RZSS).
Due to delay of the water suppression module in Fig. 1, Mzss experiences a slight magnetization recovery that can be corrected using Eq. 4 describing T1 recovery,
| Eq. 4 |
where Mmeas_z is the amplitude from measurement, and tws is the delay time of the water suppression module. It is assumed that T1 of metabolites is pre-determined from other approaches, such as inversion recovery or saturation recovery. Therefore, Mzss can be calculated.
Method
To demonstrate the spatial modulation of metabolite magnetization and its attenuation in longitudinal direction by iPFG, Bloch equation numerical simulations were performed. Code was written using MATLAB (Natick, MA). The Mzss of N-acetylaspartate (NAA) was simulated based on the protocol time series in a single repetition time (TR). For NAA, T1 and T2 literature values at 7T of 1730 ms and 170 ms, respectively, were used [18].
All scans were acquired using a 32-channel head coil on a 7T Siemens scanner. A doped water-based sphere phantom was used for phantom measurement of metabolites. Written informed consent was obtained from five healthy volunteers (four females and one male between the ages of 24–40 years).
The iPFG train applied in the measurements shown in Fig. 1 consisted of 500 sinc-Gauss pulses with gradient Gz (along the z direction) interspersed between the RF pulses. The duration of each RF pulse was 4 ms with frequency offset at 2.2 ppm for optimal determination of NAA and a constant inter-pulse delay Td of 10ms. The phases of the iPFG RF pulses alternated as base frequency shift (bfs) and bfs+180°. Base frequency shift was decided by the frequency offset used in the measurement; in our case, this was 2.2 ppm. The flip angle of the iPFG train was incremented by 10° from 0° to 60° for a total of seven measurements. Gradient amplitude and duration were Gz=2mT/m and 5ms, respectively. Spoil gradients immediately after the preparation module were set along all three directions, Gx=Gy=Gz, with amplitude = 8 mT/m and duration = 1.2 ms.
The sinc-Gaussian pulse used in Fig. 1 has a somewhat nonuniform frequency profile across the metabolites of interest. As a result, different chemical shift experiences slightly different flip angles. Although small, the flip angle at each chemical shift was corrected for data fitting based on Bloch simulation of the sinc-Gaussian pulse.
A water suppression module with an overall duration of 120 ms was applied. Nine sinc-Gauss RF pulses implemented in the water suppression module were all set to an RF duration of 9 ms with a 12 ms interpulse delay time. RF flip angle, RF phase, and spoil gradient amplitudes used after each of the pulses were (flip angle/phase/G): 80.4°/234°/Gx=32 mT/m, 80.4°/585°/Gz=32 mT/m, 152.8°/1053°/Gx=32 mT/m, 80.4/1638°/Gz=16 mT/m, 152.8°/2340°/Gy=16 mT/m, 80.4°/3159°/Gx=16 mT/m, 152.8°/4095°/Gz=40 mT/m, 152.8°/5148°/Gy=40 mT/m and 152.8°/6318°/Gx=Gy=Gz=40 mT/m. Spoil gradient durations were all set to 1.2 ms.
The PRESS sequence was implemented with an isotropic voxel (2 cm × 2 cm × 2 cm) for both in vivo and phantom experiments. The chosen location of in-vivo measurements was in the occipital (OC) lobe as illustrated in Fig. 5. TR of 8.5 s, TE1 (the first PRESS TE) = 69 ms, and TE2 (TE-TE1) = 37 ms were used in both in vivo and phantom experiments. TE1 and TE2 were chosen to minimize contamination of the glutamate (Glu) H4 resonance by glutamine (Gln) and NAA [19]. Two thousand forty-eight data points were acquired for each spectrum. Phantom and in vivo data were acquired with 12 averages and four averages, respectively. Based on previous experience, we believed that four averages would provide adequate SNR for fitting NAA, creatine (Cr), and choline (Cho). When Glu with an inherently lower signal intensity is the target metabolite, increasing the number of averages from four to 12 or more should significantly improve the fitting results. The total scan time for measuring T1/T2 using seven flip angles was approximately 18 mins for phantom and 6 mins for in vivo experiments.
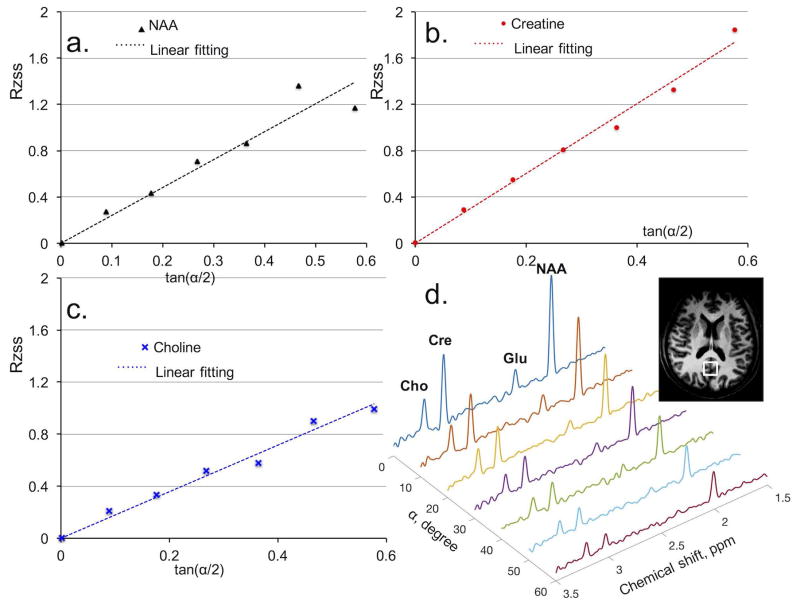
Figure 5.
In vivo metabolite T2 determination with proposed Multiple flip Angle pulse driven Ratio of longitudinal Steady States (MARZSS). (a) Linear fitting as black dotted line for N-acetylaspartate (NAA) data points, extracted with LCModel from spectra in Fig. 5d. (b) Linear fitting as red dotted line for creatine (Cr) data points, extracted from spectra in Fig. 5d. (c) Linear fitting as blue dotted line for choline (Cho) data points, extracted from spectra in Fig. 5d. (d) Signal variation with flip angle in phantom measurements of NAA, Cr, and Cho acquired with flip angle varied from 0° to 60° with 10° increments.
Conventional standard inversion recovery was implemented for independent T1 determination at the same location. With no change of water saturation or PRESS modules in the MARZSS sequence, the iPFG train was replaced with an inversion recovery pulse. Inversion recovery (TI) times of 425, 625, 825, 1125, 1225, 1425, 1625, 2725, 4725 and 6725 ms were implemented for T1 measurements. Four scans were conducted for each TI measurement, and TR was 8.5 s. For comparison purposes, a multi-TE PRESS technique for T2 quantification was also implemented at the same location.
All the metabolite signal amplitudes for MARZSS, IR, and multi-TE measurements were determined using LCModel fitting [20]. The basis set of metabolites for fitting included NAA (NAA acetyl moiety, NAA aspartyl moiety), N-acetylaspartylglutamate (NAAG acetyl moiety, NAAG aspartyl and glutamate moieties), total Cr, total Cho, Aspartate (Asp), Glu, Gln, Lactate (Lac), glutathione (GSH), and γ-aminobutyric acid (GABA).
Two-tailed, unpaired Student’s t-tests were used to compare T2s obtained via the proposed MARZSS technique and the multi-TE PRESS pulse sequence.
Results
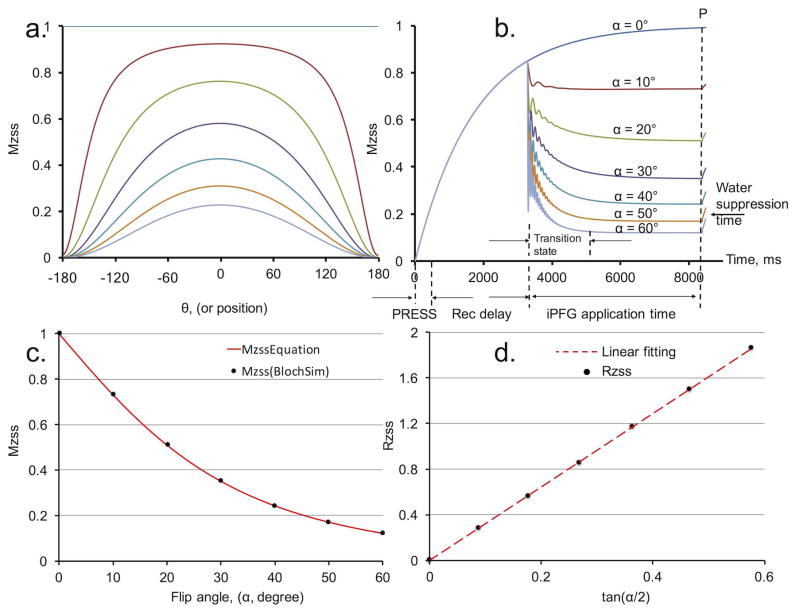
Bloch simulations were implemented to demonstrate changes in longitudinal magnetization (Mz)—both spatially at steady state and temporally at transition state—as a function of different RF flip angles (Fig. 2a and Fig. 2b, respectively). Fig. 2a illustrates the resulting spatial variations of Mz when iPFG trains with different flip angles were applied to saturate NAA to steady states. Although only one of the full spatial modulation periods was simulated in Fig. 2a, for multiple half periods signal attenuation would be identical in the voxel due to symmetry.
Figure 2.
Bloch simulation and equation validation using N-acetylaspartate (NAA) (T1, 1730 ms, T2, 170 ms). (a) Spatial variation of longitudinal magnetization at steady states in one period. From top to bottom, lines represent spatial variation induced by Multiple flip Angle pulse driven Ratio of longitudinal Steady States (MARZSS) with flip angle from 0° to 60° with 10° increments. (b) Longitudinal steady states (Mzss) temporal variation in one repetition time (TR). PRESS, PRESS sequence readout time (the small effects of two 180° PRESS pulses were omitted.); “Rec delay” is magnetization recovery delay time. (c) Mzss as a function of flip angles. Black dots were extracted from the signal curves evaluated at the dotted line P in Fig. 2b. Alternately, the black dots could be extracted from the areas under the curves in Fig. 2a. The red line is the predicted curve based on equation Eq. 2. (d) Ratio of longitudinal steady states (RZSS), in Eq. 3, as a function of tan(α/2). The red dot line is the linear fit.
In the simulation illustrated in Fig. 2a, there is only one period of modulation in the voxel, which corresponds to the gradient moment of 0.23mT/m × 5ms in the iPFG train, given the 2 cm voxel size. In practical applications, significantly larger gradient moments of 2mT/m × 5ms would be employed to avoid the potential quantification error introduced by a non-integer value of n (or a non-half-integer value due to the symmetrical shape of the even function cosθ) in Eq. 1. Approximately 17 half-periods of modulation were present in the 2 cm voxel, which minimized the quantification error introduced at the edges of voxel. The evolution of the Mz over time during an entire TR was numerically simulated using Bloch equations (Fig. 2b), where each signal point as a function of time was averaged over θ values. Fig. 2b illustrates the Mz of NAA evolving from transition states to steady states at different flip angles. Because of the 120 ms duration for the water suppression module, a slight magnetization recovery occurred between the time immediately after the iPFG train and the time before the 90° readout pulse.
Fig. 2c and 2d demonstrate the validation of Eq. 2 and Eq. 3, respectively, using Bloch numerical simulations. The black dots in Fig. 2c were extracted from steady state amplitude at the dotted line P in Fig. 2b. Alternately, the black dots could be identically extracted from integrating (summing) the areas under the symmetrical Mzss spatial variation curve in Fig. 2a. The red line in Fig. 2c illustrates the signal change as a function of flip angle calculated from Eq. 2, showing excellent agreement between the full Bloch simulation and Eq. 2. Linear fitting of the black dots based on Eq. 3 is shown in Fig. 2d. The T2 value determined from the slope was 167 ms, which is in good agreement with its true value of 170 ms. The small deviation was introduced primarily due to the approximations E1=1−Td/T1 and E2=1−Td/T2.
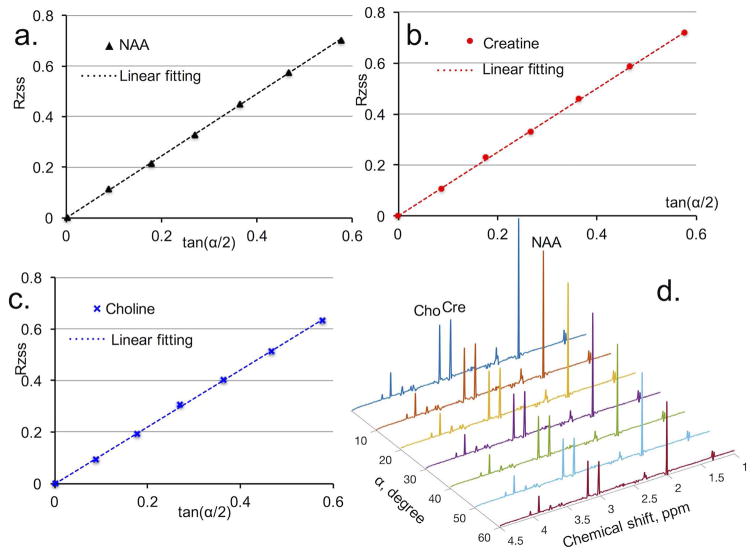
In phantom measurements, signal amplitude ratios RZSS (defined in Eq. 3) of NAA, Cr, and Cho as a function of tan(α/2) are shown in Fig. 3a, 3b, and 3c, respectively. The linear relationships between RZSS and tan(α/2) predicted by Eq. 3 were clearly confirmed by all three metabolites (Fig. 3). The T2 of NAA, Cr, and Cho were determined by least square linear fitting (the dotted lines in Fig. 3a, 3b, and 3c). The T2 results obtained from the proposed MARZSS technique are listed in Table 1 to allow comparison with T2 results obtained with the multi-TE PRESS method. Fig. 3d shows the phantom spectra acquired by the MARZSS method at different flip angles.
Figure 3.
Phantom metabolite T2 determination with proposed Multiple flip Angle pulse driven Ratio of longitudinal Steady States (MARZSS). (a) Linear fitting as black dotted line for N-acetylaspartase (NAA) data points, extracted with LCModel from spectra in Fig. 3d. (b) Linear fitting as red dotted line for creatine (Cr) data points, extracted from spectra in Fig. 3d. (c) Linear fitting as blue dotted line for choline (Cho) data points, extracted from spectra in Fig. 3d. (d) Signal variation with flip angle in phantom measurements of NAA, Cr, and Cho. Metabolite spectra were acquired with flip angle from 0° to 60° with 10° increment.
Table 1.
Comparisons between phantom metabolite T2 measurements obtained using the proposed MARZSS and conventional multi-TE PRESS methods.
| Method/Metabolite | T2, ms, NAA | T2, ms, creatine | T2, ms, choline |
|---|---|---|---|
| multi-TE PRESS | 530±37 | 310±24 | 200±23 |
| MARZSS* + PRESS** | 494±58 | 275±31 | 194±30 |
| Percent diff, % | 6.8% | 11% | 3% |
MARZSS*: preparation module; PRESS**: readout module. Metabolite T1s of the doped phantom were predetermined to be 759 ms, 449 ms, and 297 ms for NAA, creatine, and choline, respectively. Phantom was tested for 3 times and the averaged numbers were shown. NAA, N-acetylaspartate;
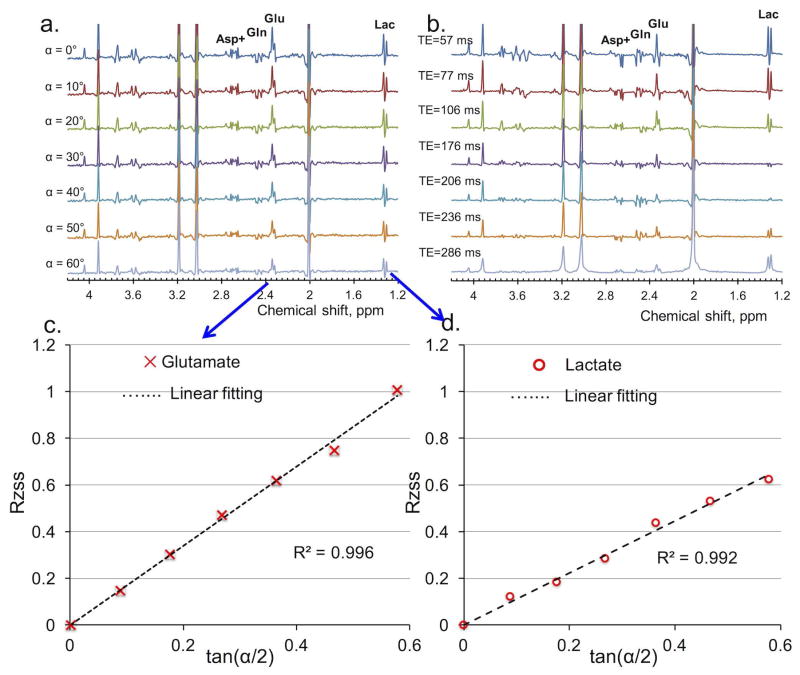
Comparisons of the spectra of metabolites with J-couplings (Asp+, partially contributed from NAA), Gln, Glu, and lactate (Lac) obtained via MARZSS versus multi-TE are demonstrated in Fig. 4a and Fig. 4b. Fig. 4a clearly shows that the proposed MARZSS technique can be implemented with no discernable modulation from J-coupling evolutions because the readout module uses a fixed TE. In contrast, strong signal modulations of metabolites of Asp+, Gln, Glu, and Lac were observed as expected because of J-coupling effects with TE variations (Fig. 4b). Changes in RZSS for Glu and Lac in Fig.4a as a function of flip angle were extracted from LCModel and plotted in Fig. 4c and Fig. 4d, respectively. T2 values for Glu and Lac in the water-based sphere phantom were determined to be 217 ms and 533 ms respectively, based on measured T1 values of 626 and 658 ms.
Figure 4.
Comparison of phantom results of J-coupled metabolites, such as aspartate (Asp+ (with contribution from N-acetylaspartate (NAA) aspartyl moiety), glutamine (Gln), glutamate (Glu), and lactate (Lac), between Multiple flip Angle pulse driven Ratio of longitudinal Steady States (MARZSS) and multi-echo time (TE) PRESS techniques. (a) Signal variation with flip angle in phantom measurements acquired by the proposed MARZSS. No discernable sensitivity to J-coupling of MARZSS technique was observed. Flip angles were varied from 0° to 60° with 10° increments (top to bottom). (b) Signal variation with TE in phantom measurements acquired by conventional multi-TE PRESS. High sensitivity to J-coupling of conventional multi-TE PRESS was observed. Echo times were 57, 77, 106, 176, 206, 236, 286 ms (top to bottom). (c) Linear fitting as black dotted line for Glu data points from the proposed MARZSS technique. Glu data were extracted with the LCModel from spectra shown in Fig. 4a. (d) Linear fitting as black dotted line for Lac data points from the proposed MARZSS technique. Lac data were extracted from spectra shown in Fig. 4a.
In vivo signal amplitude ratios (RZSS, defined in Eq. 3) of NAA, Cr, and Cho as a function of tan(α/2) acquired from one subject are shown in Fig. 5a, 5b, and 5c, respectively. The linear relationships between RZSS and tan(α/2) predicted by Eq. 3 are clearly confirmed by all three metabolites. The T2s of NAA, Cr, and Cho were determined from the slopes of the dotted straight lines in Fig. 5a, 5b, and 5c, respectively. Table 2 compares T2 results obtained from the proposed MARZSS technique with T2 results obtained via the multi-TE PRESS method. In vivo MARZSS spectra using flip angles from 0° to 60° with 10° increments are shown in Fig. 5d.
Table 2.
Comparisons between in vivo metabolite T2 measurements obtained using the proposed MARZSS and conventional multi-TE PRESS method at 7T.
| Method/Metabolites/7T | T2, ms, NAA | T2, ms, creatine | T2, ms, choline | T2, ms, glutamate |
|---|---|---|---|---|
| Multi-TE PRESS (OC) | 171±17 | 113±9 | 154±42 | N/A |
| MARZSS+PRESS (OC) | 270±36 | 185±24 | 263±53 | 155±45 |
| Percent diff, % | 58.2% | 63.7% | 70.8% | ---- |
| t-test (p) | <0.01 | <0.004 | <0.05 | ---- |
| 7T, CP-LASER (OC) [11] | 341 | 221 | N/A | N/A |
| 7T, CPMG(PA) [12] | 234 | 146 | 194 | N/A |
NAA, N-acetylaspartate; OC, occipital lobe; PA, parietal lobe; p, t-test of differences between MARZSS and multi-TE PRESS; N/A, not available. Our measurement results from multi-TE PRESS approximate values obtained in the literature at 7T [21]. The T1s of individual subjects were predetermined for calculation of T2s. The averaged in vivo T1s in the OC were 1409 ms, 1396 ms, 1160 ms and 1198 ms for NAA, creatine, choline, and glutamate, respectively.
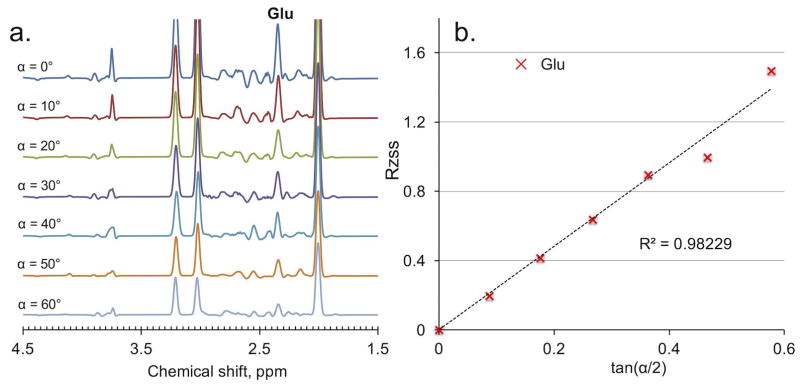
Fig. 6a illustrates the fitted in vivo spectra of Glu obtained using the MARZSS technique; here, the preparation pulses of MARZSS affected monotonically the signal attenuation of Glu. RZSS of Glu as a function of tan(α/2) was also extracted, and the result is shown in Fig. 6b. The linear relationship between RZSS and tan(α/2) predicted by Eq. 3 is clearly observable, with a fitting coefficient of determination R2 = 0.982.
Figure 6.
In vivo results of glutamate (Glu) from the Multiple flip Angle pulse driven Ratio of longitudinal Steady States (MARZSS) technique. (a) Signal variation with flip angle in in vivo measurements of Glu acquired from the proposed MARZSS. This is from fitting data shown in Fig. 5d. No discernable sensitivity to J-coupling of MARZSS technique was observed from Fig. 6a. Flip angles were varied from 0° to 60° with 10° increments (top to bottom). (b) Linear fitting as black dotted line for Glu data points, extracted with the LCModel from spectra in Fig. 6a using the proposed MARZSS technique.
Discussion
This study proposed a new technique— MARZSS—for probing metabolite T2 relaxation by generating RF-driven longitudinal steady-state magnetization without the need to vary TE. Multiple flip angle iPFG trains were used as preparation modules to generate T2/T1 weighting of steady state longitudinal magnetization. In contrast, PRESS sequence served only as a readout module. A new linear equation for quantifying longitudinal magnetization and metabolite relaxation (T2/T1) at steady state was derived from Bloch equations. The newly derived equations were validated through Bloch simulations, phantom, and in vivo experiments.
Using conventional techniques such as multi-TE PRESS, spins evolve on the transverse plane with differing evolution time between measurements (i.e., TE), which may result in confounding signal decay from diffusion, physiological motion, and/or signal modulation from J-coupling. When used as a preparation module, the proposed new technique, MARZSS, is independent from the undesirable effects noted above, which are traditionally generated by the readout sequence. Therefore, the readout sequence parameters, especially TE, can be kept consistent between measurements.
It is well known that metabolite T2s measured by multi-TE PRESS differ substantially from those obtained via CPMG-type methods (Table 2). In contrast, the proposed MARZSS technique is more similar to inversion recovery measurement, whose results are independent of the readout sequence. The three techniques for T2 measurement have significantly different characteristics.
First, it should be noted that metabolite quantification is complicated by the baseline contributions of broad resonances from macromolecules when spectra are measured at short or even medium TEs [22]. Without a priori knowledge of the concentrations or relaxation times of those macromolecules, accurate extraction of true signal amplitude of the metabolites of interest can be difficult at short TEs [23, 24]. However, using short TE data is unavoidable when measuring T2 using multi-TE PRESS or CPMG. Because the fitting results of signal amplitude acquired with short TE steps are typically less accurate than those acquired with longer TE steps due to stronger macromolecule baseline at short TE, systematic errors in metabolite T2 measurement are difficult to eliminate when using either of these two conventional techniques. In contrast, the MARZSS method proposed here completely separated relaxation weighting from readout. As a result, an optimized readout sequence with fixed TE can be used. For example, in this study, the readout sequence was previously optimized to minimize macromolecule baseline and to separate Glu H4 resonance from overlapping signals of Gln and the aspartyl moiety of NAA [21].
Second, for spin echo type sequences, metabolite transverse relaxation measurements can be affected by extra signal attenuation resulting from the diffusion effect. Variations in TE effectively alter diffusion time. In addition, the spoiling gradients used in the sequences and the intrinsic microscopic internal gradient associated with susceptibility inhomogeneity can introduce diffusion attenuation [25, 11]. Furthermore, in cases of in vivo measurements obtained in the presence of internal gradients at different TEs, the potential signal decay induced by physiological motion such as cardiac and respiration cycles can be even stronger than diffusion per se, introducing more errors. Implementing CPMG-type sequences for quantification can reduce various diffusion effects. However, in order to shorten diffusion time, the inter-pulse delay of CPMG has to be decreased while increasing the number of 180° refocusing pulses; this change may result in safety concerns about specific absorption rate in clinical applications. In addition, metabolite T2 relaxation measured using CPMG-type sequences can be contaminated by relaxation during the refocusing pulses [11, 26].
Third, for spin echo and stimulated echo sequences, the signal amplitude of coupled resonances is complicated due to scalar coupling effect. Introducing T2 relaxation by varying TE, which is necessary for both multi-TE PRESS and CPMG, introduces additional signal modulation due to scalar coupling but is again unavoidable in conventional methods. The experimenter is often forced to use multiple suboptimal TEs in order to generate different amounts of T2 weighting for T2 quantification. These suboptimal TEs can cause significant signal overlap for metabolites of interest. For example, the spectra acquired using the MARZSS technique are demonstrated in Fig. 4a, where the smooth and monotonic signal amplitude attenuation by the progressively larger saturation effects of iPFG trains can be seen. In comparison, the spectra obtained via multi-TE PRESS are shown in Fig. 4b, where the J-coupling effect interferes strongly with the exponential attenuation of transverse relaxation.
While multi-TE PRESS experiments place magnetization entirely on the transverse plane to introduce necessary T2 weighting, our pulse-driven steady state technique uses a combination of rapid refocusing with 10 ms intervals between adjacent iPFG pulses and longitudinal storage of magnetization. Given that in the MARZSS method, relaxation weighting is prepared longitudinally, we would expect this method to be substantially less susceptible to diffusion and motion effects.
Phantom measurements (listed in Table 1) found that the T2 of NAA, Cr, and Cho determined by the MARZSS technique agreed reasonably well with the conventional multi-TE PRESS technique. This is likely due to the fact that internal local susceptibility gradients (internal gradients) are not present in the phantom liquid. The free diffusion decay of metabolites is very small with multi-TE PRESS because the time difference of two spoil gradients surrounding the two 180 degree pulses is short, and because diffusion evolution times are not altered in response to different TE values in the multi-TE measurements. Therefore, with phantoms, it is reasonable that MARZSS and multi-TE T2 values would agree. However, the situation would be completely different in vivo, where internal local susceptibility gradients (internal gradients) are present [11]. TE changes in multi-TE measurements may literally cause variations in the evolution time of diffusion, thereby creating different diffusion decay rates that depend on TE. As a result, substantial, statistically significant differences were found between the two methods in in vivo experiments. Indeed, we found that the T2s of NAA, Cr, and Cho measured by the proposed MARZSS technique were significantly longer than those measured by multi-TE PRESS (see Table 2). Interestingly, the in vivo T2 values measured by the MARZSS technique—which has a mechanism of rapid partial refocusing with an inter-pulse interval of only 10 ms—were closer to values obtained using CPMG, which is known to be far less sensitive to diffusion-related effects [11, 12].
The experimental results presented here validate the concepts outlined in the Theory section, above. However, it should be noted that further optimization of this technique is possible. For example, the first measurement with flip angle of 0° is not necessary because the same data are acquired (or extracted from fitting) in the separate T1 measurements with the same readout sequence. In addition, the simulation results shown in Fig. 2b show that both recovery delay time and iPFG pulse train duration can be significantly shortened to speed up the overall experiment. Our Bloch simulation suggests that TR may be shortened to 4–5 s for flip angles larger than 15°.
This novel MARZSS technique is also associated with certain limitations. First, the accuracy of T2s depends on the measurement of T1, which introduces additional errors to the final results. However, measurement of T1 is relatively more robust, and its result is less controversial than measurement of T2 because multi-TEs are not involved in the measurements; indeed, we estimated that even when T1s were averaged across five subjects instead of using individual T1s, in most cases the deviation from T1 would be less than 10%. Second, the accuracy of B1 calibration is another concern, although multi-TE PRESS and CPMG with non-adiabatic pulses can also suffer from B1 miscalibration or B1 inhomogeneity. The effect of flip angle error can be evaluated by taking the derivative of Eq. 3: dT2/T2 = dα/sin(α). Therefore, a 10% error in flip angle causes an approximately 10% error in T2 for the range of flip angles (sin(α) ≈ α) used in this study.
Taken together, our findings suggest that the complete separation between relaxation preparation and readout opens many interesting possibilities for practical application in metabolite relaxation experiments. For example, many spectral editing experiments require a fixed TE to maximize editing yield, making it very difficult to effectively measure transverse relaxation of the edited signals using conventional multi-TE PRESS or CPMG methods. The proposed technique, however, allows the straightforward inclusion of various editing schemes (e.g, two-step J editing or multiple quantum filtering) into the readout sequence because TE can be arbitrarily selected and fixed while relaxation weighting is completely generated and quantified by the MARZSS technique. In addition, with the freedom to select an arbitrary TE for readout, TE can be set to zero or close to zero when measuring T2 relaxation. This can be very convenient for investigating MRS signals with very short T2s; examples include the ATP signals in 31P MRS and labile proton signals that are in fast exchange with water.
Conclusion
We developed a novel technique— MARZSS—for probing metabolite T2 relaxation by generating RF-driven longitudinal steady-state magnetization without the need to vary TE. An iPFG train was used to create spatial modulation for probing metabolite relaxation in vivo and a new linear equation was derived from Bloch equations. The application of this new concept was demonstrated by Bloch simulations, phantom experiments, and in vivo experiments on human brain. This new technique may have promising applications in a variety of experiments involving relaxation characterization of tissue properties.
Acknowledgments
This work was supported by the intramural research program of National Institute of Mental Health. Ioline Henter (NIMH) provided invaluable editorial assistance.
References
- 1.Tognarelli JM, Dawood M, Shariff MI, Grover VP, Crossey MM, Cox IJ, Taylor-Robinson SD, McPhail MJ. Magnetic Resonance Spectroscopy: Principles and Techniques: Lessons for Clinicians. J Clin Exp Hepatol. 2015 Dec;5(4):320–8. doi: 10.1016/j.jceh.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansen JF, Backes WH, Nicolay K, Kooi ME. 1H MR Spectroscopy of the brain: absolute quantification of metabolites 1. Radiology. 2006 Aug;240(2):318–32. doi: 10.1148/radiol.2402050314. [DOI] [PubMed] [Google Scholar]
- 3.Sibtain NA, Howe FA, Saunders DE. The clinical value of proton magnetic resonance spectroscopy in adult brain tumours. Clinical radiology. 2007 Feb 28;62(2):109–19. doi: 10.1016/j.crad.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 4.De Graaf RA. In vivo NMR spectroscopy: principles and techniques. John Wiley & Sons; 2013. Mar 21, [Google Scholar]
- 5.Dager SR, Oskin NM, Richards TL, Posse S. Research applications of magnetic resonance spectroscopy (MRS) to investigate psychiatric disorders. Topics in magnetic resonance imaging: TMRI. 2008 Apr;19(2):81. doi: 10.1097/RMR.0b013e318181e0be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du F, Cooper A, Cohen BM, Renshaw PF, Öngür D. Water and metabolite transverse T2 relaxation time abnormalities in the white matter in schizophrenia. Schizophrenia research. 2012 May 31;137(1):241–5. doi: 10.1016/j.schres.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magnetic Resonance in Medicine. 1994 Sep 1;32(3):294–302. doi: 10.1002/mrm.1910320304. [DOI] [PubMed] [Google Scholar]
- 8.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Annals of the New York Academy of Sciences. 1987 Nov 1;508(1):333–48. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 9.Frahm JA, Bruhn H, Gyngell ML, Merboldt KD, Hänicke W, Sauter R. Localized high resolution proton NMR spectroscopy using stimulated echoes: Initial applications to human brain in vivo. Magnetic resonance in medicine. 1989 Jan 1;9(1):79–93. doi: 10.1002/mrm.1910090110. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Kolkovsky AL, Mériaux S, Boumezbeur F. Metabolite and macromolecule T1 and T2 relaxation times in the rat brain in vivo at 17.2T. Magn Reson Med. 2016 Feb;75(2):503–14. doi: 10.1002/mrm.25602. [DOI] [PubMed] [Google Scholar]
- 11.Michaeli S, Garwood M, Zhu XH, DelaBarre L, Andersen P, Adriany G, Merkle H, Ugurbil K, Chen W. Proton T2 relaxation study of water, N-acetylaspartate, and creatine in human brain using Hahn and Carr Purcell spin echoes at 4T and 7T. Magnetic resonance in medicine. 2002 Apr 1;47(4):629–33. doi: 10.1002/mrm.10135. [DOI] [PubMed] [Google Scholar]
- 12.Ronen I, Ercan E, Webb A. Rapid multi-echo measurement of brain metabolite T2 values at 7 T using a single shot spectroscopic Carr-Purcell-Meiboom-Gill sequence and prior information. NMR in Biomedicine. 2013 Oct 1;26(10):1291–8. doi: 10.1002/nbm.2951. [DOI] [PubMed] [Google Scholar]
- 13.Deoni SC, Peters TM, Rutt BK. High resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magnetic resonance in medicine. 2005 Jan 1;53(1):237–41. doi: 10.1002/mrm.20314. [DOI] [PubMed] [Google Scholar]
- 14.Morris GA, Freeman R. Selective excitation in Fourier transform nuclear magnetic resonance. Journal of Magnetic Resonance (1969) 1978 Mar 31;29(3):433–62. doi: 10.1016/j.jmr.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 15.Mosher TJ, Smith MB. A DANTE tagging sequence for the evaluation of translational sample motion. Magnetic Resonance in Medicine. 1990 Aug 1;15(2):334–9. doi: 10.1002/mrm.1910150215. [DOI] [PubMed] [Google Scholar]
- 16.Li L, Miller KL, Jezzard P. DANTE prepared pulse trains: A novel approach to motion sensitized and motion suppressed quantitative magnetic resonance imaging. Magnetic resonance in medicine. 2012 Nov 1;68(5):1423–38. doi: 10.1002/mrm.24142. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Chai JT, Biasiolli L, Robson MD, Choudhury RP, Handa AI, Near J, Jezzard P. Black-blood multicontrast imaging of carotid arteries with DANTE-prepared 2D and 3D MR imaging. Radiology. 2014 Jun 11;273(2):560–9. doi: 10.1148/radiol.14131717. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Xu D, Ozturk-Isik E, Lupo JM, Chen AP, Vigneron DB, Nelson SJ. T1 and T2 metabolite relaxation times in normal brain at 3T and 7T. J Mol Imaging Dynam S. 2012;1:002. [Google Scholar]
- 19.An L, Li S, Murdoch JB, Araneta MF, Johnson C, Shen J. Detection of glutamate, glutamine, and glutathione by radiofrequency suppression and echo time optimization at 7 Tesla. Magnetic resonance in medicine. 2015 Feb 1;73(2):451–8. doi: 10.1002/mrm.25150. [DOI] [PubMed] [Google Scholar]
- 20.Provencher SW. Automatic quantitation of localized in vivo1H spectra with LCModel. NMR in Biomedicine. 2001 Jun 1;14(4):260–4. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- 21.An L, Li S, Shen J. Simultaneous determination of metabolite concentrations, T1 and T2 relaxation times. Magnetic Resonance in Medicine. 2017 Feb 1; doi: 10.1002/mrm.26612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cianfoni A, Law M, Re TJ, Dubowitz DJ, Rumboldt Z, Imbesi SG. Clinical pitfalls related to short and long echo times in cerebral MR spectroscopy. Journal of Neuroradiology. 2011 May 31;38(2):69–75. doi: 10.1016/j.neurad.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Behar KL, Rothman DL, Spencer DD, Petroff OA. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magnetic Resonance in Medicine. 1994 Sep 1;32(3):294–302. doi: 10.1002/mrm.1910320304. [DOI] [PubMed] [Google Scholar]
- 24.Gottschalk M, Lamalle L, Segebarth C. Short TE localised 1H MRS of the human brain at 3 T: quantification of the metabolite signals using two approaches to account for macromolecular signal contributions. NMR in Biomedicine. 2008 Jun 1;21(5):507–17. doi: 10.1002/nbm.1219. [DOI] [PubMed] [Google Scholar]
- 25.van der Toorn A, Dijkhuizen RM, Tulleken CA, Nicolay K. Diffusion of metabolites in normal and ischemic rat brain measured by localized 1H MRS. Magnetic resonance in medicine. 1996 Dec 1;36(6):914–22. doi: 10.1002/mrm.1910360614. [DOI] [PubMed] [Google Scholar]
- 26.Deelchand DK, Henry PG, Marjańska M. Effect of carr purcell refocusing pulse trains on transverse relaxation times of metabolites in rat brain at 9.4 Tesla. Magnetic resonance in medicine. 2015 Jan 1;73(1):13–20. doi: 10.1002/mrm.25088. [DOI] [PMC free article] [PubMed] [Google Scholar]