Abstract
BACKGROUND
Genomic testing is increasingly performed in oncology, but concerns remain regarding clinician’s ability to interpret results. We sought to determine the agreement between physicians and genomic annotators from the University of Texas MD Anderson Precision Oncology Decision Support (PODS) Team regarding actionability and the clinical utilization of test results.
METHODS
On a prospective protocol, patients underwent clinical genomic testing for hotspot mutations in 46 or 50 genes (Ampli-Seq Cancer Panel; ThermoFisher). Six months after sequencing, physicians received questionnaires for patients with a variant in an actionable gene, investigating their perceptions regarding actionability of alterations and clinical utilization of these findings. Genomic annotators independently classified these variants as: actionable, potentially actionable, unknown or not actionable.
RESULTS
Physicians completed 250 of 288 questionnaires (87% response rate). Physicians considered 168/250 patients (67%) as having an actionable alteration; 165/168 (98%) were considered actionable by PODS; three were of unknown significance. Physicians were aware of genotype-matched therapy available for 119 (71%); 48/119 (40%) received matched therapy. 46% (36/79) of patients in whom physicians regarded alterations as not actionable were classified as having an actionable/potentially actionable mutation by PODS. However, many of these were only theoretically actionable due to limited trials/therapies (e.g., KRAS).
CONCLUSIONS
Physicians are aware of recurrent mutations in actionable genes on “hot spot” panels. As larger genomic panels are used, there may be growing need for annotation of actionability. Decision support to increase awareness of genomically-relevant trials and novel treatment options for recurrent mutations (e.g., KRAS) are also needed.
Keywords: Precision medicine, genomics, somatic mutation, clinical trial, personalized therapy
INTRODUCTION
Sequencing technology is rapidly improving and costs are decreasing to the point that utilizing genomic results in routine clinical decision making is becoming practical and increasingly widespread. However, the clinical impact of genomics is limited by clinicians’ ability to interpret this growing and rapidly changing information; specifically, to determine which alternation(s) impact gene function and to identify appropriate therapy. Unfortunately, 22% of physicians at a major cancer center reported low confidence in their ability to interpret genomic results. Low physician confidence was associated with decreased anticipated utilization of genomic testing to drive clinical decision making [1].
Studies showing success using targeted therapy for the treatment of cancer have increased in the past decade. Examples of these include use of EGFR inhibitors in EGFR-mutated lung cancer [2], BRAF and MEK inhibitors in BRAF V600E-mutant unresectable or metastatic melanoma [3, 4], and HER2-targeting agents in HER2+ metastatic breast cancer [5]. These successes have fueled the drive to identify new molecular targets, which may improve outcomes for patients whose tumors harbor specific alterations. The presence of alterations in these novel targets may sensitize the tumors harboring them to treatment with targeted therapy. We have seen a rapid transition from single-gene testing into larger panel testing covering a larger number of genes and exons.
While some alterations may not affect protein function, others may change function significantly. Further, when an altered gene is felt to provide a useful target for treatment, the physician must be aware of currently available therapies and clinical trials.
The clinical impact of genomic testing may be limited by clinicians’ ability to appropriately order testing and correctly interpret results. As use of genomics in medicine continues to grow we must understand more fully how these changes are affecting physicians and the treatment that they are able to offer to their patients. We sought to understand physician perceptions regarding: 1) the clinical impact of the availability of genomic testing had and 2) the effect on patient satisfaction resulting from the addition of genomic testing (as perceived by the physician). This information may suggest areas where resources and services can be provided to support physicians in their application of targeted therapy to maximize benefit to patients.
METHODS
Patients
Cancer patients with metastatic or inoperable locally advanced or recurrent cancer, who were perceived as likely to benefit from somatic genomic characterization, were enrolled on an Institutional Review Board-approved prospective protocol for genomic profiling (NCT01772771).
Genomic Sequencing
Genomic sequencing was performed as described in Boland, et al. [6]. Briefly, Hematoxylin and eosin stained archival tissue sections with > 20% tumor cellularity were analyzed. Tumor DNA was tested in a Clinical Laboratory Improvement Amendments (CLIA) environment looking at hotspot mutations in 46 or 50 genes using an Ion Torrent Personal Genome Machine Sequencer (ThermoFisher, CA) [7]. Only those alterations designated as likely somatic were considered for this questionnaire.
Determination of Actionability
Genomic alterations were annotated to determine actionability based on known or potential functional and/or therapeutic significance of the variant, and availability of genomically-matched therapies, as established by The MD Anderson Precision Oncology Decision Support team, and previously described [8]. First, it was determined whether or not the gene and type of alteration (i.e., mutation, copy number change, or fusion) were actionable. Although a gene may be actionable if it is a biomarker of risk, or diagnostic or prognostic, we focused on genes actionable due to therapeutic implications. Several of the factors considered in making gene level call included the availability of drugs targeting the gene/pathway either directly or indirectly, published literature suggesting that the gene may play a role in driving tumorigenesis, and use of the gene as selection criteria for enrollment on a clinical trial [8]. Our actionable gene list used for this study has been previously published [9]. Once the actionability of the alteration type was determined, it was then considered whether the specific variant was actionable based upon information from published literature, a functional genomics platform, and location of the alteration within the gene and proximity to functional domains. Based upon the findings, variants are grouped into one of the following classifications of actionability: ‘yes’ (literature based, or inferred [inferred loss of function mutations in tumor suppressors), ‘potentially’, ‘unknown’, and ‘no’ [8]. For the purpose of this analysis, alterations having actionability calls of ‘yes’ or ‘potentially’ were considered to be in agreement with physicians who felt an alteration was actionable. In cases where a patient had more than one actionable alteration identified, perceptions of actionability were compared to the alteration with the highest actionability level for each patient.
Questionnaires
Questionnaires were distributed to physicians with information about their patient provided in pre-populated fields (Supplementary Text). Relevant information provided to physicians included patient medical record number, name, date the test was completed and the alterations that were identified. Questionnaires were conducted through RedCap and were sent to the physicians of these patients via a link contained within an e-mail. Questions in the questionnaire followed a pattern where the physicians were first asked regarding whether they considered the alterations identified to be actionable. They were subsequently asked if they were aware of available targeted therapy available at MD Anderson for the identified alterations and were allowed the opportunity to describe any genotype-matched treatments considered. The next set of questions included whether the patient received treatment based on the sequencing results and if so, what type of therapy was given. A drop down menu provided the options that could be selected (Clinical trial, off-label or standard of care) and a free text field allowed physicians to type the name(s) of the therapy given. The last set of questions pertained to physician perceptions of the value added to the care provided as well as to the perceived effect of genomic testing on patient satisfaction with regards to care received as a result of the genomic testing.
Results
Questionnaires were sent to 69 physicians, regarding 288 patients, in order to gain an understanding of physician perceptions regarding actionability of alterations identified in potentially actionable genes, knowledge of genotype-matched therapy available at the institution, clinical utilization of genomic testing results, and perceived value of genomic testing. Out of 288 questionnaires sent, 250 were completed (87% response rate) by 64 individual physicians from 13 different departments/disease centers. The number of questionnaires completed by each physician ranged from 1 to 29, with the median being 2 questionnaires per physician.
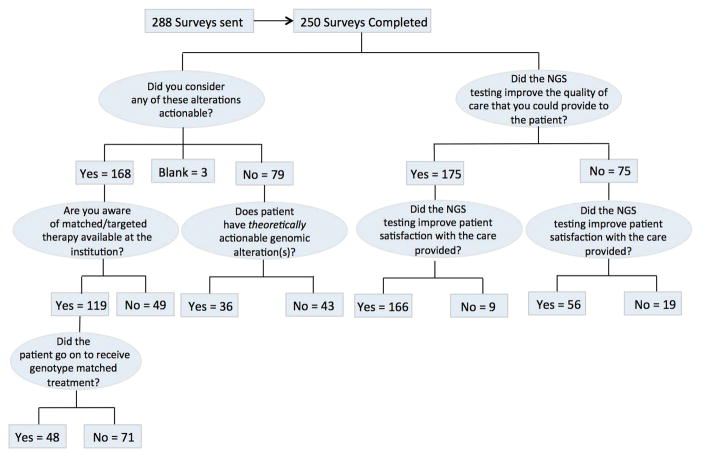
Physicians regarded 168 of the 250 patients as having an actionable alteration (Figure 1 and Supplementary Table 1. PODS annotators considered 165 out of 168 (98%) patients to have an actionable or potentially actionable alteration. Three of the 168 patients who were considered to have actionable alterations by their physicians had variants classified as “unknown significance” by PODS annotators (Supplementary Table 2).
Figure 1.
Questionnaire questions and responses. Flowchart describing the flow of questions contained within the questionnaires as well as responses given which were validated through manual review.
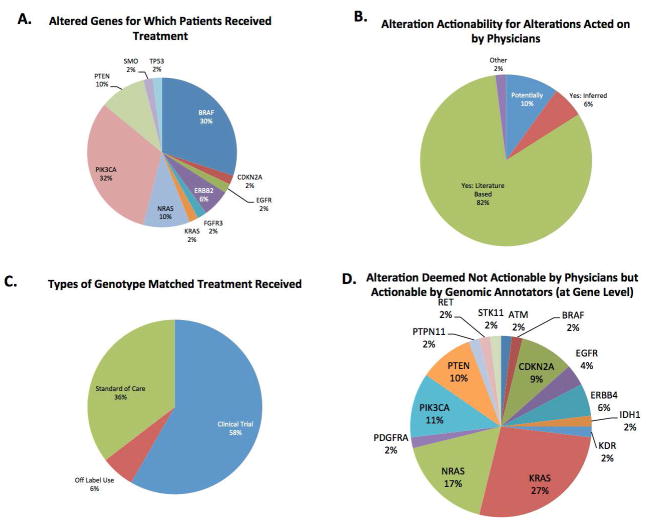
Of the 168 patients about whom physicians felt had at least one actionable alteration, we asked how often physicians were aware of genotype-matched therapy available for these alterations at the institution. 119 out of 168 (71%) were aware of genotype-matched therapy available, while 49 (29%) were not. Utilizing both physician responses provided as well as manual review, we determined that out of the 119 patients whose treating physician was aware of genotype-matched therapy at the institution, 48 (40%) went on to receive matched therapy and 71 (60%) did not. Shown in Figure 2A, the altered genes most frequently acted upon to receive targeted therapy were PIK3CA (32%), BRAF (30%), PTEN (10%), NRAS (10%), and ERBB2 (6%). Alterations that were acted upon in this patient population most frequently would have been classified as actionable based on either published literature (82%), inferred actionability (6%), or potentially actionable (10%) (Figure 2B). Shown in Figure 2C, of the patients who went onto receive genotype-matched treatment, patients most frequently went onto a clinical trial (58%) with standard of care being the second most frequent (36%), followed by off-label use of targeted therapy (6%).
Figure 2.
A. Altered genes in patients used for treatment selection with a genotype-matched therapy. B. Alteration actionability as classified by PODS for alterations subsequently acted on by physicians. C. Type of therapy received for patients with alterations receiving genotype-matched therapy. D. Alterations (gene level) which physicians deemed to be not actionable, but for which a call by the genomic annotators would have been classified as either “actionable” or “potentially actionable.”
To understand why 71 of 119 patients whose physician was aware of genotype matched therapy options did not go on to receive genotype-matched treatment, we utilized both physician comments in the questionnaires as well as manual review of patient clinic notes. Table 1 summarizes the reasons that we observed. The most frequent reason, accounting for 25 out of 71 patients was the patient’s choice to receive treatment closer to home due to unwillingness or inability to travel. Other common reasons reported were election to be treated with non-targeted therapy 13/71 (18%), ineligibility for a matched clinical trial 11/71 (15%), and poor performance status 8/71 (11%). These most common reasons are consistent with our previous findings of common barriers to clinical trial enrollment which were assessed by retrospective review rather than through a prospective questionnaire [10].
Table 1.
| Reason for Not Receiving Treatment | N=71 |
|---|---|
| Elected Local Treatment | 25 |
| Elected Non-Targeted Therapy | 13 |
| Ineligible | 11 |
| Previous Number of Treatments | 1 |
| No Measurable Disease | 3 |
| Other Co-Morbidities | 7 |
| Poor Performance Status | 8 |
| Eligible but no available openings | 2 |
| Not Financially Cleared/Insurance Declined Coverage | 3 |
| Already on Targeted Therapy | 5 |
| Not Interested in Clinical Trials | 3 |
| Stable Disease/Surveillance | 1 |
In 79 out of the 250 completed questionnaires, physicians felt that the specific alterations found in the potentially actionable genes were not truly actionable. We found that PODS would have classified at least one alteration in 36 (46%) out of these 79 patients as either actionable or potentially actionable (Figure 1 and Supplementary Table 3. For this analysis, we considered KRAS alterations present in a patient with a diagnosis of colorectal cancer to be not actionable [11]. However, KRAS alterations were categorized as theoretically actionable for other diagnoses. Figure 2D shows that differences were most frequently observed for KRAS alterations in tumor types other than colorectal cancer (27%) followed by alterations in NRAS (17%). For these 36 patients, 73% of these alterations would have been classified as actionable (either inferred or literature based) and 27% would have been classified as potentially actionable based on PODS’ definition of actionability.
For 27 (75%) of 36 patients with actionable or potentially actionable alterations that the physician deemed not actionable, there was a clinical trial open at the institution that was either selecting for alterations in an altered gene or using a drug relevant for one of the altered genes present. Full screening for eligibility could not be performed. However, based on the genotype and clinical disease type patients were considered a “match.” At the time of the questionnaire, there were very few trial options for KRAS/NRAS mutant patients, making these alterations more “theoretically” rather than “practically” actionable through targeted therapy.
Figure 1 summarizes the physician-perceived value added to the care that was provided to the patient as a result of the genomic profiling. When physicians were asked whether they felt that utilization of NGS platforms improved the “quality of care” that they were able to provide to the patient, 175 (70%) felt that it did improve quality of care, while 75 (30%) felt that it did not. When asked whether or not the physician perceived NGS as having improved patient satisfaction with efforts to personalize treatment options, 222 (89%) physicians responded that it did improve patient satisfaction and 28 (11%) felt that it did not. Notably when physicians felt that NGS improved the quality of care that they were able to provide, they were also more likely to perceive the patient as having improved satisfaction with efforts to personalize treatment options p<0.0001.
DISCUSSION
The high response rate (87%) suggests that physicians are actively engaged and willing to discuss the clinical impact of genomic testing. We observed that treating physicians and genomic annotators agreed regarding the actionability of genomic alterations. Specifically, in 67% of patients with an alteration identified in a potentially actionable gene, physicians identified at least one alteration as actionable. Within this group the physician call on actionability agreed with the PODS classifications 98% of the time.
However, of the 79 patients whose physician did not recognize any actionable alterations, 46% had an alteration that PODS classified as either “actionable” or “potentially actionable.“ This suggests an area where decision support services may have clinical utility. Another opportunity lies in the ability of decision support teams to offer treatment “match” options to physicians so that they have all of the information provided when deciding on the next best treatment option. We found that physicians were aware of genotype matched therapy for only 71% of patients whom they felt to have an actionable alteration, once again suggesting an important role for decision support to help physicians interpret results and recognize when targeted therapy options are available to their patients.
Patients whose tumors harbored alterations in BRAF, PIK3CA, PTEN, NRAS, and ERBB2 were most likely to receive targeted therapy. This is suggests that physicians may be more inclined to act upon alterations about which more information is available. Notably, several of the alterations were only theoretically actionable due to lack of trials, emphasizing the importance of having a suite of applicable clinical trials to realize the promise of targeted therapy for cancer. This discordance between theoretical and actual actionability partly suggests that studies that only look at theoretical actionability may overestimate the clinical utility of genomic testing. On the other hand, some of the alterations classified as nonactionable by physicians were potentially actionable. Both false positives and false negatives in interpretation can decrease the clinical utility of genomic testing; thus there is a need for decision support to help physicians optimally utilize targeted therapy. This challenge will become more acute as larger panels, whole exome or even genome testing becomes widely available. The importance of decision support is further supported by the previously reported lack of physician confidence to interpret genomic testing results [1].
One limitation to our questionnaire is that, in cases where a patient had more than one alteration identified in a potentially actionable gene, we could not gather alteration-specific perceptions of actionability. For example, if a patient had one alteration that PODS would have classified as “unknown actionability” and another which would been classified as “actionable”, it is impossible to know which alteration the physician was referring to as actionable unless the patient received therapy targeted to a specific alteration. Another limitation of our study is that it was conducted in a single academic center. In the community practice setting there may exist greater discordances between clinician and annotator interpretations; this may be due to a lower perception of actionability as a consequence of decreased access to investigational therapeutics at the site. To address this issue, it will be important to consider ways in which we can have a broader impact and assist physicians and treatment centers outside of MD Anderson Cancer Center by providing access to decision support on the ever changing actionability classification of genes as it relates to new scientific evidence and the offering of novel targeted therapies in the clinical trial setting. Support should also be given to ensure that physicians feel confident to suggest a visit to one of the centers where these targeted therapies are available for further evaluation of eligibility.
When physicians felt that an actionable alteration was present and were also aware of matched therapy targeting the alteration, 40% of patients went onto receive genotype-matched therapy (48/119). Notably, the type of treatment received was most commonly experimental as in a clinical trial (58%) or off-protocol use (6%), whereas standard of care made up a little over a third of genotype-matched therapy received (36%). The most common reasons patients did not receive genotype-matched treatment were similar to those previously reported [10], with the most frequent reason being patient preference of being treated closer to home (35%). Other factors limiting treatment with genotype-matched therapy was the election to be treated with alternate therapy (18%), ineligibility for clinical trial being considered (15%) and poor performance status (11%). The trial enrollment rate of 40% that we observed, when physicians both perceived as having an actionable alteration and were aware of available treatment, is in line with what others have seen. The Lung Cancer Mutation Consortium found a 28% rate of enrollment onto matched-therapy trials in patients who had an oncogenic alteration identified [12], similarly the SAFIR01 breast cancer trial found that 28% of patients with a targetable alteration went on to receive matched therapy [13].
Physicians who felt that their patient had an actionable alteration were more likely to perceive that genomic testing improved the quality of the care that they were able to provide to the patient (82% vs 43%). Interestingly however, the overall perception of improved patient satisfaction as a result of genomic testing was not different between the patients whose physician felt they had actionable alterations vs those that did not (89% for both). This suggests that whether or not the physician deems an alteration actionable, their physicians perceived that genomic testing improved patient satisfaction.
Supplementary Material
Acknowledgments
Funding: This work was supported in part by CPRIT grants RP150535 (Precision Oncology Decision Support Core) and RP170668 (Data Science and Informatics Core for Cancer Research); the Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy; NCI U01CA180964; NCATS UL1TR000371 (Center for Clinical and Translational Sciences); the Nellie B. Connally Breast Cancer Research Endowment; the MD Anderson Cancer Center Support grant (P30 CA016672); NHI/NCATS UL1TR001105 (UT-Southwestern Clinical and Translational Alliance for Research); NIH/NLM grants R01 LM011829 and R01 LM01068, and NIH/NIDCR grant R01 DE024166.
Footnotes
Disclosures: Dr. Eng reports grants from Daichi, grants from Keryx, other from Bayer, other from Sirtex, other from Genentech, outside the submitted work; Dr. Mendelsohn reports personal fees from Merrimack Pharmaceuticals, outside the submitted work; Dr. Meric reports grants from Novartis, AstraZeneca, Taiho, Genentech, Calithera, Debiopharma, Bayer, Aileron, Jounce, CytoMx, eFFECTOR, Zymeworks, PUMA, Curis, Pfizer, personal fees from Dialecta, Sumitomo Dainippon Pharma, personal fees from Inflection Biosciences, Pieris, Darwin Health, GRAIL, Clearlight Diagnostics, during the conduct of the study; Dr. Mills reports grants from Adelson Medical Research Foundation, grants, personal fees and other from AstraZeneca, grants and other from Critical Outcome Technologies, grants from Komen Research Foundation, grants from Nanostring, grants from Breast Cancer Research Foundation, grants from Karus, grants from Illumina, grants from Takeda/Millenium Pharmaceuticals, personal fees and other from Symphogen, personal fees and other from MedImmune, personal fees and other from ISIS Pharmaceuticals, grants and personal fees from Pfizer, personal fees and other from Lilly, personal fees and other from Novartis, personal fees and other from ImmunoMet, personal fees and other from Allostery, personal fees and other from Tarveda, other from Adventist Health, other from Catena Pharmaceuticals, other from Precision Medicine, other from Provista Diagnostics, other from Signalchem Lifesciences, other from Tau Therapeutics, outside the submitted work; In addition, Dr. Mills has a patent HRD assay to Myriad Genetics licensed.
Author Contributions:
Conceptualization: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Methodology: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Software: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Validation: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Formal analysis: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Investigation: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Resources: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Data curation: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Writing – original draft: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Writing – review and editing: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Visualization: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Supervision: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Project administration: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
Funding acquisition: L Brusco, C Wathoo, K Mills Shaw, V Holla, A Bailey, A Johnson, Y Khotskaya, N Sanchez, J Zeng, E Bernstam, C Eng, B Kee, R Amaria, M Routbort, G Mills, J Mendelsohn, F Meric-Bernstam
References
- 1.Gray SW, et al. Physicians’ attitudes about multiplex tumor genomic testing. J Clin Oncol. 2014;32(13):1317–23. doi: 10.1200/JCO.2013.52.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis PM, et al. The role of the epidermal growth factor receptor tyrosine kinase inhibitors as therapy for advanced, metastatic, and recurrent non-small-cell lung cancer: a Canadian national consensus statement. Curr Oncol. 2009;16(1):27–48. doi: 10.3747/co.v16i1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim G, et al. FDA approval summary: vemurafenib for treatment of unresectable or metastatic melanoma with the BRAFV600E mutation. Clin Cancer Res. 2014;20(19):4994–5000. doi: 10.1158/1078-0432.CCR-14-0776. [DOI] [PubMed] [Google Scholar]
- 4.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 6.Boland GM, et al. Clinical next generation sequencing to identify actionable aberrations in a phase I program. Oncotarget. 2015;6(24):20099–110. doi: 10.18632/oncotarget.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh RR, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15(5):607–22. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Johnson A, et al. The right drugs at the right time for the right patient: the MD Anderson precision oncology decision support platform. Drug Discov Today. 2015;2012:1433–8. doi: 10.1016/j.drudis.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meric-Bernstam F, et al. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst. 2015;107(7) doi: 10.1093/jnci/djv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meric-Bernstam F, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol. 2015;33(25):2753–62. doi: 10.1200/JCO.2014.60.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrido-Laguna I, et al. KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PLoS One. 2012;7(5):e38033. doi: 10.1371/journal.pone.0038033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kris MG, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andre F, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15(3):267–74. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.