Abstract
PREMISE OF THE STUDY
Plants will play an important role in the future of space exploration as part of bioregenerative life support. Thus, it is important to understand the effects of microgravity and spaceflight on gene expression in plant development.
METHODS
We analyzed the transcriptome of Arabidopsis thaliana using the Biological Research in Canisters (BRIC) hardware during Space Shuttle mission STS-131. The bioinformatics methods used included RMA (robust multi-array average), MAS5 (Microarray Suite 5.0), and PLIER (probe logarithmic intensity error estimation). Glycome profiling was used to analyze cell wall composition in the samples. In addition, our results were compared to those of two other groups using the same hardware on the same mission (BRIC-16).
KEY RESULTS
In our BRIC-16 experiments, we noted expression changes in genes involved in hypoxia and heat shock responses, DNA repair, and cell wall structure between spaceflight samples compared to the ground controls. In addition, glycome profiling supported our expression analyses in that there was a difference in cell wall components between ground control and spaceflight-grown plants. Comparing our studies to those of the other BRIC-16 experiments demonstrated that, even with the same hardware and similar biological materials, differences in results in gene expression were found among these spaceflight experiments.
C ONCLUSIONS
A common theme from our BRIC-16 space experiments and those of the other two groups was the downregulation of water stress response genes in spaceflight. In addition, all three studies found differential regulation of genes associated with cell wall remodeling and stress responses between spaceflight-grown and ground control plants.
Keywords: cell wall, cytoskeleton, dehydration, glycome profiling, transcriptome, heat shock, XERO1, FH2, AGP, XTH
Spaceflight is potentially stressful for biological systems. Understanding this unique environment and its impact on gene regulation is a critical step in planning for long-duration spaceflight missions (Perbal, 2008; Vandenbrink and Kiss, 2016). In support of these exploration missions, bioregenerative life support systems will use plants as a source of food, fuel, and fiber for space explorers (Mitchell, 1994; Paradiso et al., 2013). However, environmental factors such as cosmic radiation, craft vibration, and microgravity (μg) can detrimentally influence plant development (Kranz, 1986; Mitchell, 1994).
Morphological variances have been previously observed in spaceflight-grown plants relative to ground controls. These include decreased starch content and altered cell shape as a result of cell wall perturbation (Johnson et al., 2015), as well as more adventitious roots, waving of roots (Paul et al., 2012b) and a unidirectional skewing of shoots and roots (Millar et al., 2011). Consequently, the unique environment of spaceflight may negatively impact the yield of crops grown as part of the bioregenerative life support systems. Therefore, understanding the mechanisms by which plants respond to spaceflight, including alterations in gene expression, is critical for designing crops that can produce high yields in these unique environments.
Transcriptomics provides a means of screening for gene expression in biological specimens. Microarrays are used for transcriptomic spaceflight experiments because they can produce meaningful results from small sample sizes. Several recent studies have compared the transcriptomes of spaceflight-grown and Earth-grown Arabidopsis seedlings (Paul et al., 2012b, 2013; Zupanska et al., 2013; Correll et al., 2013; Fengler et al., 2015; Kwon et al., 2015). In these experiments, four different types of flight hardware were used: the European Modular Cultivation System (EMCS), Advanced Biological Research System (ABRS), Science in Microgravity box (SIMbox), and Biological Research in Canisters (BRIC). EMCS and ABRS were used in the International Space Station, SIMbox was used on the Shenzhou 8 Chinese spacecraft, and BRIC was used on the Space Shuttle.
Unfortunately, comparative analyses among these transcriptomics experiments are limited due to differences in hardware, different genotypes, varying numbers of replicates, and different procedures for handling tissues (Paul et al., 2012b). However, despite these different approaches, there are some overlapping biochemical pathways that have been identified to be differentially regulated in spaceflight. These include pathways involved in heat shock, stress response, cell wall modification, and cell defense.
Spaceflight experiments are complex, and the results obtained by plant space biologists have been variable due to differences in approaches and hardware design. For example, using the EMCS hardware on the ISS, the TROPI2 experiment investigated gene expression in Arabidopsis seedlings (Correll et al., 2013). This transcriptomics study had 280 transcripts differentially regulated at least 2-fold in spaceflight compared with the ground controls. The spaceflight hardware included an onboard centrifuge, so a comparison of 1 g spaceflight with μg spaceflight was possible. This spaceflight 1 g vs. μg comparison revealed only 27 transcripts differentially regulated 2-fold or more (P < 0.01). The transcripts that were differentially expressed included many that were responsible for cell wall functionality, cell defense, cell stress, oxygen status, cell wall development, cell polarity including auxin, and lipid metabolism, among others.
Another study used the ABRS flight hardware, which is a different hardware system from the EMCS (Paul et al., 2012a, 2013). These studies found that 408 transcripts were differentially regulated at least 1.9-fold and 58 that were at least 7-fold different in spaceflight compared with the ground control (P < 0.01). Each organ was found to have specific and unique responses to the spaceflight environment, which followed an overall change in morphology of the plant body including a change in cell wall architecture (Paul et al., 2013).
In yet another hardware scenario, SIMbox, was flown in a joint Chinese and German experiment (Fengler et al., 2015; Li et al., 2017), and the hardware included a centrifuge to provide a 1 g spaceflight control. In this project, 298 genes, with a high P-value (P < 0.1), were reported as differentially regulated 2-fold or more between space and ground control plants. Transcripts involved with cell defense response, pathogen-response, and pathogen-resistance genes were among the differentially expressed genes reported, along with calcium-signaling related MAP kinases and reactive oxygen species (Fengler et al., 2015).
Thus, with these several recent spaceflight studies, it is difficult to find similar correlations in the data among the different investigators, largely due to differences in experimental design and diverse hardware. However, one cluster of recent studies from spaceflight experiments is distinguished by similar methodology and hardware, and, therefore, these results can be thoroughly compared. In 2010, three research groups performed individual experiments in the BRIC-Petri Dish Fixation Unit (BRIC-PDFU) hardware as part of the STS-131 mission on the Space Shuttle. Each group used Arabidopsis thaliana as a model organism, and all of these experiments performed transcriptome analyses on the spaceflight samples and compared them to ground controls.
These experiments were entitled:
Actin Regulation of Arabidopsis Root Growth and Orientation During Space Flight (BRIC-16-Regulation; NASA, 2016b).
Impact of Spaceflight on Arabidopsis: Deep Sequencing and DNA Arrays as Collaborative Readouts of the Transcriptome of Arabidopsis Seedlings and Undifferentiated Cells in Space (BRIC-16-DNA; NASA, 2016c).
Investigations of the Plant Cytoskeleton in Microgravity with Gene Profiling and Cytochemistry (BRIC-16-Cytoskeleton; NASA, 2016a).
The purpose of the BRIC-16-Regulation (BRIC-16-Reg; E. Blancaflor, Principal Investigator) experiment was to compare microtubule development within the roots of wild-type and actin mutants in μg (Kwon et al., 2015; Nakashima et al., 2014). The wild type used in this experiment was Columbia (Col-0). This experiment found that there were more transcripts downregulated (148) than upregulated (26) at least 4-fold or more in spaceflight. Downregulated transcripts were mainly nuclear-encoded, while upregulated were mainly encoded in the plastid. The BRIC-16-Reg project focused on oxidative stress and cell wall structure in response to μg, as exhibited by the shorter root hairs present in spaceflight samples compared to ground controls (Kwon et al., 2015).
BRIC-16-DNA (A.-L. Paul, Principal Investigator) studied gene expression profiles from seedlings and callus tissues (Paul et al., 2012b, 2013; Zupanska et al., 2013). The wild type used in this experiment was Col-0. This experiment found that approximately 300 genes were differentially regulated more than 2-fold in whole seedlings, and another approximately 300 in tissue culture samples (P < 0.01). However, in the case of seedlings alone, only 45 were differentially regulated 5-fold or more in spaceflight vs. ground control. BRIC-16-DNA compared their work with other experiments conducted in parabolic and simulated μg environments, and concluded that heat shock proteins (HSPs) were active only in prolonged spaceflight (Zupanska et al., 2013). In addition, Paul et al. (2012b) indicated that there were no differences in gross morphology between spaceflight-grown and ground control seedlings.
Our BRIC-16-Cytoskeleton (BRIC-16-Cyt; J. Z. Kiss, Principal Investigator) studies focused on the structure of shoot statocytes and on the effects of spaceflight on plant gene expression. The wild type used in this study was Landsberg erecta (Ler-0). BRIC-16-Cyt reported a unidirectional skewing of roots and shoots was observed in μg compared with ground control plants, and adventitious roots were present in greater abundance in the spaceflight samples (Millar et al., 2011). Additionally, morphometric measurements of the endodermal cells in petioles of seedlings demonstrated a significant change in endodermal cell shape. Cells were more rounded in spaceflight vs. ground control, which was likely due to cell wall structural differences due to μg (Johnson et al., 2015).
All three of these BRIC-16 experiments were run simultaneously in the same flight hardware, with differences in sample preparation, and all three generated transcriptome data using microarrays. BRIC-16-Reg and BRIC-16-DNA used the Col-0 ecotype accession while BRIC-16-Cyt used Ler-0. Each of these groups grew seedlings on agar plates for 14 d, at which point seedlings were stabilized with RNAlater in-flight. Use of in-flight −80°C cold stowage for rapid freezing was unavailable for STS-131, so all samples were returned to Earth in RNAlater at ambient temperature.
In this paper, we present comparative analyses of our BRIC-16-Cyt spaceflight experiment with the other two BRIC-16 experiments (BRIC-16-Reg and BRIC-16-DNA). We demonstrate that even the same species grown in the same hardware can yield differing results due to specific experimental conditions including variations in genotypes, tissues used, and analytical techniques. These discrepancies led to a finding of little overlap among differentially regulated genes in these three spaceflight studies that used the same BRIC hardware system. However, transcripts within the same functional categories, including stress response, defense, auxin signaling, metabolic processes, and cell wall modification, were found to be common among these studies.
MATERIALS AND METHODS
The materials and methods for our BRIC-16-Cyt study are described in detail below. The materials and methods for the other experiments in BRIC-16-Reg and BRIC-16-DNA are described briefly here as a comparison. Additional information regarding sample preparation is presented in Kwon et al. (2015) for BRIC-16-Reg and Paul et al. (2012b) for BRIC-16-DNA.
Preflight preparation of BRIC-16-Cyt
For the BRIC-16-Cyt experiment (J. Z. Kiss, Principal Investigator), seeds of Arabidopsis thaliana (Ler-0) were surface-sterilized in a solution of 1 drop octyl phenol ethoxylate (TX-100) in 100 mL of 70% (v/v) ethanol for 5 min, followed by two, 1-min rinses in 95% (v/v) ethanol. An additional washing with 1 drop TX-100 in 100 mL water was followed by four final rinses in sterile water. Under sterile conditions within a laminar flow hood, seeds were sown in one row of 40 seeds on 13 total Petri plates (60 mm) on 1.2% (w/v) agar containing 1 mM MES buffer at pH 5.5, 1% (w/v) sucrose, one-half strength Murashige and Skoog basal salts (Millar et al., 2011).
During the sowing process of the BRIC-16-Cyt seeds, the quality of each seed was assessed using a stereomicroscope and damaged seeds were discarded. Petri plates were sealed with Parafilm M (Bemis Company, Oshkosh, Wisconsin, USA), placed in a sealed sterile box then allowed 1 day incubation in darkness at a controlled 24°C before integration into the Biological Research in Canisters-Petri Dish Fixation Unit (BRIC-PDFU) flight hardware (Kern et al., 1999; Millar et al., 2011).
Integration of biological specimens into the flight hardware was conducted in sterile conditions within a laminar flow hood by engineering staff at the NASA Space Life Sciences Laboratory (SLSL, Kennedy Space Center, Florida, USA). Directionality of each PDFU was maintained as its fluid compartment was filled with 13 mL of RNAlater (Ambion, Foster City, California, USA). Each PDFU was then tested for leaks and placed within the external BRIC shell. The assembled BRIC-PDFUs were placed within a half-sized middeck locker on the Space Shuttle Discovery 36 h before launch (see Millar et al., 2011; Kwon et al., 2015; and Paul et al., 2012b for additional details on preparation of the BRIC-PDFU spaceflight hardware).
Comparison of preflight preparation among the BRIC-16 investigative teams
All three groups used the BRIC-PDFU flight hardware. BRIC-16-Cyt was conducted using the Ler-0 ecotype, while BRIC-16-DNA and BRIC-16-Reg used the Col-0 ecotype as the wild type. All plants were grown in the dark on 60 mm petri plates and on a similar agar substrate that included MS salts. Plates from both BRIC-16-Reg and BRIC-16-Cyt were made up of 1.2% (w/v) agar with half-strength MS salts and 1% (w/v) sucrose. BRIC-16-DNA used 0.5% (w/v) Phytagel Arabidopsis Plate Media with half-strength MS salts (Paul et al., 2013; Schultz et al., 2012). Quantities of medium were equivalent for each group (6.5–6.7 mL per plate).
While all seeds were sown before loading the hardware, the patterns in which they were sown were different. BRIC-16-DNA contained seeds sown in a grid pattern, with approximately 75 seeds per plate (Paul et al., 2012b; Schultz et al., 2012), while BRIC-16-Cyt seeds were sown in a dense line of 39–42 seeds across the center of each plate (see Millar et al., 2011), and BRIC-16-Reg plates contained 150–200 seeds in a series of lines for molecular studies (Kwon et al., 2015). Seeds for the BRIC-16-Cyt were wet-sown in nanopure water, while BRIC-16-Reg seeds were dry-sown from sterile filter paper following surface-sterilization (supplemental photo from Kwon et al., 2015). Thus, sowing techniques and substrates varied among the three investigative groups.
Launch, landing, and sample processing
For all of the BRIC-16 flight samples, stabilization in RNAlater was completed in-orbit by an astronaut after 309 h of growth of the plants in darkness in the BRIC-PDFU. Following landing and after 53 h in RNA later, the BRIC-16 samples were prepared for deintegration. The three BRIC-16 investigative teams followed this timeline.
Petri plates then were removed from the flight hardware and the BRIC-16-Cyt plates with seedlings were photographed for gross morphology and germination analyses. BRIC-16-Cyt seedlings were placed in RNAse-free vials with fresh RNAlater and transported on ice to the University of Florida, Gainesville, Florida (USA). Upon arrival at the university, microarray samples were placed in a freezer at −80°C for storage until extraction for RNA.
Ground controls for all BRIC-16 samples were prepared in the same initial manner but with a 24-h delay from spaceflight samples. A ground control locker was placed within a growth chamber known as the orbital environmental simulator (OES) at the SLSL, which attempted to duplicate the temperature and humidity data collected from Discovery’s middeck, on a 24-h delay. The ground control locker was repositioned manually to mimic the directionality changes of the Shuttle’s launch sequence. Stabilization of all BRIC-16 samples was conducted in parallel to the process in-orbit, again on the same 24-h delay.
Cell wall glycomics of BRIC-16-Cyt
The cell wall glycomics analyses were performed at the University of Georgia Complex Carbohydrate Research Center (CCRC, Athens, GA). Tissue that remained in the shredder columns from the RNA extraction process from BRIC-16-Cyt samples were used to run an ELISA-based glycome analysis employing a comprehensive suite of plant cell wall glycan directed monoclonal antibodies (mAbs) (Pattathil et al., 2012, 2015, 2016).
This zero waste procedure allowed us to maximize the data produced from a minimal amount of spaceflight-grown tissue. Briefly, cell wall materials (alcohol insoluble residues; AIR) were isolated from ground and spaceflight grown plant tissues as described previously (Pattathil et al., 2012). Cell wall materials were subsequently subjected to a high alkaline extraction (4 M KOH with 1% w/v sodium borohydride) to extract most major noncellulosic cell wall glycans. The 4 M KOH extract was then neutralized, exhaustively dialyzed, and lyophilized for subsequent ELISA screens with the entire collection of cell-wall-glycan-directed mAbs (Pattathil et al., 2012). This process was extensively tested by CCRC staff with small quantities of Earth-grown Arabidopsis tissue samples before conducting these spaceflight studies, and their results were largely comparable to the results obtained with the BRIC-16-Cyt samples. Heat maps were produced using antibodies binding strength values obtained after ELISA screens (Pattathil et al., 2012). Cell walls were extracted from the shoot tissues to obtain a glycome profile, as explained above. ELISAs were performed with 0.3 μg glucose equivalent carbohydrate per-well basis in technical duplicates, and the presented data represent background-noise-subtracted means of the replicate assay. Cell-wall-glycan-directed mAbs were obtained from laboratory stocks (CCRC, JIM and MAC series) at the Complex Carbohydrate Research Center (available through Carbo-Source Services; http://www.carbosource.net) and BG1 and LAMP (LAMP2HI2H7) were from BioSupplies (Yagoona, Australia). Cell wall glycomics analyses were not conducted by the other two BRIC-16 investigative teams.
Microarray preparations for the BRIC-16-Cyt seedlings
The contents of one BRIC from spaceflight (F) and two BRICs, each from a separate ground control (G), were set aside for the molecular component of the study. Each BRIC included five PDFUs (which contained one Petri plate). To clarify the sample size: each Petri plate was sown with 40 seeds, and germination rates were 89% for flight and 90% for ground Petri plates in the BRIC-16-Cyt study, providing a total of approximately 178 seedlings from spaceflight and 360 from ground control for molecular work. Of these, half of the seedlings harvested from four spaceflight PDFUs (79 seedlings) and nine ground control PDFUs (160 seedlings) were used for the microarray analyses. Samples were transferred to RNase-free microcentrifuge tubes containing fresh RNAlater for transportation.
To process the BRIC-16-Cyt samples before the microarray studies, we thawed the samples and bisected them to separate roots from shoots. Tissues from shoots only were used for this study. Tissue of shoots was disrupted in a microcentrifuge tube using a Pellet Pestle motor (Kontes, Vineland, New Jersey, USA) and sterile RNase-free pestle tip (Stimpson et al., 2009). Extraction was conducted using the RNeasy Mini Plant Kit (Qiagen, Valencia, CA) according to the procedure outlined in the RNeasy Mini Handbook. The total RNA was provided to the University of Florida’s Interdisciplinary Center for Biotechnology Research (ICBR) Gene Expression Core Facility. There, sample was quantified using the NanoDrop Spectrophotometer ND-8000 (NanoDrop Technologies, Wilmington, Delaware, USA), and quality was determined using the RNA 6000 Pico LabChip (Agilent Technologies, Santa Clara, California, USA) and its corresponding ladder (RNA 6000 ladder, Ambion, Foster City, California, USA) containing transcript sizes of 0.2, 0.5, 1.0, 2.0, 4.0 and 6.0 kb on a Bioanalyzer (Agilent). Of the samples prepared, the three plates with the highest quality RNA were chosen from ground control and spaceflight to run the microarray in Appendix S1 (see the Supplemental Data with this article).
RNA amplification and purification of BRIC-16-Cyt were conducted by the ICBR using the Ovation Pico WTA system for biotin target labeling. Chemical and enzymatic fragmentation of cRNA was completed in a thermal cycler at 37°C for 60 min, and 70°C for 10 min, with a final holding at 4°C until sample removal. The NanoDrop spectrophotometer was used to assess the yield of the labeled cRNA. Hybridization to the microarray gene chip was conducted following the standard protocol outlined in the technical manual for Affymetrix GeneChip Expression Analysis (Affymetrix, Santa Clara, CA).
The BRIC-16-Cyt microarrays were washed and stained using the Affymetrix GeneChip Fluidics Station 450 according to the Affymetrix standard protocol outlined in the Affymetrix GeneChip Expression Analysis technical manual. Nonstringent wash buffer was used for washing. Streptavidin phycoerythrin (SAPE) was used for the staining process. Scanning was completed using the GeneChip Scanner 3000. Raw data were presented in the form of 570 nm light emission, the intensity of which is proportional to the number of targets bound at each locus of the array. Raw data from microarray studies was processed using Microarray Suite Version 5 software (Affymetrix) to convert the intensity information to quantitative values.
Analyses of microarrays
Raw microarray data were generated by each of the three investigative teams. The data obtained from BRIC-16-Reg and BRIC-16-DNA were deposited in the ArrayExpress electronic database (Kolesnikov et al., 2015). BRIC-16-Reg can be found under the identifier E-MTAB-3011, while BRIC-16-DNA is identified as E-MTAB-1009. BRIC-16-Cyt data were deposited in the NASA GeneLab under accession number GLDS-121 (https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-121/).
For analysis of the raw microarray data from all three investigative teams (BRIC-16-Cyt, BRIC-16-Reg, and BRIC-16-DNA), the Expression Console (EC) software (Affymetrix) was used for normalization. For a point of comparison, three normalization methods were separately employed for all data sets: RMA (robust multi-array average), PLIER (probe logarithmic intensity error), and MAS5 (Affymetrix MicroArray Suite version 5) (Quackenbush, 2002; Irizarry et al., 2003; McClintick and Edenberg, 2006; Seo and Hoffman, 2006; Therneau and Ballman, 2008; Qu et al., 2010). Quality control tests were run on each normalized data set. Each normalized set was further processed by running gene-level unpaired one-way between subject ANOVA analyses using the Transcription Analysis Console (TAC) (Affymetrix) (Reis et al., 2015). Specifics of this workflow were described by Ode et al. (2014). Volcano plots were generated using Excel Professional Plus 2013 (Microsoft, Redmond, Washington, USA). Hierarchical clustering of the BRIC-16-Cyt data was generated from the RMA-normalized BRIC-16-Cyt data using TAC. Gene Ontology (GO) data for each transcript was found using The Arabidopsis Information Resource (TAIR) (Rhee et al., 2003). Graphs were generated showing that the main categories were molecular function, biological process, and cellular composition for the BRIC-16-Cyt data.
Raw data for the other two investigative teams were obtained from EMBL: E-MTAB-3011 (BRIC-16-Reg) and E-MTAB-1009 (BRIC-16-DNA). For comparative transcriptomics, raw data sets from the three investigative groups (BRIC-16-Cyt, BRIC-16-Reg, BRIC-16-DNA) underwent QC testing using EC software and were processed separately using three normalization methods: MAS5, PLIER, and RMA. Normalized data sets then underwent unpaired one-way between-subject ANOVA analyses. Results of these analyses were augmented with TAIR GO data (Berardini et al., 2004), and genes were further annotated if linked to cell wall extractabilities by previous glycomics studies.
To test the hypothesis that the observed transcriptomic differences among BRIC-16-Cyt, -Reg, and -DNA were due to experimental differences (and not random due to poor RNA or experimental error), we conducted a pairwise analysis of the ground controls from each experiment. Then, we performed a pairwise analysis of the spaceflight samples from each experiment. The data were then checked for overlap between the spaceflight and ground control. This process was repeated using each normalization method (RMA, MAS5, PLIER).
To show that the number of stress genes was not abnormally high in the BRIC-16-Cyt samples as compared with the other investigative teams, we conducted a gene-level differential expression analysis: a one-way between-subject unpaired ANOVA to compare BRIC-16-Cyt ground controls with BRIC-16-DNA and BRIC-16-Reg ground control samples. The GO data were collected for the resulting gene lists and sorted.
Quantitative real-time PCR (qRT-PCR) of BRIC-16-Cyt seedlings
Shoots of seedlings from the BRIC-16-Cytoskeleton (BRIC-16-Cyt) group’s spaceflight-grown and ground control were also used for the qRT-PCR, and the following procedures took place at Miami University (Oxford, Ohio, USA). Seedlings were shipped overnight on dry ice from −80°C storage at the University of Florida and placed in −80°C storage upon arrival. Seedlings were defrosted, then dissected in RNAlater, disrupted using a pellet pestle motor and sterile pestle, and RNA was extracted from the shoots using the RNeasy Mini Plant Kit (Qiagen, Valencia, California, USA), according to the procedure outlined previously (Stimpson et al., 2009). Following extraction, excess fluid was evaporated using a centrifugal vacuum evaporator Savant Speed Vac Concentrator (Thermo Scientific, Waltham, Massachusetts, USA). Evaporated RNA was re-eluted using RNase-free water and converted to cDNA using the QuantiTect Reverse Transcription Kit (Qiagen). First, genomic DNA was removed by incubating RNA for 2 min with gDNA Wipeout Buffer in a 42°C water bath. Next, the samples underwent reverse transcription through incubation at 42°C with Quantiscript Reverse Transcriptase, Qantiscript RT Buffer, and RT Primer mix, followed by inactivation of the process by placing them in a 95°C water bath for 3 min. A negative control was run alongside spaceflight and ground samples during the reverse transcription process. This negative control lacked reverse transcriptase, but contained all other reaction products.
Real-time quantification of PCR was conducted to compare ground control and spaceflight samples using a QuantiTect SYBR Green PCR Kit (Qiagen) and a Bio-Rad iCycler (Bio-Rad; Hercules, CA) at Miami University’s Center for Bioinformatics and Functional Genomics, using the standard procedure outlined in the QuantiTect SYBR Green PCR Handbook. Primers for each gene of interest were designed using Primer 3 Plus (Untergasser, 2007; Untergasser et al., 2007; Thornton and Basu, 2011). Ubiquitin-conjugating enzyme 10 (UBC10; AT5G53300) was used as the standard. Primers are provided in Table 1. Incubation of cDNA with primers and SYBR Green Master Mix was conducted at 95°C for 15 min to activate the DNA polymerase and followed by 3-step cycling for 40 cycles. The cycles included a denaturing step for 15 s at 94 ° C, annealing for 30 s at 50 ° C, and extension at 72°C for 30 s A comparison of expression levels for each gene was conducted against our standard, UBC 10 (Stimpson et al., 2009), and negative controls— one that contained no reverse-transcriptase and one that lacked templates. Next, a melting curve analysis of the PCR products was conducted by plotting the results with the 2(−ΔΔCt) method (Livak and Schmittgen, 2001) to visualize the fold-change in RNA expression (also used by Paul et al. [2012b]). Resulting values of each were averaged, and the means were used to determine statistical significance using a Student’s t test in Excel.
TABLE 1.
Primers used for qRT-PCR by the BRIC-16-Cytoskeleton research group.
| TAIR gene ID | Gene symbol | Primer |
|---|---|---|
| AT5G53300 | UBC10 | L 5′-TCC CAA CAT TAA CAG CAA CG-3′ R 5′-CTT CGT GCA GTG GAC TCG TA-3′ |
| AT5G17850 | CAX8 | L 5′-TTG GTT AGC AGG AGG GTT TG-3′ R 5′-CAC TTG AGC TCC TTC GTT CC-3′ |
| AT1G68560 | XYL1 | L 5′-GAG ACC ATC GCA ACT CAC AA-3′ R 5′-CTG AAC CAA CCA TGG GAA CT-3′ |
| AT2G47190 | MYB2 | L 5′-GGA TGC CGA GAT TAG TGG AA-3′ R 5′-GGA GAA TTC GAA GAC GTT GC-3′ |
| AT4G24780 | EXPA10 | L 5′-TGT ACG ACC GGT AAC CCA AT-3′ R 5′-CCA CGG CGT GTC TTA AAG TT-3′ |
RESULTS
Cell wall glycomics of BRIC-16-Cyt
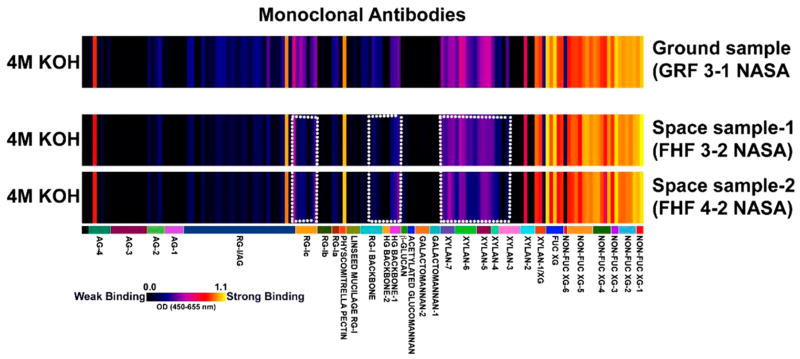
Shoot tissue samples that remained in the shredder columns after the RNA extraction process from the BRIC-16-Cyt experiments were in trace amounts. Cell wall materials isolated from these small sample amounts were subjected to a single-step strong alkaline extraction, an optimized method while working with trace amounts of starting tissue materials, to isolate most major noncellulosic matrix cell wall glycans. These extracts from ground and spaceflight samples were screened with a comprehensive suite of cell wall glycan-directed mAbs that could monitor most major noncellulosic glycan epitopes in plants. The results of this screening study are shown in Fig. 1.
FIGURE 1.
Comparative glycomics analyses of ground and spaceflight grown shoot samples of Arabidopsis thaliana. Cell-wall-glycan-directed mAbs-based ELISA screening of 4 M KOH cell wall extracts prepared from ground- and spaceflight-grown shoot samples of Arabidopsis thaliana was conducted to enable a comparative glycomics analysis. This comprehensive collection of 155 mAbs (labels in lower panel) used in the screen monitored most major classes of noncellulosic matrix cell wall glycans. The binding strengths of the mAbs that corresponded to the abundance of matrix glycan epitopes are depicted as a heatmap with a bright yellow to black color scheme in which bright yellow depicts the strongest binding and the black no binding.
The results obtained showed that despite the small amounts of samples being used to start with, the 4 M KOH extraction step could recover significant abundance of matrix cell wall glycan epitopes for enabling mAb-based comparative glycomics studies. The most abundant epitopes detected in both sample types were nonfucosylated and fucosylated xyloglucan epitopes. However, interestingly, tissues from the spaceflight-grown samples reproducibly exhibited subtle differences from that of ground samples. For instance, the abundance of xylan epitopes was reduced in the spaceflight samples (those detected by xylan-3 through -7 groups of the mAbs), and similarly, the abundance of pectic backbone epitopes, including those recognized by homogalacturonan (HG) backbone-1, HG backbone 2, rhamnogalacturonan (RG)-I backbone, and RG-Ic groups of mAbs (Fig. 1) was marginally reduced in spaceflight-grown plants in comparison to the ground control.
Overall, the results of the glycomics study of the BRIC-16-Cyt samples suggest that biosynthesis of xylan and pectic components of the walls may be impacted by microgravity, resulting in a compositional difference in the cell wall matrix. Table 2 outlines the genes involved in cell wall modification that are upregulated in spaceflight as determined by the BRIC-16-Cyt microarray, including the arabinogalactan protein AGP31 (AT1G28290), the xyloglucan XTH9 (AT4G03210), the glycosidase XTH32 (AT2G36870), and the glycosyl hydrolases GH9C2 (AT1G64390) and GH9B7 (AT1G75680) (Table 2).
TABLE 2.
Genes involved in cell wall modification category of the Gene Ontology analyses that were differentially regulated between Space Flight and Ground Control (F/G) in BRIC-16-Cyt, −5 ≤ FC ≥ 5 (P < 0.01), using three data normalization methods, RMA, MAS5, PLIER. FC = fold-change.
| Gene ID | Gene symbol | FC: RMA | FC: MAS5 | FC: PLIER |
|---|---|---|---|---|
| AT2G36870 | XTH32 | 3.19 | 7.98 | 7.02 |
| AT4G03210 | XTH9 | 2.88 | 12.92 | |
| AT1G64390 | GH9C2 | 5.33 | 5.42 | 7.64 |
| AT1G28290 | AGP31 | 22.97 | 23.44 | 22.72 |
Microarray analyses
RNA was extracted from BRIC-16-Cyt seedlings, and the quality of RNA was assessed before conducting microarrays. The first indication of poor RNA quality was provided by the Bioanalyzer data (Appendix S1), which was further assessed with a QC threshold test. The QC threshold test confirmed that the microarray data were outside bounds for all three normalization methods conducted on BRIC-16-Cyt, with a percentage spread that was ≥10% among arrays. The BRIC-16-Reg group also noted poor RNA quality (Kwon et al., 2015) and was confirmed by a QC threshold test that was ≥10% among arrays. Raw data provided by the BRIC-16-DNA group also did not pass our QC threshold tests, as their percentage present spread was also ≥10% among arrays, and their log scale factor spread was high. Since sample sizes were limited and these spaceflight experiments could not readily undergo duplication, we continued with analyses of BRIC-16-Cyt based on the successful reporting of the BRIC-16-Reg (Kwon et al., 2015) and BRIC-16-DNA results (Paul et al., 2012b).
BRIC-16-Cyt microarrays were normalized using three separate methods: RMA, MAS5, and PLIER. The BRIC-16-Cyt microarrays normalized using RMA showed that 126 transcripts were differentially expressed in spaceflight vs. ground control. Of these transcripts, 71 were upregulated while 55 were downregulated (−5 ≥ FC ≥ 5, P < 0.01). MAS5 analysis yielded 187 upregulated and 121 downregulated (−5 ≥ FC ≥ 5, P < 0.01), while PLIER analysis yielded the most, with 384 upregulated and 66 downregulated transcripts (−5 ≥ FC ≥ 5, P < 0.01) (Table 3). With all these methods, more were upregulated than downregulated overall (RMA data are displayed in Appendices S2 and S3). Of note, 71 transcripts were differentially regulated after all three normalization methods, but their fold-change values varied. Of these, only 37 transcripts were consistently upregulated, while only six were consistently downregulated among all three normalization techniques. Such differences in fold-change values among normalization methods are mentioned in the literature (Qu et al., 2010) and are to be expected. Many of the genes that were downregulated in spaceflight were annotated with gene ontology (GO) data (TAIR, 2017) for involvement in water stress response (Fig. 2).
TABLE 3.
Three normalization methods applied to the BRIC-16-Cyt microarray data. The number of differentially expressed genes in Space Flight/Ground Control (F/G) (P < 0.01) are listed.
| Normalization method | −2 ≤ Fold-change ≥ 2
|
−5 ≤ Fold-change ≥ 5
|
||
|---|---|---|---|---|
| Up | Down | Up | Down | |
| PLIER | 1125 | 242 | 384 | 66 |
| MAS5 | 492 | 275 | 187 | 121 |
| RMA | 437 | 205 | 71 | 55 |
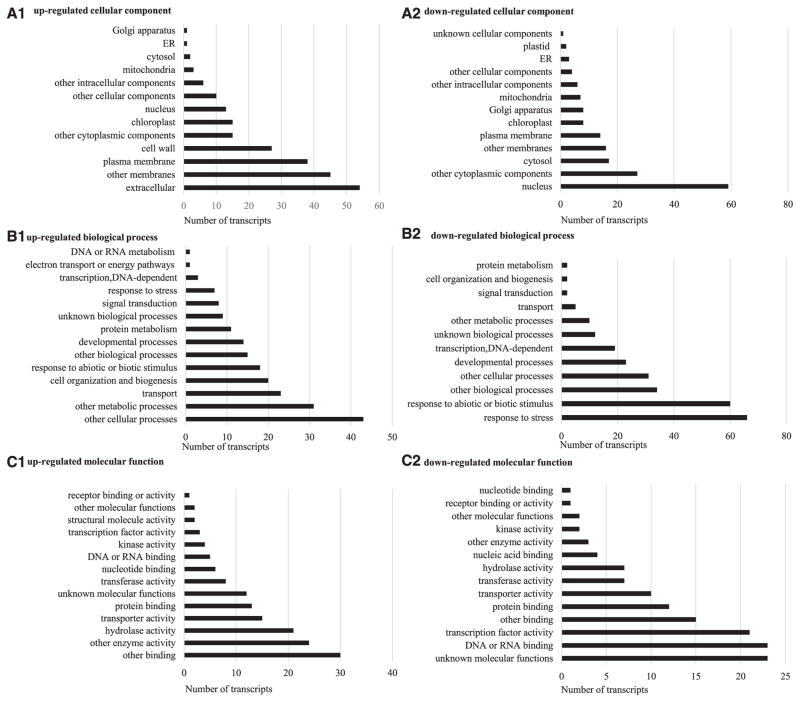
FIGURE 2.
The Arabidopsis Information Resource (TAIR) Gene Ontology data showing (A1) upregulated cellular component, (A2) downregulated cellular component, (B1) upregulated biological process, (B2) downregulated biological process, and (C1) upregulated molecular function, (C2) downregulated molecular function for the BRIC-16-Cytoskeleton group. A comparison of Space Flight/Ground Control (F/G) with RMA-normalized data, −5 ≤ fold-change ≥ 5 (P < 0.01). The most downregulated biological process was response to stress. The nucleus was the most common cellular component localized for downregulated transcripts.
GO cellular component data for our BRIC-16-Cyt experiments (Fig. 2A) indicated that most transcripts that were upregulated in spaceflight conditions were localized in the extracellular matrix, membranes, and the cell wall, while the majority of downregulated transcripts were localized in the nucleus. Biological process GO data (Fig. 2B) showed many upregulated transcripts were implicated in metabolic processes and transport, while the downregulated were often involved in stress response. GO molecular function data (Fig. 2C) indicated that the most upregulated transcripts were involved in enzyme activity, while the downregulated were implicated in some form of binding, including protein binding and DNA or RNA binding (Fig. 2C). GO data were used when identifying genes involved in hypoxia, heat shock response, cell wall organization, and water relations, as presented in the discussion.
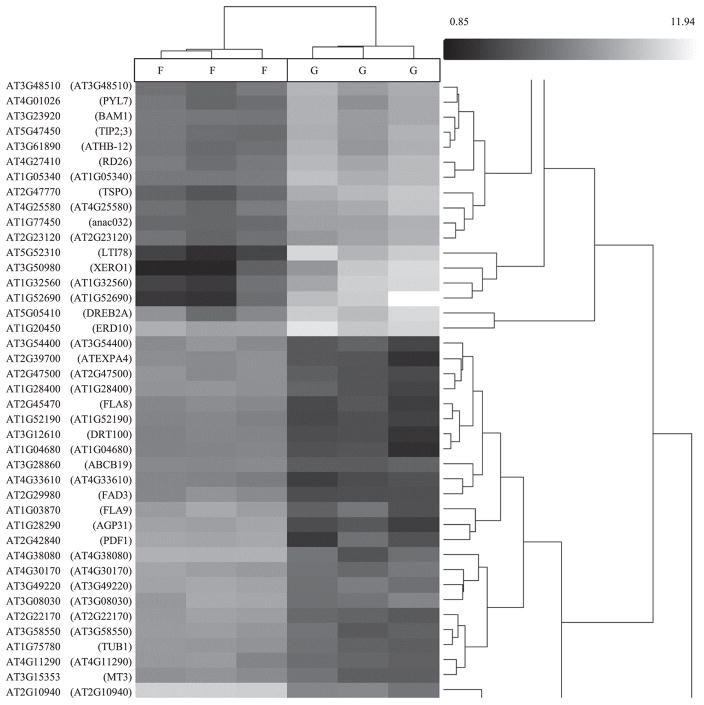
Hierarchical clustering of RMA-normalized BRIC-16-Cyt data showed that the transcriptomes of the flight samples clustered together, while the transcriptomes of the ground samples formed another distinct cluster. Within the genes, we found a clustering of dehydration-responsive genes that were downregulated including XERO1 (AT3G50980) and LTI78 (AT5G52310). Another cluster grouped the arabinogalactan protein FLA8 (aka AGP8; AT2G45470) with expansin expression, including EXPA4 (aka EXP4; AT2G39700) (Fig. 3).
FIGURE 3.
Hierarchical clustering of BRIC-16-Cytoskeleton data, normalized using RMA: spaceflight vs. ground control; −5 ≤ fold-change ≥ 5 (P < 0.01). Downregulated transcripts (black) and upregulated transcripts (white) are shown on a grayscale gradient of black to white. DRT100 (AT3G12610) is a DNA damage repair protein that is downregulated in ground control. Water stress genes, including LTI78 (AT5G52310), XERO1 (AT3G50980), are clustered, and DREB2A (AT5G05410) are clustered as downregulated in spaceflight and upregulated in ground control. To produce this heat map and dendrogram, we calculated the Euclidean distance between expression vectors to determine the similarity between two genes. Cluster-to-cluster distances were determined by complete linkage.
For cell-wall-modifying components in the BRIC-16-Cyt experiment, a number of genes were identified by all three normalization methods (RMA, MAS5, and PLIER) as differentially upregulated (F/G) 5-fold or more (P < 0.01) in the BRIC-16-Cyt study. These genes include AGP31 (AT1G28290), GH9C2 (AT1G64390), and XTH32 (AT2G36870). For xyloglucans specifically, genes that were differentially upregulated (F/G) 5-fold or more (P < 0.01) using one or more normalization method included: XTH32 (AT2G36870), XTH9 (AT4G03210), XTH8 (AT1G11545), XTH6 (AT5G65730), GH9A1 (AT5G49720), GH9B7 (AT1G75680), GH9B13 (AT4G02290), and GH9C2 (AT1G64390).
Also, genes for some heat shock proteins were found to be differentially regulated 5-fold or more (P < 0.01), including the downregulated (F/G) HSP101 (AT1G74310) and HSP90.1 (AT5G52640), and HSP40 (AT2G20550), which was upregulated (F/G) in the BRIC-16-Cyt data.
Quantitative real-time PCR (qRT-PCR) was used to verify the results of the microarray analysis. Four differentially regulated genes from the BRIC-16-Cyt microarray results: CCX2 (AT3G11490), XYL1 (AT1G68556), MYB2 (AT2G47190), EXPA10 (AT1G26770) were analyzed using qRT-PCR. The analysis showed that the expression of MYB2 was reduced in flight and EXPA10 was increased in flight, while CCX2 and XYL1 were unchanged. The results for unchanged genes are likely due to insufficient sample size, an unfortunate constraint of spaceflight studies. In the BRIC-16-Cyt microarray, MYB2 was downregulated, while EXPA10 was upregulated. These results show consistency between the two tests.
Comparison of BRIC-16 spaceflight results
Many studies that have been performed in spaceflight suggest that there is a stress response exhibited by plants (for reviews, see Musgrave, 2002; Paul et al., 2001; and Perbal, 2008). Thus, alterations in gene expression have been shown to occur between plants grown in spaceflight compared to ground controls. Here, we compared the results from three experiments that used the same hardware and the plant Arabidopsis thaliana during the same mission (STS-131), but had notable differences in sample preparation methodologies (Table 4).
TABLE 4.
Experimental methods used by the three investigative teams. Letters indicate BRIC canister identifier. Information on BRIC-16-Reg is from Kwon et al. (2015) and BRIC-16-DNA is from Paul et al. (2012b) and clarified through personal communication. AGM = Arabidopsis growth medium; MS = Murashige and Skoog medium; Noble = Noble Foundation; ICBR = Interdisciplinary Center for Biotechnology Research at the University of Florida.
| Team | BRIC identifier | Ecotype | Tissue | Sowing pattern | Medium base | Medium additives | No. of seeds | Facility | Normalization |
|---|---|---|---|---|---|---|---|---|---|
| BRIC-16-Cyt | E, F, H | Ler-0 | Shoot of seedling | 1 Central row | 1.2% AGM | MS salts, 1% sucrose | 39–42 | ICBR | RMA |
| BRIC-16-Reg | C, D, H | Col-0 | Entire seedling | Several rows | 1.2% AGM | MS salts, 1% sucrose | 150–200 | Noble | RMA |
| BRIC-16-DNA | A, B, G | Col-0 | Entire seedling | Grid pattern | 0.5% Phytagel | MS salts | 75 | ICBR | MAS5 |
Seedlings from BRIC-16-Cyt, BRIC-16-Reg, and BRIC-16-DNA grew for the same length of time, and all were stabilized in-flight using RNAlater. Off-loading from the Space Shuttle and postprocessing were conducted at the same time and in a similar fashion for each group. Whole seedlings were analyzed by the BRIC-16-DNA and BRIC-16-Reg groups, while BRIC-16-Cyt dissected the seedlings before extraction, and we used only the shoots from seedlings for our molecular analyses. Extraction of RNA and the microarrays for BRIC-16-Cyt and BRIC-16-DNA were conducted by staff at the ICBR (Gainesville, Florida, USA) and by staff at the Noble Foundation (Ardmore, Oklahoma, USA) for BRIC-16-Reg. All three groups reported relatively poor quality RNA (Appendix S1; Kwon et al., 2015; Paul et al., 2012b). RNAlater was used by all three investigative groups to stabilize the RNA between the end of the experiment and the time of extraction.
Each investigative team chose a normalization method for their microarray data that was suited to their specific data set. In our primary comparison of the three studies (Fig. 4), we used the same normalization method that was indicated by each group’s published methods. BRIC-16-Cyt and BRIC-16-Reg (Kwon et al., 2015) were normalized using RMA for the results presented here, while BRIC-16-DNA (Paul et al., 2012b) was normalized using MAS5.
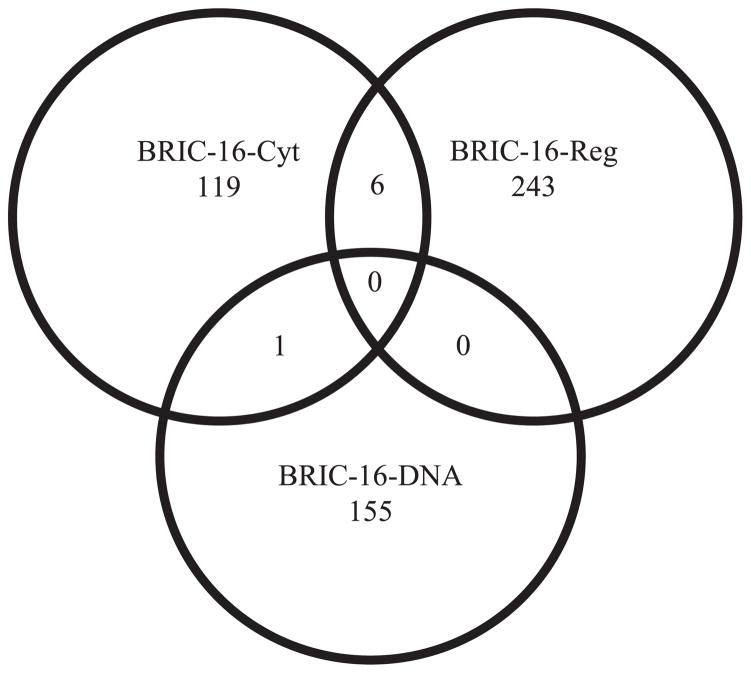
FIGURE 4.
Venn diagram showing a comparison of gene transcripts in a comparison of Space Flight/Ground Control (F/G) (downregulated and upregulated combined) in each study. BRIC-16-Cytoskeleton and BRIC-16-Regulation data were normalized using RMA, while BRIC-16-DNA microarrays were normalized using MAS5. For all groups, −5 ≤ fold-change ≥ 5 (P < 0.01). No transcripts were found in common among the three investigative teams. One transcript was in common between BRIC-16-Cyt and BRIC-16-DNA. Six transcripts were in common between BRIC-16-Cyt and BRIC-16-Reg.
The comparison of these groups, presented in a Venn diagram (Fig. 4), was conducted using each group’s reported normalization method. No transcripts were found in common among all three groups using reported normalization methods, although one transcript was in common between BRIC-16-Cyt and BRIC-16-DNA (AT3G50980), and six were in common between BRIC-16-Reg and BRIC-16-Cyt (Table 5), but these were upregulated in BRIC-16-Cyt and downregulated in BRIC-16-Reg. Overall, BRIC-16-Reg had more transcripts downregulated (Appendix S2), while BRIC-16-DNA and BRIC-16-Cyt saw more transcripts upregulated in spaceflight vs. ground control. An additional analysis was conducted on the raw data for the three groups using both normalization methods (Appendix S4). To confirm that our results were not obscured by experimental error, we conducted additional pairwise analyses between each groups’ ground controls and also between their spaceflight (not reported). The greatest similarities were found among the RMA data sets, with >8500 transcripts appearing in this pairwise comparison of 5-fold or more (P ≤ 0.01) in ground controls. This high level of overlap among the ground controls of the three investigative teams adds credibility to our results, indicating that the observed trends were not a product of experimental error.
TABLE 5.
Transcripts significantly differentially regulated by more than one investigative team. Comparison of Space Flight/Ground Control, −5 ≤ FC ≥ 5 (P < 0.01). FC = fold-change.
| Gene ID | Gene symbol/accession | Description | FC BRIC-16-DNA | FC BRIC-16-Reg | FC BRIC-16-Cyt |
|---|---|---|---|---|---|
| AT3G50980 | XERO1 | Dehydrin, water stress response | −6.07 | −268.27 | |
| AT2G23540 | GDSL | GDSL-motif esterase/acyltransferase/lipase | −8.04 | 7.65 | |
| AT1G75280 | AT1G75280 | Isoflavone reductase | −5.93 | 5.41 | |
| AT2G22510 | AT2G22510 | Hydroxyproline-rich glycoprotein family protein | −6.59 | 9.89 | |
| AT2G38390 | AT2G38390 | Peroxidase superfamily protein | −5.75 | 12.98 | |
| AT2G39700 | ATEXPA4 | Putative expansin, cell wall loosening | −5.56 | 7.78 | |
| AT4G24780 | AT4G24780 | Pectin lyase-like superfamily protein, drought response | −5.22 | 7.18 |
To verify that the reported downregulation of stress genes in spaceflight was not due to a high level of stress in the ground control samples, we conducted a gene-level differential expression analysis with a one-way between-subject unpaired ANOVA to compare BRIC-16-Cyt ground control samples with BRIC-16-DNA and BRIC-16-Reg ground control samples. In this comparison, 12% of the biological process GO terms of the BRIC-16-Cyt samples were categorized as response to stress, while only 5% of the GO terms of the combined BRIC-16-DNA and BRIC-16-Reg ground controls were categorized as response to stress. This series of comparisons indicates that more stress-response genes were present in the BRIC-16-Cyt ground controls compared with the other two investigative teams.
DISCUSSION
Here, we discuss the differences observed in the transcriptomes of the spaceflight studies, in relation to the environmental conditions of the spaceflight study. First, we consider changes in the cell wall during spaceflight, followed by convection as it relates to hypoxia in spaceflight, and then discuss the water status of plants in spaceflight. The emphasis is on the BRIC-16-Cyt results, but we compare our results with those of BRIC-16-Reg and BRIC-16-DNA throughout.
Changes in cell wall during spaceflight
Cell wall integrity is important to a plant’s overall stability. In a recent report on our spaceflight studies, we showed that alterations in the cell wall occurred when seedlings were grown in spaceflight. Specifically, the individual endodermal cells were smaller and more rounded in spaceflight than in ground controls (Johnson et al., 2015). Transcriptomic data showed that a number of genes involved with cell wall (Table 2) and cytoskeletal structure (Table 6) were differentially regulated (F/G). AGP31 (AT1G28290) is an integral cell wall protein that is often abundant within etiolated hypocotyls, which acts as a scaffolding to strengthen the cell wall (Hijazi et al., 2014). AGP31 was found to be differentially upregulated (F/G, −5 ≥ FC ≥ 5, P < 0.01) in BRIC-16-Cyt. A difference in mechanical load in μg led to structural differences within the cell wall during a spaceflight study (Klymchuk et al., 2003). BRIC-16-Reg noted a similar trend and linked these differences to the actin cytoskeleton (Nakashima et al., 2014).
TABLE 6.
Genes involved in cytoskeletal modification that were differentially regulated Space Flight/Ground Control in BRIC-16-Cyt, −5 ≤ FC ≥ 5 in at least one normalization method (P < 0.01), using various normalization methods. FC = fold-change.
| Gene ID | Gene symbol | FC: RMA | FC: MAS5 | FC: PLIER |
|---|---|---|---|---|
| AT2G39900 | WLIM2A | 4.39 | 6.01 | |
| AT3G05470 | FH2 family | 10.28 | 10.32 | |
| AT2G43800 | FH2 family | 8.74 | ||
| AT3G07540 | FH2 family | 18.66 | ||
| AT3G32400 | FH2 family | 5.45 | ||
| AT3G22790 | NET1A | 14.38 | ||
| AT3G43610 | SPC97 | 6.59 |
Cell wall structure is dependent upon the underlying cytoskeleton (reviewed by Bashline et al., 2014), and the organization of microtubules is aligned with cellulose before expansion (Sugimoto et al., 2000). BRIC-16-Cyt noted 7 differentially regulated genes relating to cytoskeleton within the shoots of seedlings grown in spaceflight (Table 6). These include four within the formin homology 2 (FH2) gene family, which are involved in cell elongation (Cao et al., 2015).
Hemicelluloses such as xylans and xyloglucans are involved in the structural integrity of cell walls (Xiao et al., 2016). Cell wall components have previously been shown to be dependent on environmental growth conditions and are specific to each plant organ, with roots and leaves differing in composition in response to moisture availability (Pattathil et al., 2016). The glycomics results of our BRIC-16-Cyt study confirmed these previous studies, as they indicated that many xylan and pectic backbone epitopes were marginally less abundant in spaceflight-grown plants vs. ground control (Fig. 1). Previous studies have shown the existence of xylan–pectin linkages in plant cell walls (Tan et al., 2013). These results suggest that the biosynthesis of such pectin–xylan networks may be altered in microgravity; however, further studies are advised. Xyloglucan-cleaving molecules were upregulated (F/G, −5 ≥ FC ≥ 5, P < 0.01) in BRIC-16-Cyt, including XTH32 (AT2G36870), XTH9 (AT4G03210), GH9C2 (AT1G64390), and GH9C2 (AT1G64390) (Table 2). These findings are in line with the results of several investigators (Hoson et al., 2002, 2003; Soga et al., 2002), who found that a breakdown of xyloglucan led to cell wall structural differences in spaceflight-grown hypocotyls. Taken together, the transcriptomic and glycomic results from this study also support the hypothesis that cell wall structural components differ between spaceflight-grown plants and ground controls.
Convection issues and hypoxia during spaceflight
Previous spaceflight studies have investigated the alteration of convection in μg (Porterfield et al., 1999) and determined that insufficient levels of oxygen, or hypoxia, were present in spaceflight-grown plants (Stout et al., 2001; Liao et al., 2004). Altered air circulation was also a factor in this study, as evidenced by differences in temperature among BRICs (Millar et al., 2011; Paul et al., 2012b). This hypoxic condition impacts the sugar and starch content of the plant, with differences among the stems, leaves, and roots (Porterfield et al., 1997). BRIC-16-Cyt and BRIC-16-Reg showed differentially regulated genes with GO data that indicated a hypoxic response. These are HSFA2 and PLA2A. HSFA2 (AT2G26150) was found to be differentially regulated in BRIC-16-Cyt, and PLA2A (AT2G26560) was differentially regulated in our analysis of BRIC-16-Reg. Both of these genes were downregulated (F/G, −5 ≥ FC ≥ 5, P < 0.01) in these studies, which indicates that in this hardware, hypoxia was less of an issue for spaceflight-grown plants than for their ground control equivalents. In flooded or otherwise hypoxic environments, plants develop adventitious roots, which immediately transport oxygen from the substrate to shoots (Ayi et al., 2016). Interestingly, in BRIC-16-Cyt, we observed an abundance of adventitious roots growing from the shoots of spaceflight samples (Millar et al., 2011). Thus, this abundance of adventitious roots could account for the reduction in shoot hypoxia.
Water and gas distribution within μg is unreliable (Hoehn et al., 2000; Stout et al., 2001); yet when water is accessible to the roots, it can travel to the rest of the plant body via capillary motion (Saint-Jalmes et al., 2007). Rounder cells observed in the shoot endodermis of spaceflight-grown seedlings (Johnson et al., 2015) may be accounted for by a higher turgor potential in the shoots along with the observed cell wall structural differences. An earlier spaceflight study noted increased turgor through vacuolation of soybean root cells (Klymchuk et al., 2003). In contrast to roots, shoot statocytes contain a large vacuole when grown in 1 g, and the vacuole pushes against the cell wall. Because roots have smaller vacuoles than shoots, alterations in gas convection and fluid dynamics that are unique to μg can impact them differently. The difference in the mechanical load of water in μg has been linked to structural and compositional differences within the cell wall (Chebli et al., 2013). This trend is further supported by the glycomic results of the BRIC-16-Cyt shoots, which indicate a decrease in the abundance of xylan, rhamnogalacturonan, and homogalacturonan epitopes in spaceflight vs. ground control (Fig. 1) and differences observed in cell shape (Johnson et al., 2015).
Water status in spaceflight
A striking abundance of genes related to water stress were highly differentially upregulated in ground control vs. spaceflight for BRIC-16-Cyt (Table 5; Appendices S2 and S4). An additional comparison of ground controls of all three spaceflight experiments indicated the relative absence of stress-response genes in the ground controls. Taken together, these results suggest that there was less water stress in spaceflight than there was on Earth. Interestingly, a water-stress-responsive transcript, XERO1, was downregulated (F/G) in both the BRIC-16-Cyt and BRIC-16-DNA studies. XERO1 (RAB18/LEA34) is a dehydrin, a late embryogenesis abundant (LEA) protein, that has been implicated in the freezing tolerance of seeds during dormancy (Hundertmark et al., 2011; Mäntylä et al., 1995) and was implicated in tissue-specific response to water deficit (Nylander et al., 2001). Like many stress-response genes, XERO1 was shown to be responsive to abscisic acid (Lång and Palva, 1992; Ghelis et al., 2000a) and dependent on calcium influx (Ghelis et al., 2000b) for activity. While many genes associated with stress response were differentially regulated in the three studies that we compared, BRIC-16-Cyt had much higher absolute (log2)-fold change values than BRIC-16-Reg and BRIC-16-DNA.
The BRIC-16-Cyt study included transcriptomic analyses on shoots alone. The BRIC-16-Reg and BRIC-16-DNA studies used complete seedlings for their transcriptome analysis. BRIC-16-DNA also used callus tissue samples, but the transcriptomes were not compared in the present analysis. In a spaceflight experiment using the ABRS hardware, Paul et al. (2013) explained the importance of separating plant organs rather than using entire seedlings for transcriptomics studies. They showed that Arabidopsis underwent organ-specific changes in response to spaceflight, with differential gene expression in leaves, hypocotyls, and roots under each condition. When organs were pooled together they provided an incomplete picture, even having a gene be oppositely regulated in different organs grown under the same conditions (Paul et al., 2013). This observation explains the comparatively high FC values with downregulation of stress-response genes in the BRIC-16-Cyt results. These results also suggest that roots may undergo greater hypoxic stress than shoots during spaceflight. Since there were abundant adventitious roots observed on the hypocotyls of spaceflight seedlings compared with ground controls, some of the differences in expression profiles may be accounted for by including root tissues (Millar et al., 2011).
CONCLUSIONS
During our analyses of the literature on spaceflight experiments, we noted that many studies conducted on different types of organisms reported on stress responses (Barrila et al., 2016; Li et al., 2017), cell expansion (Jha et al., 2016; Soga et al., 2002), cellular defense (Matía et al., 2007; Martzivanou et al., 2006), and cell polarity (Lorenzi and Perbal, 1990; Testa et al., 2014). The three BRIC-16 experiments (BRIC-16-Cyt, BRIC-16-Reg, and BRIC-16-DNA) also found many genes in these categories to be differentially regulated in Arabidopsis, with contrasting results in entire seedlings compared with shoots.
By addressing the data of three similar experiments in one spaceflight project, we acquired detailed information regarding the Arabidopsis transcriptome in microgravity. However, we noted similarities and differences in the microarray data among the three BRIC-16 studies. Minor differences in methodologies could account for the dissimilarities that we observed among the three data sets, with data analytics accounting for the majority of these differences. In addition, it is important to again note that we used shoot tissue only in our BRIC-16-Cyt studies, while the other two groups used entire seedlings. Overall, the results from this comparison support previous findings that stress response pathways and cell wall structure are altered in spaceflight.
The current study was aided by the close collaboration of three investigative groups, who participated in an experiment using identical hardware in the BRIC-16 project during the STS-131 mission of the Space Shuttle. Sharing raw data among investigative teams studying Arabidopsis has become common due to large open data sets accessible through TAIR and similar databases. In an effort to maximize collaborations, NASA has built an open bioinformatics-driven platform entitled GeneLab (Alwood et al., 2017), which brings together NASA space life science research-omics data from the diverse research programs of ISS and NASA. GeneLab (https://genelab.nasa.gov) will facilitate additional bioinformatics analyses of diverse spaceflight studies to help improve our understanding of how plants and other biological systems respond to the challenges of spaceflight.
Supplementary Material
Acknowledgments
The authors thank the anonymous reviewers and the editors of the American Journal of Botany for their additional insights and suggestions to improve this manuscript. For cooperation with this project, we thank Elison Blancaflor and Jin Nakashima from BRIC-16-Reg and Anna-Lisa Paul and Rob Ferl from BRIC-16-DNA. We also thank Richard E. Edelmann (Miami University) as well as the staff at NASA’s Kennedy Space Center, including Howard Levine, David Reed, Susan Manning-Roach, David Cox, Christopher Comstock, Kimberly Slater, and Stacy Engel. Financial support for BRIC-16-Cyt (J. Z. Kiss, Principal Investigator) was provided by NASA grant (NNX10AF44G). The authors thank Michael G. Hahn at CCRC, UGA for the glycome profiling platform. The generation of the CCRC series of plant cell wall glycan-directed monoclonal antibodies used in this work was supported by the NSF Plant Genome Program (DBI-0421683 and IOS-0923922).
LITERATURE CITED
- Alwood JS, Ronca AE, Mains RC, Shelhamer MJ, Smith JD, Goodwin TJ. From the bench to exploration medicine: NASA life sciences translational research for human exploration and habitation missions. Nature Partner Journal Microgravity. 2017;3:5. doi: 10.1038/s41526-016-0002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayi Q, Zeng B, Liu J, Li S, van Bodegom PM, Cornelissen JHC. Oxygen absorption by adventitious roots promotes the survival of completely submerged terrestrial plants. Annals of Botany. 2016;118:675–683. doi: 10.1093/aob/mcw051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrila J, Ott CM, LeBlanc C, Mehta SK, Crabbé A, Stafford P, Pierson DL, Nickerson CA. Spaceflight modulates gene expression in the whole blood of astronauts. Nature Partner Journal Microgravity. 2016;2:16039. doi: 10.1038/npjmgrav.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashline L, Lei L, Li S, Gu Y. Cell wall, cytoskeleton, and cell expansion in higher plants. Molecular Plant. 2014;7:586–600. doi: 10.1093/mp/ssu018. [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Meuller LA, et al. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiology. 2004;135:745–755. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Henty-Ridilla JL, Blanchoin L, Staiger CJ. Profilin-dependent nucleation and assembly of actin filaments controls cell elongation in. Arabidopsis Plant Physiology. 2015;170:220–233. doi: 10.1104/pp.15.01321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebli Y, Pujol L, Shojaeifard A, Brouwer I, van Loon JJWA, Geitmann A. Cell wall assembly and intracellular trafficking in plant cells are directly affected by changes in the magnitude of gravitational acceleration. PLoS One. 2013;8:e58246. doi: 10.1371/journal.pone.0058246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll MJ, Pyle TP, Millar KDL, Sun Y, Yao J, Edelmann RE, Kiss JZ. Transcriptome analyses of Arabidopsis thaliana seedlings grown in space: Implications for gravity-responsive genes. Planta. 2013;238:519–533. doi: 10.1007/s00425-013-1909-x. [DOI] [PubMed] [Google Scholar]
- Fengler S, Spirer I, Neef M, Ecke M, Nieselt K, Hampp R. A whole-genome microarray study of Arabidopsis thaliana semisolid callus cultures exposed to microgravity and in microgravity related spaceflight conditions for 5 days on board of Shenzhou 8. BioMed Research International. 2015;2015 doi: 10.1155/2015/547495. article 547495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelis T, Dellis O, Jeanette E, Bardat F, Cornel D, Miginiac E, Rona JP, Sotta B. Abscisic acid specific expression of RAB18 involves activation of anion channels in Arabidopsis thaliana suspension cells. FEBS Letters. 2000a;474:43–47. doi: 10.1016/s0014-5793(00)01574-x. [DOI] [PubMed] [Google Scholar]
- Ghelis T, Dellis O, Jeanette E, Bardat F, Miginiac E, Sotta B. Abscisic acid plasmalemma perception triggers a calcium influx essential for RAB18 gene expression in Arabidopsis thaliana suspension cells. FEBS Letters. 2000b;483:67–70. doi: 10.1016/s0014-5793(00)02088-3. [DOI] [PubMed] [Google Scholar]
- Hijazi M, Roujol D, Nguyen-Kim H, Castillo LDC, Saland E, Jamet E, Albenne C. Arabinogalactan protein 31 (AGP31), a putative network-forming protein in Arabidopsis thaliana cell walls? Annals of Botany. 2014;114:1087–1097. doi: 10.1093/aob/mcu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn A, Scovazzo P, Stodieck LS, Clawson J, Kalinowski W, Rakow A, Simmons D, et al. SAE Technical Paper 2000-01-2510. 2000. Microgravity root zone hydration systems. [Google Scholar]
- Hoson T, Soga K, Mori R, Saiki M, Nakamura Y, Wakabayashi K, Kamisaka S. Stimulation of elongation growth and cell wall loosening in rice coleoptiles under microgravity conditions in space. Plant & Cell Physiology. 2002;43:1067–1071. doi: 10.1093/pcp/pcf126. [DOI] [PubMed] [Google Scholar]
- Hoson T, Soga K, Wakabayashi K, Kamisaka S, Tanimoto E. Growth and cell wall changes in rice roots during spaceflight. Plant and Soil. 2003;255:19–26. doi: 10.1023/a:1026105431505. [DOI] [PubMed] [Google Scholar]
- Hundertmark M, Buitinik J, Leprince O, Hincha DK. The reduction of seed-specific dehydrins reduces seed longevity in Arabidopsis thaliana. Seed Science Research. 2011;21:165–173. [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scheref U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson C, Subramanian A, Edelmann RE, Kiss JZ. Morphometric analyses of petioles of seedlings grown in a spaceflight experiment. Journal of Plant Research. 2015;128:1007–1016. doi: 10.1007/s10265-015-0749-0. [DOI] [PubMed] [Google Scholar]
- Jha R, Wu Q, Singh M, Preininger MK, Han P, Ding G, Cho HC, et al. Simulated microgravity and 3D culture enhance induction, viability, proliferation and differentiation of cardiac progenitors from human pluripotent stem cells. Nature Scientific Reports. 2016;6:30956. doi: 10.1038/srep30956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern V, Sack F, White N, Anderson K, Wells W, Martin C. Spaceflight hardware allowing unilateral irradiation and chemical fixation in petri dishes. Advances in Space Research. 1999;24:775–778. doi: 10.1016/s0273-1177(99)00412-3. [DOI] [PubMed] [Google Scholar]
- Klymchuk DO, Kordyum EL, Vorobyova TV, Chapman DK, Brown CS. Changes in vacuolation in the root apex cells of soybean seedlings in microgravity. Advances in Space Research. 2003;31:2283–2288. doi: 10.1016/s0273-1177(03)00256-4. [DOI] [PubMed] [Google Scholar]
- Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, et al. ArrayExpress update—Simplifying data submissions. Nucleic Acids Research. 2015;43:D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz A. Genetic and physiological damage induced by cosmic radiation on dry plant seeds during space flight. Advances in Space Research. 1986;6:135–138. doi: 10.1016/0273-1177(86)90076-1. [DOI] [PubMed] [Google Scholar]
- Kwon T, Sparks JA, Nakashima J, Allen SN, Tang Y, Blancaflor EB. Transcriptional response of Arabidopsis seedlings during spaceflight reveals peroxidase and cell wall remodeling genes associated with root hair development. American Journal of Botany. 2015;102:21–35. doi: 10.3732/ajb.1400458. [DOI] [PubMed] [Google Scholar]
- Lång V, Palva ET. The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Molecular Biology. 1992;20:951–962. doi: 10.1007/BF00027165. [DOI] [PubMed] [Google Scholar]
- Li HS, Lu JY, Zhao H, Sun Q, Yu FT, Pan Y, Chen Y, et al. The impact of space environment on gene expression in Arabidopsis thaliana seedlings. Science China Technological Sciences. 2017;60:902–910. [Google Scholar]
- Liao J, Liu G, Monje O, Stutte GW, Porterfield D. Induction of hypoxic root metabolism results from physical limitations in O2 bioavailability in microgravity. Advances in Space Research. 2004;34:1579–1584. doi: 10.1016/j.asr.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorenzi G, Perbal G. Root growth and statocyte polarity in lentil seedling roots grown in microgravity or on a slowly rotating clinostat. Physiologia Plantarum. 1990;78:532–537. [Google Scholar]
- Mäntylä E, Lång V, Palva ET. Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LTI78 and RAB18 proteins in. Arabidopsis thaliana Plant Physiology. 1995;107:141–148. doi: 10.1104/pp.107.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martzivanou M, Babbick M, Cogoli-Greuter M, Hampp R. Microgravity-related changes in gene expression after short-term exposure of Arabidopsis thaliana cell cultures. Protoplasma. 2006;229:155–162. doi: 10.1007/s00709-006-0203-1. [DOI] [PubMed] [Google Scholar]
- Matía I, González-Camacho F, Marco R, Kiss JZ, Gasset G, van Loon J, Medina FJ. The “root” experiment of the “Cervantes” Spanish Soyuz mission: Cell proliferation and nucleolar activity alterations in Arabidopsis roots germinated in real or simulated microgravity. Microgravity Science and Technology. 2007;19:128–132. [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar KDL, Johnson CM, Edelmann RE, Kiss JZ. An endogenous growth pattern of roots is revealed in seedlings grown in microgravity. Astrobiology. 2011;11:787–797. doi: 10.1089/ast.2011.0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CA. Bioregenerative life support systems. American Journal of Clinical Nutrition. 1994;60:820S–824S. doi: 10.1093/ajcn/60.5.820S. [DOI] [PubMed] [Google Scholar]
- Musgrave ME. Seeds in space. Seed Science Research. 2002;12:1–17. [Google Scholar]
- Nakashima J, Liao F, Sparks JA, Tang Y, Blancaflor EB. The actin cytoskeleton is a suppressor of the endogenous skewing behavior of Arabidopsis primary roots in microgravity. Plant Biology. 2014;16:142–150. doi: 10.1111/plb.12062. [DOI] [PubMed] [Google Scholar]
- NASA. Biological Research in Canisters-16: Investigations of the plant cytoskeleton in microgravity with gene profiling and cytochemistry (BRIC-16-Cytoskeleton) [accessed 20 February 2017];Website. 2016a https://www.nasa.gov/mission_pages/station/research/experiments/785.html.
- NASA. Biological Research in Canisters-16: Actin regulation of Arabidopsis root growth and orientation during space flight (BRIC-16-Regulation) [accessed 20 February 2017];Website. 2016b https://www.nasa.gov/mission_pages/station/research/experiments/813.html.
- NASA. Biological Research in Canisters-16: The impact of spaceflight on Arabidopsis: deep sequencing and DNA arrays as collaborative readouts of the transcriptome of Arabidopsis seedlings and undifferentiated cells in space (BRIC-16-DNA) [accessed 20 February 2017];Website. 2016c https://www.nasa.gov/mission_pages/station/research/experiments/815.html.
- Nylander M, Svensson J, Palva ET, Welin BV. Stress-induced accumulation and tissue specific localization of dehydrins in. Arabidopsis thaliana Plant Molecular Biology. 2001;45:263–279. doi: 10.1023/a:1006469128280. [DOI] [PubMed] [Google Scholar]
- Ode A, Duda GN, Geissler S, Pauly S, Ode JE, Perka C, Strube P. Interaction of age and mechanical stability on bone defect healing: An early transcriptional analysis of fracture hematoma in rat. PLoS One. 2014;9:e106462. doi: 10.1371/journal.pone.0106462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradiso R, De Micco V, Buonomo R, Aronne G, Barbieri G, De Pascale S. Soilless cultivation of soybean for bioregenerative life-support systems: A literature review and the experience of the MELiSSA project— Food characterization Phase 1. Plant Biology. 2013;16:69–78. doi: 10.1111/plb.12056. [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Miller JS, Hahn MG. Immunological approaches to plant cell wall and biomass characterization: glycome profiling. Methods in Molecular Biology (Clifton, NJ) 2012;908:61–72. doi: 10.1007/978-1-61779-956-3_6. [DOI] [PubMed] [Google Scholar]
- Pattathil S, Hahn MG, Dale BE, Chundawat SPS. Insights into the plant cell wall structure, architecture, and integrity using glycome profiling of native and AFEX™-pre-treated biomass. Journal of Experimental Botany. 2015;66:4279–4294. doi: 10.1093/jxb/erv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Ingwers MW, Victoriano OL, Kandemkavil S, McGuire MA, Teskey RO, Aubrey DP. Cell wall ultrastructure of stem wood, roots, and needles of a conifer varies in response to moisture availability. Frontiers in Plant Science. 2016;7:882. doi: 10.3389/fpls.2016.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Amalfitano CE, Ferl RJ. Plant growth strategies are remodeled by spaceflight. BMC Plant Biology. 2012a;12:232. doi: 10.1186/1471-2229-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Daugherty CJ, Bihn EA, Chapman DK, Norwood KLL, Ferl RJ. Transgene expression patterns indicate that spaceflight affects stress signal perception and transduction in. Arabidopsis Plant Physiology. 2001;126:613–621. doi: 10.1104/pp.126.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Zupanska AK, Ostrow DT, Zhang Y, Sun Y, Li J, Shanker S, Farmerie WG, et al. Spaceflight transcriptomes: Unique responses to a novel environment. Astrobiology. 2012b;12:40–56. doi: 10.1089/ast.2011.0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Zupanska AK, Schultz ER, Ferl RJ. Organ-specific remodeling of the Arabidopsis transcriptome in response to spaceflight. BMC Plant Biology. 2013;13:112. doi: 10.1186/1471-2229-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal G. From ROOTS to GRAVI-1: Twenty five years for understanding how plants sense gravity. Microgravity Science and Technology. 2008;21:3–10. [Google Scholar]
- Porterfield D, Crispi M, Musgrave M. Changes in soluble sugar, starch, and alcohol dehydrogenase in Arabidopsis thaliana exposed to N2 diluted atmospheres. Plant & Cell Physiology. 1997;38:1354–1358. doi: 10.1093/oxfordjournals.pcp.a029129. [DOI] [PubMed] [Google Scholar]
- Porterfield D, Kuang A, Smith P, Crispi M, Musgrave M. Oxygen-depleted zones inside reproductive structures of Brassicaceae: Implications for oxygen control of seed development. Canadian Journal of Botany. 1999;77:1439–1446. [PubMed] [Google Scholar]
- Qu Y, Fei H, Chen Y. Different effects of the probe summarization algorithms PLIER and RMA on high-level analysis of Affymetrix exon arrays. BMC Bioinformatics. 2010;11:211. doi: 10.1186/1471-2105-11-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nature Genetics. 2002;32(Supplement):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Reis RS, Hart-Smith G, Eamens AL, Wilkins MR, Waterhouse PM. Gene regulation by translational inhibition is determined by Dicer partnering proteins. Nature Plants. 2015;1:14027. doi: 10.1038/nplants.2014.27. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Research. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jalmes A, Marze S, Ritacco H, Langevin D, Bail S, Dubail J, Guingot L, et al. Diffusive liquid propagation in porous and elastic materials: The case of foams under microgravity conditions. Physical Review Letters. 2007;98:058303. doi: 10.1103/PhysRevLett.98.058303. [DOI] [PubMed] [Google Scholar]
- Schultz ER, Zupanska AK, Manning-Roach S, Camacho J, Levine H, Paul A-L, Ferl RJ. Testing the bio-compatibility of aluminum PDFU BRIC hardware. Gravitational and Space Biology. 2012;26:48–63. [Google Scholar]
- Seo J, Hoffman EP. Probe set algorithms: Is there a rational best bet? BMC Bioinformatics. 2006;7:395. doi: 10.1186/1471-2105-7-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga K, Wakabayashi K, Kamisaka S, Hoson T. Stimulation of elongation growth and xyloglucan breakdown in Arabidopsis hypocotyls under microgravity conditions in space. Planta. 2002;215:1040–1046. doi: 10.1007/s00425-002-0838-x. [DOI] [PubMed] [Google Scholar]
- Stimpson AJ, Pereira RS, Kiss JZ, Correll MJ. Extraction and labeling methods for microarrays using small amounts of plant tissue. Physiologia Plantarum. 2009;135:229–236. doi: 10.1111/j.1399-3054.2008.01191.x. [DOI] [PubMed] [Google Scholar]
- Stout S, Porterfield D, Briarty L, Kuang A, Musgrave M. Evidence of root zone hypoxia in Brassica rapa L. grown in microgravity. International Journal of Plant Sciences. 2001;162:249–255. doi: 10.1086/319585. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO. New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiology. 2000;124:1493–1506. doi: 10.1104/pp.124.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAIR [The Arabidopsis Information Resource] GO annotation search, functional categorization and download. [accessed 6 February 2017];Website. 2017 https://www.arabidopsis.org/tools/bulk/go/index.jsp.
- Tan L, Eberhard S, Pattathil S, Wardre C, Glushka J, Yuan C, Hao Z, et al. An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell. 2013;25:270–287. doi: 10.1105/tpc.112.107334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa F, Palombo A, Dinicola S, D’Anselmi F, Proietti S, Pasqualato A, Masiello MG, et al. Fractal analysis of shape changes in murine osteoblasts cultured under simulated microgravity. Rendiconti Lincei. 2014;25(Supplement 1):39–47. [Google Scholar]
- Therneau TM, Ballman KV. What does PLIER really do? Cancer Informatics. 2008;6:423–431. [PMC free article] [PubMed] [Google Scholar]
- Thornton B, Basu C. Real-time PCR (qPCR) primer design using free online software. Biochemistry and Molecular Biology Education. 2011;39:145–154. doi: 10.1002/bmb.20461. [DOI] [PubMed] [Google Scholar]
- Untergasser A. Primer 3 Plus. [accessed 20 February 2017];Computer program. 2007 available at http://primer3plus.com/
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Research. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbrink JP, Kiss JZ. Space, the final frontier: A critical review of recent experiments performed in microgravity. Plant Science. 2016;243:115–119. doi: 10.1016/j.plantsci.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Zhang T, Zheng Y, Cosgrove DJ, Anderson CT. Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiology. 2016;170:234–249. doi: 10.1104/pp.15.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupanska AK, Denison FC, Ferl RJ, Paul AL. Spaceflight engages heat shock protein and other molecular chaperone genes in tissue culture cells of. Arabidopsis thaliana American Journal of Botany. 2013;100:235–248. doi: 10.3732/ajb.1200343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.