Abstract
When BDNF binds to its receptors, TrkB and p75NTR, the BDNF-receptor complex is endocytosed and trafficked to the cell body for downstream signal transduction, which plays a critical role in neuronal functions. Huntingtin-associated protein 1 (HAP1) is involved in trafficking of vesicles intracellularly and also interacts with several membrane proteins including TrkB. Although it has been known that HAP1 has functions in vesicular trafficking and receptor stabilisation, it is not yet established whether HAP1 has a role in BDNF and its receptor endocytosis. In the present study, we found that HAP1 is in an interacting complex with p75NTR, TrkB and BDNF, especially newly endocytosed BDNF. BDNF and TrkB internalisation is abolished in HAP1 knock-out (KO) cortical neurons. TrkB downstream signalling pathways such as ERK, Akt and PLCγ-1 are also impaired in HAP1 KO cortical neurons upon BDNF stimulation. Proliferation of cerebellar granule cells is also impaired in cell culture and cerebellum of HAP1 KO mice. Our findings suggest that HAP1 may play a key role in BDNF and its receptor endocytosis and may promote neuronal survival and proliferation.
Keywords: HAP1, BDNF, TrkB, P75NTR, Endocytosis, Proliferation
Introduction
Brain-derived neurotrophic factor (BDNF) is a member of neurotrophin growth factors which have pivotal roles in neuronal development and maintaining the nervous system functions in adult [1–3]. BDNF is initially synthesised as a precursor protein which is called pre-proBDNF, and further processing of the prodomain intracellularly or extracellularly produces mature BDNF (which will be called BDNF, 13 kDa) [4–7]. Recent evidence has shown that proBDNF signalling causes neuronal apoptosis and long-term depression while BDNF signalling has a critical function in neuronal survival and long-term potentiation [6, 8–10], suggesting that proBDNF and BDNF have opposite functions.
BDNF binds to tyrosine kinase B (TrkB) receptor with high affinity and to p75NTR, a pan receptor, with low affinity [11]. When neurotrophins including BDNF bind to receptors at axonal terminals, the ligand-receptor complex is endocytosed and transported retrogradely along the axon to the cell body, which results in the initiation of signalling cascades locally [12]. It has been shown that the endocytosis of BDNF/TrkB complex is clathrin dependent and is essential for outgrowth of dendrites and neuronal survival [13].
Huntingtin-associated protein 1 (HAP1) is the first identified protein that interacts with huntingtin (Htt) protein which causes Huntington disease (HD) when mutated [14]. HAP1 KO mice often do not survive long after birth and die before the third day after birth owing to hypothalamic neuronal degeneration, resulting in feeding behaviour inhibition [15]. There have been two HAP1 isoforms reported, HAP1A and HAP1B, in rodents [16, 17], and there is only one human HAP1 isoform which shows 96% identity to the rat HAP1A [17].
HAP1 is involved in retrograde and anterograde vesicular trafficking intracellularly by interacting with P150Glued [18, 19] and kinesin light chain [20], respectively. HAP1 also interacts with other proteins which are involved in vesicular transport including Duo [21], hepatocyte growth factor-regulated tyrosine kinase substrate [22], 14-3-3 protein [23] and Abelson helper integration site 1 [24]. Our previous studies have shown that HAP1 directly interacts with the prodomain of proBDNF and sortilin to prevent its lysosomal degradation and regulates the anterograde transport and release of proBDNF [25, 26].
HAP1 also interacts with several membrane receptor proteins including epidermal growth factor receptor, γ-amino butyric acid type A receptor, androgen receptor and tyrosine receptor kinase A (TrkA) and TrkB and plays a role in facilitating recycling of these receptors to the plasma membrane or stabilising them to prevent lysosomal degradation [22, 27–32]. Sheng et al. [24] demonstrated that TrkB internalisation is reduced in cultured brainstem neurons when HAP1 expression is suppressed by HAP1 siRNA in the presence of the BDNF and BDNF internalisation is reduced in the cultured brain stem cells of HAP1 KO mice [24]. However, in the study, TrkB expression level in the brainstem of HAP1 KO mice was also significantly lower than in WT mice; therefore, the reduced BDNF internalisation could be owing to reduced TrkB level. Recently, it has been also shown that HAP1 is associated with TrkB to protect TrkB from lysosomal degradation [32]. Although HAP1 is involved in signalling endosome trafficking and a few reports have demonstrated HAP1’s association with internalisation of BDNF and TrkB, it is still uncertain whether HAP1 regulates BDNF and its receptors’ endocytosis directly.
In the present study, we have investigated whether HAP1 regulates endocytosis and signalling of BDNF and its receptors, TrkB and p75NTR, using cell lines (PC12 and HEK293T) and cortical neurons from HAP1 KO mice [15]. We have found that endocytosed BDNF and HAP1 highly co-localise and BDNF endocytosis is blocked by antibodies to BDNF and p75NTR. Also, TrkB and p75NTR interact with HAP1, and these interactions are increased by BDNF stimulation. In cultured HAP1 KO cortical neurons, TrkB internalisation and downstream signalling are abolished. Moreover, the loss of HAP1 showed proliferation defect of cerebellar granule cells in response to BDNF both in vitro and in vivo.
Results
Endocytosed BDNF Highly Co-localises with HAP1
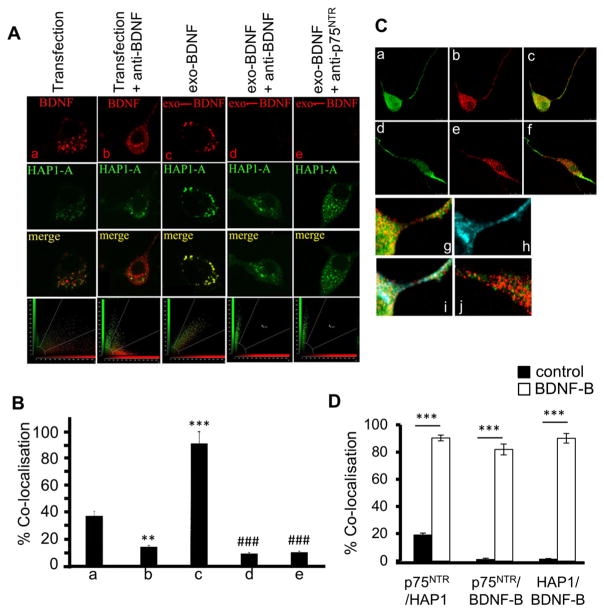
As HAP1 co-localises with proBDNF and regulates its anterograde transport and release [25], we examined whether HAP1 co-localises with BDNF and p75NTR in neuron-like PC12 cells; we could not investigate HAP1 co-localisation with TrkB in these cells, because PC12 cells do not express TrkB [33–35]. We co-transfected these cells with the two plasmids, HAP1A-GFP and BDNF-RFP, and found that HAP1A-GFP and BDNF-RFP co-localise (34%) in PC12 cells after co-transfection (Fig. 1a(a), b). This result indicates that HAP1 may have a relationship with BDNF. However, as transfected BDNF-RFP can be released and then endocytosed, it is not clear whether HAP1 co-localises with newly endocytosed BDNF or intracellular BDNF. To examine this issue, we incubated transfected cells with antibody to BDNF to neutralise secreted BDNF-RFP and found that co-localisation significantly decreased (to 13%, p < 0.01) (Fig. 1a(b), b). This result indicates that HAP1 co-localises with a large proportion of endocytosed BDNF rather than intracellular BDNF within the transfected cells. To confirm whether endocytosed BDNF highly co-localises with HAP1, PC12 cells were transfected with HAP1A-GFP only, and the cell culture medium containing RFP-BDNF was added exogenously (BDNF-exo) in the absence or presence of antibodies to BDNF or p75NTR extracellular domain [11]. While the newly endocytosed BDNF-RFP showed high co-localisation (90%, p < 0.001) with HAP1A-GFP (Fig. 1a(c), b), the co-localisation of BDNF-RFP with HAP1A-GFP was dramatically decreased (to 10%, p < 0.001) in the presence of antibody to BDNF (Fig. 1a(d), b) or p75NTR (Fig. 1a(e), b). These results demonstrate that HAP1 is clearly associated with endocytosed BDNF but not with the intracellular BDNF which is synthesised within the cells, suggesting that HAP1 may be associated with BDNF endocytosis.
Fig. 1.
HAP1 and p75NTR are highly co-localised with endocytosed BDNF. a PC12 cells were transfected with HAP1A-GFP (green)/BDNF-RFP (red) or HAP1A-GFP only. Cells were co-transfected with HAP1A-GFP/BDNF-RFP (a) or cultured in the presence of sheep anti-BDNF to neutralise the secreted BDNF after the co-transfection (b). c, d and e show that PC12 cells were transfected only with HAP1A-GFP plasmid. Two days later, cells were incubated with either cell culture medium containing RFP-BDNF (exo-BDNF) only (c), or exo-BDNF with sheep anti-BDNF antibody (d), or exo-BDNF with p75NTR antibody directed against the extracellular domain of p75NTR (e). Co-localisation is indicated in merged panels (yellow). b Graph presenting co-localisation results from a. Co-localisation of HAP1A-GFP/BDNF-RFP after co-transfection with anti-BDNF treatment was reduced (**p < 0.01 vs (a)). Addition of exo-BDNF increased the co-localisation with HAP1A-GFP (***p < 0.001 vs (a)). Addition of antibody to BDNF or p75NTR with exo-BDNF decreased co-localisation of HAP1A-GFP/exo-BDNF (###p < 0.001 vs (c)). The data are presented as mean ± SEM (n = 20 per group, one-way ANOVA). c PC12 cells were cultured with BDNF-biotin (BDNF-B) (a, b, c, g) or without BDNF-biotin (BDNF-B) (d, e, f, j) for 30 min and immunostained for HAP1 (a, d; green) and p75NTR (b, e; red); c and f merged images (yellow); g and j enlarged images of parts of c and f, respectively; h stained for -BDNF-B (blue); i merged image of h and g. d Graph presenting the co-localisation results in c. The data are presented as mean ± SEM (n = 9 per group, ***p < 0.001 vs corresponding control; Student’s t test)
As HAP1 is associated with TrkA [29] and TrkB [32], we next investigated whether p75NTR co-localises with HAP1 and endocytosed BDNF. PC12 cells were treated with or without BDNF-biotin (BDNF-B), and HAP1, p75NTR and BDNF-B were immunostained for co-localisation analysis. Whereas only a small fraction (less than 20%) of HAP1 (green) co-localises with p75NTR (red) in PC12 cells (Fig. 1c(d–f, j), d) in the absence of BDNF-B, BDNF-B induced over 90% of co-localisation (Fig. 1c(a–c, g), d; p < 0.001 vs without BDNF-B). Furthermore, internalised BNDF-B (blue) (Fig. 1c(h)) also co-localises with the HAP1/p75NTR vesicles (Fig. 1c(i)). The co-localisation of p75NTR/BDNF-B and HAP1/BDNF-B was 81% (p < 0.001) and 91% (p < 0.001), respectively. This result shows that BDNF stimulation induces the recruitment of HAP1 molecule to the complexes of p75NTR/BDNF.
BDNF Stimulates the Interactions of HAP1/TrkB and HAP1/p75NTR
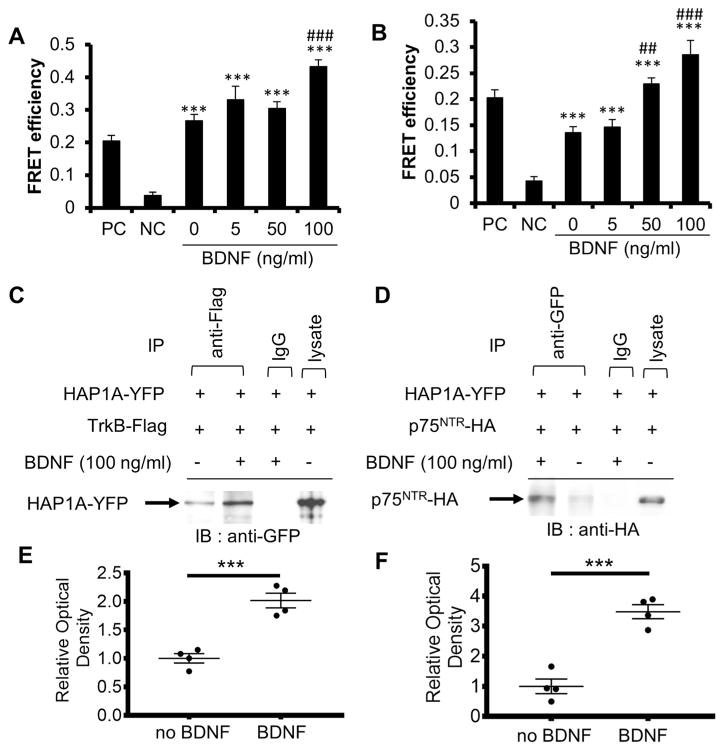
As TrkB is the receptor for BDNF [11], we performed two different experiments, FRET and Co-IP, to confirm that HAP1 interacts with TrkB and p75NTR using HEK293T cells. The result from the FRETexperiment revealed that HAP1A highly interacts with TrkB (p < 0.001) compared to negative control (pEYFP/p75NTR-CFP) and that BDNF significantly increased the HAP1A/TrkB interaction (p < 0.001) at 100 ng/ml (Fig. 2a). Compared to negative control, there was a FRET signal between HAP1A and p75NTR (p < 0.001), and BDNF also triggered significant increase in the FRET signals between HAP1A and p75NTR in a dose-dependent manner up to 100 ng/ml (50 ng/ml, p < 0.01; 100 ng/ml, p < 0.001) compared to control (0 ng/ml) (Fig. 2b). These data indicate that HAP1 interacts with both BDNF receptors TrkB and p75NTR and TrkB, and BDNF can increase such interactions. Co-IP was employed to confirm the interaction of HAP1A/TrkB and HAP1A/p75NTR. The result confirmed the interaction between BDNDF and TrkB, and addition of exogenous BDNF increased their interaction significantly by 2-fold (p < 0.001) (Fig. 2c, e). Similarly, BDNF stimulation also increased the interaction of BDNF and p75NTR significantly by 3.5-fold (p < 0.001) (Fig. 2d, f). Given that HAP1 appears to co-localise with p75NTR when TrkB is not expressed in PC12 cells, TrkB and p75NTR may be in separate complexes with HAP1 recruited in response to BDNF stimulation.
Fig. 2.
Interaction of HAP1/TrkB and HAP1/p75NTR is increased by BDNF treatment in HEK293T cells. a Graph presenting FRET acceptor bleaching analysis for HAP1A-YFP/TrkB-CFP interaction compared to NC or control (0 ng/ml BDNF). BDNF increases the HAP1A/TrkB interaction (***p < 0.001 vs NC; ###p < 0.001 vs 0 ng/ml BDNF). b Graph presenting FRET acceptor bleaching analysis for HAP1A-YFP/p75NTR-CFP interaction compared to NC or control (0 ng/ml BDNF). BDNF increases the HAP1A/p75NTR interaction (***p < 0.001 vs NC; ##p < 0.01, ###p < 0.001 vs 0 ng/ml BDNF). For a and b, data represent mean ± SEM (n = 5–11 per group, one-way ANOVA, Tukey’s post hoc test; PC positive control (p75NTR-EYFP-CFP); NC negative control (pEYFP/p75NTR-CFP). c BDNF increases the level of HAP1A-YFP pulled down by mouse anti-flag antibody (TrkB fused with Flag) as indicated. Lane 1: no BDNF; lane 2: BDNF (100 ng/ml); lane 3: mouse IgG as negative control; lane 4: cell lysate input; IP immunoprecipitation; IB immunoblot. d BDNF increases the level of p75NTR-HA pulled down by anti-GFP antibody (HAP1 fused with YFP) from lysate of HEK293T cells co-transfected with p75NTR-HA and HAP1A-YFP as indicated. Lane 1: BDNF (100 ng/ml); lane 2: no BDNF; lane 3: goat IgG as negative control; lane 4: cell lysate input; IP immunoprecipitation; IB immunoblot. e, f Each dot represents individual relative optical density to ‘no BDNF’ in c and d (mean ± SEM from 4 experiments, ***p < 0.001, Student’s t test)
HAP1 Is Required for the Endocytosis of BDNF
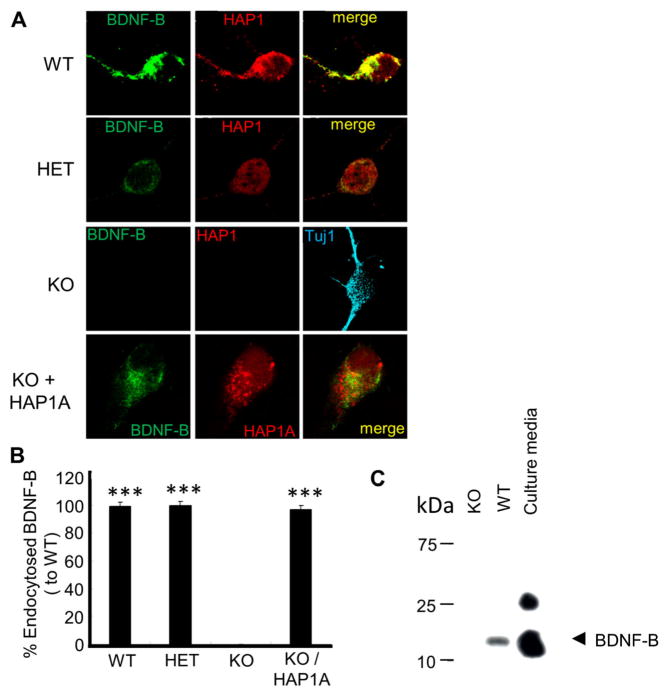
As BDNF endocytosis was reduced significantly in cultured brainstem cells of HAP1 KO [24], we therefore tested whether BDNF endocytosis could be impaired in cortical neurons of HAP1 KO mice. Cultured cortical neurons from HAP1 WT, HET or KO were treated with BDNF-B for 30 min, and the cells were then immunostained to determine endocytosed BDNF-B. We found that BDNF-B was internalised in cortical neurons from HAP1 WT and HET mice but not KO mice (p < 0.001) (Fig. 3a, b). We then transfected HAP1 KO neurons with HAP1A-CFP plasmid to examine whether the HAP1A cDNA could rescue the impaired endocytosis. Interestingly, the transfection of HAP1 KO neurons with HAP1A plasmid rescued the internalisation of BDNF-B (p < 0.001) (Fig. 3a, bottom panel, b). To support this finding, endocytosed BDNF-B was pulled down by streptavidin-agarose beads in cortical neuron lysate of HAP1 WT or KO mice after BDNF-B treatment. We found that BDNF-B was detected in only WT neuronal lysate but not in KO (Fig. 3c) neurons. Taken together, these findings propose that HAP1 has a critical role in BDNF endocytosis in cortical neurons.
Fig. 3.
Endocytosis of BDNF-B is abolished in cultured cortical neurons from HAP1 KO mice. a Cortical neurons from P1 HAP1 WT, HETor KO mice were cultured for 3 days, incubated with BDNF-B for 30 min and immunostained for BDNF-B (green), HAP1 and Tuj1 (blue). Genotypes and transfection of KO are labelled at left side of image panels. For rescue experiment, HAP1KO cortical neurons were transfected with HAP1A-CFP. Merged image is indicated (yellow). b Quantification of endocytosed BDNF-B; data are represented as the mean ± SEM (n = 50 per group, ***p < 0.001 vs KO, one-way ANOVA, Tukey’s post hoc test). c Cultured cortical neurons from HAP1 KO (lane 1) and WT mice (lane 2) were incubated with BDNF-B (100 ng/ml) for 30 min at 37 °C. Culture media containing BDNF-B (lane 3) and cell lysates were incubated with strepavidin-sepharose beads, and samples were used for SDS-PAGE and probed with streptavidin-HRP. BDNF-B can be detected in WT neurons but not HAP1 KO neurons. The high molecular weight in supernatant is the dimer of BDNF-B
HAP1 Is Required for Endocytosis of TrkB but Not p75NTR in Mouse Cortical Neurons
Previous studies showed that TrkB internalisation is reduced in neurons from the brain stem in which HAP1 expression is suppressed [24]. Our data from the current study suggest that HAP1 may be also involved in the endocytosis of BDNF receptors TrkB and p75NTR. To test the hypothesis, we determined the neuronal surface level of TrkB and p75NTR after BDNF treatment by surface biotinylation. To do this, cultured cortical neurons from WTand HAP1 KO mice were incubated with or without BDNF-containing culture medium, followed by cell surface biotinylation. We found that the surface TrkB-FL (140 kDa) level was significantly reduced (by 47%, p < 0.01 vs control) in WT neurons upon BDNF stimulation but not in HAP1 KO neurons (Fig. 4a, b). As TrkB-T1 (95 kDa) has been found to bind to BDNF and get endocytosed together with BDNF [36] and to inhibit BDNF signalling in Xenopus oocytes [37], we also examined the surface TrkB-T1 levels on BDNF stimulation. The result showed that surface TrkB-T1 levels were not changed in HAP1 WT or KO on BDNF stimulation (Fig. 4a, c). Surprisingly, no reduction of surface p75NTR level was found on BDNF stimulation of HAP1 WT or KO cortical neurons (Fig. 4a, d). Through these results, we suggest that HAP1 may play a critical role in BDNF endocytosis by regulating TrkB endocytosis but not p75NTR in cortical neurons.
Fig. 4.
Endocytosis of TrkB and p75NTR in cortical neurons upon BDNF stimulation. a Western blots showing BDNF-induced endocytosis of TrkB and p75NTR. HAP1 WT and KO cortical neurons, after 72-h culture, were stimulated with 0 (control) or 100 ng/ml BDNF for 30 min. Surface TrkB-FL, TrkB-T1 and p75NTR were detected by Western blotting using anti-TrkB and anti-p75NTR antibodies, respectively, after surface protein biotinylation and fractionation of biotinylated proteins with streptavidin-agarose. Total TrkB-FL, TrkB-T1, p75NTR and β-actin were determined using whole cell lysates by Western blot with anti-TrkB, anti-p75NTR and anti-β-actin antibody, respectively. b, c, d Each dot or square represents individual animal. Densitometric ratios of surface/total TrkB-FL levels (b), surface/total TrkB-T1 levels (c) and surface/total p75NTR levels (c) assessed by biotinylation assay. Data presented as mean ± SEM (n = 3 per group, Student’s t test). ‘WT control was normalised to 1. TrkB-FL, full-length of TrkB; TrkB-T1, truncated TrkB
TrkB Expression Is Not Reduced in Cortex of HAP1 KO Mice
It has been reported that TrkB protein expression level is reduced in cerebellum, hypothalamus and brainstem in HAP1 KO neonates [24, 32]. To investigate whether the impaired TrkB internalisation in HAP1 KO cortical neurons could be owing to the reduced TrkB expression level itself, the mRNA and protein levels of TrkB were examined in cortex and brainstem tissue of neonates (P 0) of HAP1 WT and KO, respectively. We have found that the TrkB mRNA level was reduced significantly in brainstem (p < 0.05) but not in the cortex of HAP1 KO compared to WT (Fig. 5a, c). Similarly, the protein level of TrkB was also reduced in the brainstem (p < 0.05) but not in the cortex of HAP1 KO compared to WT (Fig. 5b, d). These results confirm that impaired TrkB internalisation is caused by absence of HAP1 rather than decreased levels of TrkB, because HAP1 gene deletion does not affect TrkB mRNA and protein level in the cortex. However, HAP1 gene deletion reduced TrkB mRNA and protein levels in the brainstem, suggesting that HAP1 gene loss may affect the transcription level of TrkB in the brainstem.
Fig. 5.
Semi-quantitative RT-PCR of trkB mRNA and TrkB protein level in the cortex and brainstem tissue. The mRNA expression levels were determined in the cortex and brainstem of HAP1 WT and HAP1 KO. a trkB and gapdh RT-PCR products were subjected to agarose gel electrophoresis. Gapdh was used as internal control. b Western blot analysis of cortex and brainstem tissues from WT and HAP1 KO mice. c, d Each dot or square represents individual animal. Densitometric ratios of trkB/gapdh (c) and TrkB/β-actin (d). Mean ± SEM (n = 3 per group, Student’s t test, *p < 0.05). BS brainstem, Ctx cortex
HAP1 Is Required for BDNF-Induced Downstream Signalling
TrkB activation by BDNF triggers the downstream signalling including phosphorylation of ERK, Akt and PLCγ-1 [11]. The abolished endocytosis of BDNF and TrkB in cortical neurons from HAP1 KO mice led us to hypothesise that its downstream signal transduction pathways may also be affected. To test the hypothesis, we compared the phosphorylated ERK (pERK), Akt (pAkt) and PLCγ-1 (pPLCγ-1) under BDNF treatment between WT and HAP1 KO cortical neurons by Western blotting. We have demonstrated that the levels of pERK (p < 0.01), pAkt (p < 0.001) and pPLCγ-1 (p < 0.05) after BDNF treatment for 15 min in HAP1 WT neurons were significantly increased compared to WT control (0 ng/ml BDNF) neurons (Fig. 6a–d). However, there was no increase of phosphorylation on ERK, Akt and PLCγ-1 in HAP1 KO cortical neurons in response to BDNF treatment (Fig. 6a–d). These results suggest that HAP1 is required for the signal transduction of BDNF.
Fig. 6.
TrkB signalling is decreased in HAP1 KO cortical neurons. a Representative Western blots showing BDNF-induced TrkB downstream signal transduction proteins. HAP1 WT and KO cortical neurons were stimulated with 100 ng/ml BDNF for 30 min and then subjected to Western blot analysis with antibodies to phosphorylated ERK (pERK), Akt (pAkt) and PLCγ-1 (pPLCγ-1) and total ERK, Akt and PLCγ-1. GAPDH serves as an internal control. b, c, d Each dot or square represents individual animal. Densitometric quantification of the ratios of pERK to total ERK (b), pAkt to total Akt (c) and pPLCγ-1 to total PLCγ-1 (d). Mean ± SEM; n = 3–4 per group. *p < 0.05, **p < 0.01, *** p < 0.001; one-way ANOVA with Tukey’s post hoc test). ‘WT control was normalised to 1. All bands of Western blot were normalised with their corresponding GAPDH
Proliferation of Cerebellar Granule Cells Is Impaired in HAP1 KO Mice In Vitro and In Vivo
As BDNF plays a critical role in cellular proliferation of cerebellar granule cells, we hypothesised that neuronal proliferation in response to BDNF could be impaired in HAP1 KO mice. To test this, we observed the proliferation of cerebellar granule cells in vitro and in vivo by BrdU incorporation experiment. As BrdU is an analogue of thymidine, during the ‘S’ phase of the cell cycle, it can be incorporated into DNA instead of thymidine and can serve a proliferation marker. The in vitro experiment showed that the number of BrdU-positive cells increased 1.5 times relative to control upon BDNF treatment (at 10 and 100 ng/ml) in WT (p < 0.05 vs control) cells while there was no increased number of BrdU-positive cells in HAP1 KO cells. There was a significant difference in BDNF-treated cerebellar granule cells between WT and HAP1 KO granule cells (p < 0.01 and p < 0.001 vs KO) (Fig. 7a, b). The result of the in vivo experiment was similar to the in vitro experiment. The BrdU-positive cells (%) in BDNF-injected WT mice were increased (more than 2-fold) in comparison to PBS-injected (p < 0.01) or BDNF-injected KO (p < 0.001) granular cells (Fig. 7c, d). However, there was no difference in BrdU-positive cells (%) between PBS and BDNF-injected KO mice (Fig. 7c, d). Therefore, our results suggest that loss of Hap1 may cause defects in neuronal proliferation owing to lacking of BDNF/TrkB signalling.
Fig. 7.
BDNF induces proliferation of cerebellar granule cells in HAP1 WT but not in HAP1 KO in vivo and in vitro. a, b Isolated cells from P0 cerebellum tissue of HAP1 WT and KO mice were cultured on glass coverslips coated with poly-D-lysine (PDL) overnight and for additional 24 h with 10 μg/ml BrdU and 0 (control), 10 or 100 ng/ml BDNF containing culture medium, respectively. The cells were then fixed and stained with anti-BrdU antibody according to the procedure described in “Materials and Methods”. Scale bar, 100 μm. BrdU-positive cells (green) in the presence or absence of BDNF in WT and KO granular cells are shown in a. Quantification of BrdU-positive cells is shown in b. Data are presented as mean ± SEM; *p < 0.05 vs WT control; ##p < 0.01, ###p < 0.001 vs WT 10 ng/ml BDNF; ¥¥p < 0.01, ¥¥¥p < 0.001 vs WT 100 ng/ml BDNF; n = 3–4 coverslips per group; six areas were observed in each coverslip, and the same locations were applied to all the observed coverslips for image taking; one-way ANOVA, Tukey’s post hoc test. c, d Effect of BDNF on granule cell proliferation in WT and HAP1 KO neonatal mice in vivo. Sections were labelled with BrdU-positive cells (red colour) demonstrating proliferation of granule cells at 24 h post-BrdU injection (c, a–d). c, a–b Representative microphotographs of BrdU-positive granule cells from HAP1 KO mice treated with PBS (a) or BDNF (b). c, c, d Representative microphotographs of BrdU-positive granule cells from WT mice treated with PBS (c) or BDNF (d). Quantification of BrdU-positive cells is shown in d. Data are presented as mean ± SEM; **p < 0.05 vs WT control; ###p < 0.001, WT BDNF vs HAP1 KO BDNF; n = 6 per group; one-way ANOVA, Tukey’s post hoc test. Scale bars, 50 μm. ‘HAP1 WT BDNF was normalised as 100%
Discussion
In the present study, we found that HAP1 is co-localised with internalised mature BDNF but not mature BDNF synthesised within neurons. In addition, we also found that HAP1 is co-localised with BDNF receptors p75NTR and TrkB and the co-localisation is increased in response to BDNF stimulation in PC12 cells and neurons. In co-transfected cells, we found that HAP1 interacts with p75NTR or TrkB, respectively, as detected by co-IP and FRET analysis, and the interactions were dramatically increased in response to BDNF. Furthermore, we found that HAP1 is required for the endocytosis of BDNF and its receptors p75NTR and TrkB as the internalisation of these factors were abolished in HAP1 KO neurons. Our results also indicate that HAP1 appears essential for the activation and phosphorylation of Akt, ERK and PLCγ-1 in cortical neurons in response to BDNF. Lastly, we found that HAP1 is also required for proliferation of cerebellar granule cells in response to BDNF stimulation in vitro and in vivo. Based on these data, we conclude that HAP1 is an essential molecule for the endocytosis and signal transduction of BDNF/TrkB in neurons.
HAP1 Interacts with TrkB and p75NTR and This Interaction Is Stimulated by BDNF
We have found that exogenously added BDNF showed much higher co-localisation with HAP1A than intracellularly expressed BDNF pool in PC12 cells. Moreover, adding antibodies to BDNF and p75NTR with exogenous BDNF treatment reduced the co-localisation of HAP1A with BDNF. Immunostaining of endogenous HAP1 and p75NTR also showed that co-localisation between HAP1 and p75NTR increased by exogenous BDNF addition. These findings suggest that HAP1 may have different roles for endocytosed and intracellular BDNF. It should be noted that PC12 cells express p75NTR but not TrkB [34, 35]. Therefore, p75NTR could be the only receptor for BDNF endocytosis in PC12 cells.
Our Co-IP and FRET experiments in HEK293T cells demonstrated that the interaction of HAP1A/TrkB and HAP1A/p75NTR increased upon BDNF treatment. This result is consistent with recent findings that TrkB is in a complex with HAP1 [32]. Taken together, our data suggest that HAP1 interacts with TrkB and p75NTR and the interaction can be stimulated by BDNF.
The Role of HAP1 in Endocytosis of BDNF and Its Receptors
We have found that endocytosis of BDNF was abolished in HAP1 KO cortical neurons and the defect was rescued by HAP1A gene delivery into HAP1 KO cortical neurons. This result was also confirmed by pull-down assay of internalised biotinylated BDNF. Our results clearly demonstrate that HAP1 is essential for BDNF endocytosis. Our finding is partially consistent with a previous study which showed that the endocytosis of biotinylated BDNF was reduced in the brainstem neurons from HAP1 KO mice [24]. A few studies have shown that TrkB protein levels are significantly lower in hypothalamus [32], cerebellum and brainstem tissue [24] in HAP1 KO mice than in WT mice. RT-PCR and Western blot of TrkB in the cortex and brainstem tissue in WT and HAP1 KO confirmed that the internalisation impairment of BDNF in HAP1 KO resulted from absence of HAP1 but not from the TrkB expression level reduction.
We have found that BDNF treatment of WT cortical neurons significantly reduced cell surface TrkB but had no effect on cell surface TrkB in HAP1 KO cortical neurons. However, BDNF stimulation did not trigger endocytosis of truncated TrkB (TrkB-T1), which is consistent with a previous report [13]. It has been reported that TrkB-T1 is able to bind and become endocytosed with BDNF [36] by possibly forming a dimer with full-length TrkB, which suggests that TrkB-T1 endocytosis may be controlled by different neuronal activity [37–39]. Moreover, it has been demonstrated that TrkB-T1 internalisation begins slowly after BDNF triggering and the internalisation is most active at 90 min [40]. It is possible that our study missed the optimal time for detection of TrkB-T1 in cortical neurons during BDNF stimulation time. Moreover, it could be worthwhile examining the binding sites between HAP1-TrkB and HAP1-TrkB-T1. If TrkB-T1 does not have a binding domain with HAP1 while TrkB does, it could explain why TrkB-T1 was not endocytosed on BDNF stimulation. Interestingly, in contrast to TrkB, we failed to detect any difference in the cell surface p75NTR between WT and HAP1 KO cortical neurons in response to BDNF. One reason for this discrepancy could be due to the low level of p75NTR expression in cortical neurons [41–43].
The Role of HAP1 in TrkB Signalling and Neuronal Proliferation
Another significant finding of the present study is that HAP1 is required for the signal transduction through TrkB in response to BDNF. This study found that the phosphorylation of ERK, Akt and PLCγ-1 was increased in response to BDNF in WT cortical neurons but not in HAP1 KO neurons. It is widely accepted that BDNF/TrkB complex is critical for neuronal survival, growth and proliferation [44–48]. Therefore, our data strongly indicate that HAP1 is a critical molecule involved in the signal transduction of BDNF/TrkB. When neurotrophins including BDNF bind to their receptors, the ligand-receptor complex is endocytosed and transported retrogradely along the axon to the cell body, which results in the initiation of signalling cascades locally [12]. This signalling model is called the signalling endosome model [33, 49, 50].
As the endocytosis of BDNF and its receptors requires HAP1 and is apparently regulated by HAP1, we propose that HAP1 is a key molecule regulating trafficking and signal transduction of signalling endosomes containing BDNF. Our results also indirectly suggest that the endocytosis of neurotrophins and their receptors is an essential step for neurotrophin signalling and consistent with the signalling endosome hypothesis [33, 51, 52]. Although our findings do not exclude the possible alternative retrograde signalling mechanisms, the findings that abolishing BDNF endocytosis by HAP1 deletion diminishes TrkB downstream signalling indicate that the BDNF/TrkB signalling endosome is a main mode of signal transduction for BDNF.
We have also found that proliferation of cerebellar granule cells with BDNF treatment was impaired in HAP1 KO mice while BDNF treatment triggered the increase in proliferation of cerebellar granule cells both in vitro and in vivo. This result suggests that HAP1 is an essential intracellular molecule regulating the signal transduction of BDNF in the development of the nervous system. Our data support previous findings that the phenotype of HAP1 KO mice shares many features of BDNF KO such as postnatal lethality, and abnormal energy metabolism [15, 53, 54] as the neurotrophic function of BDNF could be defected in HAP1 KO mice. However, the finding that HAP1 is required for BDNF-induced proliferation of granule cells is in contrast to that of the recent study showing that the BDNF can rescue the deficit of progenitor cell proliferation in the hypothalamus in conditional HAP1 KO mice [32]. This discrepancy may be related to regions of the brain. In the present study, we have used new born mice as a model and focussed on cerebellar granule cells.
In addition, the findings that HAP1 is required for the endocytosis and signal transduction of BDNF and TrkB support the concept that the biological functions of BDNF require HAP1. Retrolinkin binds endophilin A1, and those proteins are involved in early endocytic trafficking of BDNF/TrkB [55]. The authors showed that suppressed Retrolinkin expression prevents TrkB internalisation under BDNF stimulation and activation of ERK, resulting in reduced dendrite growth of CNS neurons. It has been also reported that inhibition of clathrin-mediated endocytosis interferes with BDNF/TrkB internalisation and dendrites outgrowth and survival of cortical neurons through Akt activation [13]. BDNF also plays a role in cellular proliferation through Akt activation [56]. Moreover, HAP1 plays a role in retro- and anterograde trafficking to transport vesicles [20, 23, 57]. Taken together, it may be reasonable to conclude that HAP1 gene deletion impairs BDNF/TrkB internalisation and further endosomal trafficking intracellularly to the cell body to facilitate TrkB downstream signal cascade, leading to abolished neuronal proliferation.
As BDNF/TrkB endocytosis is critical for its neurotrophic function, our study warrants further investigation into roles of HAP1 in neurodegenerative disorders such as Huntington’s disease and Alzheimer’s disease and endocytosis of other growth factors.
Materials and Methods
Animals
All procedures involving animals were approved by the Animal Ethic Committee of SA Pathology (Adelaide, Australia) and the Animal Welfare Committee of Flinders University (Adelaide, Australia). All the procedures were undertaken according to the guidelines of the National Health and Medical Research Council of Australia. All animals were maintained under 12-h light/dark cycle and free access to food and water. HAP1 KO mice were generated as described previously [15]. HAP1 KO, HET (heterozygous for the HAP1 knock-out allele) or WT neonatal mice were bred from transgenic breeding pairs heterozygous for the HAP1 knock-out allele (C57/Black6 genetic background). PCR genotyping of HAP1 KO mice was carried out using primers 5′-TTTTGGAGG TCTGGTCTCGCTCTG-3′/5′-CGTCTTCCATCTTAGTGCGTTCAC-3′ for wild type (WT) and 5 ′-TTTTGGAGGTCTGGTCTCGCTCTG-3′/′ 5-CTTCATGTGGATGCTAGGGATCC-3′ for KO animals.
Plasmids
The following plasmids were used in this study: p75NTR-cyan fluorescent protein (CFP), p75NTR-yellow fluorescent protein (YFP), p75NTR-Hemagglutinin (HA) WT (all p75NTR plasmids were kindly donated by Prof. Goldstein University of California, San Diego, CA, USA), TrkB-YFP (kindly donated by Dr. Wayman, Washington State University, Pullman, WA, USA), p75NTR-CFP-YFP (kindly donated by Dr. Coulson, University of Queensland, QLD, Australia), TrkB-Flag (kindly donated by Dr. Chen, Shandong University, Shandong, China) and pEYFP-N1 was obtained from Clontech (Mountain View, CA, USA). HAP1A-green fluorescent protein (GFP), HAP1A-CFP and HAP1-YFP were derived by subcloning a PCR product generated from PRK-HAP1A using primers (forward: 5′-TAGC TAGCATGCGCCCGAAGGAC-3′ and reverse: 5′-GAGG TACCAGGGTTGATGATCGGTAGC-3′) into the NheI and KpnI sites of pEGFP-N1, pECFP-N1 and pEYFP-N1 (Clontech, Mountain View, CA, USA). To produce BDNF with red fluorescence tag, the rat cDNA sequences of mature BDNF (384 bp) were amplified from a proBDNF-EGFP construct (kindly donated by Dr. Kojima, National Institute of Advanced Industrial Science and Technology, Osaka, Japan) using the following PCR primers: forward: 5′-GCGAATT CATGCACTCCGACCCT-3′ and reverse: 5′-ATGGCGACCGGTGGATCCCT-3′ with added BamHI and EcoRI site. The amplified fragment was cut with BamHI and EcoRI and subcloned in-frame into pDsRed-Express-N1 (Clontech, Mountain View, CA, USA), and the final constructs were verified by DNA restriction enzyme digestion and DNA sequencing.
Cell Culture
HEK293T cells (ATCC, Rockville, MD, USA) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technology, VIC, Australia) containing 10% foetal bovine serum (FBS), penicillin (50 IU/ml), streptomycin (50 μg/ml) and L-glutamine (2 mM) (Life Technology, VIC, Australia). PC12 cells (ATCC, Rockville, MD, USA) were cultured in DMEM medium supplemented with horse serum (5%) (Life Technology, VIC, Australia), FBS (10%), penicillin (50 IU/ml) and streptomycin (50 μg/ml). All cells were maintained at 37 °C in a humidified incubator supplemented with 95% O2 and 5% CO2.
Biotinylation of BDNF
To analyse endocytosed BDNF in cell imaging and biochemical assay, BDNF was biotinylated as described previously [38] with minor modification. It has been shown that biotinylated BDNF has biological activity [58, 59]. BDNF (100 μg, kindly donated by Amgen, Thousand Oaks, CA, USA) was dialysed against 1 × PBS (pH 7.4) for 3 h, and PBS was changed two more times every 3 h. The dialysed BDNF was incubated with 2 mg Sulfo-NHS-SS-biotin (Thermo Fisher Scientific, Rockford, IL, USA) for 2 h on ice. Free biotin was removed by dialysis against PBS.
BDNF-Red Fluorescent Protein (RFP) Preparation
As BDNF-RFP is secreted into the cell culture medium, we used the culture medium containing BDNF-RFP for the co-localisation assay. To prepare the culture medium containing BDNF-RFP, we transfected PC12 cells with pDs-Red-BDNF plasmid for 48 h. The culture medium containing BDNF-RFP was then used immidately in the co-localisation assay.
The Co-localisation Assay
To investigate whether HAP1, BDNF and p75NTR co-localise with each other, co-localisation assay was performed. PC12 cells were seeded onto poly-L-ornithine (mol. weight 30,000–70,000, 15 μg/ml; Sigma-Aldrich, St Louis, MO, USA)-coated 13 mm coverslips (Thermo Fisher Scientific, Rockford, IL, USA) and cultured for 3 days in differentiation culture medium containing 100 ng/ml nerve growth factor (NGF) before transfection to make an in vitro neuron like model [60]. pEGFP-HAP1A only or pEGFP-HAP1A/pDs-Red-BDNF pair were mixed with NeuroPorter (Sigma-Aldrich, St Louis, MO, USA) in a ratio of 5:5 μl according to the instructions of NeuroPorter Transfection kit (Sigma-Aldrich, St Louis, MO, USA), followed by adding the mixture to the PC12 cells. The cells were cultured in NGF containing culture medium for 48 h in humidified air containing 5% CO2.
The cells were cultured with sheep anti-BDNF antibody (10 μg/ml, in-house) alone, cell culture medium containing BDNF-RFP alone and either cell culture medium containing BDNF-RFP with sheep anti-BDNF (10 μg/ml) or cell culture medium containing BDNF-RFP with rabbit anti-p75NTR antibody (10 μg/ml, a kind gift from Prof. Chao, Skirball Institute, New York, NY, USA). The cells were then fixed, and images of fluorochromes (green and red) were obtained by Leica TCS SP5 (Leica Microsystems, Mannheim, Germany) for quantitative assessment of co-localisation between HAP1A and BDNF-RFP.
For immunocytochemistry co-localisation assay, differentiated PC12 cells were treated as above, and BDNF-biotin (BDNF-B) was added at 100 ng/ml for 30 min in DMEM containing horse serum (5%) (Life Technology, VIC, Australia), FBS (10%), penicillin (50 IU/ml) and streptomycin (50 μg/ml) (Life Technology, VIC, Australia), followed by acid wash (0.25 M acetic acid, 0.25 M NaCl) for 20 min to remove surface-bound BDNF-B. Immunostaining was performed with mouse anti-HAP1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit anti-p75NTR (a gift by Prof. Chao, Skirball Institute, New York, NY, USA), followed by anti-mouse-Alexa488 and anti-rabbit-Cy3 (Life Technology, VIC, Australia) and streptavidin-Cy5 (Life Technology, VIC, Australia) for BDNF-B after fixing cells. The images of fluorochromes were obtained by Leica TCS SP5 (Leica Microsystems, Mannheim, Germany) for quantitative assessment of co-localisation of HAP1, p75NTR and BDNF-B. The cells were tested for mycoplasma and were mycoplasma-negative.
Co-immunoprecipitation (Co-IP) Assay
Co-IP assay was performed to analyse the interaction between HAP1A/TrkB and HAP1A/p75NTR as described previously [26]. HEK293T cells were co-transfected with HAP1A-YFP/TrkB-FLAG or HAP1A-YFP/p75NTR-HA pair. The cells were treated with or without BDNF at 100 ng/ml for 1 h in DMEM supplemented with 10% FBS, penicillin (50 IU/ml), streptomycin (50 μg/ml) and L-glutamine (2 mM) (Life Technology, VIC, Australia) and then lysed with RIPA buffer (50 mM Tris-HCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, pH 7.4) supplemented with protease inhibitor cocktail (Roche, Castle Hill, NSW, Australia), followed by sonication and centrifugation at 12,000g for 10 min at 4 °C. The resultant supernatants were collected, and BCA protein assay (Thermo Fisher Scientific, Rockford, IL, USA) was performed to determine protein concentrations. The supernatants (400 μg) were then incubated with 2 μg antibody (mouse anti-FLAG (Sigma-Aldrich, St Louis, MO, USA)) for TrkB-FLAG/HAP1A-YFP, goat anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for HAP1A-YFP/p75NTR-HA and mouse or goat IgG as a control for antibody specificity at 4 °C overnight with rotation at 60 rpm. Protein G beads (30 μl; Thermo Fisher Scientific, Rockford, IL, USA) were added to the mixture and rotated for further 2 h at 4 °C in order to immobilise antibody and the control IgG. The beads were washed four times with PBST (PBS, 0.1% Tween-20) and boiled in 30 μl of 2 × SDS PAGE loading buffer (0.1 M Tris-HCl, pH 6.8, 4% SDS, 10% glycerol, 10% β-mercaptoethanol, 0.004% bromophenol blue). The samples were then analysed by Western blotting to detect HAP1A-YFP with goat anti-GFP antibody and p75NTR-HA using rabbit anti-HA antibody (Sigma-Aldrich, St Louis, MO, USA), respectively. At the same time, 2 μg of the original sample supernatant was loaded as an input control.
Fluorescence Resonance Energy Transfer
Fluorescence resonance energy transfer or Förster resonance energy transfer (FRET) is a non-radiative, distance-dependent (1–10 nm), energy transfer from an excited fluorophore (donor) to another fluorophore (acceptor) and was performed as described previously [26, 61]. HEK293T cells were seeded (at 2.5 × 104) on coverslips for overnight and then transfected with HAP1A-CFP/p75NTR-YFP pair HAP1A-CFP/TrkB-YFP pair, positive control (p75NTR-CFP-YFP) or negative control (p75NTR -CFP/YFP pair). BDNF was added to the pairs of HAP1A-CFP/p75NTR-YFP and HAP1A-CFP/TrkB-YFP at various concentrations (0, 5, 50 and 100 ng/ml). A Leica SP5 spectral confocal microscope was used to perform FRET according to the manufacturer’s recommendations (Leica Microsystems, Mannheim, Germany). Briefly, acceptor bleaching was used to compare the donor fluorescence intensity in the same sample before and after photo bleaching the acceptor. If there is FRET, the intensity of the donor fluorescence increases after photo-bleaching the acceptor. The efficiency of FRET can be determined as follows: FRETeff (Dpost_Dpre)/Dpost_100%, where Dpost is the fluorescence intensity of the donor after acceptor photo bleaching and Dpre is the fluorescence intensity of the donor before acceptor photo bleaching. When FRETeff is higher than background and negative controls, the FRETeff is considered positive. The higher the FRETeff, the closer the two molecules are to each other based on the FRET principle. Each FRETeff value from the observed samples including positive and negative controls was determined according to a mean of six individual tests from different regions of interest. Three different experiments were conducted, and the results were analysed by IBM SPSS Statistics 21.0 software (IBM, supplied by University of South Australia).
Primary Neuronal Culture
Transgenic pups at postnatal day 0 (P0) to 1 (P1) were anaesthetised on ice-water slurry and killed by decapitation. After the brain meninges removal, the cortex from both hemispheres of each pup was separated from other parts of the brain and transferred into 15-ml falcon tubes containing 2 ml DMEM and centrifuged at 2000 rpm for 2 min at 4 °C. DMEM was then removed, and tissues were digested with 2 ml of 0.5% trypsin including 0.1% DNaseI (NEB, Ipswich, MA, USA) for 20 min at 37 °C with shaking every 5 min. FBS (final 15%) was added to stop the tissue digestion, and the digested tissue was triturated by passing through a 10-ml serological pipette (Thermo Fisher Scientific, Rockford, IL, USA) for 10 times. When tissue pieces settled, the supernatant was transferred into a new tube and spun at 2000 rpm for 2 min at 4 °C. The cell pellet was resuspended in 1 ml of cortical neuron culture media (Neurobasal medium containing B27 supplement (2%), L-glutamine (2 mM) and penicillin/streptomycin (100 IU/ml) (Life Technology, VIC, Australia) and β-mercaptoethanol (0.1 mM)). In order to harvest neurons after treatment for Western blotting or for immunocytochemistry assay, cortical cells were placed into poly-D-lysine and laminin-coated 6-well plate (at 2 × 106) and on poly-D-lysine-and laminin-coated coverslips (Thermo Fisher Scientific, Rockford, IL, USA) in 4- or 24-well plates (at 2.5 × 104) (Thermo Fisher Scientific, Rockford, IL, USA), respectively, and after 72-h incubation at 37 °C supplemented with 95% O2, 5% CO2 incubator, neurons were ready for treatment.
Cell Surface Biotinylation Assay
To analyse internalised TrkB and p75NTR in cultured cortical neurons, cell surface biotinylation assay was conducted as previously described [13]. Briefly, cortical neurons from P0 to P1 HAP1 WTand KO mice were isolated and cultured in 6-well plates for 72 h in cortical neuron culture media. Thereafter, the cultured neurons were washed with Neurobasal medium twice and then kept in Neurobasal medium only without B27 supplement for 2 h, followed by changing to cortical neuron culture media containing 0 (control) or 100 ng/ml of BDNF and incubation for 30 min in a 37 °C incubator supplemented with 5% CO2 and 95% O2. Culture plates were then transferred onto ice and left for 10 min. The plates were then gently rinsed three times with chilled PBS-CM (PBS containing 1 mM CaCl2, 0.5 mM MgCl2, pH 7.4), followed by incubation with 0.5 mg/ml Biotin-S-S-NHS (Thermo Fisher Scientific, Rockford, IL, USA) on ice for 60 min. The cell surface biotinylation reaction was quenched by washing twice (10 min each wash) with 100 mM glycine in PBS-CM with gentle rocking, followed by washing three times (5 min each wash) with chilled PBS-CM with gentle rocking. Neurons were then lysed with RIPA buffer containing proteinase inhibitor cocktails (Roche, Castle Hill, NSW, Australia), and the lysates were collected by cell scraper. Cell extracts were then sonicated and centrifuged at 12000 rpm for 10 min at 4 °C. The resultant supernatants were transferred to fresh tubes, and protein concentrations were determined by BCA protein assay (Thermo Fisher Scientific, Rockford, IL, USA). Total protein (40–60 μg) was mixed with streptavidin-agarose beads (Sigma-Aldrich, St Louis, MO, USA) and incubated overnight at 4 °C, and 20–30 μg of total protein was kept for total receptor detection. Next day, beads were washed 4 times with RIPA buffer, and 30 μl of 2× SDS-PAGE loading buffer (100 mM Tris-Cl (pH 6.8), 4% (w/v) SDS, 0.2% (w/v) bromophenol blue, 20% (v/v) glycerol, 200 mM DTT (dithiothreitol)) was added, followed by boiling for 5 min. The samples were then subjected to Western blot analysis with goat anti-TrkB (R&D systems, Minneapolis, MN, USA), rabbit anti-p75NTR (from Prof. Reichardt, University of California, San Francisco, CA, USA) and mouse anti-β-actin (Sigma-Aldrich, St Louis, MO, USA) to detect surface and total TrkB and p75NTR and β-actin as internal control, respectively. The band densitometry of the cell surface protein versus total protein and β-actin was determined using ImageJ software (Research Service Branch; National Institute of Health, http://rsbweb.nih.gov/ij/index.html).
Reverse Transcription (RT)-PCR
Determination of TrkB mRNA expression levels was conducted according to the standard procedures. Total RNA was extracted from the cortex and brainstem of neonatal HAP1 WT (n = 3) and KO (n = 3) mice using an RNAeasy mini kit (Qiagen, Doncaster, Vic, Australia). The concentration and purity of RNA were determined with a NanoDrop 2000 (Thermo Fisher Scientific, Rockford, IL, USA). Five hundred nanograms of total RNA was used to generate first-strand cDNA using Superscript III First-Strand Synthesis System (Life Technology, VIC, Australia). PCR was conducted with GoTaq Green Master Mix (Promega, Madison, WI, USA) with primers for mouse TrkB (forward primer, 5′-CCTCCACGGATGTTGCTGAC-3′; and reverse primer, 5′-GCAACATCACCAGCAGGCA-3′) and normalised against the housekeeping gene GAPDH (forward primer, 5′-AACATCATCCCTGCATCCAC-3′; and reverse primer, 5′-TTGAAGTCTCAGGAGACAAC-3′). RT-PCR was performed for 25 cycles in Veriti 96-well Thermal Cycler (Life Technology, VIC, Australia) at 95 °C for 30 s, 53 °C for 30 s and 72 °C for 30 s. The densitometry of the PCR bands (TrkB and GAPDH) was measured using ImageJ software (Research Service Branch; National Institute of Health, http://rsbweb.nih.gov/ij/index.html). GAPDH was used for normalising TrkB mRNA expression.
TrkB Signalling Western Blot Analysis
Cortical neurons were washed with Neurobasal medium, and the neurons were incubated in Neurobasal medium only for 2 h before treatment. Neurons were treated with 0 (control) or 100 ng/ml BDNF for 20 min at 37 °C in an incubator supplemented with 5% CO2 and 95% O2. The tissue culture plate was transferred onto ice-water slurry to stop the treatment, followed by three washes with ice-cold PBS. Ice-cold phosphorylation lysis buffer containing 0.1% SDS, 1% nondiet P-40, 0.5% sodium deoxycholate, 2 mM EDTA, 2 mM sodium orthovanadate, 1 mM phenylmethylsulphonyl fluoride (PMSF),10 mM sodium fluoride and protease inhibitor cocktail (Roche, Castle Hill, NSW, Australia) in PBS was then added to the cells. The cell lysates were sonicated for 15 s on ice-water slurry and centrifuged for 10 min at 12000 rpm at 4 °C. The supernatants were then transferred to fresh microtubes followed by determining protein concentrations. The neuron lysates were separated by 10% SDS-PAGE and transferred to Hybond-C membrane (GE Healthcare Australia, NSW, Australia) and blocked with blocking buffer (5% skim milk in Tris-buffered saline (TBS) (150 mM NaCl, 50 mM Tris-Cl, pH 7.5) for 1.5 h at 25 °C with gentle shaking. After blocking, membranes were incubated on a rocker at 4 °C with one of the following primary antibodies: rabbit anti-phospho-ERK1/2 (1:500, #4370S, rabbit anti-ERK1/2 (1:1000, #9102S), rabbit anti-PLCγ-1 (1:500, #5690S), rabbit anti-phospho-Akt1/2/3 (1:500, #9271), rabbit anti-Akt1/2/3 (1:1000, #4691) (Cell Signalling, Danvers, MA, USA), anti-phospho-PLCγ1 (1:500, #ab53125, Abcam, Cambridge, UK) and mouse anti-β-actin (1:4000. A5316, Sigma-Aldrich, St Louis, MO, USA). Thereafter, corresponding secondary antibodies linked to horse radish peroxidase were applied onto the membranes for 1 h at 25 °C. Band densities were determined using Image Quant LAS 4000 (GE Healthcare Australia, NSW, Australia). All phosphorylated protein bands were normalised to total protein and GAPDH band to yield the densitometry ratio value. For TrkB Western blot, the cortex and brainstem tissue of neonatal HAP1 WT (n = 3) and KO (n = 3) mice were homogenised in RIPA Buffer. The proteins (30 μg) were subjected to Western blot analysis as described above. Goat anti-TrkB (R&D systems, Minneapolis, MN, USA) was used, and mouse anti-β-actin (Sigma-Aldrich, St Louis, MO, USA) was used for loading control.
Proliferation Assay of Cerebellar Granule Cells In Vitro and In Vivo
For an in vitro experiment, cerebellar neurons were prepared same as cortical neuron culture method. The viable cells were counted using trypan blue (Sigma-Aldrich, St Louis, MO, USA) to exclude dead cells, and 5 × 104 cerebellar granule cells were seeded onto poly-D-lysine-coated 13 mm cover slip (Thermo Fisher Scientific, Rockford, IL, USA) and cultured. Next day, cultured cerebellar neurons were treated with cerebellar neuron culture medium containing BDNF (0, 10, 100 ng/ml) and 10 μg/ml of bromodeoxyuridine (BrdU) (Sigma-Aldrich, St Louis, MO, USA) to label cells in S phase for 24 h. Then, the cells were subjected to immunocytochemistry. Cultured cells were fixed in 4% paraformaldehyde, PBS pH 7.4 for 10 min at room temperature, followed by TBS wash. The cells were then kept in 2 N HCL at 37 °C for 1 h to expose BrdU, followed by TBS wash. Thereafter, the cells were then incubated with blocking buffer (3% bovine serum albumin (BSA) (Sigma-Aldrich, St Louis, MO, USA), TBS, 0.1% Triton X-100, 0.01% NaN3, pH 7.4) for 1 h at 25 °C, then incubated with an anti-BrdU monoclonal antibody (1:500, G3G4, DSHB, Iowa city, Iowa, USA), followed by a Cy3-conjugated goat anti-mouse IgG (1:200, C2821, Sigma-Aldrich, St Louis, MO, USA). The images were taken by CX40 fluorescence microscope (Olympus, VIC, Australia), and BrdU-positive cells were counted using ImageJ software (Research Service Branch; National Institute of Health, http://rsbweb.nih.gov/ij/index.html). Six fields were checked in each cover slip, and the same locations of the six fields were applied to the all observed cover slips. Three and four cover slips were observed for 0 and 10 and 100 ng/ml of BDNF treatment samples of each genotype, respectively.
In vivo experiment was conducted as described previously [62]. Briefly, neonatal mice were immobilised under a stereo-microscope with anaesthesia on ice. PBS (1 μl, control) or BDNF (1 μl, 5 μg) was injected into the lateral ventricle slowly (n = 6/group) at the injection target (0.5 mm lateral to the midline, 1 mm caudal to bregma, and 1.5 mm in depth). To label proliferating cells, BrdU (50 mg/kg; Sigma-Aldrich, St Louis, MO, USA) was injected subcutaneously after the lateral ventricle injection, and the animals were kept for 24 h in their home cage. Mice were perfused with cold 4% paraformaldehyde after anaesthesia. Dissected brain tissues were submerged into 30% sucrose in 0.1 M phosphate buffer for cryoprotection. Sagittal section cutting (25 μm) was performed on a microtome (Leica, Mannheim, Germany), and the cut sections were mounted on gelatin-treated slides. The sections were incubated in 0.5% Triton X-100, followed by incubation in 2 N HCl at 37 °C for 1 h to expose BrdU incorporated into DNA. Thereafter, the sections were washed three times with PBS and blocked with blocking solution containing 5% donkey serum (Sigma-Aldrich, St Louis, MO, USA) for 1 h at 25 °C. Anti-BrdU monoclonal antibody (1:500, G3G4, DSHB, Iowa city, Iowa, USA) was added and incubated overnight at 4 °C. After washing, samples were then incubated Cy3-conjugated goat anti-mouse IgG (1:200, C2821, Sigma-Aldrich, St Louis, MO, USA). The number of BrdU-positive granule cells was counted with Olympus BX50 (Olympus, VIC, Australia) with 20× objective.
Statistical Analysis
The data are presented as mean ± standard error of the mean (SEM), and it was considered significant when p < 0.05. Student’s t test or one-way ANOVA with post hoc test was used to analyse the data with intra- or intergroup. All statistical analyses were conducted using IBM Statistics 21 software (IBM, supplied by University of South Australia) or GraphPad Prism, Version 6.05 (GraphPad, supplied by University of South Australia; Graph Pad Inc., CA).
Summary Statement.
HAP1 is required for endocytosis of BDNF and its receptor TrkB in neurons, and the loss of HAP1 causes defects in endocytosis of BDNF/TrkB and neuronal proliferation.
Acknowledgments
We thank L.S. Goldstein from University of California for providing us with p75NTR-CFP, p75NTR-YFP and p75NTR-HA wt plasmids; G.A. Wayman from Washington State University for providing TrkB-YFP plasmid; E. Coulson from University of Queensland for providing p75NTR-CFP-YFP plasmid; Zheyu Chen from Shandong University for providing TrkB-Flag plasmid.
Funding This project was supported by an NHMRC grant to XFZ&XJL (480423). Yoon Lim was supported by an NHMRC Postgraduate Scholarship (GNT1017711), and LLW was supported by an IPRS Scholarship from Flinders University.
Footnotes
Author Contributions Y. L., L.L.W, S.C., Y.S., S.L.V. and M.Y. designed and performed the experiments and analysed the data. L.B., D.K. and X.J.L contributed to data interpretation and critical manuscript evaluation. X.F.Z conceived and designed the experiments, contributed to data interpretation and critical manuscript revision. Y.L. and L.B. wrote the manuscript.
Compliance with Ethical Standards
Competing Interests The authors declare that they have no competing interests.
References
- 1.Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237:1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 3.Lewin GR, Barde YA. Physiology of the neurotrophins. Annu Rev Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- 4.Mowla SJ, Farhadi HF, Pareek S, Atwal JK, Morris SJ, Seidah NG, Murphy RA. Biosynthesis and post-translational processing of the precursor to brain-derived neurotrophic factor. J Biol Chem. 2001;276:12660–12666. doi: 10.1074/jbc.M008104200. [DOI] [PubMed] [Google Scholar]
- 5.Nagappan G, Zaitsev E, Senatorov VV, Jr, Yang J, Hempstead BL, Lu B. Control of extracellular cleavage of ProBDNF by high frequency neuronal activity. Proc Natl Acad Sci U S A. 2009;106:1267–1272. doi: 10.1073/pnas.0807322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pang PT, Lu B. Regulation of late-phase LTP and long-term memory in normal and aging hippocampus: role of secreted proteins tPA and BDNF. Ageing Res Rev. 2004;3:407–430. doi: 10.1016/j.arr.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Seidah NG, Benjannet S, Pareek S, Chretien M, Murphy RA. Cellular processing of the neurotrophin precursors of NT3 and BDNF by the mammalian proprotein convertases. FEBS Lett. 1996;379:247–250. doi: 10.1016/0014-5793(95)01520-5. [DOI] [PubMed] [Google Scholar]
- 8.Lee R, Kermani P, Teng KK, Hempstead BL. Regulation of cell survival by secreted proneurotrophins. Science. 2001;294:1945–1948. doi: 10.1126/science.1065057. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, Lim Y, Li F, Liu S, Lu JJ, Haberberger R, Zhong JH, Zhou XF. ProBDNF collapses neurite outgrowth of primary neurons by activating RhoA. PLoS One. 2012;7:e35883. doi: 10.1371/journal.pone.0035883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo NH, Teng HK, Siao CJ, Chiaruttini C, Pang PT, Milner TA, Hempstead BL, Lu B. Activation of p75NTR by proBDNF facilitates hippocampal long-term depression. Nat Neurosci. 2005;8:1069–1077. doi: 10.1038/nn1510. [DOI] [PubMed] [Google Scholar]
- 11.Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 12.Seidah NG, Chretien M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 13.Zheng J, Shen WH, Lu TJ, Zhou Y, Chen Q, Wang Z, Xiang T, Zhu YC, et al. Clathrin-dependent endocytosis is required for TrkB-dependent Akt-mediated neuronal protection and dendritic growth. J Biol Chem. 2008;283:13280–13288. doi: 10.1074/jbc.M709930200. [DOI] [PubMed] [Google Scholar]
- 14.Li XJ, Li SH, Sharp AH, Nucifora FC, Jr, Schilling G, Lanahan A, Worley P, Snyder SH. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- 15.Li SH, Yu ZX, Li CL, Nguyen HP, Zhou YX, Deng C, Li XJ. Lack of huntingtin-associated protein-1 causes neuronal death resembling hypothalamic degeneration in Huntington’s disease. J Neurosci. 2003;23:6956–6964. doi: 10.1523/JNEUROSCI.23-17-06956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li SH, Gutekunst CA, Hersch SM, Li XJ. Association of HAP1 isoforms with a unique cytoplasmic structure. J Neurochem. 1998;71:2178–2185. doi: 10.1046/j.1471-4159.1998.71052178.x. [DOI] [PubMed] [Google Scholar]
- 17.Li SH, Hosseini SH, Gutekunst CA, Hersch SM, Ferrante RJ, Li XJ. A human HAP1 homologue. Cloning, expression, and interaction with huntingtin. J Biol Chem. 1998;273:19220–19227. doi: 10.1074/jbc.273.30.19220. [DOI] [PubMed] [Google Scholar]
- 18.Bartlett EJ, Brodie JD, Simkowitz P, Schlosser R, Dewey SL, Lindenmayer JP, Rusinek H, Wolkin A, et al. Effect of a haloperidol challenge on regional brain metabolism in neuroleptic-responsive and nonresponsive schizophrenic patients. Am J Psychiatry. 1998;155:337–343. doi: 10.1176/ajp.155.3.337. [DOI] [PubMed] [Google Scholar]
- 19.Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, Holzbaur EL, Ross CA. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 20.McGuire JR, Rong J, Li SH, Li XJ. Interaction of huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J Biol Chem. 2006;281:3552–3559. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]
- 21.Colomer V, Engelender S, Sharp AH, Duan K, Cooper JK, Lanahan A, Lyford G, Worley P, et al. Huntingtin-associated protein 1 (HAP1) binds to a trio-like polypeptide, with a rac1 guanine nucleotide exchange factor domain. Hum Mol Genet. 1997;6:1519–1525. doi: 10.1093/hmg/6.9.1519. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Chin LS, Levey AI, Li L. Huntingtin-associated protein 1 interacts with hepatocyte growth factor-regulated tyrosine kinase substrate and functions in endosomal trafficking. J Biol Chem. 2002;277:28212–28221. doi: 10.1074/jbc.M111612200. [DOI] [PubMed] [Google Scholar]
- 23.Rong J, Li S, Sheng G, Wu M, Coblitz B, Li M, Fu H, Li XJ. 14-3-3 protein interacts with huntingtin-associated protein 1 and regulates its trafficking. J Biol Chem. 2007;282:4748–4756. doi: 10.1074/jbc.M609057200. [DOI] [PubMed] [Google Scholar]
- 24.Sheng G, Xu X, Lin YF, Wang CE, Rong J, Cheng D, Peng J, Jiang X, et al. Huntingtin-associated protein 1 interacts with Ahi1 to regulate cerebellar and brainstem development in mice. J Clin Invest. 2008;118:2785–2795. doi: 10.1172/JCI35339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu LL, Fan Y, Li S, Li XJ, Zhou XF. Huntingtin-associated protein-1 interacts with pro-brain-derived neurotrophic factor and mediates its transport and release. J Biol Chem. 2010;285:5614–5623. doi: 10.1074/jbc.M109.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang M, Lim Y, Li X, Zhong JH, Zhou XF. Precursor of brain-derived neurotrophic factor (proBDNF) forms a complex with huntingtin-associated protein-1 (HAP1) and sortilin that modulates proBDNF trafficking, degradation, and processing. J Biol Chem. 2011;286:16272–16284. doi: 10.1074/jbc.M110.195347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kittler JT, Thomas P, Tretter V, Bogdanov YD, Haucke V, Smart TG, Moss SJ. Huntingtin-associated protein 1 regulates inhibitory synaptic transmission by modulating gamma-aminobutyric acid type a receptor membrane trafficking. Proc Natl Acad Sci U S A. 2004;101:12736–12741. doi: 10.1073/pnas.0401860101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rong J, Li SH, Li XJ. Regulation of intracellular HAP1 trafficking. J Neurosci Res. 2007;85:3025–3029. doi: 10.1002/jnr.21326. [DOI] [PubMed] [Google Scholar]
- 29.Rong J, McGuire JR, Fang ZH, Sheng G, Shin JY, Li SH, Li XJ. Regulation of intracellular trafficking of huntingtin-associated protein-1 is critical for TrkA protein levels and neurite outgrowth. J Neurosci. 2006;26:6019–6030. doi: 10.1523/JNEUROSCI.1251-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL, MacAskill AF, Rostaing P, Lumb MJ, Humbert S, Triller A, et al. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron. 2010;65:53–65. doi: 10.1016/j.neuron.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu LL, Zhou XF. Huntingtin associated protein 1 and its functions. Cell Adhes Migr. 2009;3:71–76. doi: 10.4161/cam.3.1.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang J, Yang H, Zhao T, Sun M, Xu X, Zhou XF, Li SH, Li XJ. Huntingtin-associated protein 1 regulates postnatal neurogenesis and neurotrophin receptor sorting. J Clin Invest. 2014;124:85–98. doi: 10.1172/JCI69206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe CL, Mobley WC. Signaling endosome hypothesis: a cellular mechanism for long distance communication. J Neurobiol. 2004;58:207–216. doi: 10.1002/neu.10323. [DOI] [PubMed] [Google Scholar]
- 34.Lou X, Yano H, Lee F, Chao MV, Farquhar MG. GIPC and GAIP form a complex with TrkA: a putative link between G protein and receptor tyrosine kinase pathways. Mol Biol Cell. 2001;12:615–627. doi: 10.1091/mbc.12.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saxena S, Howe CL, Cosgaya JM, Hu M, Weis J, Kruttgen A. Differences in the surface binding and endocytosis of neurotrophins by p75NTR. Mol Cell Neurosci. 2004;26:292–307. doi: 10.1016/j.mcn.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Alderson RF, Curtis R, Alterman AL, Lindsay RM, DiStefano PS. Truncated TrkB mediates the endocytosis and release of BDNF and neurotrophin-4/5 by rat astrocytes and schwann cells in vitro. Brain Res. 2000;871:210–222. doi: 10.1016/s0006-8993(00)02428-8. [DOI] [PubMed] [Google Scholar]
- 37.Eide FF, Vining ER, Eide BL, Zang K, Wang XY, Reichardt LF. Naturally occurring truncated trkB receptors have dominant inhibitory effects on brain-derived neurotrophic factor signaling. J Neurosci. 1996;16:3123–3129. doi: 10.1523/JNEUROSCI.16-10-03123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du J, Feng L, Zaitsev E, Je HS, Liu XW, Lu B. Regulation of TrkB receptor tyrosine kinase and its internalization by neuronal activity and Ca2+ influx. J Cell Biol. 2003;163:385–395. doi: 10.1083/jcb.200305134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ninkina N, Adu J, Fischer A, Pinon LG, Buchman VL, Davies AM. Expression and function of TrkB variants in developing sensory neurons. EMBO J. 1996;15:6385–6393. [PMC free article] [PubMed] [Google Scholar]
- 40.Fryer RH, Kaplan DR, Kromer LF. Truncated trkB receptors on nonneuronal cells inhibit BDNF-induced neurite outgrowth in vitro. Exp Neurol. 1997;148:616–627. doi: 10.1006/exnr.1997.6699. [DOI] [PubMed] [Google Scholar]
- 41.Kordower JH, Bartus RT, Bothwell M, Schatteman G, Gash DM. Nerve growth factor receptor immunoreactivity in the non-human primate (Cebus apella): distribution, morphology, and colocalization with cholinergic enzymes. J Comp Neurol. 1988;277:465–486. doi: 10.1002/cne.902770402. [DOI] [PubMed] [Google Scholar]
- 42.Mufson EJ, Bothwell M, Hersh LB, Kordower JH. Nerve growth factor receptor immunoreactive profiles in the normal, aged human basal forebrain: colocalization with cholinergic neurons. J Comp Neurol. 1989;285:196–217. doi: 10.1002/cne.902850204. [DOI] [PubMed] [Google Scholar]
- 43.Mufson EJ, Kordower JH. Cortical neurons express nerve growth factor receptors in advanced age and Alzheimer disease. Proc Natl Acad Sci U S A. 1992;89:569–573. doi: 10.1073/pnas.89.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barde YA. Trophic factors and neuronal survival. Neuron. 1989;2:1525–1534. doi: 10.1016/0896-6273(89)90040-8. [DOI] [PubMed] [Google Scholar]
- 45.Cheng A, Wang S, Yang D, Xiao R, Mattson MP. Calmodulin mediates brain-derived neurotrophic factor cell survival signaling upstream of Akt kinase in embryonic neocortical neurons. J Biol Chem. 2003;278:7591–7599. doi: 10.1074/jbc.M207232200. [DOI] [PubMed] [Google Scholar]
- 46.Choi SH, Li Y, Parada LF, Sisodia SS. Regulation of hippocampal progenitor cell survival, proliferation and dendritic development by BDNF. Mol Neurodegener. 2009;4:52. doi: 10.1186/1750-1326-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glass DJ, Nye SH, Hantzopoulos P, Macchi MJ, Squinto SP, Goldfarb M, Yancopoulos GD. TrkB mediates BDNF/NT-3-dependent survival and proliferation in fibroblasts lacking the low affinity NGF receptor. Cell. 1991;66:405–413. doi: 10.1016/0092-8674(91)90629-d. [DOI] [PubMed] [Google Scholar]
- 48.Meyer-Franke A, Kaplan MR, Pfrieger FW, Barres BA. Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron. 1995;15:805–819. doi: 10.1016/0896-6273(95)90172-8. [DOI] [PubMed] [Google Scholar]
- 49.Bartlett SE, Reynolds AJ, Hendry IA. Retrograde axonal transport of neurotrophins: differences between neuronal populations and implications for motor neuron disease. Immunol Cell Biol. 1998;76:419–423. doi: 10.1046/j.1440-1711.1998.00767.x. [DOI] [PubMed] [Google Scholar]
- 50.Grimes ML, Zhou J, Beattie EC, Yuen EC, Hall DE, Valletta JS, Topp KS, LaVail JH, et al. Endocytosis of activated TrkA: evidence that nerve growth factor induces formation of signaling endosomes. J Neurosci. 1996;16:7950–7964. doi: 10.1523/JNEUROSCI.16-24-07950.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bronfman FC, Tcherpakov M, Jovin TM, Fainzilber M. Ligand-induced internalization of the p75 neurotrophin receptor: a slow route to the signaling endosome. J Neurosci. 2003;23:3209–3220. doi: 10.1523/JNEUROSCI.23-08-03209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye H, Kuruvilla R, Zweifel LS, Ginty DD. Evidence in support of signaling endosome-based retrograde survival of sympathetic neurons. Neuron. 2003;39:57–68. doi: 10.1016/s0896-6273(03)00266-6. [DOI] [PubMed] [Google Scholar]
- 53.Ernfors P, Lee KF, Jaenisch R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature. 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- 54.Jones KR, Farinas I, Backus C, Reichardt LF. Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell. 1994;76:989–999. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu X, Yang Y, Xu C, Niu Y, Chen T, Zhou Q, Liu JJ. Retrolinkin cooperates with endophilin A1 to mediate BDNF-TrkB early endocytic trafficking and signaling from early endosomes. Mol Biol Cell. 2011;22:3684–3698. doi: 10.1091/mbc.E11-04-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q, Liu G, Wu Y, Sha H, Zhang P, Jia J. BDNF promotes EGF-induced proliferation and migration of human fetal neural stem/progenitor cells via the PI3K/Akt pathway. Molecules. 2011;16:10146–10156. doi: 10.3390/molecules161210146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li SH, Gutekunst CA, Hersch SM, Li XJ. Interaction of huntingtin-associated protein with dynactin P150Glued. J Neurosci. 1998;18:1261–1269. doi: 10.1523/JNEUROSCI.18-04-01261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X, Obianyo O, Chan CB, Huang J, Xue S, Yang JJ, Zeng F, Goodman M, et al. Biochemical and biophysical investigation of the brain-derived neurotrophic factor mimetic 7,8-dihydroxyflavone in the binding and activation of the TrkB receptor. J Biol Chem. 2014;289:27571–27584. doi: 10.1074/jbc.M114.562561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu Y, Ji Y, Ganesan S, Schloesser R, Martinowich K, Sun M, Mei F, Chao MV, et al. TrkB as a potential synaptic and behavioral tag. J Neurosci. 2011;31:11762–11771. doi: 10.1523/JNEUROSCI.2707-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das KP, Freudenrich TM, Mundy WR. Assessment of PC12 cell differentiation and neurite growth: a comparison of morphological and neurochemical measures. Neurotoxicol Teratol. 2004;26:397–406. doi: 10.1016/j.ntt.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 61.Pollok BA, Heim R. Using GFP in FRET-based applications. Trends Cell Biol. 1999;9:57–60. doi: 10.1016/s0962-8924(98)01434-2. [DOI] [PubMed] [Google Scholar]
- 62.Xu ZQ, Sun Y, Li HY, Lim Y, Zhong JH, Zhou XF. Endogenous proBDNF is a negative regulator of migration of cerebellar granule cells in neonatal mice. Eur J Neurosci. 2011;33:1376–1384. doi: 10.1111/j.1460-9568.2011.07635.x. [DOI] [PubMed] [Google Scholar]