Abstract
Agrobacterium tumefaciens is a widely used microbial tool in plant molecular biology to transfer DNA into plant cells and produce, e.g., stable or transient transformants or induce gene silencing. In our study, we present a simplified version of electrocompetent cell preparation that is not only time and cost efficient, but it requires minimal handling of bacterial cells. Liquid cultures are normally used to prepare competent Agrobacterium cells. To overcome the difficulties of working with liquid cultures, we propose suspending bacterial cells directly from overnight agar plate cultures. In addition, we optimized several parameters to simplify the procedure and maximize the number of transformants (e.g., Agrobacterium strains, number of washing steps, amount of required plasmid DNA, electroporation parameters, type of incubation media, or incubation time). This optimized, simple, and fast protocol has proved to be efficient enough to obtain transformed colonies with low amounts (as little as 1 ng) of plasmid DNA. In addition, it also enabled us to introduce ligated plasmids directly into Agrobacterium omitting the E. coli transformation step and accelerating the cloning procedure further.
Keywords: Agrobacterium, Electroporation, Plate method, Direct transformation
Introduction
Agrobacterium tumefaciens (Rhizobium radiobacter) is an important species of phytopathogenic bacteria that can cause crown gall disease in hundreds of eudicot plants by inducing tumors (Agrios 1997). Tumors in plant tissues are triggered by DNA transfer. During this process, the T-DNA fragment(s) are excised from the Ti (tumor-inducing) plasmid of Agrobacterium and are exported into plant cells by a type IV bacterial secretion system. The T-DNA can incorporate into the genome of host cells and the genes of this DNA insert (including tumor-inducing genes) are expressed by the compromised host cells (Valentine 2003). Besides the phytopathological importance of this bacterium species, it has become an important biotechnological tool of gene transfer into plant cells. Agrobacterium-mediated transformation is applicable for stable and transient gene delivery or for the introduction of gene silencing constructs into plant cells. Binary Ti vector plasmid systems are widely used for gene transfer experiments. In this system, the Agrobacterium strain contains a disarmed Ti plasmid with essential vir genes as a resident plasmid and another plasmid that carries the desired transgene on T-DNA between the right (RB) and left (LB) borders (Hellens et al. 2000; Lee et al. 2008). The advantage of this system is that DNA manipulation can be performed with a smaller size T-DNA containing plasmid that replicates both in Agrobacterium and in Escherichia coli.
The most efficient transformation method of A. tumefaciens is electroporation that uses electrical field to increase the permeability of the bacterial cell membrane and to enhance the uptake of DNA (Mattanovich et al. 1989; Mersereau et al. 1990; Kotnik 2013). Based on the published A. tumefaciens electroporation protocols (Mattanovich et al. 1989; Mersereau et al. 1990; De la Riva et al. 1991; Main et al. 1995; Lin 1995; den Dulk-Ras et al. 1995; McCormac et al. 1998; Hitzeroth et al. 2016), we have improved steps of the original protocol to obtain a simple method that requires minimal handling of bacterial cells but efficiently transforms A. tumefaciens for routine applications.
Materials and methods
Bacterial strains and plasmid
A. tumefaciens strains: MOG301 (Hood et al. 1993), EHA 105 (Hood et al. 1993) and LBA 4404 (Hoekema et al. 1983) were grown on agar solidified Luria broth (LB) (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl, and 15 g/L agar) supplemented with 50 μg/mL rifampicin (Serva). After electroporation, the bacteria were grown in SOC liquid medium (20 mM glucose, 20 g/L tryptone, 5 g/L yeast extract, 10 mM NaCl, 2.5 mM MgCl2, and 10 mM MgSO4), or in LB liquid medium.
For electroporation experiments, the pTRV2 in pCAMBIA3301 plasmid (Arabidopsis Biological Resource Center, STOCK:CD3-1043) was used.
Optimized protocol of electrocompetent cell preparation and electroporation
Freshly growing cells (1–2 days old) of the chosen Agrobacterium strain were spread on LB agar plates (diameter = 90 mm, supplemented with the required antibiotics) and incubated overnight (~ 16 h) at 27 °C to produce bacterial lawn that covered the surface of the plate completely. If a higher volume of electrocompetent cell suspension was required, cells were collected from more than one plate.
Bacterial cells were carefully washed off the plate with 4 mL ice cold 10% (v/v) sterile glycerol. Cells growing on the surface of the plate were scraped with an inoculation loop avoiding damages of the agar medium and were suspended in the glycerol solution. The bacterial suspension was then transferred into two sterile 2 mL centrifuge tubes.
Suspensions in the two tubes were centrifuged at 14,000 rpm (18,000 g) for 1 min at 4 °C; the supernatant was discarded.
1 mL ice cold 10% (v/v) sterile glycerol was added to each tube containing the bacterial pellet. The tubes were thoroughly vortexed afterwards to resuspend the cells and step 3 was repeated one more time.
After the two centrifugation steps, the supernatant was removed and discarded again and the bacterial pellets in the two tubes were resuspended in 200–200 μl ice cold 10% (v/v) sterile glycerol each and combined in one tube (yielding 400 μl in total).
The tube with the Agrobacterium cell suspension was kept on ice until electroporation. (The unused electrocompetent cells can be aliquoted and frozen in liquid nitrogen and stored at − 70 °C.)
For electroporation 70–80 μl of the ice cold suspension of electrocompetent bacterial cells was mixed with 1–3 μl plasmid DNA (1–100 ng) in a sterile centrifuge tube.
This mixture was loaded into a chilled electroporation cuvette (gap = 2 mm) and placed into the cuvette holder.
The electroporator was used with the following parameters: 2.5 kV, 25 μF capacitance, and 400 Ohm resistance.
One mL SOC medium was added immediately to the electroporation cuvette and the resulting bacterial suspension was transferred into a 15 mL centrifuge tube, and the tube was incubated at 27 °C for 1 h with rotating. (1 h of incubation is usually long enough to obtain a desired number of transformant cells, but to reach an even higher number of transformants, the incubation time can be extended to 3 h.)
When the incubation was over 100 μl from each suspension of electroporated cells was spread onto LB plates which contained the required antibiotics. The plates were incubated for 2 days at 27 °C and successfully transformed colonies were verified by specifically designed PCR amplifications.
Transformation efficiency calculations
After the 1–3 h-long incubation time, the efficiency of transformation was calculated. For this calculation, the electroporated cell suspension was diluted 100 times (performing two subsequent 10× dilution steps) with sterile SOC medium to get a final volume of 1 mL diluted suspension. Then, 100 μl aliquots of the diluted bacterial cell suspension were spread onto rifampicin (50 μg/mL) and kanamycin (50 μg/mL) containing LB plates. The plates were incubated at 27 °C for 2 days and bacterial colonies were counted. The efficiency was calculated by counting the number of the colonies and presented as CFU (colony-forming unit)/μg DNA.
Direct electroporation of ligation product into A. tumefaciens
A 350 bp cDNA fragment was amplified with Phusion High-Fidelity DNA Polymerase (Thermo Scientific) from pGEM-T Easy plasmid (Promega) that contained the cDNA of Arabidopsis thaliana At4g10540 gene. The sequences of the used primers were the following: Fw 5′-TGA AGA TCT ATG AAG AGT TGC AGA ACC TTA A-3′ and Rev 5′-CCA CTA GTC CGA GTT GTA TCT AGT TGG-3′. The amplified PCR product was purified using High Pure PCR Product Purification Kit manufactured by Roche. The purified PCR product and the pCAMBIA1302 plasmid were digested with BcuI/BglII enzymes in FastDigest buffer (Thermo Scientific) and then were purified again by High Pure PCR Product Purification Kit. The digested insert and plasmid (40 ng) were ligated by T4 DNA Ligase (Thermo Scientific) adjusting a molar ratio of 3:1. After ligation, the ligase enzyme was inactivated at 65 °C for 10 min. One μl aliquots of the ligation reaction mixture were used for electroporation.
Statistical analysis
Two or three independent biological experiments with three replicates for each were conducted. Experimental data were analyzed using one-way ANOVA and posthoc tests. The mean differences were considered statistically significant at P ≤ 0.05 level. Statistical analysis was done with IBM SPSS Statistics 20. Software.
Results and discussion
Agrobacterium-mediated transformation is a widely used technique in molecular biotechnology labs working with plants. To simplify the procedure of the preparation of competent cells, we replaced the liquid culture-based preparation method with solid plate based one that was previously used effectively with other bacterial species (Gonzales et al. 2013). There are several advantages of our plate-based method described in “Materials and methods” over liquid culture-based methods. For instance, it requires less equipment and media or a reduced number of inoculation and centrifugation steps (lowering the risk of contamination). Moreover, by selecting the appropriate number of plates, the amount of competent cells produced becomes easily controlled and calculable. In our first transformation attempts, we removed bacterial cells from plates (that were incubated at 27 °C overnight) with 10% sterile glycerol and washed four times with the same glycerol solution before electroporation. The final concentration of bacterial cells was about 1.5 × 1011 cells/mL as determined by serial dilution. This concentration is comparable to concentrations published before, where bacterial cells were collected from liquid cultures (Mersereau et al. 1990; De la Riva et al. 1991; den Dulk-Ras et al. 1995). Because the first results showed that this method is efficient enough for routine plasmid transfer into Agrobacterium, we optimized and simplified further the electrocompetent cell preparation and transformation protocol.
Effect of the number of washing steps on transformation
To make the transformation procedure easier and more rapid, we minimized the number of washing steps that refines the quality of the bacterial cell suspension. For testing the minimal number of washing steps required for efficient plasmid transfer, we washed the bacteria in parallel experiments 1–3 times with 10% glycerol. After electroporation, the efficiency of transformation was determined. The results confirmed that even one washing step is sufficient for successful electroporation as no statistically significant difference was observed in the number of developing transformant colonies with the introduction of a second or third washing step (data not shown).
Effect of electroporation parameters on the efficiency of transformation
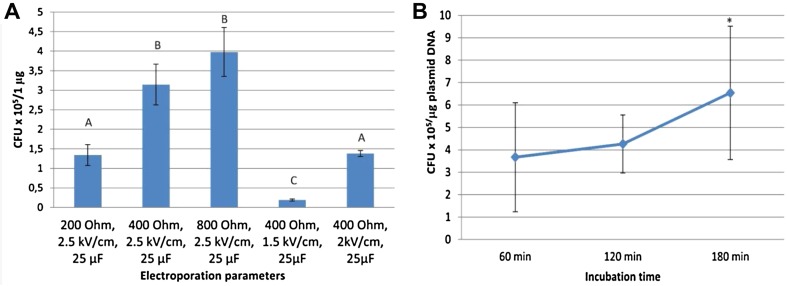
In a series of experiments, we evaluated the effect of different electroporation parameters on the transformation efficiency of our electrocompetent cells (A. tumefaciens MOG 301) by changing one parameter at a time. We tested three different voltage settings (1.5, 2, and 2.5 kV/cm) and three different pulse controller settings (200, 400, and 800 Ohm), while the capacitor setting was constant (25 μF). Figure 1a shows that the mean of the efficiency of transformation was the highest in the case of an 800 Ohm and 2.5 kV/cm setting, although no statistically significant difference could be observed in the efficiency of transformation between 800 and 400 Ohm resistance at 2.5 kV/cm. It was also apparent that an increase in the voltage setting (from 1.5 to 2.0 or 2.5 kV/cm) can improve the transformation efficiency.
Fig. 1.
a Effect of different electroporation parameters on transformation efficiency of A. tumefaciens MOG301 electrocompetent cells. The mean differences are statistically significant at 0.05 level, denoted by different letters. Statistical analysis was performed using one-way ANOVA and LSD posthoc test. The results show the average of three experiments and three replicates for each. b Effect of incubation time of bacterial suspension on the number of transformants. After electroporation, bacterial suspension was diluted with 1 mL SOC media and incubated for 60–180 min. The number of transformants were calculated after dilution and plating. Data represent the mean of two independent biological experiments with three replicates for each. The mean difference is statistically significant at 0.05 level. Statistical analysis was performed using one-way ANOVA and Tukey’s posthoc test. Asterisk indicates statistically significant differences between 60, 120, and 180 min incubation times
Comparing the effect of SOC and LB incubation media on transformation efficiency
After electroporation, bacterial cells were immediately suspended in liquid medium in 15 mL centrifuge tubes and incubated at 27 °C for 3 h. To test the influence of the type of liquid incubation media on transformation efficiency of our electrocompetent cells, 1 mL SOC or LB liquid media were added to the bacterial suspensions after electroporation. Our results showed that SOC incubation resulted in higher transformation efficiency, since approximately 1.5 times higher transformant cells were obtained after SOC than after LB medium incubation.
Influence of incubation time of bacterial suspension after electroporation on the number of transformants
After electroporation, bacterial cell suspensions were incubated at 27 °C with rotating. To evaluate the effect of the incubation time on transformation efficiency and make the transformation protocol shorter without drastic decrease in the number of transformants, the incubation time was changed between 1 and 3 h. Our results showed that longer incubation time resulted in significantly more transformant colonies (Fig. 1b.). However, even shorter incubation resulted in sufficient number of transformants for routine plasmid transfer. Therefore, the incubation period can be shortened to 1 h in 1 mL SOC liquid medium if it is necessary.
Influence of using different Agrobacterium strains on the efficiency of transformation
The efficiency of our electroporation protocol was tested on three different A. tumefaciens strains (MOG301, EHA 105, and LBA 4404) to see if this protocol can be used widely to introduce plasmids into various Agrobacterium strains. These strains belong to two different genotypes, because MOG 301 and EHA 105 possess C58 (Hood et al. 1993), while LBA4404 has TiAch5 chromosomal background (Hellens et al. 2000). The transformation was successful for all three strains, but the effectiveness of the transformation process greatly varied between the three strains (Table 1). The highest transformation efficiency was obtained with A. tumefaciens MOG301 (2.7x105 CFU/μg plasmid DNA). In conclusion, our results showed that the simplified electroporation method can be used for Agrobacterium strains with different genotype backgrounds.
Table 1.
Transformation efficiency of different Agrobacterium strains
| Agrobacterium strains | Efficiency (CFU × 105/μg plasmid DNA) |
|---|---|
| MOG301 | 2.7 ± 0.29a |
| EHA 105 | 1.25 ± 0.15b |
| LBA 4404 | 0.044 ± 0.011c |
These results show the average of two experiments ± SE with three replicates for each. The mean differences are statistically significant at 0.05 level, denoted by different letters. Statistical analysis was performed using one-way ANOVA and Tukey’s posthoc test
Low amount of plasmid DNA is enough for efficient transformation
In some cases, the amount of plasmid DNA is a limiting factor of efficient electroporation. To find the lowest plasmid concentration that can be transformed into our competent cells, we added various amounts of plasmid (1, 20, 50, and 100 ng) to A. tumefaciens MOG301 competent cells. The electroporation results showed that transformation was efficient even if 1 ng plasmid DNA was used for electroporation. This low quantity of plasmid DNA was sufficient for successful electroporation and produced 3.6 × 103 CFU/μg DNA transformant cells. This result implies that our competent cells and electroporation protocol are suitable to introduce low amount of DNA into Agrobacterium.
Efficient direct electroporation of ligation product into A. tumefaciens
During the standard cloning procedure, if one wants to clone a DNA insert into an Agrobacterium binary plasmid, the ligation product should first be transformed into E. coli after the ligation reaction is completed. Then, using the E. coli colonies that develop on selective medium, the incorporation of the insert must be checked by PCR, and after picking the positive colonies, plasmid DNA needs to be purified. This time-consuming procedure normally precedes the transformation of A. tumefaciens cells. To accelerate this process, we examined if our electrocompetent A. tumefaciens cells are suitable for direct transformation of ligation products. To test this shortened protocol, we ligated a 350 bp Arabidopsis cDNA PCR product into pCAMBIA1302 plasmid and attempted to electroporate into A. tumefaciens MOG301 cells. One μl heat-treated (65 °C 10 min) ligation product was added to the suspension of competent cells. This direct transformation procedure was repeated twice and 200–400 colonies were obtained after the electroporation procedure when the whole amount of bacterial suspension (1 mL) was spread on the surface of selective LB medium-containing plates. We checked the incorporation efficiency of the insert into pCAMBIA1302 by specific PCR reactions. In the first experiment, the efficiency of incorporation was 93% (14 out of the tested 15 colonies showed successful plasmid incorporation), and in the second experiment, it was 73% (with 11 successfully transformed colonies out of the 15 tested ones). These results confirmed that the competent cells prepared by our protocol are useful for direct transformation of ligated plasmid products, and thus, our improved method may considerably simplify the cloning procedure into A. tumefaciens.
Conclusions
The advantage of the described plate-based electrocompetent cell preparation method is that it needs minimal handling of bacterial cells, less equipment (e.g., there is no need for shaking incubators or large capacity centrifuges) and requires only limited amount of laboratory consumables.
In addition, we simplified the whole process to shorten time by avoiding some unessential washing steps and decreasing the length of incubation that follows the electroporation step. The whole refined process from electrocompetent cell preparation to spreading the electroporated cell suspension onto the plate takes only approximately 90 min. This time is considerably short especially in comparison with methods that use diluted liquid culture for electrocompetent cell preparation (e.g., De la Riva et al. 1991; den Dulk-Ras et al. 1995).
The efficiency of plasmid transfer of our improved plate-based method is about 2–5 × 105 CFU/μg plasmid DNA. There are other electrocompetent preparation methods that result in higher transformation efficiency (e.g., Mersereau et al. 1990; De la Riva et al. 1991); nevertheless, our plate based method is fully sufficient for routine plasmid transformation (and other advantages of our method thoroughly compensate for this lower yield).
Usefulness of our optimized protocol was also corroborated by the fact that plasmid DNA even in low amount (1 ng) could have been consistently transferred into Agrobacterium. Moreover, it was shown that competent cells prepared by our protocol were suitable for direct transformation of ligated products into Agrobacterium. On one hand, direct transformation is able to accelerate the cloning procedure, and on the other hand, it can also be advantageous if a plasmid construct that contains a special insert would be instable in E. coli. This procedure was also shown to be effective for routine preparation of competent cells using different Agrobacterium strains.
Acknowledgements
Research in the authors’ laboratories has been funded by the Hungarian Scientific Research Fund (OTKA K 104730, OTKA PD 109050), the Bolyai Scholarship (BO 609 12, BO 00348/09), and the Széchenyi 2020 Programme, the European Regional Development Fund and the Hungarian Government (GINOP-2.3.2-15-2016-00061) which are gratefully acknowledged.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest in the publication.
References
- Agrios GN. Bacterial galls. San Diego: Academic Press; 1997. pp. 438–441. [Google Scholar]
- De la Riva G, Van Montagu M, Inzé D, Dhaese P. High-efficiency transformation of Agrobacterium tumefaciens with plasmid DNA by electroporation. Biotecnol Aplicada. 1991;8:345–351. [Google Scholar]
- den Dulk-Ras A, Hooykaas PJ. Electroporation of Agrobacterium tumefaciens. Methods Mol Biol. 1995;55:63–72. doi: 10.1385/0-89603-328-7:63. [DOI] [PubMed] [Google Scholar]
- Gonzales MF, Brooks T, Pukatzki SU, Provenzano D. Rapid protocol for preparation of electrocompetent Escherichia coli and Vibrio cholerae. J Vis Exp. 2013;80:e50684. doi: 10.3791/50684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R, Mullineaux P, Klee H. Technical Focus: a guide to Agrobacterium binary Ti vectors. Trends Plant Sci. 2000;5:446–451. doi: 10.1016/S1360-1385(00)01740-4. [DOI] [PubMed] [Google Scholar]
- Hitzeroth II, van Zyl AR. Transient expression of viral proteins in plants using Agrobacterium tumefaciens. Methods Mol Biol. 2016;1404:581–595. doi: 10.1007/978-1-4939-3389-1_38. [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. doi: 10.1038/303179a0. [DOI] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993;2:208–218. doi: 10.1007/BF01977351. [DOI] [Google Scholar]
- Kotnik T. Lightning-triggered electroporation and electrofusion as possible contributors to natural horizontal gene transfer. Phys Life Rev. 2013;10:351–370. doi: 10.1016/j.plrev.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Lee LY, Gelvin SB. T-DNA binary vectors and systems. Plant Physiol. 2008;146:325–332. doi: 10.1104/pp.107.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ. Electrotransformation of Agrobacterium. Methods Mol Biol. 1995;47:171–178. doi: 10.1385/0-89603-310-4:171. [DOI] [PubMed] [Google Scholar]
- Main GD, Reynolds S, Gartland JS. Electroporation protocols for Agrobacterium. Methods Mol Biol. 1995;44:405–412. doi: 10.1385/0-89603-302-3:405. [DOI] [PubMed] [Google Scholar]
- Mattanovich D, Rüker F, Machado AC, Laimer M, Regner F, Steinkellner H, Himmler G, Katinger H. Efficient transformation of Agrobacterium spp. by electroporation. Nucleic Acids Res. 1989;17:6747. doi: 10.1093/nar/17.16.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac AC, Elliott MC, Chen DF. A simple method for the production of highly competent cells of Agrobacterium for transformation via electroporation. Mol Biotechnol. 1998;9:155–159. doi: 10.1007/BF02760816. [DOI] [PubMed] [Google Scholar]
- Mersereau M, Pazour GJ, Das A. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene. 1990;90:149–151. doi: 10.1016/0378-1119(90)90452-W. [DOI] [PubMed] [Google Scholar]
- Valentine L. Agrobacterium tumefaciens and the plant: the David and Goliath of modern genetics. Plant Physiol. 2003;133:948–955. doi: 10.1104/pp.103.032243. [DOI] [PMC free article] [PubMed] [Google Scholar]