Abstract
The applicability of near-infrared (NIR) and mid-infrared (MIR) spectroscopy combined with chemometrics was explored in this study to develop rapid, low-cost and non-destructive spectroscopic methods for classification and quantification of aflatoxins in brown rice. A total of 132 brown rice samples within the aflatoxin concentration range of 0–2435.8 μg/kg were prepared by artificially inoculated with A. flavus and A. parasiticus strains of fungus. For the classification of samples at varying levels of aflatoxin B1, the linear discriminant analysis model obtained correct classification rate of 96.9 and 90.6% for NIR and MIR spectroscopy, respectively. For the simultaneous determination of aflatoxins B1, B2, G1, G2 and the total aflatoxins, partial least squares regression also showed good predictive accuracy for both NIR (rv = 0.936–0.973, RPD = 2.5–4.0) and MIR spectroscopy (rv = 0.922–0.970, RPD = 2.5–4.0). The overall results indicated that the two spectroscopic techniques offered the feasibility to be used as alternative tools for rapid detection of various aflatoxin contaminations in grain.
Electronic supplementary material
The online version of this article (10.1007/s13197-018-3033-1) contains supplementary material, which is available to authorized users.
Keywords: Aflatoxins, Near-infrared (NIR), Mid-infrared (MIR), Brown rice, Chemometrics
Introduction
It is well known that toxigenic fungi are ubiquitous in nature and can occur regularly in various grains. The secondary metabolites of those fungi, namely mycotoxins, can elicit a wide range of toxic activities that adversely affect the health of both humans and animals when they consume the grain (and their products) contaminated by mycotoxins (Gnonlonfin et al. 2013). Among the thousands of existing mycotoxins, the most commonly known are the aflatoxins, which are mainly produced by A. flavus and A. parasiticus strains (Wu et al. 2016). Aflatoxins consist of a group of approximately 20 related fungal metabolites, but only four are naturally found in food products. These are aflatoxin B1, B2, G1 and G2. Aflatoxin B1 (AFB1) is the most commonly found one and also the most toxic and carcinogenic, hence it was classified as a Group I carcinogen by the International Agency for Research on Cancer (IARC) (Binder 2007). Considering aflatoxin hazard to health, many countries have established mandatory sanitary regulations of aflatoxin level in food and agricultural products. The European Commission (EU) has set the maximum limit for AFB1 contamination in food products less than 5 ppb [Regulation (EU) No. 165/2010]. The Food and Drug Administration (FDA) in the United States has also set the limiting value of aflatoxin content at 20 ppb for food and 300 ppb for feed (USFDA 2009).
However, the precise determination of aflatoxins in grain is very complicated, because they are often present in quite small quantities. Therefore, sophisticated methods and instrumentations are usually required. Thin layer chromatography (TLC) was first applied to measure aflatoxins (Marutoiu et al. 2004). Currently, the most commonly used methods are based on gas chromatography (GC), high performance liquid chromatography (HPLC), liquid chromatography–mass spectrometry (LC–MS) (Huang et al. 2014). Other methodologies, such as enzyme-linked immunosorbent assays (ELISA) (Liu et al. 2013), immuno-affinity (Abd-Elghany and Sallam 2015) or fluorescence (Hruska et al. 2014) were also applied in some applications. Although these methods may have a high accuracy, difficulties can be encountered in sample preparation, including the isolation of aflatoxins from complex biological matrices. The sample preparation procedure often makes most of the analytical methods laborious, expensive and less effective.
Considering large amounts of cereals are consumed by animals and humans every day, it is necessary to develop analytical methods able to detect mycotoxin contamination in real time and with minimum cost. Spectroscopic techniques, such as near-infrared (NIR) and mid-infrared (MIR) spectroscopy, are excellent candidates for quality detection of agricultural products (Liu et al. 2015; Wang et al. 2015). These techniques are based on the measurement of the wave-absorption frequencies of chemical bonds in functional groups, such as C–C, C–H, O–H, C=O and N–H, that are closely related to the chemical composition and structure of samples. The main difference between NIR and MIR is that absorption in MIR range corresponds to fundamental frequency of molecular vibrations, whereas absorption in NIR corresponds to overtones and combinations of vibrations (Bellon-Maurel and McBratney 2011). With the combination of chemometrics, they can provide both qualitative and quantitative information without any complicated sample preparation.
NIR and MIR spectroscopy are now used in a remarkably wide range of analytical applications, for quantitative analysis of nutritive parameters such as water, protein, fat, starch and amino acids in grains and seeds (Ferreira et al. 2014). More recently, the potential of these two techniques to predict changes due to fungal infection during storage or processing have been examined (Hossain and Goto 2014). Berardo has applied NIRS to predict the percentage of Fusarium verticillioides infection in maize kernels as well as the concentration of ergosterol and fumosin B1 in meals (Berardo et al. 2005). Similar studies were conducted by using MIR spectroscopy to detect Fusarium graminearum infection on corn with respect to the content of deoxynivalenol (Kos et al. 2002) and ergosterol (Kos et al. 2003). In the same way, promising results were also obtained when NIR or MIR methodology were applied to discriminate samples into different grades with regarded to mycotoxins concentration (Kaya-Celiker et al. 2014) as well as to detect and estimate aflatoxin, deoxynivalenol, ochratoxin and fumonisin in milled and single kernels of many grain products (Fernández-Ibañez et al. 2009; Gaspardo et al. 2012).
Although studies can be found in the literature on the use of NIR and MIR methods for mycotoxin determination in grain, there is still a lack of information on the characterization of aflatoxin contamination in rice products. In addition, most previous reports were usually focused on the detection of single species of toxin (AFB1 or total aflatoxins), but little for simultaneous analysis of aflatoxin B1, B2, G1 and G2. A study could be found on the determination of aflatoxin B1, B2, G1 and G2 in peanut using MIR spectroscopy (Mirghania et al. 2001). However, only the solvent-extract of ground peanut cake rather than the peanut itself was employed. Therefore, the objective of this work was to explore NIR and MIR spectroscopy as alternative tools for rapid qualitative and quantitative evaluation of aflatoxin B1, B2, G1 and G2 and total aflatoxins in real brown rice samples. The performance of these two spectroscopic methods were also evaluated and compared. This study could provide useful information to the simultaneous detection of several different mycotoxins based the two methods, and to determine the most suitable method for quality control of grain.
Materials and methods
Aspergillus spp. spore suspension preparation
Two strains (A. flavus 3.17; A. parasiticus 3.395) known of aflatoxin production were purchased from Beina Chuanglian Biotechnology Research Institute (Beijing, China). Cultures of the strains were incubated at 28 °C and 85% relative humidity (RH) for 7 days on the Salt Czapek-Dox Medium to produce large numbers of spores. After incubation, spores were harvested by depositing sterile distilled water on the plate surface, slowly rubbing with a sterile stainless steel inoculation loop. Subsequently, the suspension obtained was filtered through the sterile cheesecloth for further use. The concentration of spore was determined using standard pour plate method, which were around 6.03 × 106 CFU/mL for the two strains (adjusted by using of sterile water).
Grain sample inoculation
Fresh brown rice samples with plastic vacuum packaging were purchased from a retail store in Heilongjiang Province, China. The samples were firstly irradiated by gamma-rays from Co-60 source at dose of 12 kGy, which was sufficient to kill fungi on or within the grain kernels (Kirkin et al. 2014). After irradiation, a total of 4 kg samples were aseptically transferred into two sterile stainless steel containers (2 kg/container) with agitators. With the continuously stirring, one container was sprayed with 40 mL spores suspension of A. flavus 3.17. The other container was inoculated with the same volume of A. parasiticus 3.395. Inoculated samples were then placed in sterile transparent PVC bags for 24 h at 4 °C to ensure water equilibrium. The inoculum concentration was determined by standard pour plate method, which was found at around 1.02 × 105 CFU/g. Subsequently, the samples of each bag were equally divided into 8 sub-groups (250 g/group). Finally, a total of 16 sample groups (8 × 2 bags) were obtained. The 16 samples were then placed in incubators at 28 °C, 85% RH for 30 days for aflatoxin production.
HPLC analysis of aflatoxins
Reference analysis for aflatoxins (B1, B2, G1, G2) in blank (control) and aflatoxin-contaminated brown rice samples was carried out using a Waters Alliance 2695 HPLC system (Waters, Milford, MA, USA), which was combined with an fluorescence detector. Before analysis, all samples were crushed into homogenized power using a No. 40 mesh sieve, and then stored at − 18 °C. Analytical procedures could be summarized briefly as follow: (1) a 50-g ground sample was mixed with 200 mL of acetonitrile/water (84/16, v/v) and shaken at 200 rpm for 30 min, (2) 8 mL of the filtrate was passed through a Mycosep 226 column (Romer Labs. Inc., MO, USA), (3) 2 mL of the eluate was evaporated to dryness under a stream of nitrogen gas at 50 °C, (4) The remaining residue was dissolved in n-hexane (200 μL) and trifluoroacetic acid (100 μL) for pre-column derivatization for 15 min at 40 °C, and (5) the dried residue was dissolved in 1 mL of acetonitrile/water (15/85, v/v) and filtered through a 0.45 μm syringe filter, and then injected into the HPLC automatically. The injection volume was 10 μL. The excitation and emission wavelengths were 380 and 420 nm, respectively. Each sample was measured in triplicate, and external standard method was used. The coefficients of determination (R2) of the method were > 0.997 for calibration curves of all aflatoxin standards.
Sample preparation for NIR and MIR analysis
HPLC results indicated that aflatoxins were not detected in blank (control) samples, and high aflatoxin concentrations were detected in all 16 contaminated samples. In order to prepare sample sets of calibration with varying concentrations of aflatoxin of interest, highly contaminated samples were later blended with blank samples on a weight basis. All samples were first crushed into homogenized power using a No. 40 mesh sieve, and then stored at − 18 °C. In order to reduce spectral interference caused by water, all contaminated and blank samples were dried at 40 °C for 6 h to make the moisture content in samples below 15% before blending. Specifically, 10 g of ground grain taken from each contaminated sample were blended with a blank sample according to the ratios of 1:2, 1:4, 1:6, 1:8, 1:10, 1:12, and 1:14 (m/m), respectively. A total of 112 contaminated samples (16 sub-groups × 7 levels) were then obtained. Finally, 128 samples were prepared, including 16 blank samples and 112 contaminated samples, which covered the concentrations of total aflatoxins ranging from 0 to 2406.4 μg/kg.
Spectra acquisition
NIR spectra of 128 samples were acquired using a FT-NIR spectrometer (MB3600, ABB-Bomem, QC, Canada) in diffuse reflection mode in the range of 4000–12,000 cm−1. The spectrometer was equipped with an interferometer, a long life light source and an InGaAs detector. Around 8 g of sample was uniformly packed in a glass bottle and directly placed on the sample holder for measurement. Measurements were conducted with the resolution of 4 cm−1 and 64 scans to ensure an adequate signal-to-noise ratio. Each sample was scanned in triplicate and air background was taken each hour.
MIR spectra were collected using a Bruker Tensor 27 FT-IR spectrometer (Bruker Optik, Ettlingen, Germany), which was equipped with a horizontal ZnSe crystal attenuated total reflectance (ATR) accessory (Pike Technologies, Madison, WI, USA) and a deuterated triglycine sulphate (DTGS) detector. The measurements were directly carried out by placing about 1 g ground sample on the ZnSe surface. The spectra were collected in the region of 600–4000 cm−1, by accumulating 64 scans with the resolution of 4 cm−1. Three replicate measurements were taken for each sample and the average spectrum was saved for further analysis. The background spectrum (air) was taken before every sample scanning. The ZnSe crystal was cleaned with 70% ethanol and dried by medical absorbent cotton after each measurement.
Multivariate calibration
All statistical analyses were conducted using TQ Analyst (version 6.2.1, Thermo Electron Corp., Madison, WI, USA) and Matlab 8.4 (The Mathworks Inc., USA) with a PLS toolbox 8.0 (Eigenvector Research Inc., USA) in this study. The goals of statistical analyses were: (a) select effective wavelength range, (b) extract feature information from NIR and MIR spectra, (c) discriminate brown rice samples with varying levels of aflatoxin, and (d) predict aflatoxin concentration in samples. Principal component analysis (PCA), linear discriminant analysis (LDA), stepwise multiple linear regression (SMLR) and partial least squares regression (PLSR) have been proven to be effective in many applications (Shi et al. 2013), and were therefore used in the present study to qualitative and quantitative evaluation of aflatoxins in brown rice samples.
PCA was primarily used to transform large amounts of original variables into a few new principal components (PCs), which accounted as much as possible for the variability in the raw data (Bro and Smilde 2014). LDA is a supervised technique that is widely recognized in classification problems. It is based on the determination of linear discriminant functions, which maximizes the ratio of between-class variance by minimizing the within-class variances (Khanmohammadi et al. 2013). SMLR is an efficient and rapid algorithm which selects characteristic wavelengths of sample spectra, determined by F test and predicted residual sum of squares (PRESS), as input variables of models. PLSR is a regression method which has no restriction in using the number of variables that can be selected for the calibration to make the model suitable to extract the maximum information (Shi et al. 2013).
Before modeling, spectral pretreatment methods, including smoothing, multiplicative scatter correction (MSC) and 1st/2nd derivative were tried and the best results were shown. During model development, the data were divided into a calibration set (2/3) and a validation set (1/3) by using the Kennard-Stone (KS) algorithm (Rajer-Kanduc et al. 2003). In order to evaluate the performance of calibration models, the correct classification rate, the correlation coefficient of calibration, and the validation (rc/rv), root mean-square error of cross-validation and prediction (RMSECV/RMSEP) were determined and discussed. The value of residual predictive deviation (RPD: the ration of the standard deviation of the reference value in prediction to the RMSEP) was also applied to evaluate the ability of the calibration model in predicting the chemical composition in samples.
Results and discussion
Aflatoxin data
The descriptive statistics of aflatoxin B1, B2, G1, G2, and total aflatoxins (AFs) for calibration and validation sample sets are summarized in Table 1. It could be observed that the samples showed a wide range in aflatoxin concentration. High SD (standard deviation) values were obtained for AFB1, G1 and AFs. The data manifested that the range of aflatoxin concentration analyzed covered almost all possible aflatoxin contaminated brown rice found in agriculture and industry (Elzupir et al. 2015). For each parameter, the range of calibration set covered the larger scale and the concentrations were generally evenly distributed in both calibration and validation data sets, which was beneficial for developing stable calibration models.
Table 1.
Descriptive statistics of aflatoxin data for calibration and validation sample sets
| Parameter (μg/kg) | Calibration sets (n = 86) | Validation sets (n = 42) | ||||
|---|---|---|---|---|---|---|
| Range | Mean | SD | Range | Mean | SD | |
| AFB1 | 0–890.20 | 274.04 | 279.03 | 0–884.19 | 267.38 | 282.98 |
| AFB2 | 0–55.97 | 15.68 | 16.39 | 0–47.01 | 14.73 | 15.28 |
| AFG1 | 0–1416.90 | 385.90 | 404.00 | 0–1358.47 | 373.91 | 400.64 |
| AFG2 | 0–102.40 | 25.64 | 27.31 | 0–77.95 | 23.64 | 24.46 |
| AFs | 0–2406.40 | 694.53 | 724.54 | 0–2363.59 | 672.65 | 716.49 |
Spectral features analysis
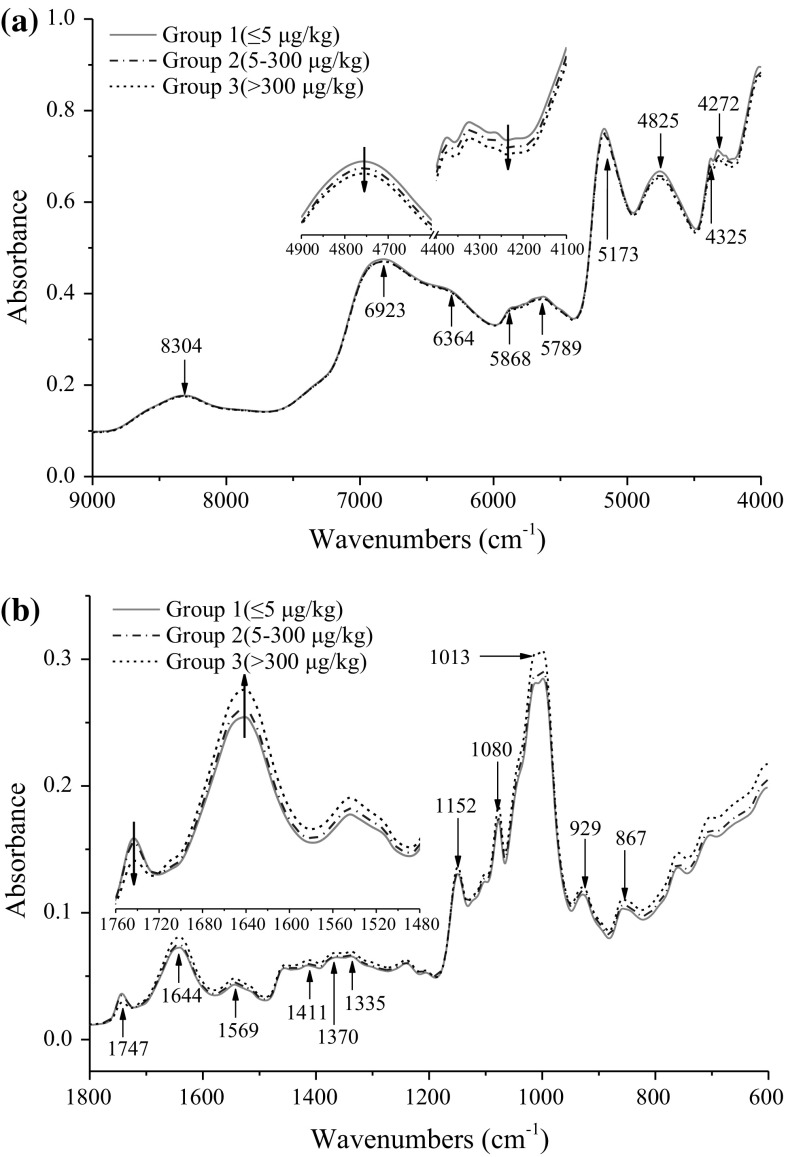
Using the permissible levels of aflatoxin in grains issued by EU and FDA as the reference, the samples were divided into three groups based on AFB1 concentration: group 1: ≤ 5 μg/kg (infected free); group 2: 5–300 μg/kg (slightly infected); group 3: > 300 μg/kg (highly infected). In order to examine different spectral features related to changes of aflatoxin concentration in samples, the variations of average NIR spectra from 9000 to 4000 cm−1 and MIR spectra from 1800 to 600 cm−1 for the three groups were investigated (Fig. 1), after scatter correction by applying MSC as pre-mathematical treatment. Other regions were not used in further analysis due to interference caused by water absorption or little useful information contained. Compared to MIR spectra (Fig. 1b), no obvious differences, resulting from the effect of aflatoxin contamination levels on brown rice, was observed among NIR spectra from visual inspection (Fig. 1a). The main absorption bands were found at 5173 and 4825 cm−1, which might be related to O–H stretching of water and carbohydrate. The absorption bands at 8304, 6923, 5868 and 5789 cm−1 were relevant to C–H stretching and deformation (Tripathi and Mishra 2009). The band at 4272 cm−1 could be assigned to C–H stretching in fatty acids and sugars (Xu et al. 2009). After a careful examination, spectral changes associated with aflatoxin concentration could be found in some regions, particularly in the range of 5000–4000 cm−1. The absorbance of group 1 was a little higher than that of group 2 and 3, as shown in Fig. 1a.
Fig. 1.
Average NIR (a) and MIR (b) absorbance spectra of ground brown rice samples with different levels of AFB1 (group 1: ≤ 5 μg/kg; group 2: 5–300 μg/kg; group 3: > 300 μg/kg)
More significant and pronounced difference for MIR spectra among aflatoxin contamination groups could be observed in the whole range. The absorption of group 3 was slightly higher than that of group 1 and 2 from 1720 to 600 cm−1, but lower in the range of 1800–1720 cm−1. The main absorption bands were located at 1013, 1080 and 1052 cm−1, which were related to O–H, C–O and C–H groups in organic acids and carbohydrate (Oliveira et al. 2014). The absorption difference at 1747 and 1644 cm−1 among three groups might be related to fungal infection in brown rice (Abramović et al. 2007). Small bands at 1569–1335 cm−1 were associated with C–O stretching and C–H bending, which were considered to be significant for identifying aflatoxin contamination (Kaya-Celiker et al. 2014). In fact, NIR and MIR bands assignments for aflatoxin are relatively difficult for direct identification, mainly because the peaks are sometimes contributed and interfered from predominant variations in chemical and physical properties of samples. Therefore, effective feature extraction for spectral differences was very crucial for classification and quantification of aflatoxin levels in brown rice samples using chemometric methods.
Development and validation of LDA models for classification
In this step, in order to detect minor differences among different spectra, the first 10 PCs extracted from NIR (9000–4000 cm−1) and MIR (1800–600 cm−1) spectral data, which covered most of the variance (> 98%), were employed to construct LDA models for discrimination. Leave-one-out (full) cross-validation was employed for evaluation of performance of LDA models. Table 2 presented the overall classification results for AFB1 concentration levels based on NIR and MIR spectra. Generally, good classification accuracy was achieved (> 90.0%) in both calibration and validation. For NIR spectra in calibration, only 2 samples were misclassified, which resulted in a correct classification rate of 98.4%. Meanwhile, in leave-one-out cross-validation, 96.9% of the samples were correctly classified. The LDA model did not misclassify any samples with high AFB1 concentration (> 5 μg/kg) as aflatoxin negative. This is of practical significance for using NIR to detect mycotoxins because misclassification (not detecting) of aflatoxin contamination could lead to human exposure to aflatoxins in real-life. For MIR spectra, the LDA yielded slightly lower classification accuracy as compared to NIR. Specifically, 12 samples were misclassified in cross-validation, and the correct classification rate was 90.6%. Two samples with AFB1 concentration above 5 μg/kg were wrongly classified as aflatoxin negative. This could be interpreted by less sample consumption and smaller scanning area than NIR to yield equivalent results. Moreover, the classification of samples based on the concentration of AFB2, G1, G2, and AFs were also carried out and they provided quite similar results to that of AFB1. Overall, the models performance could be further promoted, and change into more robust by collecting more calibration samples naturally contaminated with aflatoxin.
Table 2.
LDA classification results of samples with different levels of AFB1 obtained by NIR and MIR spectroscopy
| AFB1 | NIR | MIR | ||||||
|---|---|---|---|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Accuracy (%) | Group 1 | Group 2 | Group 3 | Accuracy (%) | |
| Calibration | ||||||||
| Group 1 | 16 | 0 | 0 | 100 | 16 | 0 | 0 | 100 |
| Group 2 | 0 | 64 | 0 | 100 | 1 | 60 | 3 | 93.8 |
| Group 3 | 0 | 2 | 46 | 95.8 | 0 | 6 | 42 | 87.5 |
| Total | 98.4 | 92.2 | ||||||
| Validation | ||||||||
| Group 1 | 16 | 0 | 0 | 100 | 15 | 1 | 0 | 93.8 |
| Group 2 | 0 | 63 | 1 | 98.4 | 2 | 59 | 3 | 92.2 |
| Group 3 | 0 | 3 | 45 | 93.8 | 0 | 6 | 42 | 87.5 |
| Total | 96.9 | 90.6 | ||||||
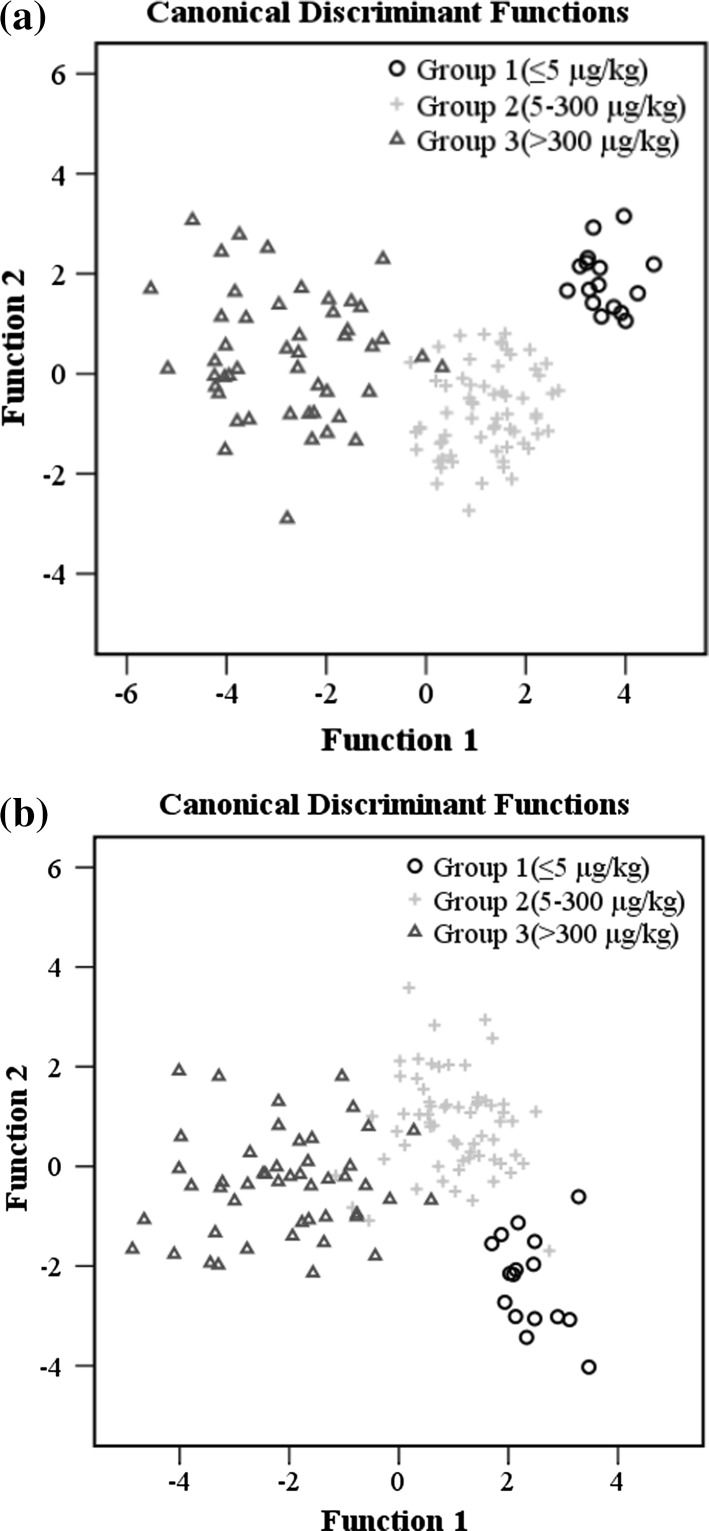
Because brown rice samples were not adequately separated according to predefined aflatoxin groups on the PCA score plot (data not shown), LDA scatter plots of two discrimination functions for samples with various AFB1 concentrations are shown in Fig. 2. The function variable is the combination of original variables that can enable us to discriminate among the groups (Shen et al. 2012). Brown rice samples with different levels of AFB1 could be successfully separated by NIR spectroscopy on the plot while the same group was clustered closely together. Towards the classification based on MIR spectroscopy, some overlapping between group 2 and 3 was observed. This study seems to indicate that NIR method might be slightly superior to MIR in terms of correct rate in classifying aflatoxin contaminated samples, which could be in part attributed to better reproducibility and repeatability of NIR spectrum regardless of broader bands and lower resolution. Overall, the clustering of samples on plots was consistent with the results of correct classification rates for the LDA models. Regions and degree of overlap among aflatoxin groups appeared to be related to the misclassification rate of relevant groups. On the whole, the result indicated that samples with varying levels of aflatoxins contamination could be properly distinguished by LDA based on NIR and MIR spectroscopy.
Fig. 2.
LDA score plots of samples with different levels of AFB1 based on NIR (a) and MIR (b) spectroscopy
Development and validation of PLSR models for quantification
As indicated above, eighty-six (86) samples were selected for training the model, while the remaining 42 samples were used for model validation. Both PLSR and SMLR algorithms were employed for establishment of aflatoxin quantification models. In comparison with SMLR, the PLSR models yielded higher predictive precision, better regression quality and lower error rate in general (data not shown). A summary of the statistics of calibration, cross-validation, and external validation based on PLSR algorithm is thus shown in Table 3.
Table 3.
A summary of PLSR calibration and validation results for the quantification of aflatoxins obtained by NIR and MIR spectroscopy
| Chemo-metrics | r c | RMSECV (μg/kg) | r v | RMSEP (μg/kg) | RPD | Factors | Validation sets | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Slope | Intercept | Bias (μg/kg) | ||||||||
| AFB1 | NIR | 0.981 | 105.0 | 0.973 | 70.4 | 4.0 | 10 | 1.010 | 21.98 | 24.786 |
| MIR | 0.980 | 100.0 | 0.970 | 70.8 | 4.0 | 8 | 0.931 | 39.75 | 21.368 | |
| AFB2 | NIR | 0.958 | 6.4 | 0.969 | 4.7 | 3.3 | 8 | 1.080 | 0.74 | 1.930 |
| MIR | 0.982 | 5.9 | 0.965 | 4.7 | 3.3 | 8 | 1.029 | 1.50 | 16.668 | |
| AFG1 | NIR | 0.941 | 180.0 | 0.936 | 144.0 | 2.8 | 8 | 0.934 | 54.01 | 29.642 |
| MIR | 0.932 | 198.0 | 0.922 | 155.0 | 2.6 | 5 | 0.816 | 83.26 | 14.709 | |
| AFG2 | NIR | 0.939 | 13.1 | 0.945 | 9.8 | 2.5 | 8 | 1.077 | 1.61 | 3.449 |
| MIR | 0.909 | 14.2 | 0.932 | 9.9 | 2.5 | 4 | 0.954 | 5.09 | 4.024 | |
| AFs | NIR | 0.950 | 299.0 | 0.951 | 231.0 | 3.1 | 8 | 0.967 | 77.53 | 55.720 |
| MIR | 0.975 | 302.0 | 0.948 | 232.0 | 3.1 | 8 | 0.915 | 111.00 | 54.190 | |
For PLSR models, high correlation and good predictive accuracy were obtained in calibration and validation based on NIR spectra (Table 3). In calibration, good calibration statistics were obtained for AFB1, B2, G1, G2 and AFs (rc > 0.930). However, relatively high values of RMSECV for AFB1, G1 and AFs were observed, which might be related to the wide range of reference values. The chemical composition with a small range of variation usually leads to a low RMSECV value, and vice versa (ElMasry et al. 2012). In validation, high values of rv and slope were obtained for all models, which indicated good regression quality for aflatoxins quantification. The AFB1 model showed the best accuracy and robustness in prediction (rv = 0.970, RPD = 4.0). The AFB2 and AFs models also yielded high rv (0.951–0.969) and RPD (3.1–3.3) values. Referring to the criteria used by other authors, RPD value greater than 3.0 is considered to be indicative of excellent prediction, whereas between 2.5 to 3.0 denotes a good prediction (Chang et al. 2001). Therefore, the above three models are considered to be suitable for quantify the aflatoxin concentrations. The model performance for AFG1 and AFG2 was slightly lower (rv = 0.936–0.945, RPD = 2.5–2.8), which presented general robustness and might be adequate for screening purpose.
The detailed statistics of calibration and validation based on MIR in the range of 1800–600 cm−1 was also given in Table 3. It could be observed that PLSR models also yielded better regression quality and lower error than those of SMLR models. For PLSR models, similar liner correlation (rc > 0.900) were obtained for AFB1, B2, G1, G2 and AFs in calibration. The models yielded higher RMSECV values for AFG1, G2 and AFs compared to those for NIR, but slightly lower values for AFB1 and AFB2. In validation, the models for AFB1, B2, G2 and AFs presented good accuracy (rv = 0.948–0.970, slope = 0.915–1.029). Quantification of AFG1 were slightly less accurate (rv = 0.922, slope = 0.816). RPD values obtained for AFB1, B2, G2 and AFs models were the same as those for NIR (RPD = 2.5–4.0). But the RPD value obtained for AFG1 was slightly low (RPD = 2.6). Attending to criterion of RPD values described above, PLSR models might be effective for screening of samples contaminated with aflatoxin. Figure 3 showed the correlation between the values determined by HPLC reference analysis and the values predicted by NIR and MIR spectroscopy on the validation sample set. The performance of NIR spectroscopy was found to be slightly superior to MIR in this work. However, it should be much cautious to decide which method was better. MIR spectroscopy, with higher sensitivity and resolution, might present better results if more considerations could be paid on sample preparation, scanning area, feature extraction and analysis efficiency. To investigate the correlation between the spectra and aflatoxin concentration, regression coefficients of the PLSR models were analyzed and were shown in supplementary data. It was found that the curves for all aflatoxins displayed the same pattern, which might be due to similar chemical structure of aflatoxins. In addition, the wavelengths with high regression coefficient were found to be consistent with the main absorption bands observed in original spectra. The result indicated that these wavelength regions played important roles for the development of calibration models.
Fig. 3.
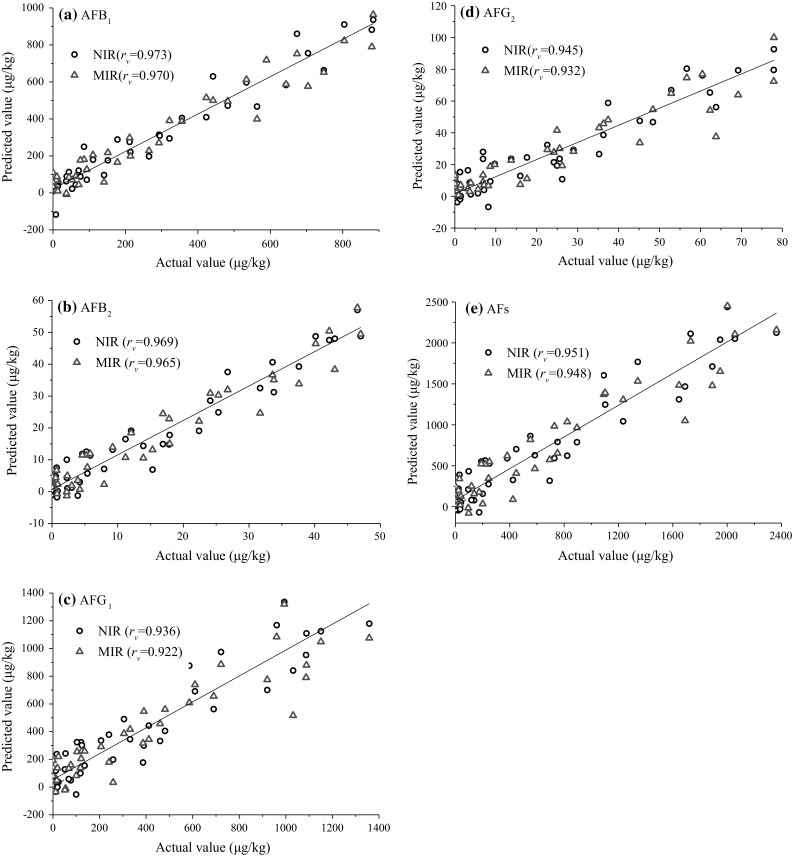
Linear regression plots of NIR and MIR predicted versus reference aflatoxin values of the external validation sample set
On the whole, both NIR and MIR models displayed good prediction capability for detection and screening of aflatoxin contamination in rice samples. However, the accurate prediction of very low aflatoxin concentration in some brown rice samples due to lager relative errors obtained, similar to the result reported in the literature (Lee et al. 2015). The reasons could be (1) associated with inhomogeneous particle size and aflatoxin distribution over this sample batch, (2) in part explained by inability of infrared spectra to detect internal compounds, inherently weak and missing vibrations of chemical groups in spectroscopy. Therefore, in order to improve predictive accuracy and minimize the prediction error of spectral models, representation and repeatability of spectrum need to be further promoted by means of larger and multiple scanning area of sample, homogenization of particle size, multiple location of sample collection, as well as consistent status of sample placement and detection. In addition, in compared to previous studies, which evaluated the feasibility for aflatoxin qualitative and quantitative analysis at similar or lower concentration ranges in some agricultural products using infrared spectroscopy, higher or comparable correct discrimination rate and predictive precision were obtained in this work (Kaya-Celiker et al. 2014; Lee et al. 2015). Besides, this study indicated that NIR and MIR methods offered some feasibility for simultaneous detection of aflatoxin B1, B2, G1 and G2 in brown rice, which also provided meaningful theoretical references for simultaneous analysis of several different mycotoxins in food and agricultural products.
Conclusion
The present study indicated that both NIR and MIR spectroscopy had the potential as rapid and non-destructive tools to quantify and/or discriminate aflatoxin contamination in brown rice over standard wet chemical methods, although certain spectral stability and detection limit need to be further improved. The LDA and PLSR models yielded a robust and satisfactory predictive ability for simultaneous detection of several aflatoxins. Both NIR and MIR spectroscopy appeared to have quite similar performance in quantification of aflatoxins with good regression quality. However, the problem of spectral change caused by inhomogeneous aflatoxin distribution, differences of sampling and sample batch as well as variation of surrounding environment should be solved or reduced, as the development of new sample scanning techniques and implementation of more efficient algorithms. Besides, in order to verify and evaluate the practicability of NIR and MIR methods, more studies focused on on-line detection of aflatoxin in agricultural products need to be investigated further. With the development of infrared spectrometer and chemometrics, NIR and MIR methods would be potentially powerful tools for real-time monitoring and rapid screening of high-throughput samples contaminated with aflatoxin in grain supply chains, which could greatly help improve the quality and safety of our food supply.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors gratefully acknowledge the financial support provided by the National Natural Science Foundation of China (No. 31772061 and 31301482), Key Research and Development Program of Zhejiang Province (No. 2018C02050), Jiangsu Agriculture Science and Technology Innovation Fund (CX(17)1003) and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13197-018-3033-1) contains supplementary material, which is available to authorized users.
References
- Abd-Elghany SM, Sallam KI. Rapid determination of total aflatoxins and ochratoxins A in meat products by immuno-affinity fluorimetry. Food Chem. 2015;179:253–256. doi: 10.1016/j.foodchem.2015.01.140. [DOI] [PubMed] [Google Scholar]
- Abramović B, Jajić I, Abramović B, Ćosić J, Jurić V. Detection of deoxynivalenol in wheat by Fourier transform infrared spectroscopy. Acta Chim Slov. 2007;54:859–867. [Google Scholar]
- Bellon-Maurel V, McBratney A. Near-infrared (NIR) and mid-infrared (MIR) spectroscopic techniques for assessing the amount of carbon stock in soils–critical review and research perspectives. Soil Biol Biochem. 2011;43:1398–1410. doi: 10.1016/j.soilbio.2011.02.019. [DOI] [Google Scholar]
- Berardo N, Pisacane V, Battilani P, Scandolara A, Pietro A, Marocco A. Rapid detection of kernel rots and mycotoxins in maize by near-infrared reflectance spectroscopy. J Agric Food Chem. 2005;53:8128–8134. doi: 10.1021/jf0512297. [DOI] [PubMed] [Google Scholar]
- Binder EM. Managing the risk of mycotoxins in modern feed production. Anim Feed Sci Tech. 2007;133:149–166. doi: 10.1016/j.anifeedsci.2006.08.008. [DOI] [Google Scholar]
- Bro R, Smilde AK. Principal component analysis. Anal Methods. 2014;6:2812–2831. doi: 10.1039/C3AY41907J. [DOI] [Google Scholar]
- Chang CW, Laird D, Mausbach MJ, Hurburgh CR. Near-infrared reflectance spectroscopy-principal components regression analysis of soil properties. Soil Sci Soc Am J. 2001;65:480–490. doi: 10.2136/sssaj2001.652480x. [DOI] [Google Scholar]
- ElMasry G, Sun DW, Allen P. Near-infrared hyperspectral imaging for predicting colour, pH and tenderness of fresh beef. J Food Eng. 2012;110:127–140. doi: 10.1016/j.jfoodeng.2011.11.028. [DOI] [Google Scholar]
- Elzupir AO, Alamer AS, Dutton MF. The occurrence of aflatoxin in rice worldwide: a review. Toxin Rev. 2015;34:37–42. doi: 10.3109/15569543.2014.984229. [DOI] [Google Scholar]
- Fernández-Ibañez V, Soldado A, Martínez-Fernández A, De Roza-Delgado B. Application of near infrared spectroscopy for rapid detection of aflatoxin B1 in maize and barley as analytical quality assessment. Food Chem. 2009;113:629–634. doi: 10.1016/j.foodchem.2008.07.049. [DOI] [Google Scholar]
- Ferreira DS, Galao OF, Pallone JAL, Poppi RJ. Comparison and application of near-infrared (NIR) and mid-infrared (MIR) spectroscopy for determination of quality parameters in soybean samples. Food Control. 2014;35:227–232. doi: 10.1016/j.foodcont.2013.07.010. [DOI] [Google Scholar]
- Gaspardo B, Del Zotto S, Torelli E, Cividino SR, Firrao G, Della Riccia G, Stefanon B. A rapid method for detection of fumonisins B1 and B2 in corn meal using Fourier transform near infrared (FT-NIR) spectroscopy implemented with integrating sphere. Food Chem. 2012;135:1608–1612. doi: 10.1016/j.foodchem.2012.06.078. [DOI] [PubMed] [Google Scholar]
- Gnonlonfin GJ, Hell K, Adjovi Y, Fandohan P, Koudande DO, Mensah GA, Sanni A, Brimer L. A review on aflatoxin contamination and its implications in the developing world: a sub-saharan african perspective. Crit Rev Food Sci Nutr. 2013;53:349–365. doi: 10.1080/10408398.2010.535718. [DOI] [PubMed] [Google Scholar]
- Hossain MZ, Goto T. Near- and mid-infrared spectroscopy as efficient tools for detection of fungal and mycotoxin contamination in agricultural commodities. World Mycotoxin J. 2014;7:507–515. doi: 10.3920/WMJ2013.1679. [DOI] [Google Scholar]
- Hruska Z, Yao H, Kincaid R, Brown R, Cleveland T, Bhatnagar D. Fluorescence excitation-emission features of aflatoxin and related secondary metabolites and their application for rapid detection of mycotoxins. Food Bioprocess Tech. 2014;7:1195–1201. doi: 10.1007/s11947-014-1265-2. [DOI] [Google Scholar]
- Huang LC, Zheng N, Zheng BQ, Wen F, Cheng JB, Han RW, Xu XM, Li SL, Wang JQ. Simultaneous determination of aflatoxin M1, ochratoxin A, zearalenone and α-zearalenol in milk by UHPLC–MS/MS. Food Chem. 2014;146:242–249. doi: 10.1016/j.foodchem.2013.09.047. [DOI] [PubMed] [Google Scholar]
- Kaya-Celiker H, Mallikarjunan PK, Schmale D, Christie ME. Discrimination of moldy peanuts with reference to aflatoxin using FTIR-ATR system. Food Control. 2014;44:64–71. doi: 10.1016/j.foodcont.2014.03.045. [DOI] [Google Scholar]
- Khanmohammadi M, Garmarudi AB, Guardia MDL. Feature selection strategies for quality screening of diesel samples by infrared spectrometry and linear discriminant analysis. Talanta. 2013;104:128–134. doi: 10.1016/j.talanta.2012.11.032. [DOI] [PubMed] [Google Scholar]
- Kirkin C, Mitrevski B, Gunes G, Marriott PJ. Combined effects of gamma-irradiation and modified atmosphere packaging on quality of some spices. Food Chem. 2014;154:255–261. doi: 10.1016/j.foodchem.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Kos G, Lohninger H, Krska R. Fourier transform mid-infrared spectroscopy with attenuated total reflection (FT-IR/ATR) as a tool for the detection of Fusarium fungi on maize. Vib Spectrosc. 2002;29:115–119. doi: 10.1016/S0924-2031(01)00196-5. [DOI] [Google Scholar]
- Kos G, Lohninger H, Krska R. Development of a method for the determination of Fusarium fungi on corn using mid-infrared spectroscopy with attenuated total reflection and chemometrics. Anal Chem. 2003;75:1211–1217. doi: 10.1021/ac0260903. [DOI] [PubMed] [Google Scholar]
- Lee KM, Davis J, Herrman TJ, Murray SC, Deng Y. An empirical evaluation of three vibrational spectroscopic methods for detection of aflatoxins in maize. Food Chem. 2015;173:629–639. doi: 10.1016/j.foodchem.2014.10.099. [DOI] [PubMed] [Google Scholar]
- Liu BH, Hsu YT, Lu CC, Yu FY. Detecting aflatoxin B1 in foods and feeds by using sensitive rapid enzyme-linked immunosorbent assay and gold nanoparticle immunochromatographic strip. Food Control. 2013;30:184–189. doi: 10.1016/j.foodcont.2012.07.008. [DOI] [Google Scholar]
- Liu TB, Zhou Y, Zhu YB, Song MJ, Li BB, Shi Y, Gong JY. Study of the rapid detection of γ-aminobutyric acid in rice wine based on chemometrics using near infrared spectroscopy. J Food Sci Technol. 2015;52(8):5347–5351. doi: 10.1007/s13197-014-1576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marutoiu C, Puiu S, Moise MI, Soran L, Marutoiu OF, Bobos L. Optimization of the separation of some aflatoxins by thin-layerchromatography planar. J Chromatogr A. 2004;17:372–374. [Google Scholar]
- Mirghania MES, Mana YBC, Jinapb S, Baharina BS, Bakara J. A new method for determining aflatoxins in groundnut and groundnut cake using Fourier transform infrared spectroscopy with attenuated total reflectance. J Am Oil Chem Soc. 2001;78:985–992. doi: 10.1007/s11746-001-0376-y. [DOI] [Google Scholar]
- Oliveira GAD, Castilhos FD, Bureau S. Comparison of NIR and MIR spectroscopic methods for determination of individual sugars, organic acids and carotenoids in passion fruit. Food Res Int. 2014;60:154–162. doi: 10.1016/j.foodres.2013.10.051. [DOI] [Google Scholar]
- Rajer-Kanduc K, Zupan J, Majcen N. Separation of data on the training and test set for modelling: a case study for modelling of five colour properties of a white pigment. Chemom Intell Lab. 2003;65:221–229. doi: 10.1016/S0169-7439(02)00110-7. [DOI] [Google Scholar]
- Regulation (EU) No 165/2010. Commission regulation (EC) No 165/2010 of 26 February (27.2.2010). Amending regulation (EU) No 1881/2006 setting maximum levels for certain contaminants in foodstuffs. Off J Eur Union L50/8eL50/12
- Shen F, Li F, Liu D, Xu H, Ying Y, Li B. Ageing status characterization of Chinese rice wines using chemical descriptors combined with multivariate data analysis. Food Control. 2012;25:458–463. doi: 10.1016/j.foodcont.2011.11.019. [DOI] [Google Scholar]
- Shi T, Cui L, Wang J, Fei T, Chen Y, Wu G. Comparison of multivariate methods for estimating soil total nitrogen with visible/near-infrared spectroscopy. Plant Soil. 2013;366:363–375. doi: 10.1007/s11104-012-1436-8. [DOI] [Google Scholar]
- Tripathi S, Mishra HN. A rapid FT-NIR method for estimation of aflatoxin B1 in red chili powder. Food Control. 2009;20:840–846. doi: 10.1016/j.foodcont.2008.11.003. [DOI] [Google Scholar]
- USFDA (2009) Action levels for aflatoxins in animal feeds. In: Vol. Sec. 683.100 (CPG7126.33)
- Wang YW, Ding W, Kou LP, Li L, Wang C, Jurick WM., II A non-destructive method to assess freshness of raw bovine milk using FT-NIR spectroscopy. J Food Sci Technol. 2015;52(8):5305–5310. doi: 10.1007/s13197-014-1574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LX, Ding XX, Li PW, Du XH, Zhou HY, Bai YZ, Zhang LX. Aflatoxin contamination of peanuts at harvest in China from 2010 to 2013 and its relationship with climatic conditions. Food Control. 2016;60:117–123. doi: 10.1016/j.foodcont.2015.06.029. [DOI] [Google Scholar]
- Xu H, Liu ZC, Cai WS, Shao XG. A wavelength selection method based on randomization test for near-infrared spectral analysis. Chemom Intell Lab. 2009;97:189–193. doi: 10.1016/j.chemolab.2009.04.006. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.