Abstract
Cephalopods are primarily active predators throughout life. Flying squids (family Ommastrephidae) represents the most widely distributed and ecologically important family of cephalopods. While the diets of adult flying squids have been extensively studied, the first feeding diet of early paralarvae remains a mystery. The morphology of this ontogenetic stage notably differs from other cephalopod paralarvae, suggesting a different feeding strategy. Here, a combination of Laser Capture Microdissection (LCM) and DNA metabarcoding of wild-collected paralarvae gut contents for eukaryotic 18S v9 and prokaryotic 16S rRNA was applied, covering almost every life domain. The gut contents were mainly composed by fungus, plants, algae and animals of marine and terrestrial origin, as well as eukaryotic and prokaryotic microorganisms commonly found in fecal pellets and particulate organic matter. This assemblage of gut contents is consistent with a diet based on detritus. The ontogenetic shift of diet from detritivore suspension feeding to active predation represents a unique life strategy among cephalopods and allows ommastrephid squids to take advantage of an almost ubiquitous and accessible food resource during their early stages. LCM was successfully applied for the first time to tiny, wild-collected marine organisms, proving its utility in combination with DNA metabarcoding for dietary studies.
Introduction
Cephalopods are active carnivorous predators, with only a few exceptions: Nautilus spp. are mainly scavengers and opportunistic predators (e.g.,1,2), the vampire squid Vampyroteuthis infernalis is a detritivore3, and the mesopelagic Ram’s horn squid Spirula spirula feeds mainly on detritus and zooplankton4. The remaining 845 species described to date1,5 are active predators6 and their diets are mainly known from studies on their subadult and adult forms. Cephalopods can hatch as large juveniles similar to the adult in morphology, habitat and feeding habits, or may have a less developed planktonic form, known as paralarvae, usually with a different lifestyle than the adults7,8. The behavior and diet of cephalopod hatchlings reported to date has demonstrated their active predatory habits from hatching (e.g.,9,10), however, this knowledge is mainly based on coastal shallow-water species, due to availability for sampling and laboratory maintenance11.
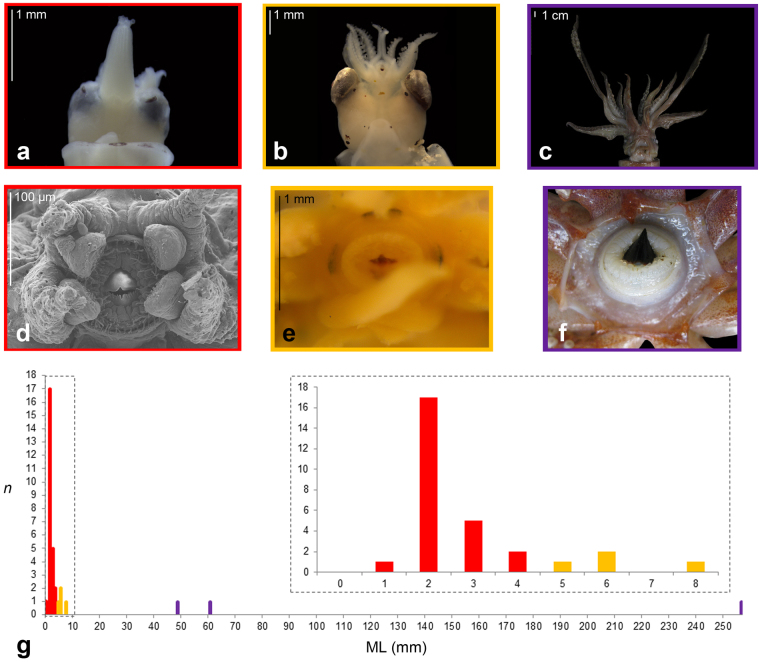
The squid Family Ommastrephidae is currently formed by 22 oceanic species, and represents one of the most widely distributed and ecologically important families of cephalopods12. Due to their huge biomass in the oceanic realm, they support some of the largest invertebrate fisheries13 and represent nearly 50% of the total cephalopod biomass fished worldwide14. The characteristic paralarva of ommastrephids, known as rhynchoteuthion, is characterized by the fusion of both tentacles into a proboscis (Fig. 1a), the function of which is unknown15. Along the ontogeny of the squid, the proboscis starts to split16 (Fig. 1b) and eventually becomes two independent raptorial tentacles (Fig. 1c), used for prey capture. Newly hatched paralarvae are provided with numerous filamentous buccal papillae around the mouth17 (Fig. 1d), which become less abundant as the paralarvae grow until they totally disappear17 (Fig. 1e), coinciding with the split of the proboscis into raptorial tentacles16 (Fig. 1b,e). The function of these papillae is also unknown. For clarity, throughout the current work, the paralarvae prior to losing the buccal papilla are referred to as “early paralarvae” and after as “late paralarvae”.
Figure 1.
(a–f) Morphology of ommastrephid squids. (a) Early paralarva (individual E100) showing an unsplit proboscis. (b) Todarodes sagittatus late paralarva (individual E5) with the proboscis beginning to split. (c) Adult Ommastrephes sp. individual E3 with the two raptorial tentacles. (d) SEM frontal photomicrograph of a Illex coindetii early paralarva obtained by in vitro fertilization (after Fernández-Álvarez et al.,15), showing the buccal papillae around the mouth. (e) Buccal area of a Todarodes sagittatus late paralarva (individual E7). (f) Buccal area of a T. sagittatus subadult. (g) Histogram representing the size classes used in this study, vertical axis represents the number of individuals, the horizontal axis represents the mantle length (mm); red bars represent early paralarvae, while yellow and violet bars represent late paralarvae, and subadults and adults, respectively.
The diet of both subadult and adult ommastrephids has been extensively studied (e. g.,18,19), however, the diet of the early paralarvae remains unknown20,21. All attempts of ommastrephid paralarval rearing have been unsuccessful as paralarvae would not ingest any offered prey22,23. Studies on wild caught ommastrephid paralarvae did not provide recognizable prey20,21 until the proboscis began to split, the filamentous buccal papillae disappeared and the remains of crustaceans and cephalopods appeared in their stomach contents20,24. Interestingly, Vidal and Haimovici24 observed a great diversity of microorganisms (dinoflagellates, flagellates, ciliates, cysts and bacteria) on the paralarva mucus cover, on the proboscis suckers and in the digestive tracts of the early ommastrephid paralarvae. They suggested that this mucus may act as a substrate for microbial growth that paralarvae may use this as food and ingest it with the aid of the proboscis. Other authors suggested that ommastrephid paralarvae feed on suspended particles by using the mucus cover of the body25, but they did not provide further evidence.
In recent years, studies using molecular tools for identifying gut contents have become more common26, especially since Next-Generation Sequencing (NGS) methods became more affordable. Based on this approach, DNA metabarcoding of gut samples is a powerful approach to identify prey remains27–29. Particularly, the high number of reads that NGS platforms produce allows the detection of DNA traces or underrepresented prey, highly improving the understanding of the diet of the focal species28. Despite these advantages, co-amplification of the target species (self-contamination, hereafter) is an important problem. The key factor is to avoid amplification of the target species, which may be the major27,29 or only component of the gut content reads30. A number of methods have been selected to overcome this problem, such as the use of primers specific to the prey species31. However, this method may only serve to increase the previously extant bias in our knowledge (or belief) about the predator diet26 and it cannot be applied when no previous knowledge is available, as in the case of ommastrephid paralarvae. PCR enrichment methods are based on a combination of amplified products with restriction enzymes32, DNA blockers33 or peptide nucleic acid clamps34,35. Nevertheless, Piñol et al.27 stressed the fact that such blocking molecules are not necessary given the huge number of sequence reads obtained by NGS platforms, which are sufficient to study the diet of focal species even if its DNA co-amplifies.
A critical step that can help diminish self-contamination of the target species is to decrease the amount of predator tissue as much as possible during gut content dissections or extractions. Although this step seems straightforward in large animals, it is not easy to achieve in some tiny organisms, such as small larvae or juveniles of marine animals, which may measure from <1 mm to a few centimeters. Until now, the best dissection method applied to tiny wild-collected marine organisms is syringing of the gut contents36. However, the Laser-Capture Microdissection (LCM) method allows the selection of particular tissues or cells from histological sections37 and thus, it is a promising method for gut content extraction from tiny animals. Nevertheless, for dietary studies LCM has only tentatively been applied for aquaria-reared cod larvae with a previously known diet, and specific prey primers were applied38. Here, we applied LCM gut content dissections in combination with DNA metabarcoding for the first time to assess the first feeding diet of wild-collected ommastrephid squid paralarvae.
Results
Taxonomic assignment of eukaryotic reads
Self-contamination reads represented 88.5% of the 2,587,082 reads obtained for 18S v9. Supplementary Fig. S1 represents the percentage of self-contamination and gut contents of each sample successfully sequenced for this molecular marker. For the early paralarvae (n = 25), self-contamination was 78 ± 30% of the reads (range: 0–100%). Four gut samples (E3 to E7) failed to provide any 18S v9 reads matching the SILVA database by the closed-reference approach in Qiime. Whereas the adult individual E3 did not provide any read, the late T. sagittatus paralarvae E5 to E7 provided 4,814–208,655 identical sequences that were discarded by the software because they did not match any sequences in the database. A subsequent analysis revealed that these reads matched the 18S v9 Sanger sequenced T. sagittatus sequence MF980452, resulting in a self-contamination value of 100%. The LCM-dissected individual E0 was the only late paralarva whose gut contents were successfully sequenced for 18S v9, with a self-contamination percentage of 87.6%. The subadult individuals E1 and E2 were successfully amplified for 18S v9 and showed self-contamination values of 51.0 and 96.7%, respectively.
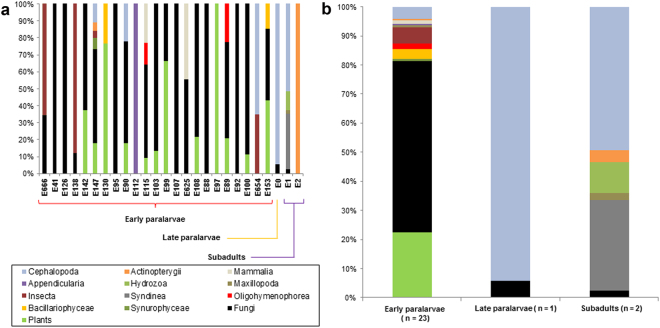
After cleaning the self-contamination reads, 299,509 total reads of eukaryotes remained, resulting in 11,519 ± 9,331 (range 1,089–31,566) reads per sample. A total of 59 molecular operational taxonomic units (MOTUs) were identified in the gut contents of all the samples. The percentages of each gut content item of each sample and size class are represented in Fig. 2a and b, respectively, and are summarized by class size in Table 1. The raw gut content reads are available in Supplementary Table S1. Early paralarvae shows 3 ± 2.2 (range 1–11, n = 23) MOTUs, the late paralarva E0 showed 3 MOTUs, and the 2 subadults, 1 and 9 different MOTUs. For the early paralarvae, 22.3% of the gut content reads was composed of plants and 59% was fungi. Animals accounted for 12.6% of the reads, with insects (5.5%) and cephalopods (4.2%) being the most represented groups. The protist groups Chromista and Ciliophora were also present in this size class. The most represented group for the late paralarva and subadults was Metazoa, representing 94 and 66%, respectively. In both the late paralarva and subadults, cephalopods were the most represented group (87.8 and 49.3%, respectively). Parasitic dinoflagellates of the Class Syndinea were only present in the subadult squid E1, representing 31% of the reads of this size class.
Figure 2.
Percentage (%) of eukaryotic 18S v9 reads in the gut contents of each sample (a) and grouped by size class (b). The taxonomic assignments are at the Class level except plants and fungi, which were collapsed. Self-contamination reads were excluded. Individuals are ordered by mantle length.
Table 1.
18S v9 eukaryotic MOTUs detected in the gut contents of ommastrephid squids as a percentage (%) and clustered by size categories.
| Kingdom | Phyllum | Class | Early paralarvae (n = 23) | Late paralarva (n = 1) | Subadults (n = 2) | ||
|---|---|---|---|---|---|---|---|
| Reads (%) | Count | Reads (%) | Reads (%) | Count | |||
| Plantae | Magnoliophyta | N/A | 3.38 | 2 | |||
| Eudicotyledoneae | 0.69 | 1 | |||||
| Magnoliopsida sp. 1 | 0.25 | 1 | |||||
| Magnoliopsida sp. 2 | 0.82 | 2 | |||||
| Magnoliopsida sp. 3 | 9.95 | 3 | |||||
| Magnoliopsida sp. 4 | 1.97 | 2 | |||||
| Magnoliopsida sp. 5 | 1.01 | 1 | |||||
| Magnoliopsida sp. 6 | 0.47 | 1 | |||||
| Monocotyledoneae sp. 1 | 0.71 | 1 | |||||
| Monocotyledoneae sp. 2 | 1.40 | 1 | |||||
| Rosopsida | 1.69 | 3 | |||||
| Fungi | N/A | N/A | 2.71 | 1 | |||
| Ascomycota | Dothideomycetes sp. 1 | 5.10 | 1 | ||||
| Dothideomycetes sp. 2 | 0.33 | 1 | |||||
| Eurotiomycetes sp. 1 | 4.89 | 1 | |||||
| Eurotiomycetes sp. 2 | 1.05 | 1 | |||||
| Eurotiomycetes sp. 3 | 0.29 | 1 | |||||
| Eurotiomycetes sp. 4 | 2.33 | 1 | 2.52 | 1 | |||
| Pezizomycetes | 0.33 | 1 | |||||
| Pleosporomycetidae sp. 1 | 0.42 | 1 | |||||
| Pleosporomycetidae sp. 2 | 0.64 | 1 | |||||
| Saccharomycetes | 12.59 | 2 | |||||
| Basidiomycota | Agaricomycetes sp. 1 | 4.55 | 2 | ||||
| Agaricomycetes sp. 2 | 0.27 | 1 | |||||
| Agaricomycetes sp. 3 | 0.23 | 1 | |||||
| Agaricomycetes sp. 4 | 1.05 | 1 | |||||
| Basidiomycetes | 1.30 | 1 | |||||
| Microbotryomycetes sp. 1 | 0.49 | 1 | |||||
| Microbotryomycetes sp. 2 | 2.42 | 1 | 5.68 | ||||
| Microbotryomycetes sp. 3 | 0.64 | 1 | |||||
| Microbotryomycetes sp. 4 | 3.05 | 4 | |||||
| Tremellomycetes sp. 1 | 8.60 | 5 | |||||
| Tremellomycetes sp. 2 | 0.56 | 1 | |||||
| Tremellomycetes sp. 3 | 2.04 | 1 | |||||
| Tremellomycetes sp. 4 | 0.64 | 1 | |||||
| Entomophthoromycota | Entomophthoraceae | 2.51 | 1 | ||||
| Chromista | Ochrophyta | Synurophyceae | 0.62 | 1 | |||
| Bacillariophyceae sp. 1 | 1.78 | 1 | |||||
| Bacillariophyceae sp. 2 | 1.73 | 1 | |||||
| Protista | Ciliophora | Oligohymenophorea sp. 1 | 1.50 | 1 | |||
| Oligohymenophorea sp. 2 | 0.42 | 1 | |||||
| Dinoflagellata | Syndinea sp. 1 | 2.53 | 1 | ||||
| Syndinea sp. 2 | 4.17 | 1 | |||||
| Syndinea sp. 3 | 3.14 | 1 | |||||
| Syndinea sp. 4 | 21.28 | 1 | |||||
| Metazoa | Arthropoda | Insecta sp. 1 | 3.83 | 1 | |||
| Insecta sp. 2 | 0.41 | 1 | |||||
| Insecta sp. 3 | 1.21 | 2 | |||||
| Maxillopoda sp. 1 | 0.71 | 1 | |||||
| Maxillopoda sp. 2 | 2.32 | 1 | |||||
| Chordata | Appendicularia | 0.55 | 1 | ||||
| Actinopterygii sp. 1 | 4.13 | 1 | |||||
| Actinopterygii sp. 2 | 0.46 | 1 | |||||
| Mammalia | 1.16 | 2 | |||||
| Cnidaria | Hydrozoa | 10.64 | 1 | ||||
| Mollusca | Cephalopoda sp. 1 (Ommastrephes sp.) | 6.53 | 6.59 | 1 | |||
| Cephalopoda sp. 2 (Illex sp.) | 42.68 | 1 | |||||
| Cephalopoda sp. 3 (Sthenoteuthis sp.) | 3.27 | 2 | |||||
| Cephalopoda sp. 4 (Eucleoteuthis luminosa/Dosidicus gigas) | 1.00 | 2 | 87.78 | ||||
| Total reads | 261,611 | 11,541 | 26,357 | ||||
Taxonomic assignments are at a Class level, with the exception of Cephalopoda, which are identified at a genus level. The count number indicates the number of individuals of each class size category with reads for the gut content item. N/A, not applicable.
Taxonomic assignment of prokaryotic reads
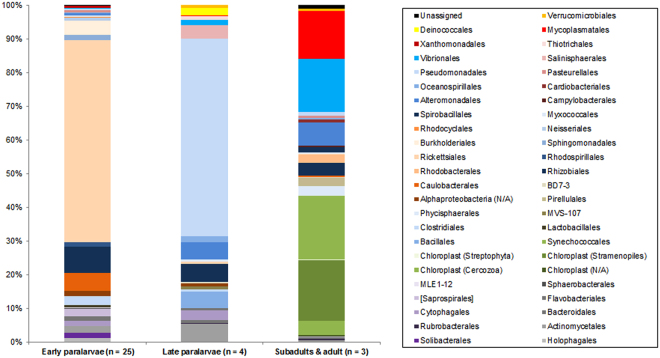
A total of 453,883 prokaryotic reads were obtained from the gut contents resulting in 14,183 ± 28,280 (range 12–124,004) reads per sample. A total of 141 different MOTUs were identified, with three of them unassigned to any taxonomic level (2,608 reads in total). The percentages of each gut content item at the Order level are represented in Supplementary Fig. S2 for each sample. Percentages of each bacterial Order grouped by size class are represented in Fig. 3 and in Table 2. The raw data are available in Supplementary Table S2. For early paralarvae (n = 25), the most represented group was the Class Proteobacteria (86% of the reads). The Proteobacteria Order Rickettsiales represented 60% of the reads. Some bacterial groups commonly found in Particulate Organic Material (POM), such as Cytophagia, Deltaproteobacteria, Flavobacteria and Firmicutes39 were present in early paralarvae gut content. The autotrophic component (Cyanobacteria and chloroplasts) represented 0.3% of the reads of early paralarvae. The Phylum Acidobacteria was only present in early paralarvae (2.6%), while Planctomycetes, present in the other two size categories, was absent in early paralarvae. For late paralarvae (n = 4), the Class Proteobacteria was the most represented group (80%) and the autotrophic component represented 0.05% of the reads. For subadults and the adult (n = 3), Cyanobacteria and chloroplasts were the most represented groups (42%), while Proteobacteria accounted for 35% of the reads and the parasitic Mycoplasmatales for 14%. Since the small size of unicellular Cyanobacteria prevent its selected ingestion by subadults and adults, these sequences can only be explained by the ingestion of food items enriched with these organisms, suggesting predation over herbivores.
Figure 3.
Percentage (%) of the prokaryotic 16S reads in the gut contents grouped by size class. The taxonomic assignments are at the Order level. Chloroplast sequences are eukaryotic chloroplasts amplified with this molecular marker. N/A, not applicable (the finest identification was at the class level).
Table 2.
16S prokaryotic MOTUs detected in the gut contents of ommastrephid squids as a percentage (%) and sorted by size category.
| Kingdom | Phyllum | Class | Order | Early paralarvae (n = 25) reads (%) | Late paralarvae (n = 4) reads (%) | Subadults & adult (n = 3) reads (%) |
|---|---|---|---|---|---|---|
| Bacteria | Acidobacteria | Holophagae | Holophagales | 1.0354 | ||
| Solibacteres | Solibacterales | 1.6018 | ||||
| Actinobacteria | Actinobacteria | Actinomycetales | 2.0923 | 5.3699 | 0.5777 | |
| Rubrobacteria | Rubrobacterales | 0.2250 | 0.2582 | |||
| Bacteroidetes | Bacteroidia | Bacteroidales | 0.0005 | 0.9424 | ||
| Cytophagia | Cytophagales | 1.4985 | 2.7949 | |||
| Flavobacteriia | Flavobacteriales | 1.3095 | 0.7464 | 0.4268 | ||
| [Saprospirae] | [Saprospirales] | 2.2642 | 0.0017 | 0.3750 | ||
| Chloroflexi | Sphaerobacteridae | Sphaerobacterales | 0.1870 | |||
| Cyanobacteria | 4C0d-2 | MLE1-12 | 0.2908 | |||
| Chloroplast | N/A | 0.3389 | ||||
| Cercozoa | 0.0005 | 0.0051 | 4.1795 | |||
| Stramenopiles | 0.0020 | 0.0119 | 18.0005 | |||
| Streptophyta | 0.0010 | 0.2289 | ||||
| Synechococcophycideae | Synechococcales | 0.0039 | 0.0375 | 18.9647 | ||
| Firmicutes | Bacilli | Bacillales | 4.7189 | |||
| Lactobacillales | 0.6359 | 0.1409 | ||||
| Clostridia | Clostridiales | 2.7018 | 0.6919 | |||
| Planctomycetes | C6 | MVS-107 | 0.8316 | |||
| Phycisphaerae | Phycisphaerales | 2.8827 | ||||
| Planctomycetia | Pirellulales | 0.0034 | 2.3244 | |||
| Proteobacteria | Alphaproteobacteria | N/A | 1.5773 | 0.8862 | ||
| BD7-3 | 0.0005 | 0.6374 | 0.1713 | |||
| Caulobacterales | 5.2989 | 0.0017 | 0.6002 | |||
| Rhizobiales | 7.6903 | 5.1518 | 3.6583 | |||
| Rhodobacterales | 0.0005 | 0.0136 | 2.4391 | |||
| Rhodospirillales | 1.3247 | |||||
| Rickettsiales | 60.1709 | 0.7601 | 0.1990 | |||
| Sphingomonadales | 1.5759 | |||||
| Betaproteobacteria | Burkholderiales | 4.1440 | 0.0068 | 0.1461 | ||
| Neisseriales | 0.8959 | |||||
| Rhodocyclales | 0.2286 | |||||
| Deltaproteobacteria | Myxococcales | 0.5189 | 0.7004 | 0.3734 | ||
| Spirobacillales | 1.4712 | |||||
| Epsilonproteobacteria | Campylobacterales | 0.0017 | 0.5651 | |||
| Gammaproteobacteria | Alteromonadales | 0.5488 | 4.9660 | 6.8196 | ||
| Cardiobacteriales | 0.0005 | 0.0034 | 0.9438 | |||
| Oceanospirillales | 0.3099 | 1.7860 | 0.4667 | |||
| Pasteurellales | 0.6124 | 0.0205 | 0.7228 | |||
| Pseudomonadales | 0.4773 | 58.7110 | 0.9014 | |||
| Salinisphaerales | 4.0406 | |||||
| Vibrionales | 0.1601 | 1.4997 | 15.8191 | |||
| Thiotrichales | 1.2066 | |||||
| Xanthomonadales | 0.6271 | |||||
| Tenericutes | Mollicutes | Mycoplasmatales | 0.0029 | 0.1432 | 14.3777 | |
| [Thermi] | Deinococci | Deinococcales | 2.2206 | |||
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | 0.8453 | 0.4907 | ||
| Unassigned | N/A | N/A | N/A | 0.2095 | 0.0170 | 1.1365 |
| Total reads | 204270 | 58679 | 190934 |
Taxonomic assignments are at the Order level. N/A, not applicable.
Discussion
The mixture of continental (insects, plants and freshwater algae) and exclusively marine animal DNA (appendicularians and cephalopods) in combination with single cell organisms (cyanobacteria, diatoms and ciliophorans), other organisms often associated with organic material degradation (fungi) and bacteria typically associated with POM and fecal pellets, strongly suggest that ommastrephid squids are detritivores during their early planktonic life. Similar assemblages of general gut content composition and protists taxa have been reported in other marine suspension feeders during their larval life, such as eel and spiny lobsters larvae34,35,40–42. Interestingly, these gut contents have not been previously reported in the literature for any other cephalopod paralarvae6,11,21,24,29. The detritus-based diet of early ommastrephid paralarvae is an unexpected finding, since posterior ontogenetic stages are voracious predators6,12. A first feeding diet based on detritus represents a unique life strategy among predatory cephalopods and is potentially one of the reasons for the ecological success of the Family Ommastrephidae in the oceanic realm. This ontogenetic shift in the diet agrees with the change in stable isotopic composition found between ommastrephid early paralarvae and adults in a previous work43. A detritivore diet would allow ommastrephid squids to take advantage of an almost ubiquitous and accessible food resource during their early stages, such that they do not directly compete with conspecifics of later ontogenetic stages for the same prey (even if they do predate on different ontogenetic stages of a particular species) or with other cephalopod paralarvae. Since detritus is almost ubiquitous44–46, competition for trophic resources between early ommastrephid paralarvae would also be minimal. The new knowledge provided in this work can be applied in the future to the development of experimental culture protocols for ommastrephid hatchlings obtained by in vitro fertilization47 or aquaria spawning48.
Identifying the diet of wild cephalopod paralarvae by DNA sequencing is a poorly studied topic. Only two previous studies exist, both working with coastal species whose paralarvae have a very different morphology and ecology, the common octopus Octopus vulgaris and the midsize squid Alloteuthis media29,49. As far as we know, no previous attempts to study the diet of ommastrephid squids by NGS sequencing have been made. Since no reliable knowledge on the diet of early ommastrephid paralarvae was available, a mixed approach based on sequencing the hypervariable eukaryotic 18S v9 and prokaryotic 16S rRNA was performed here, covering almost every life domain. This combination provided a good snapshot of the diet of early ommastrephid paralarvae. Although more specific eukaryotic metabarcodes are available, the spectrum of taxonomic groups they are able to amplify is usually narrower50. Thus, if one of these molecular markers was selected, many eukaryotic MOTUs would not be detected and the study may be critically biased, providing very different results and possibly misleading conclusions.
It should be noted that the bacteria found did not only come from the diet, since gut microbiomes of marine animals are formed by an enormous diversity of bacteria51. The cephalopod gut microbiome is poorly understood at present, but has recently gained attention in efforts to overcome mortality problems in laboratory reared paralarvae52. To the best of our knowledge, there is no previous work dealing with the gut microbiome of ommastrephid squids, either for paralarvae or later ontogenetic stages. The absence of this information precluded us from reliably distinguishing the bacteria that came from the diet from those that are common residents in the gut microbiome of squids. Similarly, some prokaryotic MOTUs may represent parasites, such as seven MOTUs of Mycoplasmataceae, which represented an important part of the subadult and adult reads (14%, Table 2, Supplementary Table S2). Although the prokaryotic data generated here (Table 2, Supplementary Table S2; Fig. 3 and Supplementary Fig. S2) are in the context of a dietary study, the results provided may aid in the understanding of the gut microbiome of ommastrephids when more directed studies are carried out and may bring to light the possible pathogens that infect these oceanic cephalopods.
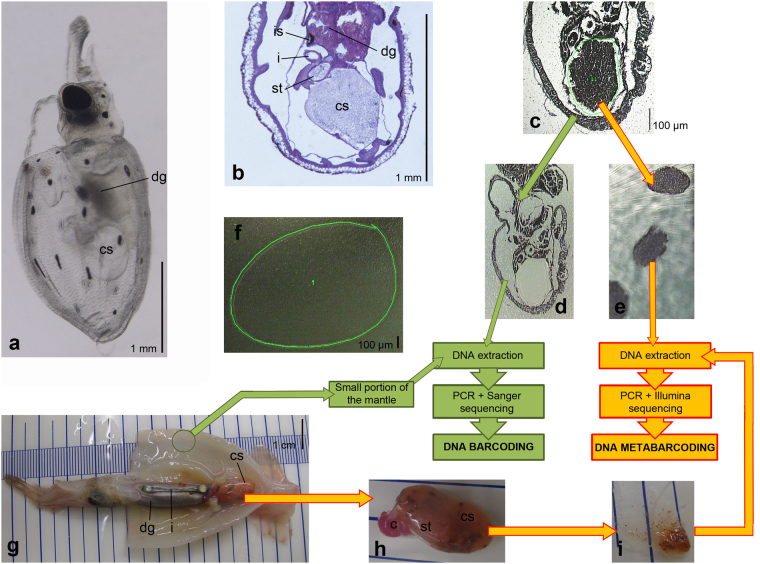
In this study, gut contents were successfully LCM-isolated from histological paralarvae sections (Fig. 4) and NGS sequencing was carried out with small portions of gut contents (Table 3) obtaining low values of self-contamination (Supplementary Fig. S1). Three late paralarvae were not LCM-processed (E5-E7, Table 3) and the whole digestive system was used in the DNA extraction. Despite the fact that these paralarvae revealed conspicuous gut contents in the digestive system during dissection, no reads were retrieved from the first bioinformatic analysis. A posterior bioinformatic analysis showed 100% self-contamination of T. sagittatus, a species not present in the database used (SILVA) (see in Material and Methods). Thus, the only possible explanation is that paralarvae tissues strongly prevail in the PCR product producing 100% of the reads during NGS sequencing. This indicates the importance of avoiding the inclusion of gut tissues from the focal species when performing dietary analyses. The low self-contamination reads obtained in this study for LCM-dissected paralarvae are unusual in the literature of dietary metabarcoding studies of tiny organisms with universal primers and without PCR enrichment methods, which usually show self-contamination values above 90% (e. g.,27,29). This is the first time LCM has been applied on wild-collected samples in a dietary study and our results are promising for applying this methodology to other tiny animals, even when universal primers are used without PCR enrichment methods.
Figure 4.
Diagram of the lab workflow. (a–f) LCM gut content extraction (late paralarva E0 and early paralarvae, Table 3). (g–i) Direct dissection of gut contents (subadult and adult individuals E1 to E3 and late paralarvae E5 to E7, Table 3). (a) Lateral view of a live hatchling of the ommastrephid squid Todaropsis eblanae, obtained by in vitro fertilization (after Fernández-Álvarez et al.,15). (b) Histological sagittal section of a T. eblanae paralarvae, showing the structure of the digestive system. (c) Sagittal section of the early paralarva E41 (Dosidicus gigas) mounted on the PEN slide during a LCM session; the green line encircles the area selected for laser cutting. (d) Same section as in subfigure C with the caecum sac contents LCM-excised. (e) Cuts of LCM-isolated gut contents of several sections of the paralarva E41. (f) PEN slide without tissues (blank), the green line shows the portion selected for laser cutting. (g) Subadult individual E2 (Sthenoteuthis pteropus) with the mantle opened to show the internal organs. (h) Caecum sac and caecum of individual E2. (i) Isolated gut contents by direct dissection. Abbreviations: c, caecum; cs, caecum sac; dg, digestive gland; i, intestine; is, ink sac; st, stomach.
Table 3.
Individuals studied ordered by mantle length (ML).
| Labcode | ML (mm) | Species | LCM dissected area (µm2) | Approximate weight of dissected gut content (g) | Observations |
|---|---|---|---|---|---|
| Early paralarvae | |||||
| E666 | 0.69 | Dosidicus gigas 1 | 315,441 | N/A | Paralarva stage 1 |
| E41 | 1.02 | Dosidicus gigas 1 | 1,375,292 | N/A | Paralarva stage 1 |
| E126 | 1.13 | SD complex2 | 846,826 | N/A | Paralarva stage 1 |
| E138 | 1.14 | SD complex2 | 55,824 | N/A | Paralarva stage 1 |
| E142 | 1.21 | Sthenoteuthis oualanensis 1 | 1,978,216 | N/A | Paralarva stage 1 |
| E147 | 1.29 | SD complex2 | 453,431 | N/A | Paralarva stage 1 |
| E130 | 1.39 | Sthenoteuthis oualanensis 1 | 219,731 | N/A | Paralarva stage 1 |
| E95 | 1.4 | SD complex2 | 2,509,496 | N/A | Paralarva stage 1 |
| E90 | 1.49 | Dosidicus gigas 1 | 3,033,288 | N/A | Paralarva stage 1 |
| E112 | 1.55 | SD complex2 | 1,893,110 | N/A | Paralarva stage 1 |
| E115 | 1.59 | SD complex2 | 1,181,529 | N/A | Paralarva stage 1 |
| E103 | 1.64 | Sthenoteuthis oualanensis 1 | 6,503,474 | N/A | Paralarva stage 1 |
| E99 | 1.67 | SD complex2 | 328,239 | N/A | Paralarva stage 1 |
| E107 | 1.74 | SD complex2 | 831,532 | N/A | Paralarva stage 1 |
| E625 | 1.88 | Dosidicus gigas 1 | 3,161,446 | N/A | Paralarva stage 1 |
| E108 | 1.9 | SD complex2 | 2,166,047 | N/A | Paralarva stage 1 |
| E88 | 1.91 | SD complex2 | 1,328,333 | N/A | Paralarva stage 1 |
| E97 | 1.91 | SD complex2 | 483,834 | N/A | Paralarva stage 1 |
| E89 | 2.06 | SD complex2 | 3,263,717 | N/A | Paralarva stage 1 |
| E626 | 2.15 | Dosidicus gigas 1 | 970,475 | N/A | Paralarva stage 1 |
| E92 | 2.17 | SD complex2 | 919,236 | N/A | Paralarva stage 1 |
| E100 | 2.29 | SD complex2 | 2,432,780 | N/A | Paralarva stage 1 |
| E654 | 2.75 | SD complex2 | 4,625,858 | N/A | Paralarva stage 2 |
| E153 | 3.23 | SD complex2 | 3,310,476 | N/A | Paralarva stage 1 |
| E510 | 3.75 | Dosidicus gigas 1 | 6,975,999 | N/A | Paralarva stage 1 |
| Late paralarvae | |||||
| E6 | 4.8 | Todarodes sagittatus 1 | N/A | No data | Paralarva stage 3 |
| E7 | 5.2 | Todarodes sagittatus 1 | N/A | No data | Paralarva stage 3 |
| E5 | 5.9 | Todarodes sagittatus 1 | N/A | No data | Paralarva stage 3 |
| E0 | 7.7 | Sthenoteuthis pteropus 1 | 3,300,000 | N/A | Paralarva stage 3 |
| Subadults and adult | |||||
| E1 | 49 | Sthenoteuthis pteropus 1 | N/A | 0.009 | Subadult |
| E2 | 61 | Sthenoteuthis pteropus 1 | N/A | 0.045 | Subadult |
| E3 | 257 | Ommastrephes sp.1,3 | N/A | 1.643 | Adult male |
| Extraction blanks | |||||
| B1 | N/A | N/A | 5,415,922 | N/A | Extraction blank 1 |
| B2 | N/A | N/A | 6,343,695 | N/A | Extraction blank 2 |
LCM, Laser Capture Microdissection; N/A, not applicable. Paralarvae stages after Shea15.
1DNA barcoded individual.
2Sthenoteuthis/Dosidicus species complex: there are no known morphological differences between the two species until S. oualanensis paralarvae develop their photophores (ca. 4 mm ML).
3Ommastrephes bartramii is a species complex according to Fernández-Álvarez et al.65 although the genus is currently considered monotypic12. We avoided providing a species-level identification until its taxonomic status is resolved.
Material and Methods
Sample collection
In total, 32 individuals were analyzed: 25 early paralarvae, 4 late paralarvae, 2 subadults and an adult (Table 3, Supplementary Table S3). Further information on sample collection and species identification is available in Supplementary Methods.
Gut contents extraction
Two methods were developed to extract the gut contents of the individuals according to their size: LCM for small paralarvae and direct dissections for larger squids (Fig. 4). Further information is available in Supplementary Methods.
DNA extraction
The DNA from the gut samples obtained by LCM dissections was extracted using the QIAamp DNA Investigator Kit (Qiagen) following the corresponding manufacturer’s protocols, and “Isolation of Total DNA from Tissues” for the remaining samples (Table 3). The samples were eluted twice in 30 µl and the second elution was stored at −20 °C as a back-up. Ambient contamination was avoided as much as possible working in the “Ancient DNA lab” of the Museo Nacional de Ciencias Naturales (MNCN-CSIC, Madrid, Spain), isolated from the other rooms and provided with UV light sterilization. Beyond the usual measures to avoid contamination in a molecular systematics lab, additional measures to avoid ambient contamination were: (1) the whole laboratory was cleaned and UV-sterilized before starting the work, and (2) no additional people were working in the same lab during the DNA extraction session.
For molecular identification of the individuals, the remaining tissues on the PEN slides (Fig. 4d) of the LCM-dissected paralarvae and a small portion of the mantle (Fig. 4g) of the remaining squids were dissected with a sterile blade. DNA was extracted using the BioSprint 15 DNA kit (Qiagen), following the manufacturer’s protocol.
DNA metabarcoding of gut contents
DNA extractions of gut contents were used to construct two libraries, for eukaryotic and prokaryotic DNA identification, covering almost all life domains. This strategy was selected due to of the absence of reliable knowledge of the actual diet of ommastrephid paralarvae. Further information on library preparation and sequencing is available in Supplementary Methods.
Bioinformatic analysis
The quality of the FASTQ files was checked using the software FastQC53 and the Illumina-specific adapters were trimmed by running the cut adapter tool implemented in Trimmomatic54. The sequences were quality-filtered (minimum Phred quality score of 20) and labeled using Qiime 1.9.055. The paired-end assembly of forward (R1) and reverse (R2) reads was executed in FLASH56 implemented in Qiime. The mismatch resolution in the overlapping region was accomplished by keeping the base with the higher quality score. Artifacts such as point mutations and chimaeras were detected and deleted using the UCHIME algorithm57 implemented in VSEARCH58. Using the final list of representative sequences, each molecular operational taxonomic unit (MOTU) was searched against the reference database SILVA59 v. 128 (September, 2016) and the last available version (May, 2013) of Greengenes60 for the 18S v9 and 16S databases, respectively. The 18S v9 reads were clustered into MOTUs using the closed-reference approach with the UCLUST algorithm61 in Qiime with a similarity threshold of 100%. Sequences that did not provide a 100% match were discarded. The 16S reads were clustered using the open-reference approach with a similarity threshold of 97% and reads that did not hit the reference sequence collection were subsequently clustered de novo. After this step, singletons and sequences representing less than 0.005% of the total number of sequences of each dataset were excluded. Sequences were compared with the GenBank database by BLAST62. For 16S, 30% of the reads remain unidentified using only Greengenes as a database. Thus, BLAST hits were applied to identify at the lowest taxonomic level possible following the same criteria: (1) sequences with <90% for identity or coverage were not considered; (2) 97% similarity is considered the species-level threshold; (3) when more than one sequence has the same identity value, the one identified to the lowest taxonomic level was selected; (4) if the GenBank identification differs at the genus level to that of Greengenes, the latter is applied. Before applying the BLAST identifications, only 0.6% of the sequences were unidentified.
In DNA metabarcoding studies, the mistagging phenomenon has been reported63,64, in which a low percentage of the reads of a sample can be assigned to another as the result of the misassignment of the indices during library preparation, sequencing, and/or demultiplexing steps. To correct for this phenomenon, for 18S v9 the low abundance MOTUs of each sample were removed by applying a threshold based on the presence of mistagging in the PCR negative control (i.e., the higher number of reads for a particular sequence in the PCR blank), resulting in a particular threshold for each sequence. As a result, for 18S v9 no sequences were assigned to three of the late paralarvae (individuals E5 to E7) and the adult (individual E3). For 16S, the 0.005% threshold was selected according to the presence of low abundance MOTUs in the whole dataset. Although extraction measures for avoiding ambient contamination were applied, some MOTUs were present in the extraction blanks. Any sequence present in at least one of these blank samples was taken as ambient contamination and removed from the study. For 18S v9, the identifications were performed at the Class level, since some Orders of some of the Classes (e.g., Mammalia and Actinopterigii) could not be reliably assessed with this region. It should also be noted that several related species may share the same metabarcode50 and thus, the number of actual eukaryotic species inside the gut may be larger than the number of detected MOTUs. Taxonomic assignments of 16S reads were considered as species-level identifications. Rarefaction plots of each sample were constructed showing the rarefied number of MOTUs defined at 100 and 97% similarity thresholds for 18S v9 and 16S, respectively (Supplementary Fig. S3).
The selected primers for 18S v9 can amplify ommastrephid squid DNA. Thus, although the dissection methodology was performed carefully, DNA of the paralarvae or the squids may be present among the gut contents and must be considered as self-contamination. In a first step, all the 18S v9 ommastrephid sequences available in GenBank were downloaded. An additional Todarodes sagittatus sequence obtained with the primers Euk-B and 18_v9_Con following the PCR conditions explained above and Sanger sequenced (GenBank accession number MF980452) was added. All of these sequences were aligned and the p-distance percentages were calculated to determine if there were sufficient differences to distinguish these species with this molecular marker (Supplementary Table S4). For those paralarvae successfully identified with DNA barcoding (Table 3), the reads that matched their identification at a genus level were regarded as self-contamination and the others as a component of the diet. For paralarvae that were not molecularly identified and had reads for only one ommastrephid species, the reads were considered as self-contamination reads. If an unidentified paralarva had sequences for two genera, the most represented was regarded as self-contamination and the other as gut contents. The self-contamination component of the reads is expressed as a percentage of the individual reads in relation to the total number of reads obtained for the sample.
Once the self-contamination reads were identified, they were discarded and the remaining reads were analyzed. The percentage of each gut content item was calculated in relation to the number of total sequences of each sample without the self-contamination reads.
Data Availability
DNA sequences from ommastrephid 18S v9 and 16S gut content and COI sequences of barcoded ommastrephids were deposited in GenBank with the accession numbers MF980393-MF980451, MF980453-MF980593 and MF980594-MF980608, respectively. All data generated and analyzed during this study are included here or as Supplementary Information files.
Electronic supplementary material
Acknowledgements
We thank Ángel Soria (RV Hesperides) for the collection of individual E3; María Ángeles Artaza (Vall d’Hebron Institut de Recerca, VHIR) for her help during the LCM sessions; Iván Acevedo, Violeta López-Márquez, Paula C. Rodríguez-Flores and Mar Soler-Hurtado (MNCN-CSIC) for their help in the molecular systematics lab; and Mireia Mestre and Marta Albo-Puigserver (ICM-CSIC) for her help with prokaryotic and detritus literature, respectively. NGS sequencing and some of the bioinformatics analyses were carried out in AllGenetics (A Coruña, Spain). Francisco Olivas performed the curation of the morphological vouchers under the Biological Reference Collections (CBR, ICM). Seven individuals used here were collected on board the RV Hesperides during the research project MAFIA (CTM2012-39587-C04-03), funded by the Spanish Ministry of Economy and Competitiveness (MINECO). FAFA was supported by the MINECO grants BES-2013-063551 and EEBB-C-16-00694. RV was supported by the Spanish Ministry of Education and Culture (MECD) grant PRX17/00090. This study was funded by the research projects AGL2012-39077 and CTM2014-57949-R from the MINECO.
Author Contributions
F.A.F.-A., A.M., R.G.-J. and R.V. conceived and designed the project; C.A.S.-Z. and R.V. contributed samples, A.M. contributed reagents and F.A.F.-A. performed laboratory work and data analysis, with assistance from R.G.-J. in DNA extractions. F.A.F.-A. wrote the first draft of the manuscript and prepared figures, and all authors contributed substantially to revisions.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21501-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Saunders W. The role and status of Nautilus in its natural habitat: evidence from deep- water remote camera photosequences. Paleobiology. 1984;10:469–486. doi: 10.1017/S0094837300008472. [DOI] [Google Scholar]
- 2.Dunstan, A., Bradshaw, C. J. A. & Marshall, J. Nautilus at Risk - Estimating Population Size and Demography of Nautilus pompilius. PLoS ONE6 (2011). [DOI] [PMC free article] [PubMed]
- 3.Hoving HJT, Robison BH. Vampire squid: Detritivores in the oxygen minimum zone. Proc. Royal Soc. B. 2012;279:4559–4567. doi: 10.1098/rspb.2012.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohkouchi N, Tsuda R, Chikaraishi Y, Tanabe K. A preliminary estimate of the trophic position of the deep-water ram’s horn squid Spirula spirula based on the nitrogen isotopic composition of amino acids. Mar. Biol. 2013;160:773–779. doi: 10.1007/s00227-012-2132-1. [DOI] [Google Scholar]
- 5.Hoving HJT, et al. The Study of Deep-Sea Cephalopods. Adv. Mar. Biol. 2014;67:235–359. doi: 10.1016/B978-0-12-800287-2.00003-2. [DOI] [PubMed] [Google Scholar]
- 6.Villanueva R, Perricone V, Fiorito G. Cephalopods as predators: a short journey among behavioural flexibilities, adaptions and feeding habits. Front. Physiol. 2017;8:598. doi: 10.3389/fphys.2017.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young RE, Harman RF. “Larva,” “paralarva” and “subadult” in cephalopod terminology. Malacologia. 1988;29:201–207. [Google Scholar]
- 8.Villanueva R, Vidal EAG, Fernández-Álvarez FÁ, Nabhitabhata J. Early mode of life and hatchling size in cephalopod molluscs: influence on the species distributional ranges. PLoS ONE. 2016;11:e0165334. doi: 10.1371/journal.pone.0165334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen DS, Dykhuizen G, Hodge J, Gilly WF. Ontogeny of copepod predation in juvenile squid (Loligo opalescens) Biol. Bull. 1996;190:69–81. doi: 10.2307/1542676. [DOI] [PubMed] [Google Scholar]
- 10.Sugimoto, C. & Ikeda, Y. Comparison of the ontogeny of hunting behavior in pharaoh cuttlefish (Sepia pharaonis) and oval squid (Sepioteuthis lessoniana). Biol. Bull. 225 50–59, 10.1086/BBLv225n1p50. [DOI] [PubMed]
- 11.Iglesias, J., Fuentes, L. & Villanueva, R. Cephalopod Culture. (Springer, 2014).
- 12.Jereb, P. & Roper, C. F. E. Cephalopods of the world. An annotated and illustrated catalogue of cephalopod species known to date. Vol. 2. (FAO, 2010).
- 13.Arkhipkin, A. I. et al. World Squid Fisheries. Rev. Fish. Sci. & Aquacul. 23, 10.1080/23308249.2015.1026226 (2015).
- 14.FAO. FAO Yearbook2014. Fishery and aquaculture statistics. (FAO, 2016).
- 15.Fernández-Álvarez FÁ, Martins CPP, Vidal EAG, Villanueva R. Towards the identification of the ommastrephid squid paralarvae (Mollusca: Cephalopoda): morphological description of three species and a key to the north-east Atlantic species. Zool. J. Linnean Soc. 2017;180:268–287. [Google Scholar]
- 16.Shea EK. Ontogeny of the fused tentacles in three species of ommastrephid squids (Cephalopoda, Ommastrephidae) Invertebr. Biol. 2005;124:25–38. doi: 10.1111/j.1744-7410.2005.1241-04.x. [DOI] [Google Scholar]
- 17.Shigeno S, Kidokoro H, Goto T, Tsuchiya K, Segawa S. Early ontogeny of the Japanese common squid Todarodes pacificus (Cephalopoda, Ommastrephidae) with special reference to its characteristic morphology and ecological significance. Zool. Sci. 2001;18:1011–1026. doi: 10.2108/zsj.18.1011. [DOI] [Google Scholar]
- 18.Markaida U, Salinas-Zavala CA, Rosas-Luis R, Gilly WF, Booth JAT. Food and Feeding of jumbo squid Dosidicus gigas in the Central Gulf of California during 2005–2007. Cal. Coop. Ocean Fish. 2008;49:90–103. [Google Scholar]
- 19.Rosas-Luis R, Villanueva R, Sánchez P. Trophic habits of the ommastrephid squid Illex coindetii and Todarodes sagittatus in the northwestern Mediterranean Sea. Fish. Res. 2014;152:21–28. doi: 10.1016/j.fishres.2013.10.009. [DOI] [Google Scholar]
- 20.Uchikawa K, Sakai M, Wakabayashi T, Ichii T. The relationship between paralarval feeding and morphological changes in the proboscis and beaks of the neon flying squid Ommastrephes bartramii. Fish. Sci. 2009;75:317–323. doi: 10.1007/s12562-008-0036-2. [DOI] [Google Scholar]
- 21.Camarillo-Coop S, Salinas-Zavala CA, Lavaniegos BE, Markaida U. Food in early life stages of Dosidicus gigas (Cephalopoda: Ommastrephidae) from the Gulf of California, Mexico. Journal of the Marine Biological Association of the United Kingdom. 2013;93:1903–1910. doi: 10.1017/S0025315413000398. [DOI] [Google Scholar]
- 22.Yatsu A, Tafur R, Maravi C. Embryos and rhynchoteuthion paralarvae of the jumbo flying squid Dosidicus gigas (Cephalopoda) obtained through artificial fertilization from Peruvian waters. Fisheries Science. 1999;65:904–908. doi: 10.2331/fishsci.65.904. [DOI] [Google Scholar]
- 23.Staaf DJ, et al. Natural egg mass deposition by the Humboldt squid (Dosidicus gigas) in the Gulf of California and characteristics of hatchlings and paralarvae. J. Mar. Biol. Assoc. U. K. 2008;88:759–770. doi: 10.1017/S0025315408001422. [DOI] [Google Scholar]
- 24.Vidal EAG, Haimovici M. Feeding and the possible role of the proboscis and mucus cover in the ingestion of microorganisms by rhynchoteuthion paralarvae (Cephalopoda: Ommastrephidae) Bull. Mar. Sci. 1998;63:305–316. [Google Scholar]
- 25.O’Dor RK, Helm P, Balch N. Can rhynchoteuthions suspension feed? (Mollusca: Cephalopoda) Vie Milieu. 1985;35:267–271. [Google Scholar]
- 26.O’Rorke R, Lavery S, Jeffs A. PCR enrichment techniques to identify the diet of predators. Mol. Ecol. Resour. 2012;12:5–17. doi: 10.1111/j.1755-0998.2011.03091.x. [DOI] [PubMed] [Google Scholar]
- 27.Piñol J, San Andres V, Clare EL, Mir G, Symondson W. A pragmatic approach to the analysis of diets of generalist predators: The use of next-generation sequencing with no blocking probes. Mol. Ecol. Resour. 2014;14:18–26. doi: 10.1111/1755-0998.12156. [DOI] [PubMed] [Google Scholar]
- 28.Albaina A, Aguirre MDA, Santos M, Estonba A. 18S rRNA V9 metabarcoding for diet characterization: a critical evaluation with two sympatric zooplanktivorous fish species. Ecol. Evol. 2016;6:1809–1824. doi: 10.1002/ece3.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olmos-Perez L, Roura A, Pierce GJ, Boyer S, Gonzalez AF. Diet Composition and Variability of Wild Octopus vulgaris and Alloteuthis media (Cephalopoda) Paralarvae: a Metagenomic Approach. Front. Physiol. 2017;8:321. doi: 10.3389/fphys.2017.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falk BG, Reed RN. Challenges to a molecular approach to prey identification in the Burmese python, Python molurus bivittatus. PeerJ. 2015;3:e1445. doi: 10.7717/peerj.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarman S, Deagle B, Gales NJ. Group-specific polymerase chain reaction for DNA-based analysis of species diversity and identity in dietary samples. Mol. Ecol. 2004;13:1313–1322. doi: 10.1111/j.1365-294X.2004.02109.x. [DOI] [PubMed] [Google Scholar]
- 32.Dunshea G. DNA-based diet analysis for any predator. PLoS ONE. 2009;4:e5252. doi: 10.1371/journal.pone.0005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vestheim H, Jarman SN. Blocking primers to enhance PCR amplification of rare sequences in mixed samples - a case study on prey DNA in Antarctic krill stomachs. Front. Zool. 2008;5:12. doi: 10.1186/1742-9994-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Rorke R, et al. Determining the diet of larvae of western rock lobster (Panulirus cygnus) using high-throughput DNA sequencing techniques. PLoS ONE. 2012;7:e42757. doi: 10.1371/journal.pone.0042757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Rorke R, Lavery SD, Wang M, Nodder SD, Jeffs AG. Determining the diet of larvae of the red rock lobster (Jasus edwardsii) using high-throughput DNA sequencing techniques. Mar. Biol. 2013;161:551–563. doi: 10.1007/s00227-013-2357-7. [DOI] [Google Scholar]
- 36.O’Rorke R, Jeffs AG, Fitzgibbon Q, Chow S, Lavery S. Extracting DNA from whole organism homogenates and the risk of false positives in PCR based diet studies: A case study using spiny lobster larvae. J. Exp. Mar. Bio. Ecol. 2013;441:1–6. doi: 10.1016/j.jembe.2013.01.003. [DOI] [Google Scholar]
- 37.Bonner RF, et al. Cell sampling – laser capture microdissection: molecular analysis of tissue. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 38.Maloy AP, Culloty SC, Bolton-Warberg M, Fitzgerald R, Slater JW. Molecular identification of laser-dissected gut contents from hatchery-reared larval cod, Gadus morhua: a new approach to diet analysis. Aquacul. Nutr. 2011;17:536–541. doi: 10.1111/j.1365-2095.2010.00836.x. [DOI] [Google Scholar]
- 39.Mestre M, Borrull E, Sala M, Gasol JM. Patterns of bacterial diversity in the marine planktonic particulate matter continuum. ISME J. 2017;11:999–1010. doi: 10.1038/ismej.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chow S, et al. Investigation on natural diets of larval marine animals using peptide nucleic acid-directed polymerase chain reaction clamping. Mar. Biotechnol. 2010;13:305–313. doi: 10.1007/s10126-010-9301-3. [DOI] [PubMed] [Google Scholar]
- 41.Govoni JJ. Feeding on protists and particulates by the leptocephali of the worm eels Myrophis spp. (Teleostei: Anguilliformes: Ophichthidae), and the potential energy contribution of large aloricate protozoa. Sci. Mar. 2010;74:339–344. doi: 10.3989/scimar.2010.74n2339. [DOI] [Google Scholar]
- 42.Riemann L, et al. Qualitative assessment of the diet of European eel larvae in the Sargasso Sea resolved by DNA barcoding. Biol. Lett. 2010;6:819–822. doi: 10.1098/rsbl.2010.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parry M. Trophic variation with length in two ommastrephid squids, Ommastrephes bartramii and Sthenoteuthis oualaniensis. Mar. Biol. 2008;153:249–256. doi: 10.1007/s00227-007-0800-3. [DOI] [Google Scholar]
- 44.Polis G, Strong D. Food web complexity and community dynamics. Am. Nat. 1996;147:813–846. doi: 10.1086/285880. [DOI] [Google Scholar]
- 45.Moore JC, et al. Detritus, trophic dynamics and biodiversity. Ecol. Lett. 2004;7:584–600. doi: 10.1111/j.1461-0248.2004.00606.x. [DOI] [Google Scholar]
- 46.Hagen EM, et al. A meta-analysis of the effects of detritus on primary producers and consumers in marine, freshwater, and terrestrial ecosystems. Oikos. 2012;121:1507–1515. doi: 10.1111/j.1600-0706.2011.19666.x. [DOI] [Google Scholar]
- 47.Villanueva R, et al. A laboratory guide to in vitro fertilization of oceanic squids. Aquac. 2012;342-343:125–133. doi: 10.1016/j.aquaculture.2012.02.025. [DOI] [Google Scholar]
- 48.Puneeta P, Vijai D, Yoo HK, Matsui H, Sakurai Y. Observations on the spawning behavior, egg masses and paralarval development of the ommastrephid squid Todarodes pacificus in a laboratory mesocosm. J. Exp. Biol. 2015;218:3825–3835. doi: 10.1242/jeb.127670. [DOI] [PubMed] [Google Scholar]
- 49.Roura Á, González ÁF, Redd K, Guerra Á. Molecular prey identification in wild Octopus vulgaris paralarvae. Mar. Biol. 2012;159:1335–1345. doi: 10.1007/s00227-012-1914-9. [DOI] [Google Scholar]
- 50.Leray M, Knowlton N. Censusing marine eukaryotic diversity in the twenty-first century. Philos. Trans. R. Soc. Lond. B. 2016;371:20150331. doi: 10.1098/rstb.2015.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nayak SK. Role of gastrointestinal microbiota in fish. Aquac. Res. 2010;41:1553–1573. doi: 10.1111/j.1365-2109.2010.02546.x. [DOI] [Google Scholar]
- 52.Roura A, Doyle SR, Nande M, Strugnell JM. You are what you eat: a genomic analysis of the gut microbiome of captive and wild Octopus vulgaris paralarvae and their zooplankton prey. Front. Physiol. 2017;8:362. doi: 10.3389/fphys.2017.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrews, S. FastQC: a quality control tool for high throughput sequence data http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
- 54.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ Prepr. 2016;4:e2409v2401. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 62.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 63.Esling P, Lejzerowicz F, Pawlowski J. Accurate multiplexing and filtering for high-throughput amplicon-sequencing. Nucleic Acids Res. 2015;43:2513–2524. doi: 10.1093/nar/gkv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartram J, et al. Accurate sample assignment in a multiplexed, ultrasensitive, high-throughput sequencing assay for minimal residual disease. J. Mol. Diagn. 2016;18:494–506. doi: 10.1016/j.jmoldx.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Fernández-Álvarez, F. Á. et al. The genus Ommastrephes d’Orbigny, 1834: a single species or more than one hidden behind a single name?in CIAC 2015 (Hakodate, Japan, 2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences from ommastrephid 18S v9 and 16S gut content and COI sequences of barcoded ommastrephids were deposited in GenBank with the accession numbers MF980393-MF980451, MF980453-MF980593 and MF980594-MF980608, respectively. All data generated and analyzed during this study are included here or as Supplementary Information files.