Abstract
Obesity is characterized by excess body fat accumulation due to an increase in the size and number of differentiated mature adipocytes. Adipocyte differentiation is regulated by genetic and environmental factors, and its inhibition could represent a strategy for obesity prevention and treatment. The current study was designed with two aims: (i) to evaluate the changes in the expression of adipogenic markers (C/EBPα, PPARγ variant 1 and variant 2, and GLUT4) in 3T3-L1 murine preadipocytes at four stages of the differentiation process and (ii) to compare the effectiveness of sulforaphane, genistein, and docosahexaenoic acid in reducing lipid accumulation and modulating C/EBPα, PPARγ1, PPARγ2, and GLUT4 mRNA expression in mature adipocytes. All bioactive compounds were shown to suppress adipocyte differentiation, although with different effectiveness. These results set the stage for further studies considering natural food constituents as important agents in preventing or treating obesity.
1. Introduction
Obesity is the main dysfunction of adipose tissue and is associated with premature death and the development of chronic diseases such as cardiovascular diseases (CVD), type 2 diabetes, hypertension, and certain cancers [1]. In particular, a chronic inflammation in the absence of overt infection or autoimmune process is a puzzling phenomenon linked to obesity [2].
Environment, lifestyle, and genetic susceptibility certainly contribute to the increased risk of obesity, one of the easiest to be recognized and the most difficult to treat medical conditions [3]. Antiobesity drugs lack physiology specificity and have side effects [4].
Obesity is characterized by an excess accumulation of white adipose mass, resulting from both the increase in adipocyte cell size and the development of mature cells from undifferentiated precursors. Particularly, de novo generation of fat cells plays a key role in the development of obesity.
Discovering compounds able to regulate the size, number, and function of adipocytes and understanding their mechanisms of action could greatly contribute to obesity prevention and treatment. In this light, natural compounds represent a potential novel strategy, already exploited for preventing other metabolic disorders [5]. Bioactive compounds have been shown to exert specific effects on the biochemical and metabolic functions of adipocytes [6–8], in particular inhibition of preadipocyte differentiation, lipolysis stimulation, and induction of apoptosis of existing adipocytes [9], therefore contributing to a possible decrease in adipose tissue mass [10].
The aim of the current study was to compare the antiadipogenic effect of three bioactive compounds, namely, docosahexaenoic acid (DHA), genistein (GEN), and sulforaphane (SFN). DHA (C22:6 n-3) is an n-3 polyunsaturated fatty acid (PUFA) abundant in fatty fish. It is considered effective in the prevention of many chronic diseases, mainly CVD [11]. GEN (4,5,7-trihydroxyisoflavone), the most abundant isoflavone found in soybeans, has received particular attention for its structural similarity to estrogen and its high affinity to the estrogen receptor. It is a well-known antioxidant, chemopreventive, and anti-inflammatory agent [12, 13]. SFN, an isothiocyanate compound, is a constituent of cruciferous vegetables such as broccoli sprouts, Brussels sprouts, and cabbage. SFN is known to have antioxidant, immunomodulatory, anticancer, and antidiabetic properties [14, 15].
In some previous earlier studies [16–19], all tested bioactive compounds have been shown to be antiadipogenic in the 3T3-L1 cell line. Notwithstanding, to our knowledge, their effectiveness has never been compared in the same experimental conditions. Although in vitro studies always need confirmation in vivo, the selection of the most active bioactive could be useful to formulate functional foods contributing to the development of new strategies to prevent obesity.
3T3-L1 cells constitute the most frequently used preadipocyte model, sharing many properties with normal adipocytes [20]. Their differentiation into mature adipocytes involves the exposure of a confluent, quiescent population of cells to a variety of effectors that activate a complex cascade of genes [21].
It is well documented that adipogenesis is finely controlled by key transcription factors such as peroxisome proliferator-activated receptor-γ (PPARγ) and CCAAT-enhancer-binding protein-α (C/EBPα). PPARγ and C/EBPα regulate the expression of various genes involved in lipogenesis, lipolysis, and insulin sensitivity, such as the one encoding for glucose transporter type 4 (GLUT4) [22, 23]. In the first part of the study, changes in the expression of PPARγ, C/EBPα, and GLUT4 genes were evaluated in the murine 3T3-L1 cell line at various stages of the differentiation process.
In the second part of the study, preadipocytes were supplemented during and after differentiation with DHA, GEN, and SFN, and both lipid accumulation and the mRNA expression of PPARγ, C/EBPα, and GLUT4 were evaluated to evidence and compare their potential inhibitory activity on adipogenesis.
2. Materials and Methods
Dulbecco's modified Eagle's medium (DMEM)/F12 GlutaMAX I was purchased from Invitrogen (Darmstadt, Germany), donor bovine serum (DBS) was from Gibco Life Technologies (Darmstadt, Germany), fetal bovine serum (FBS GOLD) was from PAA Laboratories (Pasching, Austria), and TRIzol Reagent was from Ambion, Life Technologies (Darmstadt, Germany). All other chemicals were purchased from Sigma-Aldrich (Schnelldorf, Germany) and were of the highest analytical grade.
2.1. Cell Culture and Differentiation
3T3-L1 mouse preadipocytes were obtained from the American Type Culture Collection (ATCC) and maintained at 37°C in a humidified atmosphere containing 95% air and 5% CO2; preadipocytes were subcultured every three days when 80% confluent or less into a new 175 cm2 flask. Cells were cultured in DMEM/F12 GlutaMAX I with the addition of D-glucose (3151 mg/L f.c.) (GM) containing 10% DBS and 1% penicillin/streptomycin (PS). Cells were seeded in 12-well plates or a 25 cm2 flask at a concentration of 50,000 cells/mL. Three days after seeding, cells were stimulated to differentiate with GM supplemented with 10% FBS, 1% PS, insulin (10 μg/mL), dexamethasone (1 μM), isobutylmethylxanthine (0.2 mM), and rosiglitazone (10 μM) (differentiation medium). After further 3 days (differentiation), cells were maintained in GM with FBS, PS, and insulin (postdifferentiation medium) for another 5 days (postdifferentiation) when approximately 90% of the cells displayed the characteristic lipid-filled adipocyte phenotype.
2.2. Bioactive Supplementation
DHA, GEN, and SFN were added to the differentiation and postdifferentiation medium at three different final concentrations (10, 25, or 50 μM). The SRB assay was performed in preliminary experiments, evidencing no cytotoxicity for any of the tested concentrations of each bioactive.
The treatment with bioactives began three days after seeding and lasted until the end of postdifferentiation (eleven days from seeding). All bioactive compounds were purchased from Sigma-Aldrich (Schnelldorf, Germany). Each compound was dissolved in dimethyl sulfoxide (DMSO). Unsupplemented control cells (CTR) received a corresponding amount of DMSO (<0.5% final concentration). The medium was changed every two days during postdifferentiation.
2.3. Lipid Staining
The effect of the bioactive compounds on adipogenesis was evaluated morphologically by staining accumulated lipids with Oil Red O [24] as previously described [25]. Briefly, cells were fixed with 4% formalin solution in phosphate-buffered saline (PBS) for two hours, washed with water, rinsed with isopropanol 60%, and stained with Oil Red O for 30 minutes at room temperature. After washing with distilled water for 3 times, the lipid droplets were quantified by dissolving Oil Red O in isopropanol 100% and measuring the optical density at 500 nm.
The lowest bioactive concentrations able to influence lipid accumulation (10 μM GEN, 10 μM SFN, 25 μM DHA) were then used in gene expression experiments.
2.4. Gene Expression Analysis
Unsupplemented, control cells were collected at four different steps of the differentiation protocol: one day after seeding (T1); three days after seeding (postconfluent cells), before the beginning of differentiation (T2); six days after seeding (end of the differentiation), before the addition of the postdifferentiation medium (T3); and eleven days after seeding, at the end of postdifferentiation (T4). Cells were collected at the different time points, and total RNA was extracted as described below.
In experiments evaluating bioactives' effect, 10 μM GEN, 10 μM SFN, or 25 μM DHA was added to the differentiation and postdifferentiation media as described above. At the end of the postdifferentiation period (T4), cells were collected, and total RNA was extracted with TRIzol Reagent following the manufacturer's protocol. Contaminating DNA was eliminated by DNase treatment (DNA-free Kit from Ambion, Life Technologies, Darmstadt, Germany). RNA quantity and quality, respectively, were assessed by spectrophotometric analyses at 260/230 nm using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and by the microfluidics-based Bioanalyzer platform with an RNA Nano 6000 Chip (Agilent Technology, Waldbronn, Germany).
cDNA was synthesized from 0.5 μg or 1 μg of DNase-treated total RNA using SuperScript II reverse transcriptase (Invitrogen, Darmstadt, Germany) according to the manufacturer's instructions. Quantitative real-time PCR (qPCR) was performed using the Universal ProbeLibrary system (Roche, Mannheim, Germany) on a Roche LightCycler 480 real-time PCR system (Roche, Mannheim, Germany). The cycling program for analysis was 15 min at 95°C followed by 45 cycles of 10 s at 95°C, 20 s at 55°C, and 10 s at 72°C with the primer pairs and the respective monocolor hydrolysis probes indicated in Table 1. The expression levels of target mRNAs were normalized to three reference genes: β-actin (ACTB), hypoxanthine phosphoribosyltransferase 1 (HPRT1), and TATA-box-binding protein (TBP).
Table 1.
Primer sequences used in qPCR.
| Gene | Forward primer | Reverse primer | Probe number |
|---|---|---|---|
| Target genes | |||
| PPARγ1 | GAAAGACAACGGACAAATCACC | GGGGGTGATATGTTTGAACTTG | 7 |
| PPARγ2 | TGCTGTTATGGGTGAAACTCTG | CTGTGTCAACCATGGTAATTTCTT | 2 |
| C/EBPα | AAACAACGCAACGTGGAGA | GCGGTCATTGTCACTGGTC | 67 |
| GLUT4 | GACGGACACTCCATCTGTTG | GCCACGATGGAGACATAGC | 5 |
| Reference genes | |||
| ACTB | GTGGGAGAGCAAGGAAGAGA | CACTCTTGGCCCAGTCTACG | 56 |
| HPRT1 | TCCTCCTCAGACCGCTTTT | CCTGGTTCATCATCGCTAATC | 95 |
| TBP | CGGTCGCGTCATTTTCTC | GGGTTATCTTCACACACCATGA | 107 |
2.5. Statistical Analysis
Gene expression data were analyzed using DataAssist software version 3.01 (Applied Biosystems, Foster City, CA, USA) and expressed as the mean fold change in relative expression compared with the untreated control cells, which were normalized to one. Average fold change and standard deviation (SD) were obtained from three biological replicate samples per condition.
All data were analyzed by one-way ANOVA, followed by Dunnett's or Tukey's tests. Statistical analysis of the data was performed using the GraphPad Prism 5 software (San Diego, CA, USA).
3. Results
3.1. Characterization of Preadipocyte Differentiation
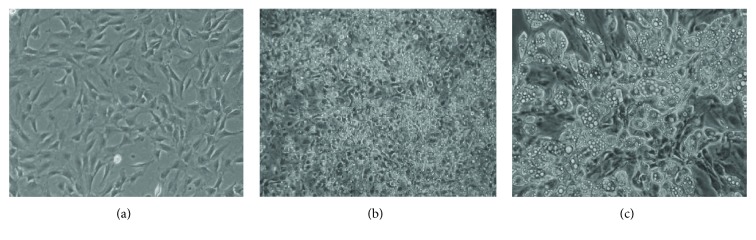
During differentiation (T1–T4), preadipocytes acquired the characteristics of mature adipocytes. At three days after seeding (T2), nondifferentiated cells showed typical fibroblastoid morphology, while at the end of the differentiation process (T4), cells had abundant intracytoplasmic lipid accumulation, showing typical white adipocyte morphology (Figure 1).
Figure 1.
Morphological changes among (a) preadipocytes three days after seeding (T2), (b) adipocytes at the end of the differentiation (T3), and (c) adipocytes at the end of postdifferentiation (T4). Images showing different cell morphologies were captured at the different steps using a Leica DM IL microscope (Wetzlar, Germany), with 10x magnification.
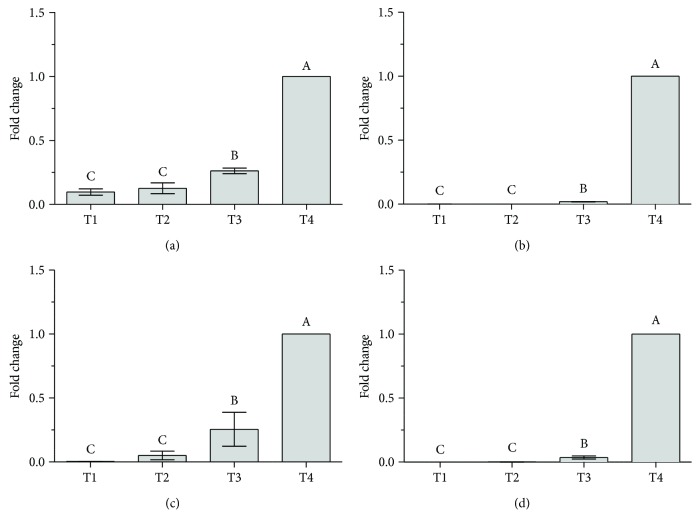
To characterize the differentiation process, PPARγ1, PPARγ2, C/EBPα, and GLUT4 gene expression was evaluated at four different stages of adipocyte differentiation: one day after seeding (T1), three days after seeding (T2), at the end of the differentiation (T3), and at the end of postdifferentiation (T4).
The expression of selected genes was very low and similar at T1 and T2, while it significantly increased at T3. For all analyzed genes, a prominent increase in mRNA levels was observed in mature adipocytes (T4) (Figure 2).
Figure 2.
(a) PPARγ1, (b) PPARγ2, (c) C/EBPα, and (d) GLUT4 mRNA expression at 4 different stages of adipocyte differentiation. T1: one day after seeding; T2: three days after seeding, before the beginning of differentiation; T3: at the end of the differentiation, before the addition of the postdifferentiation medium; T4: at the end of postdifferentiation (mature adipocytes). Data are expressed as the mean fold change relative to the mature cells (T4), normalized to one. Statistical analysis was by one-way ANOVA (p < 0.001 for all panels) followed by Tukey's HSD test. The expression is significantly different between groups marked with different letters (at least p < 0.05).
3.2. Effects of Bioactive Compound Supplementation
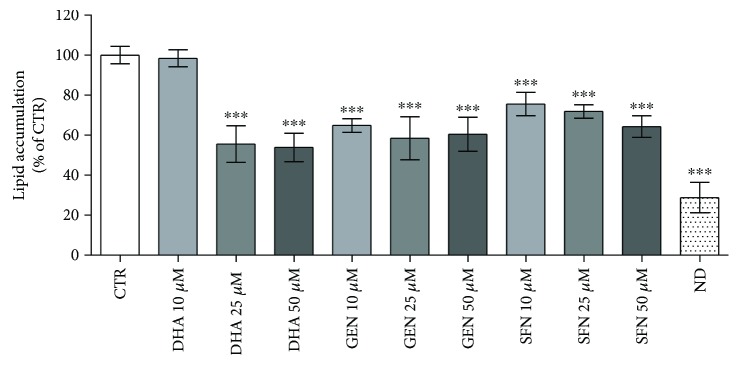
The antiadipogenic potential of DHA, GEN, and SFN was first investigated evaluating their influence on lipid accumulation. Preadipocytes were supplemented with different concentrations (10, 25, and 50 μM) of the test compounds during the differentiation and postdifferentiation periods, as described above, and lipid accumulation was detected by Oil Red O staining. All bioactive compounds markedly reduced lipid droplet formation compared to controls. GEN and SFN were effective at the lowest concentration used for supplementation (10 μM), while a higher DHA concentration (25 μM) was required to reduce lipid accumulation (Figure 3).
Figure 3.
Lipid accumulation in supplemented and control cells. Data are expressed as a percentage relative to unsupplemented control cells (CTR), assigned as 100%. Statistical analysis was performed by one-way ANOVA (p < 0.001) followed by Dunnett's test: ∗∗∗p < 0.001 versus CTR. ND: nondifferentiated cells, before the beginning of the differentiation process.
The lowest bioactive concentrations causing a significant decrease in lipid accumulation were used to verify the modification in the mRNA levels of the adipogenesis marker genes after differentiation.
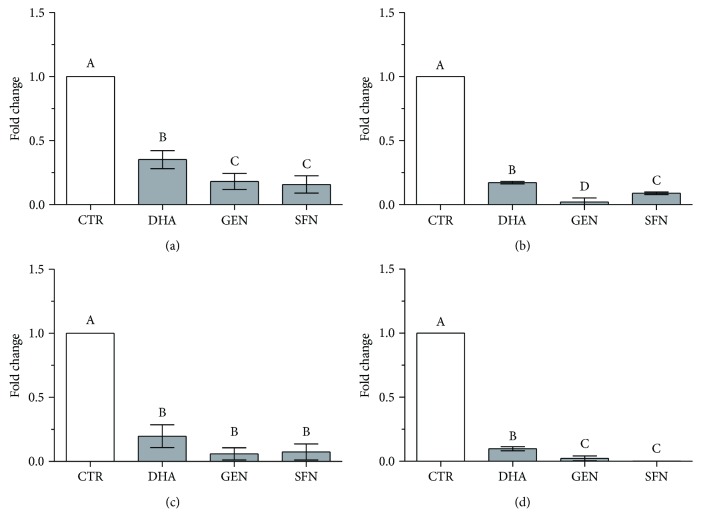
At T4, all bioactive compounds significantly reduced the transcript levels of PPARγ1, PPARγ2, C/EBPα, and GLUT4. The effect of GEN and SFN on PPARγ and GLUT4 expression appeared stronger than the DHA effect did (Figure 4).
Figure 4.
Modulatory effect of GEN, SFN, and DHA on (a) PPARγ1, (b) PPARγ2, (c) C/EBPα, and (d) GLUT4 mRNA expression. Data are expressed as the mean fold change relative to the unsupplemented control cells (CTR) at T4, normalized to one. Statistical analysis was by one-way ANOVA (p < 0.001 for all panels) followed by Tukey's HSD test. The expression is significantly different between groups marked with different letters (at least p < 0.05).
4. Discussion
Adipose tissue has an important function in the energy balance by regulating lipid metabolism, glucose homeostasis, and adipokine secretion. Thus, its dysfunction is critical in developing metabolic diseases [26]. Indeed, the incidence of metabolic syndrome, a combination of cardiometabolic risk determinants, is increasing worldwide largely as a consequence of the continued obesity epidemic [27].
In general, obesity is related to the extent of adipocyte differentiation, intracellular lipid accumulation, and lipolysis [17]. The process of adipocyte differentiation requires the activation of numerous transcription factors which are in charge of the coordinated induction and silencing of more than 2000 genes [28]. Several transcriptional regulators, including C/EBP and PPARγ, play a pivotal role in this process.
The master regulator PPARγ is both necessary and sufficient for adipogenesis [29, 30]. PPARγ has two isoforms, the ubiquitary PPARγ1 and the adipose tissue-specific PPARγ2. Both isoforms are strongly induced during preadipocyte differentiation [25], and our data confirm that PPARγ1 induction foreruns PPARγ2 induction [31].
C/EBPβ and C/EBPδ are overexpressed in the earlier phases of differentiation and have been shown to play a role in PPARγ induction [30]. C/EBPα is involved in the maintenance of the terminally differentiated adipocyte phenotypes [28, 32, 33]. In agreement, we observed a lower C/EBPα expression at T1 and T2 than at T3 and T4.
In adipocytes, C/EBPα regulates the expression of the gene encoding for GLUT4, the major insulin-responsive glucose transporter in adipose tissue as well as in skeletal and cardiac muscles [34]. Accordingly, in the present study, GLUT4 expression paralleled C/EBPα expression.
Overall, our results confirm that differentiation of 3T3-L1 cells includes distinguishable multiple stages [35, 36].
Our results evidence that all tested bioactives efficiently block adipocyte differentiation. At T4, the expression of all the tested genes was significantly lower in supplemented cells than in unsupplemented ones and comparable to the expression level observed in unsupplemented cells at the first stages of differentiation.
The antiadipogenic effect of DHA, GEN, and SFN has been already reported in previous earlier studies [16–19]. Our study is not simply a confirmation that the tested bioactives act mainly through modification of the adipocyte life cycle [8], but mainly a representation of the first study comparing the effectiveness of DHA, GEN, and SFN in the same experimental conditions. Although GEN and SFN appeared effective at lower concentrations than DHA did, it is worth noting that in vivo the latter is absorbed and delivered to peripheral cells in its parent form. GEN and SFN are extensively metabolized, and they are detectable at very low concentrations in the bloodstream [37–39]. On the contrary, the DHA concentration used in this study for cell supplementation is easily reachable in vivo in the human plasma [40–42].
5. Conclusions
Our results represent an additional step in the evaluation of the antiadipogenic effects of three natural bioactive molecules, DHA, GEN, and SFN. Although in vitro all tested bioactive compounds appeared to be putative contributors to the prevention and treatment of obesity, their in vivo metabolism suggests that mainly DHA could potentially be used for the formulation of new functional food products devoted to a new dietetic natural strategy for overweight counteraction.
Further investigations are needed to verify whether the antiadipogenic properties evidenced in vitro do translate into in vivo efficacy in humans and to sort out the pathway(s) responsible for the beneficial effects. Moreover, the compounds here considered have been studied as discrete molecules and not as part of a food, ignoring both the matrix effect and the eventual synergistic or enhanced activities between the selected compounds and other food components or other bioactive molecules [43]. This issue also deserves future attention.
Acknowledgments
The authors wish to thank Professor Peter Arner (Karolinska Institutet, Stockholm, Sweden) and his research group for their kind help to Veronica Valli in setting 3T3-L1 cell cultures and Karin Klimo for her skillful technical assistance. This paper is based on the fourth chapter of the PhD thesis of Veronica Valli. The research leading to these results has received funding from the European Union Seventh Framework Program (FP7/2007-2013) under Grant Agreement no. 311876: Pathway-27.
Disclosure
Veronica Valli's current address is Sociedad Española de Colorantes Naturales y Afines (SECNA), Polígono 33, Parcela 254, El Muladar, 46370 Chiva, Valencia, Spain.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
Veronica Valli participated in the study design and conducted all the experiments; Katharina Heilmann gave a substantial contribution in performing qPCR assays; Francesca Danesi performed the gene expression data normalization and carried out the statistical analysis; Alessandra Bordoni is the coordinator of the Pathway-27 European project and supervised the study; and Clarissa Gerhäuser conceived, designed, and supervised the experiments and wrote the manuscript together with Veronica Valli and Alessandra Bordoni. All authors contributed actively to the revision of the manuscript.
References
- 1.Friedman J. M. Obesity in the new millennium. Nature. 2000;404(6778):632–634. doi: 10.1038/35007504. [DOI] [PubMed] [Google Scholar]
- 2.Vandanmagsar B., Youm Y. H., Ravussin A., et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nature Medicine. 2011;17(2):179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rayalam S., Yang J. Y., Della-Fera M. A., Park H. J., Ambati S., Baile C. A. Anti-obesity effects of xanthohumol plus guggulsterone in 3T3-L1 adipocytes. Journal of Medicinal Food. 2009;12(4):846–853. doi: 10.1089/jmf.2008.0158. [DOI] [PubMed] [Google Scholar]
- 4.Bray G. A., Tartaglia L. A. Medicinal strategies in the treatment of obesity. Nature. 2000;404(6778):672–677. doi: 10.1038/35007544. [DOI] [PubMed] [Google Scholar]
- 5.Hwang J. T., Kim S. H., Lee M. S., et al. Anti-obesity effects of ginsenoside Rh2 are associated with the activation of AMPK signaling pathway in 3T3-L1 adipocyte. Biochemical and Biophysical Research Communications. 2007;364(4):1002–1008. doi: 10.1016/j.bbrc.2007.10.125. [DOI] [PubMed] [Google Scholar]
- 6.González-Castejón M., Rodriguez-Casado A. Dietary phytochemicals and their potential effects on obesity: a review. Pharmacological Research. 2011;64(5):438–455. doi: 10.1016/j.phrs.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Andersen C., Rayalam S., Della-Fera M. A., Baile C. A. Phytochemicals and adipogenesis. BioFactors. 2010;36(6):415–422. doi: 10.1002/biof.115. [DOI] [PubMed] [Google Scholar]
- 8.Moseti D., Regassa A., Kim W. K. Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules. International Journal of Molecular Sciences. 2016;17(1, article 124) doi: 10.3390/ijms17010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baile C. A., Yang J. Y., Rayalam S., et al. Effect of resveratrol on fat mobilization. Annals of the New York Academy of Sciences. 2011;1215(1):40–47. doi: 10.1111/j.1749-6632.2010.05845.x. [DOI] [PubMed] [Google Scholar]
- 10.Yang J. Y., Della-Fera M. A., Rayalam S., et al. Enhanced inhibition of adipogenesis and induction of apoptosis in 3T3-L1 adipocytes with combinations of resveratrol and quercetin. Life Sciences. 2008;82(19-20):1032–1039. doi: 10.1016/j.lfs.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Lorente-Cebrián S., Costa A. G., Navas-Carretero S., Zabala M., Martínez J. A., Moreno-Aliaga M. J. Role of omega-3 fatty acids in obesity, metabolic syndrome, and cardiovascular diseases: a review of the evidence. Journal of Physiology and Biochemistry. 2013;69(3):633–651. doi: 10.1007/s13105-013-0265-4. [DOI] [PubMed] [Google Scholar]
- 12.Polkowski K., Mazurek A. P. Biological properties of genistein. A review of in vitro and in vivo data. Acta Poloniae Pharmaceutica. 2000;57(2):135–155. [PubMed] [Google Scholar]
- 13.Vitale D. C., Piazza C., Melilli B., Drago F., Salomone S. Isoflavones: estrogenic activity, biological effect and bioavailability. European Journal of Drug Metabolism and Pharmacokinetics. 2013;38(1):15–25. doi: 10.1007/s13318-012-0112-y. [DOI] [PubMed] [Google Scholar]
- 14.Yeh C. T., Yen G. C. Chemopreventive functions of sulforaphane: a potent inducer of antioxidant enzymes and apoptosis. Journal of Functional Foods. 2009;1(1):23–32. doi: 10.1016/j.jff.2008.09.002. [DOI] [Google Scholar]
- 15.Dinkova-Kostova A. T., Kostov R. V. Glucosinolates and isothiocyanates in health and disease. Trends in Molecular Medicine. 2012;18(6):337–347. doi: 10.1016/j.molmed.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Wang A. S., Xu C. W., Xie H. Y., et al. DHA induces mitochondria-mediated 3T3-L1 adipocyte apoptosis by down-regulation of Akt and ERK. Journal of Functional Foods. 2016;21:517–524. doi: 10.1016/j.jff.2016.01.001. [DOI] [Google Scholar]
- 17.Lee J. H., Moon M. H., Jeong J. K., et al. Sulforaphane induced adipolysis via hormone sensitive lipase activation, regulated by AMPK signaling pathway. Biochemical and Biophysical Research Communications. 2012;426(4):492–497. doi: 10.1016/j.bbrc.2012.08.107. [DOI] [PubMed] [Google Scholar]
- 18.Choi E. J., Jung J. Y., Kim G. H. Genistein inhibits the proliferation and differentiation of MCF-7 and 3T3-L1 cells via the regulation of ERα expression and induction of apoptosis. Experimental and Therapeutic Medicine. 2014;8(2):454–458. doi: 10.3892/etm.2014.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murali G., Desouza C. V., Clevenger M. E., Ramalingam R., Saraswathi V. Differential effects of eicosapentaenoic acid and docosahexaenoic acid in promoting the differentiation of 3T3-L1 preadipocytes. Prostaglandins, Leukotrienes, and Essential Fatty Acids. 2014;90(1):13–21. doi: 10.1016/j.plefa.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Iwashita K., Yamaki K., Tsushida T. Effect of flavonoids on the differentiation of 3T3-L1 adipocytes. Food Science and Technology Research. 2001;7(2):154–160. doi: 10.3136/fstr.7.154. [DOI] [Google Scholar]
- 21.Prusty D., Park B. H., Davis K. E., Farmer S. R. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor γ (PPARγ) and C/EBPα gene expression during the differentiation of 3T3-L1 preadipocytes. Journal of Biological Chemistry. 2002;277(48):46226–46232. doi: 10.1074/jbc.M207776200. [DOI] [PubMed] [Google Scholar]
- 22.Ntambi J. M., Young-Cheul K. Adipocyte differentiation and gene expression. The Journal of Nutrition. 2000;130(12):3122S–3126S. doi: 10.1093/jn/130.12.3122S. [DOI] [PubMed] [Google Scholar]
- 23.Lowe C. E., O'Rahilly S., Rochford J. J. Adipogenesis at a glance. Journal of Cell Science. 2011;124(16):2681–2686. doi: 10.1242/jcs.079699. [DOI] [PubMed] [Google Scholar]
- 24.Rydén M., Dicker A., Götherström C., et al. Functional characterization of human mesenchymal stem cell-derived adipocytes. Biochemical and Biophysical Research Communications. 2003;311(2):391–397. doi: 10.1016/j.bbrc.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Ghini V., Di Nunzio M., Tenori L., et al. Evidence of a DHA signature in the lipidome and metabolome of human hepatocytes. International Journal of Molecular Sciences. 2017;18(2, article 359) doi: 10.3390/ijms18020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trayhurn P., Bing C., Wood I. S. Adipose tissue and adipokines – energy regulation from the human perspective. The Journal of Nutrition. 2006;136(7):1935S–1939S. doi: 10.1093/jn/136.7.1935S. [DOI] [PubMed] [Google Scholar]
- 27.Bruce K. D., Hanson M. A. The developmental origins, mechanisms, and implications of metabolic syndrome. The Journal of Nutrition. 2010;140(3):648–652. doi: 10.3945/jn.109.111179. [DOI] [PubMed] [Google Scholar]
- 28.Li H. X., Xiao L., Wang C., Gao J. L., Zhai Y. G. Epigenetic regulation of adipocyte differentiation and adipogenesis. Journal of Zhejiang University Science B. 2010;11(10):784–791. doi: 10.1631/jzus.B0900401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farmer S. R. Transcriptional control of adipocyte formation. Cell Metabolism. 2006;4(4):263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen E. D., Hsu C.-H., Wang X., et al. C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes & Development. 2002;16(1):22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho Y.-W., Hong S., Jin Q., et al. Histone methylation regulator PTIP is required for PPARγ and C/EBPα expression and adipogenesis. Cell Metabolism. 2009;10(1):27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamm J. K., el Jack A. K., Pilch P. F., Farmer S. R. Role of PPARγ in regulating adipocyte differentiation and insulin-responsive glucose uptake. Annals of the New York Academy of Sciences. 1999;892(1 THE METABOLIC):134–145. doi: 10.1111/j.1749-6632.1999.tb07792.x. [DOI] [PubMed] [Google Scholar]
- 33.Otto T. C., Lane M. D. Adipose development: from stem cell to adipocyte. Critical Reviews in Biochemistry and Molecular Biology. 2005;40(4):229–242. doi: 10.1080/10409230591008189. [DOI] [PubMed] [Google Scholar]
- 34.Yokomori N., Tawata M., Onaya T. DNA demethylation during the differentiation of 3T3-L1 cells affects the expression of the mouse GLUT4 gene. Diabetes. 1999;48(4):685–690. doi: 10.2337/diabetes.48.4.685. [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto H., Kogo Y., Ohgane J., et al. Sequential changes in genome-wide DNA methylation status during adipocyte differentiation. Biochemical and Biophysical Research Communications. 2008;366(2):360–366. doi: 10.1016/j.bbrc.2007.11.137. [DOI] [PubMed] [Google Scholar]
- 36.Tang Q. Q., Otto T. C., Lane M. D. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(1):44–49. doi: 10.1073/pnas.0137044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasper A. V., Al-Janobi A., Smith J. A., et al. Glutathione S-transferase M1 polymorphism and metabolism of sulforaphane from standard and high-glucosinolate broccoli. The American Journal of Clinical Nutrition. 2005;82(6):1283–1291. doi: 10.1093/ajcn/82.6.1283. [DOI] [PubMed] [Google Scholar]
- 38.Hosoda K., Furuta T., Ishii K. Metabolism and disposition of isoflavone conjugated metabolites in humans after ingestion of kinako. Drug Metabolism and Disposition. 2011;39(9):1762–1767. doi: 10.1124/dmd.111.038281. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z., Kulkarni K., Zhu W., Hu M. Bioavailability and pharmacokinetics of genistein: mechanistic studies on its ADME. Anti-Cancer Agents in Medicinal Chemistry. 2012;12(10):1264–1280. doi: 10.2174/187152012803833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jans J. J., de Sain-van der Velden M. G., van Hasselt P. M., et al. Supplementation with a powdered blend of PUFAs normalizes DHA and AA levels in patients with PKU. Molecular Genetics and Metabolism. 2013;109(2):121–124. doi: 10.1016/j.ymgme.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Lara J. J., Economou M., Wallace A. M., et al. Benefits of salmon eating on traditional and novel vascular risk factors in young, non-obese healthy subjects. Atherosclerosis. 2007;193(1):213–221. doi: 10.1016/j.atherosclerosis.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 42.Di Nunzio M., Valli V., Tomas-Cobos L., Murgui-Bosch L., Danesi F., Bordoni A. Is cytotoxicity a determinant of the different in vitro and in vivo effects of bioactives? BMC Complementary and Alternative Medicine. 2017;17(1, article 453) doi: 10.1186/s12906-017-1962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danesi F., Govoni M., D'Antuono L. F., Bordoni A. The molecular mechanism of the cholesterol-lowering effect of dill and kale: the influence of the food matrix components. Electrophoresis. 2016;37(13):1805–1813. doi: 10.1002/elps.201600033. [DOI] [PMC free article] [PubMed] [Google Scholar]