Antibiotic resistance is a major challenge for the effective treatment of infectious diseases. Identifying adaptive mechanisms that bacteria use to survive low levels of antibiotic stress is important for understanding pathways to antibiotic resistance. Furthermore, little is known about the effects of individual bacterial interactions on multispecies communities. This work demonstrates that subinhibitory amounts of some antibiotics produced by streptomycetes induce active motility in B. subtilis, which may alter species interaction dynamics among species-diverse bacterial communities in natural environments. The use of antibiotics at subinhibitory concentrations results in many changes in bacteria, including changes in biofilm formation, small-colony variants, formation of persisters, and motility. Identifying the mechanistic bases of these adaptations is crucial for understanding how bacterial communities are impacted by antibiotics.
KEYWORDS: Bacillus subtilis, Streptomyces venezuelae, antibiotics, chloramphenicol, competition, hormesis, ribosome, sliding motility
ABSTRACT
Competitive interactions between bacteria reveal physiological adaptations that benefit fitness. Bacillus subtilis is a Gram-positive species with several adaptive mechanisms for competition and environmental stress. Biofilm formation, sporulation, and motility are the outcomes of widespread changes in a population of B. subtilis. These changes emerge from complex, regulated pathways for adapting to external stresses, including competition from other species. To identify competition-specific functions, we cultured B. subtilis with multiple species of Streptomyces and observed altered patterns of growth for each organism. In particular, when plated on agar medium near Streptomyces venezuelae, B. subtilis initiates a robust and reproducible mobile response. To investigate the mechanistic basis for the interaction, we determined the type of motility used by B. subtilis and isolated inducing metabolites produced by S. venezuelae. Bacillus subtilis has three defined forms of motility: swimming, swarming, and sliding. Streptomyces venezuelae induced sliding motility specifically in our experiments. The inducing agents produced by S. venezuelae were identified as chloramphenicol and a brominated derivative at subinhibitory concentrations. Upon further characterization of the mobile response, our results demonstrated that subinhibitory concentrations of chloramphenicol, erythromycin, tetracycline, and spectinomycin all activate a sliding motility response by B. subtilis. Our data are consistent with sliding motility initiating under conditions of protein translation stress. This report underscores the importance of hormesis as an early warning system for potential bacterial competitors and antibiotic exposure.
IMPORTANCE Antibiotic resistance is a major challenge for the effective treatment of infectious diseases. Identifying adaptive mechanisms that bacteria use to survive low levels of antibiotic stress is important for understanding pathways to antibiotic resistance. Furthermore, little is known about the effects of individual bacterial interactions on multispecies communities. This work demonstrates that subinhibitory amounts of some antibiotics produced by streptomycetes induce active motility in B. subtilis, which may alter species interaction dynamics among species-diverse bacterial communities in natural environments. The use of antibiotics at subinhibitory concentrations results in many changes in bacteria, including changes in biofilm formation, small-colony variants, formation of persisters, and motility. Identifying the mechanistic bases of these adaptations is crucial for understanding how bacterial communities are impacted by antibiotics.
INTRODUCTION
Bacteria have various mechanisms to maintain fitness under conditions of competitive stress. Examples of competitive fitness mechanisms include type VI secretion systems or contact-dependent inhibition (1–3) and chemical mechanisms as exemplified by antibiotics and other specialized metabolites (4–8). Resistance to a specific challenge also promotes competitive fitness through chemical or genetic modifications to a target or a toxin (9–11). Additionally, adaptations to the physiology of cells within a population or community may alter susceptibility to various competitive stresses. For instance, bacteria may induce biofilm formation (8), enter a persister state (12), or activate a specialized form of metabolism in response to competitors (13–16). One adaptive mechanism available to many species is motility, which imparts to bacteria the ability to physically relocate in the event of a competitive challenge (17–21). In some cases, the response may be chemotactic, manifesting as avoidance of a toxic substance through receptor activation of motility controls. Other sensing or stress mechanisms that activate mobility are not well defined. In one example, swimming and swarming motility are enhanced when Pseudomonas aeruginosa is exposed to the antibiotic tobramycin, but the underlying mechanism is unknown (22). How bacteria sense and respond to antibiotic stress is of particular interest for understanding the development of antibiotic resistance. Indeed, a connection between motility and antibiotic resistance, where resistance is elevated in some motile populations of bacteria, has been found (23, 24).
Bacillus subtilis serves as a model for motility of Gram-positive bacteria. B. subtilis has three described mechanisms of motility: swimming, swarming, and sliding (25–29). Swimming and swarming motility are driven by the action of flagella, which provide propulsion to the bacteria. Swimming B. subtilis use multiple, peritrichous flagella to move as single cells through aqueous media. When the surrounding medium is sufficiently viscous, B. subtilis cells join into rafts that use swarming motility to migrate across surfaces under the power of flagella extending from multiple cells (26). The third type of movement, sliding, is flagellum-independent motility driven by growth. Sliding is currently understood to depend upon multiple factors, including potassium, production of the lipopeptide surfactin, exopolysaccharides (EPS), and extracellular proteins BslA and TasA (29–31). At the vanguard of a sliding population, combinations of surfactin-producing cells and EPS-producing cells cooperate to generate “van Gogh” bundles characteristic of sliding on specialized media (30). The coordinated activities of cell subpopulations within a colony indicates orchestration of multiple events to promote cooperative sliding. Some regulatory functions that contribute to sliding mobility have been described previously (31), but the overall process is less clearly understood than either swimming or swarming motilities. Additionally, other competitive functions may be coordinately controlled with mobilization of cells. In combination with resistance functions, a mobilized bacterial population potentially possesses multiple advantages for competitive fitness.
Here we describe a competitive interaction between Streptomyces venezuelae and B. subtilis. We observed that, under conditions of coculture with S. venezuelae, B. subtilis activates a motile response. First, we identified the type of motility as sliding. Second, we extracted an inducer of sliding motility from agar plates of S. venezuelae and, to our surprise, identified the inducer as the antibiotic chloramphenicol (Cm). At subinhibitory concentrations, many antibiotics possess stimulatory activity, triggering a response in exposed bacteria. This phenomenon, known as hormesis, has been studied for many species and antibiotics (32, 33). Prior studies have shown that subinhibitory concentrations of antibiotics induce responses in exposed bacteria, including changes in transcription, biofilm formation, persistence, and altered virulence (5, 22, 33–37). While tobramycin was previously seen to enhance motility of P. aeruginosa, induction of motility in an otherwise nonmotile population has rarely been reported (19, 20, 22, 34, 38). In addition to chloramphenicol, we found that other antibiotics that target the ribosome also induce motility. Targeted analysis of genes associated with translation stress and antibiotic resistance suggested that the sliding response occurs when ribosome function is perturbed. On the basis of these observations, we suggest that B. subtilis engages a programed motile response to competitive stress that results from subinhibitory antibiotic interference with protein translation.
RESULTS
Competitive interaction with Streptomyces venezuelae induced mobilization of Bacillus subtilis NCIB 3610.
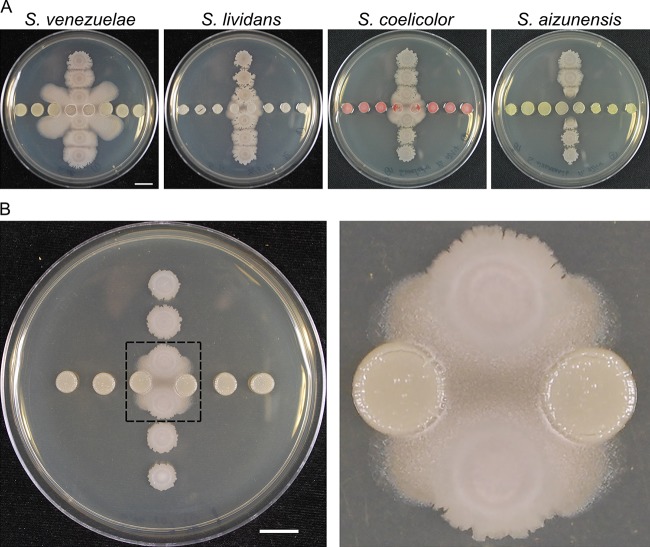
To identify patterns of interaction of B. subtilis NCIB 3610 with Streptomyces species, we plated pairs of the two species on rich agar media in a cross-wise pattern. The spotting pattern enables assessment of differential interactions determined by the proximity of competing species. Streptomyces venezuelae reproducibly induced proximal spots of B. subtilis to initiate a migration across the agar surface (Fig. 1A). In contrast, other species (e.g., Streptomyces lividans and Streptomyces coelicolor) also induce mobilization but do so with delayed timing and to a lesser extent than S. venezuelae (38). In some cases (e.g., Streptomyces aizunensis and Streptomyces sp. strain Mg1), mobilization is not observed, either due to lack of induction or because the lysis observed upon coculture disrupts mobilization (19) (Fig. 1A). On the basis of the pattern and the robust reproducibility of B. subtilis mobility induced by S. venezuelae, this interaction was investigated further.
FIG 1 .
S. venezuelae induces B. subtilis mobilization. (A) Different species of Streptomyces were cultured with B. subtilis to identify patterns of interaction. Streptomyces species were spotted in the horizontal line, and B. subtilis was in the vertical line. Pictures were taken at h 40. (B) S. venezuelae (horizontal spots) induced proximal B. subtilis (vertical spots) to migrate across the agar surface, while this migration was not observed in the distal spots. The right panel presents an enlarged view, highlighting the mobile region inside the dashed box. The picture was taken at h 18. Bars, 1 cm.
Two features evident in the observed pattern indicated a complex interspecies interaction. First, the initial migration of B. subtilis across the agar surface is oriented toward the competitor S. venezuelae (Fig. 1B) (see Movie S1 in the supplemental material). The surface characteristics change for the B. subtilis mobile population, which acquires a rough appearance in comparison to the parent spot. The difference in colony texture indicates a major transition in cellular organization, reminiscent of swarming motility or biofilms (26, 39). Second, as the mobilized population progresses outward toward adjacent S. venezuelae patches, it appears to be repelled (Fig. 1A) (Movie S1). The observed patterns of migration toward S. venezuelae suggested that B. subtilis responds to the presence of diffusible substances produced by S. venezuelae. On the basis of the observed interaction pattern, we sought, first, to define the type of motility used by B. subtilis and, second, to identify inducing substances produced by S. venezuelae.
Competitive interaction between B. subtilis and S. venezuelae. Spots of each bacterial species on agar media were captured by time-lapse video over 72 h and reveal the pattern of sliding motility exhibited by B. subtilis. Initially, B. subtilis moved toward the proximal S. venezuelae spots (up to ~36 h). Continued culture showed that the sliding population of B. subtilis progressed outward and that the population deflected away from the S. venezuelae population (up to 72 h). The agar plate was 8.4 cm in diameter. Download MOVIE S1, MOV file, 5 MB (5.1MB, mov) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
S. venezuelae induces flagellum-independent sliding motility in B. subtilis.
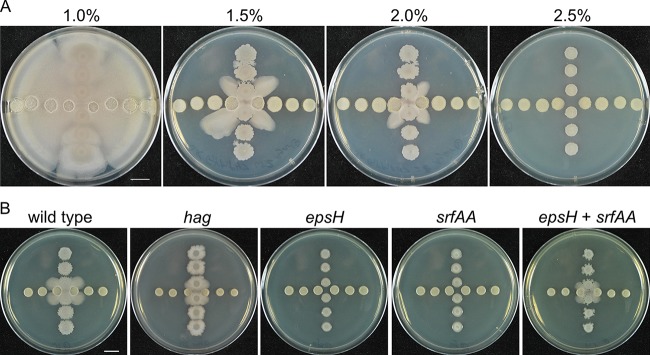
To understand the molecular basis for migration of B. subtilis, multiple approaches were used to identify the type of motility induced by S. venezulae. Bacillus subtilis migration depends upon the viscosity of the surrounding medium. For instance, increasing agar concentrations limit the type of motility available. Agar concentrations above 0.3% (wt/vol) and 1% (wt/vol) prevent swimming and swarming motilities, respectively (25). To characterize the motility of B. subtilis in response to S. venezuelae, we spotted both species onto solid media of different agar concentrations. The motile response to S. venezuelae persisted at agar concentrations of up to 2% (wt/vol), limiting the possibility of identifying swimming or swarming as a basis for motility (Fig. 2A). A third type of motility, sliding, has been demonstrated on specialized media with agar or agarose concentrations typically less than 1% (wt/vol) (29–31, 40). However, because the induced migration of B. subtilis was observed at up to 2% (wt/vol) agar, a concentration which has not been tested in sliding motility experiments, additional experiments were performed to determine the type of motility.
FIG 2 .
Identification of S. venezuelae-induced mobility as sliding. S. venezuelae was spotted in the horizontal line in both panels A and B. Pictures were taken at h 48. (A) The mobilization induced by S. venezuelae was observed at up to 2% agar. (B) Different B. subtilis mutants were cultured with S. venezuelae. The mobility of hag mutant was induced but was not observed in either epsH mutants or srfAA mutants. However, when epsH and srfAA were mixed, the mixture was able to mobilize upon challenge with S. venezuelae. Pictures were taken at h 24. Bars, 1 cm.
To identify genetic requirements for motility, we tested mutant strains that are defective for different types of motility. We first used a strain deficient in the flagellin protein (Δhag) which is incapable of producing flagella (41, 42). Although the strain with the Δhag mutation displayed defects in colony morphology and motility, possibly due to overproduction of surfactin, induction of migration by S. venezuelae was clearly observed using this strain (26) (Fig. 2B). Therefore, the motility observed includes a flagellum-independent component. Sliding motility depends on extracellular polysaccharides (EPS) and surfactin (28–30). Mutant strains that are unable to produce a poly-N-acetylglucosamine component of EPS (epsH) or surfactin (srfAA) were unable to migrate in response to S. venezuelae (43) (Fig. 2B). Because EPS and surfactin are both extracellular products, the single mutant strains were combined to test for extracellular complementation (30). When the B. subtilis epsH and srfAA mutant strains were mixed and competed with S. venezuelae, the migration was restored. Consistent with those results, we identified disruptions in eps and srf genes in a transposon mutagenesis screen for B. subtilis strains that failed to exhibit migration (see Table S3 and Text S1 in the supplemental material). Together, these results strongly suggest that B. subtilis sliding motility is induced by S. venezuelae.
Supplemental Methods. Download TEXT S1, DOCX file, 0.1 MB (99.6KB, docx) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of an inducing metabolite produced by S. venezuelae.
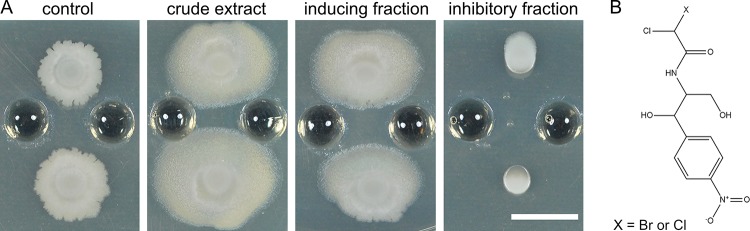
The observed patterns of migration suggested that S. venezuelae produces a substance or substances that induce sliding by B. subtilis. One hypothesis is that a metabolite or enzyme secreted by S. venezuelae activates a specific response in B. subtilis cells, leading to the observed sliding motility. To identify an inducer substance, we extracted agar media after culturing S. venezuelae in isolation. Concentrated crude extracts were then added to wells adjacent to B. subtilis colonies to determine whether inducing activity was present (Fig. 3A). Comparison to a medium-only control revealed robust inducing activity in the crude extract, which was subsequently fractionated using solid-phase extraction first and then high-performance liquid chromatography (HPLC) (see Fig. S1 in the supplemental material). We then collected time-based HPLC fractions and screened for activity on agar plates. The inducing activity was abundant in a single fraction (Fig. 3A). The active fraction was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify candidate inducer metabolites (Fig. S2A and B). An abundant signal identified by MS1 and MS2 analysis was consistent with that of monobromamphenicol, a variant of chloramphenicol where one chlorine atom is replaced by a bromine atom (44) (Fig. 3B).
FIG 3 .
Identification of monobromamphenicol as a sliding inducer. (A) Crude extract from S. venezuelae agar plates was loaded into the wells near B. subtilis and induced robust sliding motility compared with the medium-only control. All time-based HPLC fractions were collected and tested for activity. One fraction had the sliding inducing activity, and one fraction had the growth inhibitory activity. Pictures were taken at h 24. (B) The inducing fraction was brominated chloramphenicol (X = Br [monobromamphenicol]). The inhibitory fraction was chloramphenicol (X = Cl). Bar, 1 cm.
HPLC trace of 40% methanol fraction from crude extracts. Crude extract was further fractionated. The active 40% methanol fraction was applied to HPLC for further separation at a wavelength of 254 nm. The peak corresponding to the inhibitory fraction is labeled with a star. The peak corresponding to the inducing fraction is labeled with a pound sign. Download FIG S1, TIF file, 0.2 MB (189.9KB, tif) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of monobromamphenicol and chloramphenicol by HPLC-MS/MS. (A) The mass and the isotope profile are consistent with those of monobromamphenicol. Different forms of parent ions are labeled in the MS1 spectrum. (B) The identity of monobromamphenicol was further confirmed by analysis of the fragment ions in the MS2 spectrum. (C) The mass and the isotope profile are consistent with those of chloramphenicol. Different forms of parent ions are labeled in the MS1 spectrum. (D) The identity of chloramphenicol was further confirmed by analysis of the fragment ions in the MS2 spectrum. Download FIG S2, TIF file, 0.5 MB (575.9KB, tif) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
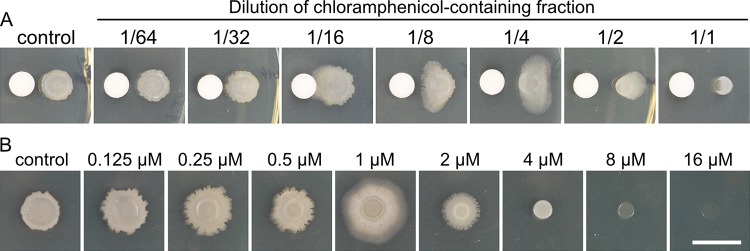
Streptomyces venezulae is well known as a producer of chloramphenicol and is the species from which the antibiotic was originally identified (45, 46). Brominated derivatives have been produced synthetically and by feeding bromine to cells but are not described as natural products of S. venezuelae biosynthesis (44, 47). Possible explanations for the observed activity are that monobromamphenicol is a minor biosynthetic product of S. venezuelae and that chloramphenicol was present at greater abundance in a separate fraction. We identified an inhibitory fraction among the HPLC fractions collected. The inhibitory fraction contained chloramphenicol as detected by LC-MS/MS (Fig. S2C and D). We considered the possibility that chloramphenicol is primarily responsible for inducing B. subtilis sliding motility but that, upon concentrating the crude extract, the more abundant chloramphenicol achieved an inhibitory concentration while monobromamphenicol reached a stimulatory concentration. To determine whether chloramphenicol induces sliding mobility, the chloramphenicol-containing fraction was serially diluted, and each dilution was tested for activity with B. subtilis (Fig. 4A). At a concentration approximately 8-fold lower than that of the parent fraction, chloramphenicol induced a sliding response by B. subtilis that was similar to the response seen upon challenge with S. venezuelae (Fig. 1B). Concentration-dependent differences in activity are described as hormesis, a phenomenon typically characterized by stimulatory effects of an agent at low doses and inhibitory or toxic effects of the same agent at higher concentrations (32, 33). To determine the corresponding concentration of chloramphenicol that is active for sliding induction, a commercially available source of pure chloramphenicol was serially diluted and added directly into the agar media. We observed the maximal sliding response by B. subtilis at 0.3 µg/ml chloramphenicol, which corresponds to an approximate concentration of 1 µM (Fig. 4B; see also Movie S2). These results demonstrate that subinhibitory amounts of chloramphenicol induce a widespread change in a population of B. subtilis, leading to mobilization of the colony.
FIG 4 .
Chloramphenicol induced B. subtilis sliding at subinhibitory concentrations. (A) The chloramphenicol fraction was 2-fold serially diluted, and 10 μl of each dilution was applied onto a filter paper disc 0.6 cm away from B. subtilis. The control was the 40% (vol/vol) methanol solvent. (B) Pure chloramphenicol was serially diluted and added to the agar plate. At 1 µM, the maximal sliding response was induced. The control was the plate without chloramphenicol. Pictures were taken at h 24. Filter disc diameter, 0.6 cm. Bar, 1 cm.
Induction of B. subtilis sliding motility by a subinhibitory concentration of chloramphenicol. (A) A B. subtilis population cultured on medium without chloramphenicol for 72 h at 30°C. (B) A B. subtilis population cultured on the same medium as that described for panel A with supplementation of 0.3 µg/ml (~1 µM) chloramphenicol. The chloramphenicol induced migration in the form of sliding motility. The agar plates were 8.4 cm in diameter. Download MOVIE S2, MOV file, 1.8 MB (1.8MB, mov) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibiotics that block translation induce B. subtilis sliding motility.
To determine whether the sliding response was specific to chloramphenicol, we selected 14 antibiotics to test for induction primarily on the basis of their different mechanisms of action. Serial dilutions of each antibiotic were spotted on filter discs placed adjacent to B. subtilis. In addition to chloramphenicol, three other antibiotics induced sliding mobility. The inducing antibiotics were tetracycline, erythromycin, and spectinomycin, which all target the ribosome and block protein translation (Table 1) (48–51). Interestingly, no aminoglycoside antibiotic tested resulted in activation of sliding mobility by B. subtilis, indicating that errors in translation do not trigger the sliding response. These results led us to conclude that B. subtilis responds to some types of translation inhibitors at subinhibitory concentrations by activating sliding motility.
TABLE 1 .
Four of 14 tested antibiotics induced slidinga
| Antibiotic | Mobility inducer | Target |
|---|---|---|
| Chloramphenicol | Yes | 50S |
| Spectinomycin | Yes | 30S |
| Erythromycin | Yes | 50S |
| Tetracycline | Yes | 30S |
| Apramycin | No | 30S |
| Kanamycin | No | 30S |
| Gentamicin | No | 30S |
| Lincomycin | No | 50S |
| Hygromycin | No | 30S |
| Phleomycin | No | DNA |
| Novobiocin | No | DNA gyrase |
| Ampicillin | No | Transpeptidase |
| Rifamycin | No | RNA polymerase |
In each case, the tested concentrations ranged from inhibitory levels to levels having no detectable effect.
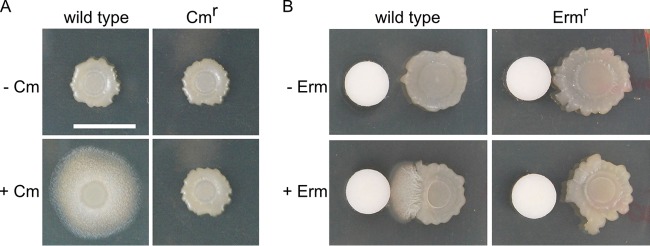
To determine whether the sliding response was dependent upon interaction of the antibiotics with the ribosome, as opposed to an unidentified cellular target, we investigated the effect of antibiotic resistance on sliding. First, a chloramphenicol-resistant (Cmr) B. subtilis strain, which expressed chloramphenicol acetyltransferase, was used to determine whether chemical modification of the antibiotic disrupted sliding. Acetylation of chloramphenicol interferes with binding of the drug to the ribosome (52–54). The Cmr strain did not induce sliding when challenged with chloramphenicol (Fig. 5A). Correspondingly, when wild-type B. subtilis was treated with chloramphenicol acetate at a concentration equivalent to the concentration at which chloramphenicol induced sliding, there was no response (Fig. S3A). However, at elevated (4-fold-greater) levels, chloramphenicol acetate induced sliding activity, indicating that the resistance was overcome with greater amounts of the modified antibiotic. Second, to determine whether direct modification of the ribosome prevented sliding mobility, an erythromycin-resistant (Ermr) B. subtilis strain was treated with inducing concentrations of erythromycin. The Ermr strain expressed a methyltransferase that specifically methylates 23S rRNA, which blocks erythromycin binding (55, 56). In comparison to the wild-type strain results, the Ermr B. subtilis strain did not induce sliding in response to erythromycin (Fig. 5B). Collectively, these results suggest that, when present at subinhibitory concentrations, antibiotics that induce sliding motility target the ribosome and presumably cause protein translation stress.
FIG 5 .
The ribosome plays a key role in antibiotic-induced sliding. (A) Wild-type strain NCIB 3610 and chloramphenicol (Cm)-resistant strain Cmr were spotted on the agar plate in the absence (-) or presence (+) of Cm (0.3 µg/ml). (B) Wild-type strain NCIB 3610 and erythromycin (Erm)-resistant strain Ermr were spotted on the agar plate in the absence or presence of Erm (10 µl of 12.5 µg/ml solution). Pictures were taken at h 24. Filter disc diameter, 0.6 cm. Bar, 1 cm.
Chloramphenicol acetate and lincomycin are inactive for sliding induction. (A) Different amounts of Cm and Cm acetate were spotted on filter discs adjacent to B. subtilis colonies. Cm acetate did not induce sliding at an amount (625 ng) equivalent to the amount of Cm that induced sliding. However, Cm acetate induced sliding in a greater amount (2,500 ng), which was equivalent to the amount of Cm that inhibited growth of B. subtilis. The solvent control for both Cm and Cm acetate was 10% ethanol (in H2O). Pictures were taken at h 24. Filter disc diameter, 0.6 cm. (B) Different amounts of lincomycin (indicated in micrograms per milliliter) were spotted on filter discs adjacent to B. subtilis colonies. The negative-control solvent used in the assay was 10% ethanol, and the positive control was 625 ng Cm. Pictures were taken at h 24. Filter disc diameter, 0.6 cm. Download FIG S3, TIF file, 2.5 MB (2.6MB, tif) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Induction of bmrCD by a subinhibitory concentration of chloramphenicol is consistent with translation stress.
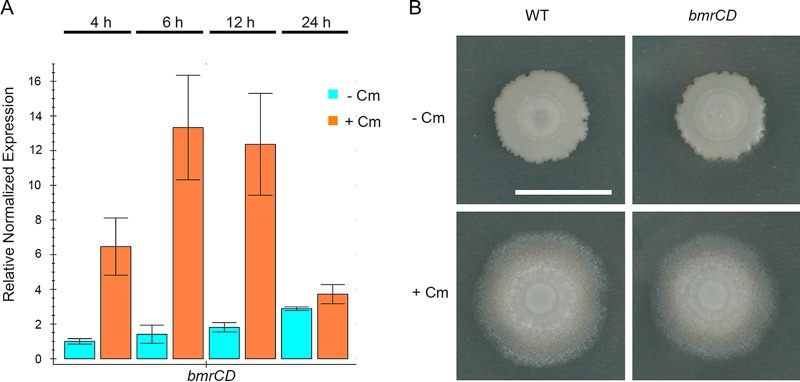
Treatment of B. subtilis with chloramphenicol and other translation inhibitors at subinhibitory concentrations has been shown to affect gene expression (35, 57, 58). Levels of expression of several genes changed due to chloramphenicol exposure (35). Expression of the bmrCD genes, which encode a multidrug efflux transporter, was subsequently shown to be dependent upon the activity of an upstream open-reading frame named bmrB (57). The mechanism of expression control couples efficient translation of BmrB to transcription of downstream bmrCD, where disruption of translation by inhibitory antibiotics causes enhanced production of BmrCD. However, those prior studies investigated laboratory strains B. subtilis 168 and 1A757 in liquid cultures where sliding would not be observed. To determine whether undomesticated B. subtilis NCIB 3610 would activate bmrCD expression in our sliding assays, the transcript abundance of bmrCD was monitored using quantitative reverse transcription-PCR (qRT-PCR). Transcripts of bmrCD were elevated a maximum of 12-fold over the untreated control abundance during the initial 12 h of the experiment (Fig. 6A). The peak abundance of bmrCD transcript occurred between 6 and 12 h. However, after 24 h, when sliding motility was clearly observed, the bmrCD transcript abundance was restored to nearly wild-type levels. This pattern of bmrCD expression is consistent with a transient expression pattern observed previously (57). The elevated expression of bmrCD indicated that the presence of chloramphenicol at a subinhibitory concentration was stressing protein translation, in accordance with the coupled transcription-translation of bmrBCD.
FIG 6 .
bmrCD is related to translation stress but is not required for sliding. (A) Quantitative RT-PCR of bmrCD transcript of the wild-type (WT) strain in the absence (-) and presence (+) of chloramphenicol (Cm) at the indicated time points, 4 h, 6 h, 12 h, and 24 h. Quantification cycle (Cq) values were normalized to Cq values for gyrB. Fold expression values are reported relative to the value for the 4-h sample in the absence of Cm. (B) The WT NCIB 3610 strain and a bmrCD deletion strain were spotted on the agar plate in the absence or presence of Cm (0.3 µg/ml). Pictures were taken at h 24. Bar, 1 cm.
The induced transcription of bmrCD is not limited to chloramphenicol. Multiple antibiotics, all targeting the ribosome, were shown to also lead to elevated bmrCD expression when used at subinhibitory concentrations (57). Intriguingly, lincomycin was previously shown to induce bmrCD expression strongly at subinhibitory concentrations but did not induce sliding at any concentration tested in our assays (Fig. S3B). This observation indicated independence of sliding motility and the effects of translation stress on expression of the BmrCD multidrug efflux pump. To determine whether bmrCD induction is required for sliding motility, bmrC, bmrD, and bmrCD mutant strains were challenged with a subinhibitory chloramphenicol concentration. Despite the absence of BmrCD, the sliding response was intact for the mutant strains (Fig. 6B; see also Fig. S4A). Therefore, the bmrCD genes are not required for sliding motility. To determine whether disrupting regulation of bmrCD would perturb chloramphenicol-induced sliding, we generated a markerless deletion of the bmrB open reading frame (ORF), placing the bmrCD genes directly under the transcriptional control of the bmrB promoter. When exposed to a subinhibitory chloramphenicol concentration, the bmrB mutant strain maintained the sliding response, further supporting the conclusion that the bmrCD genes are not involved in sliding motility (Fig. S4A). Additionally, the mutant strains were not hypersensitive to chloramphenicol, either for sliding or for growth (Fig. 6B; see also Fig. S4B). These observations suggest that, while elevated bmrCD expression indicates translation stress, as-yet-unidentified events are the drivers of antibiotic-induced sliding motility.
Phenotypic effects of bmrBCD disruptions on sliding and sensitivity. (A) Wild-type (WT) B. subtilis NCIB3610 and bmrB, bmrC, and bmrD knockout strains were spotted on the agar plate in the absence or presence of Cm. Pictures were taken at h 24. Bar, 1 cm. (B) Growth curves of the wild-type (WT) and bmrB, bmrC, bmrD, and bmrCD knockout strains in response to different concentrations (0, 1, 2, 4, 8, and 16 μM) of Cm in the period of 18 h. Download FIG S4, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Through tracking changes in colony morphology and mobility during competition between two species of bacteria, we observed that S. venezuelae induces sliding motility in B. subtilis. We found that exposure to low doses of monobromamphenicol and chloramphenicol induced mobilization of the B. subtilis population. Subsequently, we found that multiple translation-inhibiting antibiotics induced B. subtilis sliding. The observed pattern of interaction is indicative of antibiotic hormesis. In this instance, exposure to low doses of translation inhibitory molecules triggers a mobilization of a B. subtilis population. The activation of sliding motility may provide a substantial competitive advantage to B. subtilis, enabling the cells to relocate rapidly and avoid inhibitory doses of antibiotics. Streptomycetes produce many translation-inhibiting antibiotics, consistent with our observation that sliding is frequently observed using pairings of Streptomyces spp. with B. subtilis NCIB 3610 (Fig. 1A).
Perception of low doses of toxic or growth-inhibitory substances provides an opportunity for bacteria to activate protective responses. For instance, biofilms provide a specialized niche for inhabitant bacteria, which alter their physiology and expression of resistance functions, and persisters are protected due to their paused growth and metabolism. Two described examples of antibiotic-protective responses are the formation of biofilms and the formation of persister cells, which lend adaptive resistance to the target organism (8, 12, 34, 59, 60). In both cases, the outcomes are cells that become recalcitrant in the presence of antibiotics. Upon exposure to subinhibitory levels of translation stress, the outcome for B. subtilis is strikingly different. The cells engage a growth-dependent type of mobility, which provides a means to physically relocate a subpopulation. Thus, instead of preventing growth to avert antibiotic stress, B. subtilis activates growth-dependent mobilization. Induced motility in response to antibiotics has rarely been described. Tobramycin was shown previously to enhance the swarming motility of P. aeruginosa (22). In contrast, exposure to several antibiotics was found to diminish motility in multidrug-resistant Salmonella enterica serovar Typhimurium (5). The observed effects of antibiotics suggest that enhanced motility plays an important role in physiological adaptations of bacteria to antibiotic exposure.
Bacillus subtilis displayed counterintuitive directionality with respect to its reaction to chloramphenicol in our assays, which may have additional benefits suggested by the migration pattern relative to S. venezuelae. As observed in still and video images of the interaction, the initial response of B. subtilis is movement toward the colony of S. venezuelae. One speculative idea is that the apparent directionality is a product of the assay format, where cells on the proximal side of a patch are first to respond and expand outward. The outward expansion leads to rapid colonization of the agar surface, including the original spot of S. venezuelae (Fig. 7). Although further evidence is required, the interaction pattern suggests that an early expansion of B. subtilis populations results in suppression of continued growth of the streptomycete, thereby preventing further production of chloramphenicol. Following several more hours of culture, the B. subtilis outward migration extends toward more distantly situated spots of S. venezuelae. However, the migratory population is repelled from the S. venezuelae spots (Fig. 7). One possible explanation is that additional growth of the streptomycetes results in production of growth-inhibitory amounts of chloramphenicol or other antibiotics. If the patterns do indeed reflect responses to changing antibiotic concentrations, the competitive fitness advantage to early activation of sliding mobility would be dually protective, providing an early opportunity to overtake the competitor and an escape mechanism if antibiotic concentrations reach inhibitory levels.
FIG 7 .

Summary model for concentration-dependent effects of chloramphenicol on B. subtilis. The competitive culture format for S. venezuelae and B. subtilis suggests a model for the spatial and temporal effects of population growth on production and diffusion of chloramphenicol in the agar medium. (A) Early (~24 h) development of the S. venezuelae strain (light green spots) results in low concentrations (yellow) of chloramphenicol in the medium, sufficient for stimulating sliding motility in the proximal B. subtilis strain (light tan shapes). (B) Continued growth (~48 h) and, presumably, chloramphenicol biosynthesis by the proximal S. venezuelae spot are impeded by the migratory population of B. subtilis. During this time, the more distal spots of S. venezuelae grow to a greater extent and produce higher yields of chloramphenicol. The concentration of chloramphenicol (and possibly other, unidentified metabolites) becomes sufficient (red) to impede growth and progression of the sliding population of B. subtilis, which is therefore prevented from contacting the S. venezuelae population. The unaffected populations of B. subtilis (not mobilized by chloramphenicol exposure) are visible as dark tan spots.
The mechanism by which subinhibitory antibiotics induce mobilization is likely linked to protein translation. The mechanism of action for each of the inducing antibiotics is that of blocking translation. Intriguingly, the effect is not limited to a single site of action, such as the peptidyl transfer site or the exit tunnel (54, 61). Instead, the mechanisms of activation converge on blockage of progression of translation, as opposed to misincorporation of amino acids or damage to other cellular structures (62–64). This connection is illustrated by the transcriptional activation of bmrCD by subinhibitory concentrations of chloramphenicol and other antibiotics. Stalling in translation of BmrB permits the transcription of the bmrCD genes (57). Because the bmrCD genes are not required for mobilization, sliding induction must require other changes in B. subtilis exposed to inducing antibiotics. Further pursuit of changes in transcriptome, proteome, and metabolome analyses will likely uncover key factors that lead from translation stress to sliding mobility for B. subtilis.
A growing body of evidence demonstrates the many mechanisms by which bacteria detect antibiotics in the environment and initiate protective responses. These responses include formation of biofilm and persisters, enhanced virulence, motility, and other physiological adaptations. The consequences of bacterial adaptation to low doses of antibiotics are likely to have a substantial impact on bacterial communities. Adaptive changes provide opportunities for bacteria to acquire specific mechanisms of resistance to a given antibiotic or class of antibiotics (23, 65, 66). In addition, adaptive changes that influence specialized metabolism, virulence, and mobility are likely to affect interactions in ways that ripple outward to impact other species in a community and even plant and animal host organisms.
MATERIALS AND METHODS
Strains, primers, antibiotics, and growth media.
The strains of Bacillus subtilis used in this study are listed in Table S1 in the supplemental material. Bacillus subtilis mutant strains in the strain 168 (originally from Bacillus Genetic Stock Center [BGSC]) or strain PY79 background were transduced into NCIB 3610 by SPP1 phage transduction using standard procedures (67). Plasmid pDR244 was used to generate markerless deletions in the mls-marked B. subtilis NCIB 3610 strains by looping out a loxP-flanked macrolide, lincosamide, and streptogramin B (MLS) resistance cassette. To obtain a bmrCD double-knockout strain with kanamycin resistance, long-flanking region homology (LFH) PCR was used. Primers bmrC-up1000-fwd and bmrC-up1000-rev were used to amplify the bmrC upstream 1-kb region, and primers bmrD-down1000-fwd and bmrD-down1000-rev were used to amplify the bmrD downstream 1-kb region. Primers kan-fwd and kan-rev were used to amplify the kanamycin cassette. The primers are listed in Table S2. All antibiotics were purchased from Sigma. B. subtilis strains were cultured at 37°C in lysogeny broth (LB) and were inoculated onto GYM7 plates (0.4% [wt/vol] d-glucose, 0.4% [wt/vol] yeast extract, 1.0% [wt/vol] malt extract, 1.5% [wt/vol] agar, 100 mM MOPS [morpholinepropanesulfonic acid], 2.5 mM KH2PO4, 2.5 mM K2HPO4, pH 7.0) and grown to an optical density at 600 nm (OD600) of 1. Streptomyces spore stocks were maintained in water at 4°C. Additional details of the methods used are provided in Text S1 in the supplemental material.
Bacterial strains used in this study. Download TABLE S1, DOCX file, 0.1 MB (95.5KB, docx) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S2, DOCX file, 0.05 MB (52.7KB, docx) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transposon mutagenesis results. Download TABLE S3, DOCX file, 0.03 MB (33.6KB, docx) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Coculture assays and motility assays.
Coculture assays were performed as previously described (19). Briefly, 2.5 μl of Streptomyces spores (107 spores/ml) was spotted in the horizontal line and grown for 12 h at 30°C. A 1.5-μl volume of B. subtilis was then spotted 6 mm from a Streptomyces sp. in the vertical line. For motility assays, 1.5 μl of B. subtilis was spotted 6 mm from wells or filter discs on the agar plate.
Sliding inducer extraction and identification.
S. venezuelae was cultured on the top layer of GYM7 plates separated from the bottom layer by a sheet of cellophane. The top layer (5 ml) along with the cellophane was removed after 5 days of S. venezuelae growth. Metabolites were extracted from the lower layer (20 ml) by freezing the agar and separating aqueous media by filtration through a 60-ml syringe containing a layer of Miracloth (EMD Millipore). The squeezed extracts were pooled and then lyophilized. The crude extract was suspended in one-fifth of the original volume in H2O. The crude extracts were initially fractionated by the use of an SPE C18 column (Sigma). To extract the mobility inducer, 3 ml of crude extract was applied to the 3-ml SPE C18 column (Supelco). The column was washed with 6 ml of 10% (vol/vol) methanol, followed by elution with a 20% (vol/vol) stepwise gradient (from 20% to 100%) of methanol/H2O. Methanol in all fractions was removed using a rotary evaporator. The concentrated fractions were suspended in 200 µl H2O. All fractions were tested for mobility-inducing activity by spotting 10 µl on a well or a filter disc 6 mm away from B. subtilis colonies, and the mobility induction was observed after 24 h. The 40% (vol/vol) methanol fraction was active, and multiple 40% (vol/vol) extracts were pooled for further analysis. The 40% (vol/vol) methanol fraction was further fractionated by HPLC (Agilent 1200) using a semipreparative C18 column (Phenomenex) (10 by 250 mm, 5-μm particles). An isocratic method was used (30% [vol/vol] solvent A, 70% [vol/vol] solvent B. 20 min in total) with a flow rate of 4 ml/min. Solvent A was acetonitrile. Solvent B was 0.1% (vol/vol) formic acid–H2O. For each injection, 100 µl pooled active fraction was applied. Time-based fractions from HPLC were collected and tested for mobility-inducing activity. Those active fractions were analyzed by LC-MS/MS. Specifically, LC-MS/MS was performed with an Agilent 1260 HPLC system coupled with a binary pump and a 1200 series diode array detector UV light-visible light (UV-Vis) detector (compounds were detected at 254 nm, 340 nm, and 420 nm) followed by a MicroTOF-Q Ⅱ mass spectrometer (Bruker Daltonics) using an electrospray ionization (ESI) source. Separation was performed with a Supelcosil LC-18 column (Supelco) (15 cm by 3 mm, 3-µm particles). LC conditions were as follows: t = 0 min, 100% A; t = 2 min, 100% A; t = 12 min, 30% A; t = 20 min, 30% A; t = 25 min, 100% A; t = 35 min, 100% A; t = 40 min, 100% A. The flow rate was 400 µl/min. Solvent A was 5 mM ammonium acetate buffer (pH 6.6). Solvent B was 75% (vol/vol) methanol and 25% H2O. A mass spectrometer was calibrated with a diluted sodium acetate solution, and six m/z values (158.9641, 362.9263, 498.9012, 566.8886, 634.8760, and 770.8509) were used for the calibration. The mass spectrometer was operated in positive mode in a mass range from 50 to 1,500 Da. The ion source temperature was maintained at 200°C with 8 eV of ionization energy and 4,500 V of capillary voltage. Helium was used as the collision gas.
RNA extraction.
Wild-type B. subtilis NCIB 3610 was grown to the early stationary phase (OD600 = 1) and was inoculated on GYM7 plates with or without 1 μM chloramphenicol, followed by incubation at 30°C. B. subtilis colonies at 4 h, 6 h, and 12 h and the outer region of colonies at 24 h were scraped after treatment with 3 ml of stabilization mixture (2-ml RNAprotect Bacteria Reagent [Qiagen] with 1-ml Tris-buffered saline [TBS] buffer) on each plate. The bacterial suspension was transferred to a 15-ml conical tube, subjected to 5 s of vortex mixing, and incubated at room temperature for 5 min. Aliquots (500 µl) were transferred to individual 2-ml Eppendorf tubes. Cell pellets were collected by centrifugation at 17,900 × g for 10 min. RNA was isolated as previously described (19). Briefly, cells were lysed with lysis buffer (15 mg/ml lysozyme, 5 mg/ml proteinase K, 100 mM Tris HCl−50 mM EDTA buffer, pH 8.0) and subjected to vigorous vortex mixing for 45 min at ambient temperature. A 1-ml volume of Trizol reagent (Sigma) was added to each sample. RNA was precipitated using standard procedures. RNA samples were cleaned with a Turbo DNA-free kit (Applied Biosystems).
Quantitative RT-PCR (qRT-PCR).
qRT-PCR was performed as described previously (38). Briefly, 50 ng of total RNA was used as the template for cDNA synthesis with a High-Capacity RNA-to-cDNA kit (Thermo Fisher Scientific). A SsoAdvanced Universal SYBR green Supermix kit (Bio-Rad) and a CFX96 Touch real-time PCR thermocycler (Bio-Rad) were used to perform quantitative PCR as previously described (38). gyrB was used as the reference gene. Target abundance was normalized to gyrB, and the fold change value was calculated by comparison to the untreated sample at 4 h. Each experiment was repeated three times.
ACKNOWLEDGMENTS
We thank Larry Dangott, Tadhg Begley, and Yindrila Chakrabarty for the training and use of the mass spectrometer. We thank Daniel Ziegler (BGSC) for providing B. subtilis strains. We thank B. Christopher Hoefler for experimental advice and Reed Stubbendieck for helpful comments on the manuscript.
This work was supported by the National Science Foundation (NSF-CAREER award MCB-1253215) to P.D.S. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 2.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. 2011. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol 193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornforth DM, Foster KR. 2013. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol 11:285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 5.Brunelle BW, Bearson BL, Bearson SMD. 2015. Chloramphenicol and tetracycline decrease motility and increase invasion and attachment gene expression in specific isolates of multidrug-resistant Salmonella enterica serovar Typhimurium. Front Microbiol 5:801. doi: 10.3389/fmicb.2014.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller S, Strack SN, Hoefler BC, Straight PD, Kearns DB, Kirby JR. 2014. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl Environ Microbiol 80:5603–5610. doi: 10.1128/AEM.01621-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traxler MF, Kolter R. 2015. Natural products in soil microbe interactions and evolution. Nat Prod Rep 32:956–970. doi: 10.1039/c5np00013k. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira NM, Oliveria NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, Foster KR. 2015. Biofilm formation as a response to ecological competition. PLoS Biol 13:e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoefler BC, Gorzelnik KV, Yang JY, Hendricks N, Dorrestein PC, Straight PD. 2012. Enzymatic resistance to the lipopeptide surfactin as identified through imaging mass spectrometry of bacterial competition. Proc Natl Acad Sci U S A 109:13082–13087. doi: 10.1073/pnas.1205586109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molina L, Constantinescu F, Michel L, Reimmann C, Duffy B, Défago G. 2003. Degradation of pathogen quorum-sensing molecules by soil bacteria: a preventive and curative biological control mechanism. FEMS Microbiol Ecol 45:71–81. doi: 10.1016/S0168-6496(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 11.Kelsic ED, Zhao J, Vetsigian K, Kishony R. 2015. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature 521:516–519. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 13.Oh D-C, Kauffman CA, Jensen PR, Fenical W. 2007. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod 70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 14.Oh D-C, Jensen PR, Kauffman CA, Fenical W. 2005. Libertellenones A−D: induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg Med Chem 13:5267–5273. doi: 10.1016/j.bmc.2005.05.068. [DOI] [PubMed] [Google Scholar]
- 15.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. 2013. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 4:e00459-13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeckh V, Scherlach K, Nützmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A 106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flannagan RS, Valvano MA, Koval SF. 2004. Downregulation of the motA gene delays the escape of the obligate predator Bdellovibrio bacteriovorus 109J from bdelloplasts of bacterial prey cells. Microbiology 150:649–656. doi: 10.1099/mic.0.26761-0. [DOI] [PubMed] [Google Scholar]
- 18.An D, Danhorn T, Fuqua C, Parsek MR. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci U S A 103:3828–3833. doi: 10.1073/pnas.0511323103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stubbendieck RM, Straight PD. 2015. Escape from lethal bacterial competition through coupled activation of antibiotic resistance and a mobilized subpopulation. PLoS Genet 11:e1005722. doi: 10.1371/journal.pgen.1005722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones SE, Ho L, Rees CA, Hill JE, Nodwell JR, Elliot MA. 2017. Streptomyces exploration is triggered by fungal interactions and volatile signals. Elife 6:e21738. doi: 10.7554/eLife.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 22.Linares JF, Gustafsson I, Baquero F, Martinez JL. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A 103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai S, Tremblay J, Déziel E. 2009. Swarming motility: a multicellular behaviour conferring antimicrobial resistance. Environ Microbiol 11:126–136. doi: 10.1111/j.1462-2920.2008.01747.x. [DOI] [PubMed] [Google Scholar]
- 24.Butler MT, Wang Q, Harshey RM. 2010. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci U S A 107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol 49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 27.Calvio C, Celandroni F, Ghelardi E, Amati G, Salvetti S, Ceciliani F, Galizzi A, Senesi S. 2005. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J Bacteriol 187:5356–5366. doi: 10.1128/JB.187.15.5356-5366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinsinger RF, Kearns DB, Hale M, Fall R. 2005. Genetic requirements for potassium ion-dependent colony spreading in Bacillus subtilis. J Bacteriol 187:8462–8469. doi: 10.1128/JB.187.24.8462-8469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinsinger RF, Shirk MC, Fall R. 2003. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol 185:5627–5631. doi: 10.1128/JB.185.18.5627-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Gestel J, Vlamakis H, Kolter R. 2015. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol 13:e1002141. doi: 10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grau RR, de Oña P, Kunert M, Leñini C, Gallegos-Monterrosa R, Mhatre E, Vileta D, Donato V, Hölscher T, Boland W, Kuipers OP, Kovács ÁT. 2015. A duo of potassium-responsive histidine kinases govern the multicellular destiny of Bacillus subtilis. mBio 6:e00581. doi: 10.1128/mBio.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yim G, Wang HH, Davies J. 2007. Antibiotics as signalling molecules. Philos Trans R Soc Lond B Biol Sci 362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies J, Spiegelman GB, Yim G. 2006. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 35.Lin JT, Connelly MB, Amolo C, Otani S, Yaver DS. 2005. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob Agents Chemother 49:1915–1926. doi: 10.1128/AAC.49.5.1915-1926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones C, Allsopp L, Horlick J, Kulasekara H, Filloux A. 2013. Subinhibitory concentration of kanamycin induces the Pseudomonas aeruginosa type VI secretion system. PLoS One 8:e81132. doi: 10.1371/journal.pone.0081132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bleich R, Watrous JD, Dorrestein PC, Bowers AA, Shank EA. 2015. Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. Proc Natl Acad Sci U S A 112:3086–3091. doi: 10.1073/pnas.1414272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vargas-Bautista C, Rahlwes K, Straight P. 2014. Bacterial competition reveals differential regulation of the pks genes by Bacillus subtilis. J Bacteriol 196:717–728. doi: 10.1128/JB.01022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fall R, Kearns DB, Nguyen T. 2006. A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol 6:31. doi: 10.1186/1471-2180-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joys TM, Frankel RW. 1967. Genetic control of flagellation in Bacillus subtilis. J Bacteriol 94:32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iino T. 1977. Genetics of structure and function of bacterial flagella. Annu Rev Genet 11:161–182. doi: 10.1146/annurev.ge.11.120177.001113. [DOI] [PubMed] [Google Scholar]
- 43.Roux D, Cywes-Bentley C, Zhang YF, Pons S, Konkol M, Kearns DB, Little DJ, Howell PL, Skurnik D, Pier GB. 2015. Identification of poly-N-acetylglucosamine as a major polysaccharide component of the Bacillus subtilis biofilm matrix. J Biol Chem 290:19261–19272. doi: 10.1074/jbc.M115.648709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bald R, Erdmann VA, Pongs O. 1972. Irreversible binding of chloramphenicol analogues to E. coli ribosomes. FEBS Lett 28:149–152. doi: 10.1016/0014-5793(72)80698-7. [DOI] [PubMed] [Google Scholar]
- 45.Ehrlich J, Bartz QR, Smith RM, Joslyn DA, Burkholder PR. 1947. Chloromycetin, a new antibiotic from a soil actinomycete. Science 106:417. doi: 10.1126/science.106.2757.417. [DOI] [PubMed] [Google Scholar]
- 46.Ehrlich J, Gottlieb D, Burkholder PR, Anderson LE, Pridham TG. 1948. Streptomyces venezuelae, n. sp., the source of chloromycetin. J Bacteriol 56:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith CG. 1958. Effect of halogens on the chloramphenicol fermentation. J Bacteriol 75:577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 49.Weisblum B. 1995. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 51.Pioletti M, Schlünzen F, Harms J, Zarivach R, Glühmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J 20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw WV, Brodsky RF. 1968. Characterization of chloramphenicol acetyltransferase from chloramphenicol-resistant Staphylococcus aureus. J Bacteriol 95:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leslie AG, Moody PC, Shaw WV. 1988. Structure of chloramphenicol acetyltransferase at 1.75-A resolution. Proc Natl Acad Sci U S A 85:4133–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulkley D, Innis CA, Blaha G, Steitz TA. 2010. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci U S A 107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong P, Pratt SD, Edalji RP, Walter KA, Holzman TF, Shivakumar AG, Katz L. 1995. Substrate requirements for ErmC’ methyltransferase activity. J Bacteriol 177:4327–4332. doi: 10.1128/jb.177.15.4327-4332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skinner R, Cundliffe E, Schmidt FJ. 1983. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem 258:12702–12706. [PubMed] [Google Scholar]
- 57.Reilman E, Mars RAT, van Dijl JM, Denham EL. 2014. The multidrug ABC transporter BmrC/BmrD of Bacillus subtilis is regulated via a ribosome-mediated transcriptional attenuation mechanism. Nucleic Acids Res 42:11393–11407. doi: 10.1093/nar/gku832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dar D, Shamir M, Mellin JR, Koutero M, Stern-Ginossar N, Cossart P, Sorek R. 2016. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352:aad9822. doi: 10.1126/science.aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bui LMG, Conlon BP, Kidd SP. 2017. Antibiotic tolerance and the alternative lifestyles of Staphylococcus aureus. Essays Biochem 61:71–79. doi: 10.1042/EBC20160061. [DOI] [PubMed] [Google Scholar]
- 60.Kaplan JB. 2011. Antibiotic-induced biofilm formation. Int J Artif Organs 34:737–751. [DOI] [PubMed] [Google Scholar]
- 61.Dunkle JA, Xiong L, Mankin AS, Cate JHD. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci U S A 107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davies J, Gorini L, Davis BD. 1965. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol Pharmacol 1:93–106. [PubMed] [Google Scholar]
- 63.Davis BD, Chen LL, Tai PC. 1986. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc Natl Acad Sci U S A 83:6164–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hol FJH, Hubert B, Dekker C, Keymer JE. 2016. Density-dependent adaptive resistance allows swimming bacteria to colonize an antibiotic gradient. ISME J 10:30–38. doi: 10.1038/ismej.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 67.Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol 14:1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Competitive interaction between B. subtilis and S. venezuelae. Spots of each bacterial species on agar media were captured by time-lapse video over 72 h and reveal the pattern of sliding motility exhibited by B. subtilis. Initially, B. subtilis moved toward the proximal S. venezuelae spots (up to ~36 h). Continued culture showed that the sliding population of B. subtilis progressed outward and that the population deflected away from the S. venezuelae population (up to 72 h). The agar plate was 8.4 cm in diameter. Download MOVIE S1, MOV file, 5 MB (5.1MB, mov) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental Methods. Download TEXT S1, DOCX file, 0.1 MB (99.6KB, docx) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HPLC trace of 40% methanol fraction from crude extracts. Crude extract was further fractionated. The active 40% methanol fraction was applied to HPLC for further separation at a wavelength of 254 nm. The peak corresponding to the inhibitory fraction is labeled with a star. The peak corresponding to the inducing fraction is labeled with a pound sign. Download FIG S1, TIF file, 0.2 MB (189.9KB, tif) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of monobromamphenicol and chloramphenicol by HPLC-MS/MS. (A) The mass and the isotope profile are consistent with those of monobromamphenicol. Different forms of parent ions are labeled in the MS1 spectrum. (B) The identity of monobromamphenicol was further confirmed by analysis of the fragment ions in the MS2 spectrum. (C) The mass and the isotope profile are consistent with those of chloramphenicol. Different forms of parent ions are labeled in the MS1 spectrum. (D) The identity of chloramphenicol was further confirmed by analysis of the fragment ions in the MS2 spectrum. Download FIG S2, TIF file, 0.5 MB (575.9KB, tif) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Induction of B. subtilis sliding motility by a subinhibitory concentration of chloramphenicol. (A) A B. subtilis population cultured on medium without chloramphenicol for 72 h at 30°C. (B) A B. subtilis population cultured on the same medium as that described for panel A with supplementation of 0.3 µg/ml (~1 µM) chloramphenicol. The chloramphenicol induced migration in the form of sliding motility. The agar plates were 8.4 cm in diameter. Download MOVIE S2, MOV file, 1.8 MB (1.8MB, mov) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chloramphenicol acetate and lincomycin are inactive for sliding induction. (A) Different amounts of Cm and Cm acetate were spotted on filter discs adjacent to B. subtilis colonies. Cm acetate did not induce sliding at an amount (625 ng) equivalent to the amount of Cm that induced sliding. However, Cm acetate induced sliding in a greater amount (2,500 ng), which was equivalent to the amount of Cm that inhibited growth of B. subtilis. The solvent control for both Cm and Cm acetate was 10% ethanol (in H2O). Pictures were taken at h 24. Filter disc diameter, 0.6 cm. (B) Different amounts of lincomycin (indicated in micrograms per milliliter) were spotted on filter discs adjacent to B. subtilis colonies. The negative-control solvent used in the assay was 10% ethanol, and the positive control was 625 ng Cm. Pictures were taken at h 24. Filter disc diameter, 0.6 cm. Download FIG S3, TIF file, 2.5 MB (2.6MB, tif) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phenotypic effects of bmrBCD disruptions on sliding and sensitivity. (A) Wild-type (WT) B. subtilis NCIB3610 and bmrB, bmrC, and bmrD knockout strains were spotted on the agar plate in the absence or presence of Cm. Pictures were taken at h 24. Bar, 1 cm. (B) Growth curves of the wild-type (WT) and bmrB, bmrC, bmrD, and bmrCD knockout strains in response to different concentrations (0, 1, 2, 4, 8, and 16 μM) of Cm in the period of 18 h. Download FIG S4, TIF file, 2.6 MB (2.7MB, tif) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains used in this study. Download TABLE S1, DOCX file, 0.1 MB (95.5KB, docx) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S2, DOCX file, 0.05 MB (52.7KB, docx) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transposon mutagenesis results. Download TABLE S3, DOCX file, 0.03 MB (33.6KB, docx) .
Copyright © 2018 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.