Abstract
Background: Key genes related to cell cycle and apoptosis pathways play critical roles in bladder cancer. Single nucleotide polymorphisms (SNPs) in the 3’-untranslated regions (3’-UTR) of genes may impact microRNA (miRNA)-messenger RNA (mRNA) binding capacity and alter gene expression to contribute to the susceptibility of cancers. However, an association of genetic variations in cell cycle and apoptosis pathways with bladder cancer risk, has not been reported . Materials and Methods: We selected SNPs in the 3’-UTR of cell cycle and apoptosis pathways genes and genotyped them with a case-control study consisting of 578 bladder cancer patients and 1,006 cancer-free subjects. Dual luciferase reporter gene assay was performed to validate the biological function of important SNPs. Results: We found that 5 SNPs might change the binding ability of miRNA to their target genes, among which PPP3CC rs7431 A>G located in the 3’-untranslated regions with the minimum p-value (p=5.75×10–4). Analysis revealed that the rs7431 disrupted miR-212 and miR-132 targeting sites. Logistic regression revealed a significantly decreased risk of bladder cancer associated with the PPP3CC rs7431 A>G polymorphism with an odds ratio (OR) of 0.76 [95% confidence interval (CI)=0.66-0.89, p=5.75×10–4]. Luciferase report assay showed that both miR-212 and miR-132 could lead to significantly increased PPP3CC expression levels in the construct with the G allele compared to the A allele. Conclusion: PPP3CC rs7431 may alter miRNA binding ability of miR-212 and miR-132, and thus decrease bladder cancer risk.
Keywords: 3’-UTR, bladder cancer, cell-cycle and apoptosis pathways, miRNA, SNP
Bladder cancer is one of the most frequent malignancies of the urinary tract worldwide (1). In China, the incidence rates of bladder cancer have increased gradually from 2000 to 2011 (2). The overall incidence was 7.68 per 100,000 and mortality rates were 1.99 per 100,000 in the population according to the National Central Cancer Registry (NCCR) of China 2015 annual report (3). Despite the relatively low incidence of bladder carcinoma compared to other cancer types, the high local recurrence rate requires regular medical examination associated with a significant impact on healthcare resources and the patient’s quality of life (4). Evidence showed that bladder cancer is a complex disease caused by both genetic and environmental risk factors, including cigarette smoking, occupational exposure to carcinogens (5). Even following the same exposure to environmental carcinogenic factors, only a small number of the exposed individuals eventually occur cancers, suggesting that genetic susceptibility in critical genes may play an important role in the etiology of bladder cancer.
Cell-cycle and apoptosis pathways are related to the occurrence and development of tumors (6). Cell cycle is the series of events that happen in a cell contributing to its division and duplication of its DNA to produce two daughter cells (7), and cell apoptosis is a process of programmed cell death that occurs in multicellular organisms (8). Both of the pathways play critical roles in regulating cellular transformation and maligancy. Defective processes of cell cycle and apoptosis affected by key genes have been implicated in bladder cancer (6).
The regulation of cell cycle and apoptosis can be controlled by epigenetics. As an important aspect of epigenetics, microRNAs (miRNAs) are a kind of small noncoding and single-stranded RNA molecules (~22 nt), which normally regulate gene expression through binding the 3’-untranslated regions (3’-UTR) of messenger RNA (mRNA) of target gene at the post-transcriptional level (9). It has been suggested that miRNAs play critical roles in regulating a variety of biological processes, such as organ development, tumorigenesis, cell differentiation, cell proliferation and apoptosis (10). Several studies showed that miRNAs could act as oncogenes and tumor suppressors to influence the occurrence and development of tumor by targeting 3’-UTR of important genes (11,12).
Single nucleotide polymorphisms (SNPs) are one of the most frequent variations in the human genome. Accumulating evidence have demonstrated that an increasing number of 3’-UTR SNPs located in miRNA binding sites impact miRNA binding ability and affect tumor development (13-15). Therefore, genetic variants in the miRNAs binding sites of key target genes involved in cell cycle and apoptosis pathways may impact miRNA-mRNA binding and alter gene expression. However, the function between genetic variants in miRNA binding sites and bladder cancer risk is still unknown.
Cell-cycle and apoptosis are regulated by epigenetic miRNA and genetic factors that occur in gene 3’-UTR SNPs. We investigated the association between genetic variations of key genes involved in cell cycle and apoptosis pathways and bladder cancer risk with a case-control study including 578 cases and 1,006 controls in the a Chinese population.
Materials and Methods
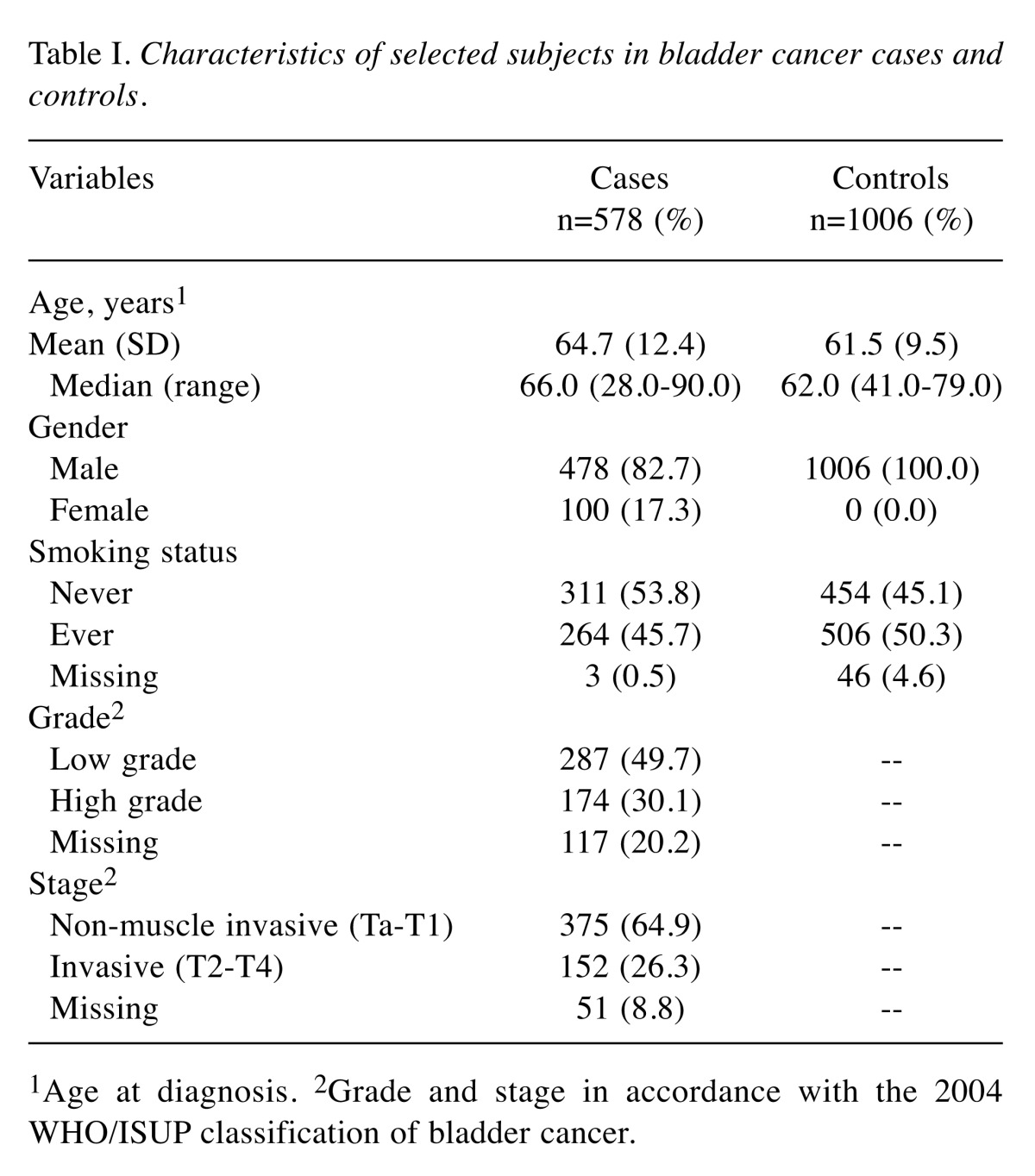
Study population. This study was approved by the Institutional Review Board of Nanjing Medical University (Nanjing, China). The recruitment of all individuals was described previously (16). Briefly, 578 cases were collected from Nanjing beginning in May 2003, and 1,006 controls shared with ChinaPCa (17). The bladder cancer patients were histologically diagnosed by at least two pathologists, and detailed clinical data were collected by local urologists. Pathological grade and stage were consistent with the 2004 WHO/ISUP classification of bladder cancer. Individuals who smoked daily for >1 year were considered smokers, and the others were considered non-smokers. All participants were provided written informed consent form for their samples and data to be used.
SNPs selection and genotyping. The candidate genes involved in the cell cycle and apoptosis pathways were found using KEGG pathway maps (www.kegg.jp/kegg/pathway.html), and all the SNPs in the 3’-UTR of these genes were selected based on 1000 Genomes Project data. Subsequently, SNPs were filtered if they (a) exhibited genotype call rate of <90%; (b) showed a minor allele frequency (MAF) of <0.05 in either cases or controls; or (c) deviated from Hardy-Weinberg equilibrium (HWE) in the controls at p<0.001.
The genomic DNA was extracted from peripheral blood lymphocytes. The selected SNPs were genotyped in 578 cases and 1,006 controls using the Illumina Infinium® BeadChip platform (Illumina, San Diego, CA, USA). Genotype calling was conducted by the GenTrain version 1.0 clustering algorithm in GenomeStudio v2011.1 (Illumina, San Diego, CA, USA).
Bioinformatics analysis of miRNA-mRNA binding ability. The online software miRNASNP (http://www.bioguo.org/miRNASNP/) was used to predict whether 3’-UTR SNPs would change the miRNAs binding ability to the target genes.
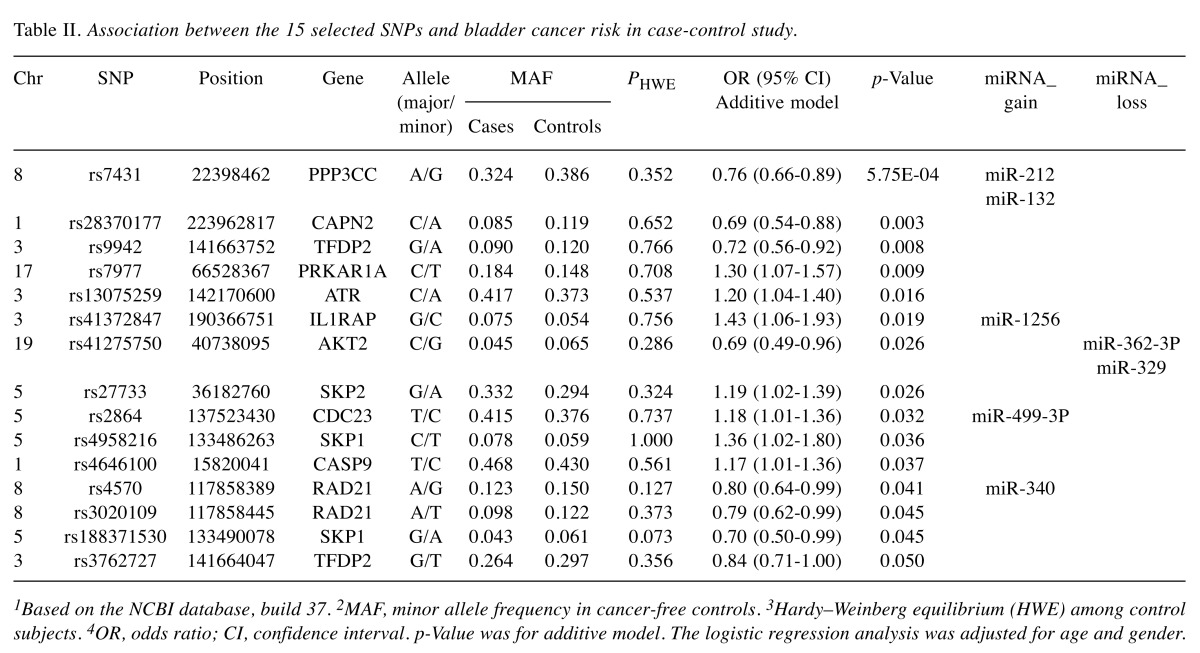
Construction of reporter plasmids and luciferase reporter assay. To construct luciferase reporter plasmids, PPP3CC 3’-UTR fragments (A allele or G allele for SNP rs7431) were gene synthesized and inserted at the XhoI/NotI site, downstream of the Renilla luciferase gene in the psiCHECK™-2 vector (Promega, Madison, WI, USA) by Generay Company (Shanghai, China). The nucleotide sequences of the constructed plasmids were confirmed by DNA sequencing method.
In this study, we chose T24 and J82 cell lines for the further luciferase assay. Both cell lines were grown at 37˚C in the presence of 5% CO2 in a humidified incubator. T24 and J82 cells were seeded on 24-well plates and then each transient co-transfected reaction contained 0.8 μg constructed vectors, either with A allele or with G allele and 20 pmol chemically synthesized miR-212 or miR-132 or negative control miRNA with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), respectively. Luciferase activity was determined with a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) after 48 h. Relative protein levels were expressed as Renilla luciferase normalized against Firefly luciferase signals. Independent triplicate experiments were performed for each plasmid construct.
Statistical analysis. HWE of the controls’ genotype frequencies was tested using a goodness-of-fit χ2 test. Associations between each SNP and risk of bladder cancer were estimated with odds ratios (ORs) and 95% confidence intervals (CIs) derived from logistic regression analysis under an additive model. All statistical tests were two-sided and a p-value <0.05 was used as the criterion of statistical significance. The Student’s t-test was used to analyze the results of miRNA or mRNA expression in bladder cancer and normal tissues. The miRNA expression levels in bladder cancer tissues and normal tissues were downloaded from the publicly available Gene Expression Omnibus (GEO) database (GSE68594). Statistical analyses were conducted with Statistical Analysis System software (v.9.1 SAS Institute Inc., Cary, NC, USA)
Results
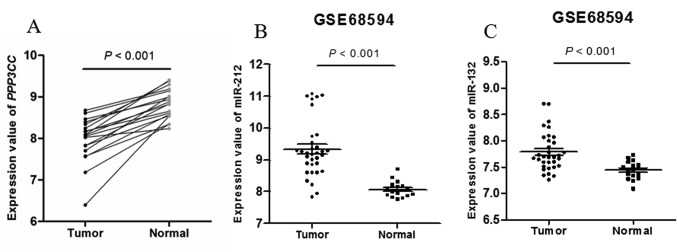
Association between 15 selected SNPs with bladder cancer risk. There were 230 genes involved in the cell cycle and apoptosis pathways. We obtained 2,562 SNPs in the 3’-UTR of these candidate genes based on the 1000 Genomes Project and then 597 SNPs were selected after quality control. We detected these 597 SNPs genotype distribution in a case-control study and the distribution of the selected characteristics between case and control subjects is summarized in Table I. Finally, we chose 15 significantly different (p<0.05) candidate SNPs and the genotype distributions were shown in Table II. The genotype frequencies of these 15 SNPs in controls were in agreement with HWE (p>0.001). Among them, we found that the SNP rs7431 A>G with the minimum p-value performed a significantly decreased risk of bladder cancer with an OR of 0.76 (95%CI=0.66-0.89, p=5.75×10–4).
Table I. Characteristics of selected subjects in bladder cancer cases and controls.
1Age at diagnosis. 2Grade and stage in accordance with the 2004 WHO/ISUP classification of bladder cancer.
Table II. Association between the 15 selected SNPs and bladder cancer risk in case-control study.
1Based on the NCBI database, build 37. 2MAF, minor allele frequency in cancer-free controls. 3Hardy–Weinberg equilibrium (HWE) among control subjects. 4OR, odds ratio; CI, confidence interval. p-Value was for additive model. The logistic regression analysis was adjusted for age and gender.
PPP3CC 3’-UTR rs7431 A>G affects PPP3CC expression by regulating the hsa-miR-212 and hsa-miR-132 binding. We used bioinformatics analysis to determine whether the genetic variants would change the binding ability of miRNA. As a result, we found 5 SNPs might change the binding ability to 3’-UTR of the target genes among 15 candidate SNPs (Table II), and rs7431 had both the minimum p-value and the biological effect. To explore whether the SNP rs7431 may influence PPP3CC regulation mediated by miR-212 and miR-132, we constructed luciferase reporter gene assay by using psiCHECK™-2 vector (Figure 1a). Based on the bioinformatics analysis, the PPP3CC rs7431 was located on the target site of miR-212 and miR-132 (Figure 1b). As shown in Figure 1c, both of miR-212 and miR-132 can significantly reduce luciferase activities in T24 cells transfected with rs7431A/G allelic reporter constructs compared to negative control miRNAs. Moreover, vectors within the rs7431 G allele had an increased luciferase activity in adding miR-212 (p<0.001) and miR-132 (p=0.001) respectively compared with those within the rs7431 A allele in T24 cells. The similar result was also observed in J82 cells (p=0.001 and p=0.002, respectively). Our findings suggested that both miR-212 and miR-132 could bind and positively regulate the transcription of PPP3CC which was more positively influenced by the variant G allele in vitro.
Figure 1. Characterization and functional analyses of the 3’-UTR of PPP3CC. (A) Schematic representation of reporter plasmids containing the PPP3CC 3’-UTR, which was inserted downstream of Renilla luciferase gene in the psiCHECK™-2 vector. (B) Complementarity between miR-212/miR-132 and the PPP3CC 3’-UTR site targeted. The SNP rs7431 was located within the ‘seed region’ of the miR-212/miR-132 binding site. (C) The effect of SNP rs7431 on the interaction between the PPP3CC 3’-UTR and miR-212/ miR-132 in T24 and J82 cells. The luciferase activity of each construct was normalized against the negative control miRNA (NC) transient transfected with constructed vectors with A allele.
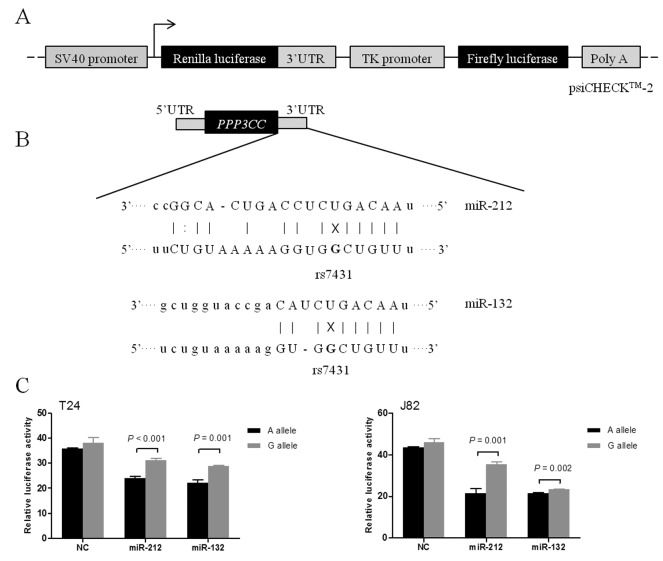
From the independent TCGA data, we found the mRNA expression levels of PPP3CC significantly decreased in 19 tumor tissues compared with their adjacent normal tissues (p<0.001) (Figure 2a). On the contrary, the expression of miR-212/miR-132 significantly increased in tumor tissues compared with normal tissues (p<0.001; Figure 2b and c) by GSE68594.
Figure 2. Expression of PPP3CC and miR-212/ miR-132 in bladder cancer and normal tissues. (A) PPP3CC mRNA expression levels were analyzed in paired bladder tissues from 19 subjects from TCGA data. The p-values were determined using the paired t-test. The expression levels of (B) miR-212 and (C) miR-132 were evaluated using the GSE68594 database.
Discussion
In the present study, we conducted a case-control study to investigate the association between SNPs in the 3’-UTR of genes involved in the cell cycle and apoptosis pathways and bladder cancer risk. We identified that rs7431 on the miR-212 and miR-132 binding site in PPP3CC 3’-UTR was associated with the decreased risk of bladder cancer. The Dual luciferase reporter gene assay indicated that PPP3CC rs7431 A to G substitution may decrease the binding ability of miR-212 and miR-132 with PPP3CC 3’-UTR. This is the first study to evaluate the possible association of the cell cycle and apoptosis associated genes 3’-UTR SNPs with bladder cancer.
Apoptosis was an initiative cell death process that is controlled by genes (18). It was also a therapeutic target of cancer (19). Genetic variations in apoptosis-regulating genes may play key roles in alterations in cellular apoptotic or anti-apoptotic signaling (20). Genetic variants in apoptosis-associated FAS and FASL genes affected risk of bladder cancer in a Turkish population (21). Disorders in the process of apoptosis may have a direct or indirect relationship with the occurrence of a tumor. However, the precise mechanism of apoptosis was not fully understood. PPP3CC, also called CNA3, CALNA3 or PP2Bgamma, was involved in the apoptosis process and it was localized on chromosome 8. Previous studies have shown that low expression of PPP3CC was highly significantly related to the cancer recurrence (22). In our study, we found a significantly decreased risk of bladder cancer associated with the PPP3CC rs7431 A>G polymorphism under an additive model.
Evidence demonstrated that SNPs in the 3’-UTRs of several genes were related to diseases by affecting miRNA-regulated gene expression (15,23,24). For example, Song et al. found that the rs6976789 variant allele strengthened miR-148a binding to the 3’-UTR of SCRN1 gene, thus decreased the ability of SCRN1 with gastric cancer risk (25). In this study, we identified PPP3CC rs7431 located in miR-212 and miR-312 binding sites. Luciferase assays indicated that PPP3CC rs7431 A to G substitution can decrease the binding activity of miR-212 and miR-132 with PPP3CC 3’-UTR, and increased PPP3CC expression. This finding indicated that the SNP rs7431 may strengthen the ability of PPP3CC to decrease the bladder cancer risk. Although the exact mechanism about the PPP3CC rs7431 polymorphism with bladder cancer risk remained to be explained, this difference could be partially due to miR-212 and miR-132 binding abilities.
This study was the first time to evaluate the PPP3CC rs7431 with bladder cancer risk. However several following limitations should be figured out. We did not validate the result in an extra population. Although this SNP located in the miRNA binding site, we did not carry on more work on this SNP. Further functional studies are needed in the future.
In conclusion, we found that genes involved in the cell cycle and apoptosis pathways were associated with bladder cancer. Our results suggested that the PPP3CC rs7431 A>G polymorphism was located in miR-212 and miR-132 binding sites and decreased the bladder cancer risk in a Chinese population. All these indicated that PPP3CC and its genetic variations may be a potential biomarker for risk assessment. Further larger prospective studies are still needed to verify our findings.
Funding
National Natural Science Foundation of China (81473050), Jiangsu Provincial “333” project, Beijing Municipal Administration of Hospitals’ Youth Programme (QML20150101), the Priority Academic Program Development of Jiangsu (Public Health and Preventive Medicine), Natural Science Foundation of Jiangsu Province (BK20151603), Nanjing Science and Technology Program (201605009) and Collaborative Innovation Center for Cancer Personalized Medicine.
Conflicts of Interest
The Authors declare no conflicts of interest.
Acknowledgements
The Authors are deeply grateful for the participation of all subjects contributing to this research.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Pang C, Guan Y, Li H, Chen W, Zhu G. Urologic cancer in China. Jpn J Clin Oncol. 2016;46(6):497–501. doi: 10.1093/jjco/hyw034. [DOI] [PubMed] [Google Scholar]
- 4.Weilandt M, Koch A, Rieder H, Deenen R, Schwender H, Niegisch G, Schulz WA. Target genes of recurrent chromosomal amplification and deletion in urothelial carcinoma. Cancer Genomics Proteomics. 2014;11:141–153. [PubMed] [Google Scholar]
- 5.Kiriluk KJ, Prasad SM, Patel AR, Steinberg GD, Smith ND. Bladder cancer risk from occupational and environmental exposures. Urol Oncol. 2012;30:199–211. doi: 10.1016/j.urolonc.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Da Silva GN, Evangelista AF, Magalhaes DA, Macedo C, Bufalo MC, Sakamoto-Hojo ET, Passos GA, Salvadori DM. Expression of genes related to apoptosis, cell cycle and signaling pathways are independent of TP53 status in urinary bladder cancer cells. Mol Biol Rep. 2011;38:4159–4170. doi: 10.1007/s11033-010-0536-x. [DOI] [PubMed] [Google Scholar]
- 7.Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennetts PS, Pierce JD. Apoptosis: understanding programmed cell death for the CRNA. Aana J. 2010;78:237–245. [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 12.Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci USA. 2007;104:13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song F, Zheng H, Liu B, Wei S, Dai H, Zhang L, Calin GA, Hao X, Wei Q, Zhang W, Chen K. An miR-502-binding site single-nucleotide polymorphism in the 3’-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin Cancer Res. 2009;15:6292–6300. doi: 10.1158/1078-0432.CCR-09-0826. [DOI] [PubMed] [Google Scholar]
- 14.Ratner E, Lu L, Boeke M, Barnett R, Nallur S, Chin LJ, Pelletier C, Blitzblau R, Tassi R, Paranjape T, Hui P, Godwin AK, Yu H, Risch H, Rutherford T, Schwartz P, Santin A, Matloff E, Zelterman D, Slack FJ, Weidhaas JB. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–6515. doi: 10.1158/0008-5472.CAN-10-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB. A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Li Z, Chu H, Lv Q, Ye D, Ding Q, Xu C, Guo J, Du M, Chen J, Song Z, Yin C, Qin C, Gu C, Zhu Y, Xia G, Liu F, Zhang Z, Yuan L, Fu G, Hu Z, Tong N, Shen J, Liu K, Sun J, Sun Y, Li J, Li X, Shen H, Xu J, Shi Y, Zhang Z. Genome-Wide Association Study of Bladder Cancer in a Chinese Cohort Reveals a New Susceptibility Locus at 5q12.3. Cancer Res. 2016;76:3277–3284. doi: 10.1158/0008-5472.CAN-15-2564. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Mo Z, Ye D, Wang M, Liu F, Jin G, Xu C, Wang X, Shao Q, Chen Z, Tao Z, Qi J, Zhou F, Wang Z, Fu Y, He D, Wei Q, Guo J, Wu D, Gao X, Yuan J, Wang G, Xu Y, Wang G, Yao H, Dong P, Jiao Y, Shen M, Yang J, Ou-Yang J, Jiang H, Zhu Y, Ren S, Zhang Z, Yin C, Gao X, Dai B, Hu Z, Yang Y, Wu Q, Chen H, Peng P, Zheng Y, Zheng X, Xiang Y, Long J, Gong J, Na R, Lin X, Yu H, Wang Z, Tao S, Feng J, Sun J, Liu W, Hsing A, Rao J, Ding Q, Wiklund F, Gronberg H, Shu XO, Zheng W, Shen H, Jin L, Shi R, Lu D, Zhang X, Sun J, Zheng SL, Sun Y. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet. 2012;44:1231–1235. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Hao XU. Effects of exercise on apoptosis (A review) J Phys Educ. 2002 [Google Scholar]
- 19.Yeh CC, Kuo HM, Li TM, Lin JP, Yu FS, Lu HF, Chung JG, Yang JS. Shikonin-induced apoptosis involves caspase-3 activity in a human bladder cancer cell line (T24) In Vivo. 2007;21:1011–1019. [PubMed] [Google Scholar]
- 20.Portt L, Norman G, Clapp C, Greenwood M, Greenwood MT. Anti-apoptosis and cell survival: a review. Biochim Biophys Acta. 2011;1813:238–259. doi: 10.1016/j.bbamcr.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Verim L, Timirci-Kahraman O, Akbulut H, Akbas A, Ozturk T, Turan S, Yaylim I, Ergen A, Ozturk O, Isbir T. Functional genetic variants in apoptosis-associated FAS and FASL genes and risk of bladder cancer in a Turkish population. In Vivo. 2014;28:397–402. [PubMed] [Google Scholar]
- 22.Hornstein M, Hoffmann MJ, Alexa A, Yamanaka M, Muller M, Jung V, Rahnenfuhrer J, Schulz WA. Protein phosphatase and TRAIL receptor genes as new candidate tumor genes on chromosome 8p in prostate cancer. Cancer Genomics Proteomics. 2008;5:123–136. [PubMed] [Google Scholar]
- 23.Shi Y, Hu Z, Wu C, Dai J, Li H, Dong J, Wang M, Miao X, Zhou Y, Lu F, Zhang H, Hu L, Jiang Y, Li Z, Chu M, Ma H, Chen J, Jin G, Tan W, Wu T, Zhang Z, Lin D, Shen H. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011;43:1215–1218. doi: 10.1038/ng.978. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Li F, Mao Y, Zhou H, Sun J, Li R, Liu C, Chen W, Hua D, Zhang X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132:641–648. doi: 10.1007/s00439-013-1275-6. [DOI] [PubMed] [Google Scholar]
- 25.Song P, Zhu H, Zhang D, Chu H, Wu D, Kang M, Wang M, Gong W, Zhou J, Zhang Z, Zhao Q. A genetic variant of miR-148a binding site in the SCRN1 3’-UTR is associated with susceptibility and prognosis of gastric cancer. Sci Rep. 2014;4:7080. doi: 10.1038/srep07080. [DOI] [PMC free article] [PubMed] [Google Scholar]