Key Points
Question
Do circulating innate immune monocytes in patients with amyotrophic lateral sclerosis have an inflammatory gene expression profile?
Findings
This study found that amyotrophic lateral sclerosis monocytes express a proinflammatory gene profile. More inflammation-related differentially expressed genes are observed in monocytes of rapidly progressing patients than slowly progressing patients with amyotrophic lateral sclerosis.
Meaning
The gene expression profile of circulating monocytes would be of significant value in defining the potential role of monocytes in pathogenesis and pathoprogression of amyotrophic lateral sclerosis and providing a scientific basis for therapeutic strategies of immunomodulation.
Abstract
Importance
Amyotrophic lateral sclerosis (ALS) is a common adult-onset neurodegenerative disease characterized by selective loss of upper and lower motor neurons. Patients with ALS have persistent peripheral and central inflammatory responses including abnormally functioning T cells and activated microglia. However, much less is known about the inflammatory gene profile of circulating innate immune monocytes in these patients.
Objective
To characterize the transcriptomics of peripheral monocytes in patients with ALS.
Design, Setting, and Participants
Monocytes were isolated from peripheral blood of 43 patients with ALS and 22 healthy control individuals. Total RNA was extracted from the monocytes and subjected to deep RNA sequencing, and these results were validated by quantitative reverse transcription polymerase chain reaction.
Main Outcomes and Measures
The differential expressed gene signatures of these monocytes were identified using unbiased RNA sequencing strategy for gene expression profiling.
Results
The demographics between the patients with ALS (mean [SD] age, 58.8 [1.57] years; 55.8% were men and 44.2% were women; 90.7% were white, 4.65% were Hispanic, 2.33% were black, and 2.33% were Asian) and control individuals were similar (mean [SD] age, 57.6 [2.15] years; 50.0% were men and 50.0% were women; 90.9% were white, none were Hispanic, none were black, and 9.09% were Asian). RNA sequencing data from negative selected monocytes revealed 233 differential expressed genes in ALS monocytes compared with healthy control monocytes. Notably, ALS monocytes demonstrated a unique inflammation-related gene expression profile, the most prominent of which, including IL1B, IL8, FOSB, CXCL1, and CXCL2, were confirmed by quantitative reverse transcription polymerase chain reaction (IL8, mean [SE], 1.00 [0.18]; P = .002; FOSB, 1.00 [0.21]; P = .009; CXCL1, 1.00 [0.14]; P = .002; and CXCL2, 1.00 [0.11]; P = .01). Amyotrophic lateral sclerosis monocytes from rapidly progressing patients had more proinflammatory DEGs than monocytes from slowly progressing patients.
Conclusions and Relevance
Our data indicate that ALS monocytes are skewed toward a proinflammatory state in the peripheral circulation and may play a role in ALS disease progression, especially in rapidly progressing patients. This increased inflammatory response of peripheral immune cells may provide a potential target for disease-modifying therapy in patients with ALS.
This study characterizes the transcriptomics of peripheral monocytes in patients with amyotrophic lateral sclerosis.
Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease. At sites of motor neuron injury in the central nervous system (CNS), neuroinflammation is a prominent pathological finding in autopsy tissues that is characterized by microglial activation and perivascular infiltration of monocytes and T cells. Positron emission tomographic images of brains from living patients with ALS demonstrate widespread microglial activation. Moreover, increased microglial activation in these patients’ motor cortices correlates with the severity of upper motor neuron clinical signs. Similar results have been obtained from mutant superoxide dismutase (mSOD1) transgenic animal models of ALS. Activated microglia were observed before disease onset and increased as disease progressed. Furthermore, replacing mSOD1 CNS microglia with wild-type microglia or removing mSOD1 from microglia prolonged survival of ALS mice. These data suggest that microglia play an active role in motor neuron degeneration of patients with ALS as well as transgenic mice. Because monocytes are the peripheral blood homologue of CNS microglia, these data raise the question as to whether monocytes similarly contribute to ALS pathology.
Because direct sampling of microglia from patients with ALS is not feasible, peripheral monocyte/macrophages may provide evidence of systemic innate immune system activation in patients. In this study, using high-throughput unbiased deep RNA sequencing (RNA-seq), we sought to demonstrate that ALS monocytes are skewed toward a proinflammatory state in the peripheral circulation and determine whether they represent a potential target for disease modification.
Methods
Participants
This study was approved by the Houston Methodist Hospital institutional review board. All patients and healthy control individuals signed informed consent before peripheral blood samples were drawn. Patients were seen at the Muscular Dystrophy Association/ALS Clinic at Houston Methodist Hospital Neurological Institute (Houston, Texas) and evaluated for functional status using the Appel ALS (AALS) score (range, 30–164) as well as the revised ALS Functional Rating Scale. None of the patients and healthy control individuals had evidence of ongoing infectious/inflammatory diseases. Progression rate was determined as the change of AALS score from the initial clinical visit to the collection time. Rapidly progressing patients were defined as those progressing at a rate of greater than or equal to 1.5 AALS points/month, and slowly progressing patients were defined as those progressing at a rate of less than 1.5 AALS points/month. The demographics between the patients with ALS (mean [SD] age, 58.8 [1.57] years; 55.8% were men and 44.2% were women; and 90.7% were white, 4.65% were Hispanic, 2.33% were black, and 2.33% were Asian) and control individuals were similar (mean [SD] age, 57.6 [2.15] years; 50.0% were men and 50.0% were women; and 90.9% were white, none were Hispanic, none were black, and 9.09% were Asian).
Monocyte Isolation
Human monocytes were freshly isolated from peripheral blood of 43 patients with ALS and 22 healthy control individuals. According to the manufacturer’s instructions, human Pan Monocyte Isolation Kit (Miltenyi Biotec) was used to obtain monocytes by negative selection. The purity of pan monocytes was 91% to 96% as determined by flow cytometry.
Quantitative Reverse Transcription Polymerase Chain Reaction
RNA was extracted and purified from ex vivo monocytes using Direct-zol RNA MiniPrep Kit (Zymo Research) according to manufacturer’s instructions. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using a 1-step RT-PCR kit with SYBR Green on an iQ5 Multicolor Real-time PCR detection System (Bio-Rad Laboratories). Primers and conditions of PCR were shown in Table 1. Primer efficiency was assessed by analyzing a serial dilution of RNA. The relative expression level of each mRNA was calculated using the ∆∆Ct method normalizing to β-actin and relative to the control samples. The presence of 1 product of the correct size was verified by both 2% agarose gel electrophoresis and melt curve analyses containing a single melt curve peak.
Table 1. Primer Sets and Temperature for Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction.
| Genes | Primer Sets | Temperature, °C |
|---|---|---|
| IL1B | Forward: 5′-TTC GAC ACA TGG GAT AAC GAG G-3′ Reverse: 5′-TTC GAC ACA TGG GAT AAC GAG G-3′ |
53.8 |
| NLRP3 | Forward: 5′-CGT GAG TCC CAT TAA GAT GGA GT-3′ Reverse: 5′-CCC GAC AGT GGA TAT AGA ACA GA-3′ |
58 |
| IL8 | Forward: 5′-AGC TCT GTG TGA AGG TGC AGT-3′ Reverse: 5′-AAT TTC TGT GTT GGC GCA GTG-3′ |
55.4 |
| CD83 | Forward: 5′-ATG GAG ACA CCC CAG GAA GAC-3′ Reverse: 5′-TCA GGG AAT AGG GCC TTT CA-3′ |
63.1 |
| FOSB | Forward: 5′-AGC AGC AGC TAA ATG CAG GA-3′ Reverse: 5′-TTC GTA GGG GAT CTT GCA GC-3′ |
65 |
| SOCS3 | Forward: 5′-GCT CCA AGA GCG AGT ACC AG-3′ Reverse: 5′-CTG TCG CGG ATC AGA AAG GT-3′ |
61.3 |
| CXCL1 | Forward: 5′-GCA GGG AAT TCA CCC CAA GA-3′ Reverse: 5′-GAT GCA GGA TTG AGG CAA GC-3′ |
57.6 |
| CXCL2 | Forward: 5′-GAT TTC ACA GTG TGT GGT CAA C-3′ Reverse: 5′-TCG AAA CCT CTC TGC TCT AAC A-3′ |
57.6 |
| β-ACTIN | Forward: 5′-GCA TCC ACG AAA CTA CCT TCA-3′ Reverse: 5′-GCA GTG ATC TCC TTC TGC ATC-3′ |
60 |
RNA Sequencing and Data Analysis
Monocyte-derived RNA was assayed for integrity quality control using 2100 Bioanalyzer (Agilent Technologies). RNA samples with an RNA integrity number of at least 8.0 were subjected to deep RNA-seq. Sequencing libraries were prepared by the Illumina TruSeq RNA Library Prep, version 2, Protocol D, using 500-ng total RNA (Illumina). The qualities of the libraries were assessed using 2100 Bioanalyzer with a DNA1000 assay. Libraries were quantified by qPCR using the KAPA Library Quantification kit for Illumina sequencing platforms (KAPA Biosystems). Quality control of library pools to establish equimolarity of individual libraries was done by Illumina MiSeq (SR 1 × 50 bp). Libraries were sequenced using an Illumina HiSeq1500 (PE 2 × 100 bp). The quality control of FASTQ files, alignment with Hg19, and quality control of BAM files were performed using ArrayStudio, version 7.1.1.51 (Omicsoft). Gene transcripts were quantified using DESeq2, version 3.3 (Bioconductor) analysis with raw RNA-seq read count data. DESeq2 is more accurate for discovery of differentially expressed genes (DEGs) than methods using fragments per kilobase of transcript per million mapped reads or reads per kilobase of transcript per million mapped reads; the normalization of DESeq2 is more precise than other normalization methods. DESeq2 analysis is based on the hypothesis that most genes are not differentially expressed; the median of this ratio for each sample provides an estimate of the correction factor that should be applied to all read counts of this sample to fulfill the hypothesis. The function analysis of DEGs was performed using Ingenuity Pathway Analysis (Qiagen).
Enzyme-Linked Immunosorbent Assay
Human interleukin (IL) 8 enzyme-linked immunosorbent assay Ready-SET-Go kit (eBioscience) and Quantikine enzyme-linked immunosorbent assay kits for human IL-1β, IL-6, tumor necrosis factor (TNF) α, or interferon (IFN) γ (R&D Systems) were used to determine cytokine concentrations in serum samples of patients with ALS and healthy control individuals according to manufacturer’s instructions.
Statistical Analysis
The DEGs between control individuals and patients with ALS were analyzed using analysis of variance for more than 2 groups or t test for 2 groups. Data are expressed as mean (SE), and P less than .05 was considered significant. The P value was 2-sided.
Results
Gene Expression of Monocytes of Patients With ALS
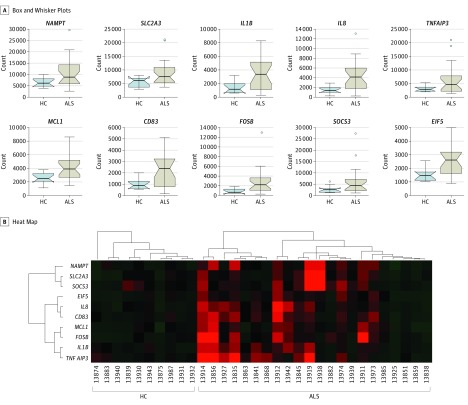
To determine the gene expression profile of ALS monocytes in an unbiased manner, we subjected the first set of monocyte RNA samples from 23 patients with ALS and 10 healthy control individuals to deep RNA-seq. Based on selection criteria of fold change greater than 2, P < .01, and false discovery rate less than 0.25, RNA-seq results revealed 233 DEGs from monocytes of patients with ALS compared with the monocytes of healthy control individuals (eTable 1 in the Supplement). Of these DEGs, Figure 1A shows the top 10 upregulated genes that had the largest differences on medium of gene expression levels between patients with ALS and healthy control individuals. The heat map of these 10 genes showed a distinct expression pattern of monocytes of patients with ALS compared with healthy control individuals’ monocytes (Figure 1B). Most importantly, except for EIF5 (a GTPase-activating protein), 9 of these 10 top DEGs, including IL1B, IL8, NAMPT, SLC2A3, FOSB, and CD83, are involved in proinflammatory responses of monocytes.
Figure 1. Comparison of Differentially Expressed Genes (DEGs) in Monocytes of Patients With Amyotrophic Lateral Sclerosis (ALS) and Healthy Control Participants by Deep RNA Sequencing.
Box and whisker plots (A) and heat map (B) illustrate top 10 upregulated DEGs that have the largest differences of medium expression between the ALS group (n = 23) and healthy control group (HC, n = 10). In box and whisker plots, upper ends of boxes represent the third quartiles of corresponding groups; lower ends of boxes represent the first quartiles of groups; middle lines indicate mediums; upper whiskers indicate the maximum; and lower whiskers indicate the minimum. Outlier data are shown as circles plotted beyond the whiskers.
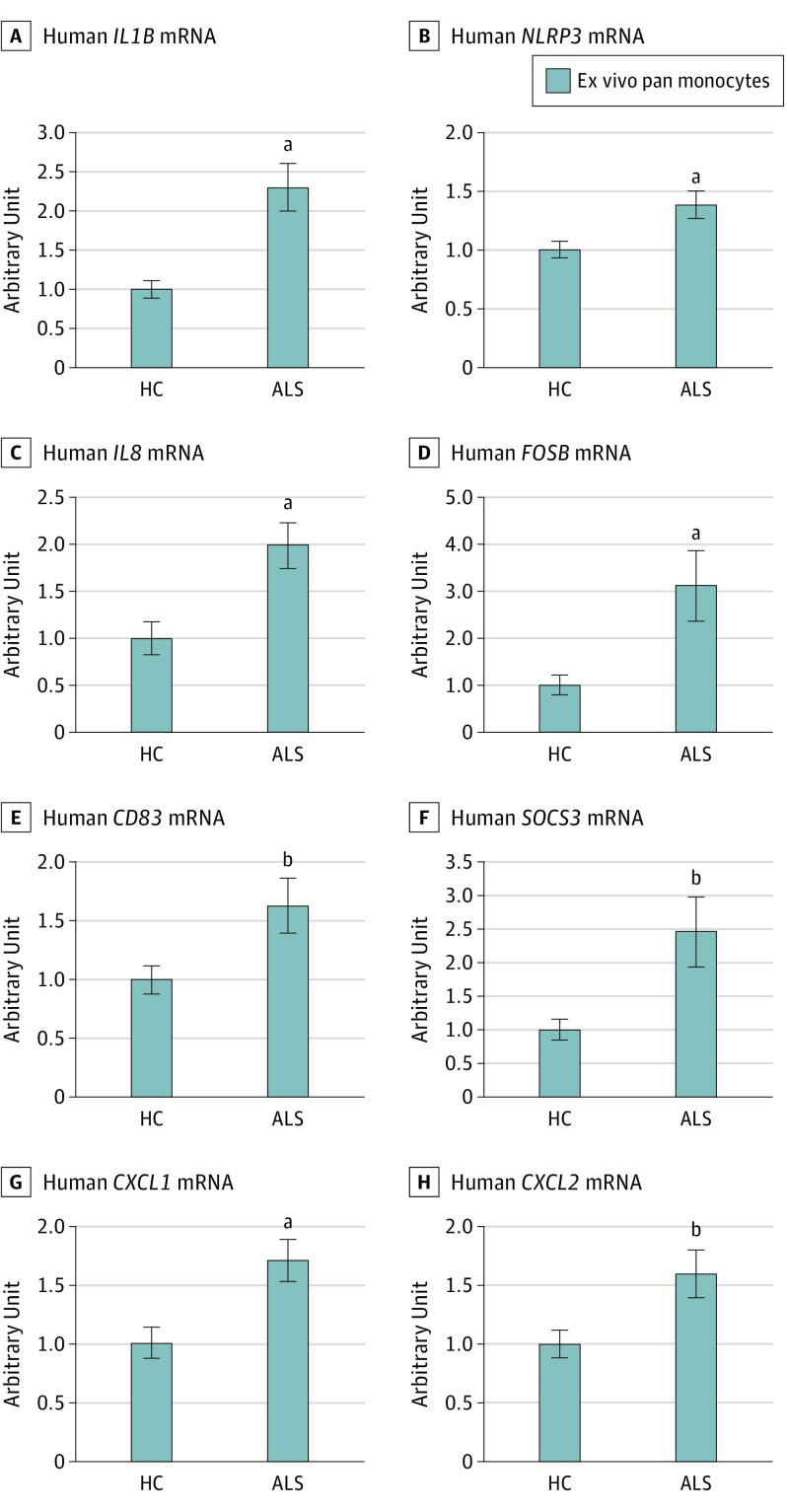
To validate RNA-seq data, we prepared monocyte RNA samples from additional patients with ALS and healthy control individuals and measured the mRNA level of a proinflammatory cytokine IL-1β by qRT-PCR. As shown in Figure 2A, monocytes of patients with ALS (n = 43; mean [SE], 2.3 [0.30]) expressed more IL1B message than monocytes isolated from control individuals (n = 22; mean [SE], 1.0 [0.11]; P < .001). Expression of NLRP3, the major component of NLRP3 inflammasome responsible for IL-1β production, was also upregulated in monocytes of patients with ALS (n = 43) compared with control monocytes (n = 22; mean [SE], 1.00 [0.07]; P = .007; Figure 2B). Furthermore, we chose IL8, FOSB, CD83, SOCS3, CXCL1, and CXCL2 from the inflammation-related DEGs and assessed their mRNA expression levels by qRT-PCR. Monocytes of patients with ALS (n = 43) had higher IL8, FOSB, CD83, SOCS3, CXCL1, and CXCL2 expression levels than monocytes from healthy control individuals (n = 22, Figure 2C-H) (IL8, mean [SE], 1.00 [0.18]; P = .002; FOSB, 1.00 [0.21]; P = .009; CD83, 1.00 [0.12]; P = .02; SOCS3, 1.00 [0.16]; P = .01; CXCL1, 1.00 [0.14]; P = .002; and CXCL2, 1.00 [0.11]; P = .01). These data confirmed RNA-seq results, indicating that inflammation-associated genes were upregulated in ALS monocytes.
Figure 2. Expression of Inflammation-Related Differentially Expressed Genes Verified by Quantitative Reverse Transcription Polymerase Chain Reaction .
Upregulation of IL1B (A), NLRP3 (B), IL8 (C), FOSB (D), CD83 (E), SOCS3 (F), CXCL1 (G), and CXCL2 (H) in monocytes of patients with amyotrophic lateral sclerosis (ALS) (n = 43) were verified by quantitative reverse transcription polymerase chain reaction. Arbitrary unit of each mRNA was normalized to β-actin and relative to the healthy control (HC) samples (n = 22). Error bars indicate the standard error.
aP < .01 vs HC (IL1B, P < .001; NLRP3, P = .007; IL8, P = .002; FOSB, P = .009; and CXCL1, P = .002).
bP < .05 vs HC (CD83, P = .02; SOCS3, P = .01; and CXCL2, P = .01).
Both qRT-PCR and deep RNA-seq data were generated from pan monocytes by negative selection, which allowed us to obtain untouched pan monocytes. We also performed RNA-seq on monocytes isolated by a positive selection method using CD14 microbeads (Miltenyi Biotec). Principal component analysis indicated that overall RNA messages of negatively selected pan monocyte from patients with ALS were more distinguishable from healthy control individuals than CD14+ ALS monocytes vs CD14+ control monocytes by positive selection (eFigure 1 in the Supplement). CD14+ ALS monocytes exhibited fewer DEGs when compared with control monocytes isolated by the same positive selection method (data not shown). The attachment of CD14 antibody on monocytes may activate them and influence gene expression of these cells, while pan monocytes by negative selection had no bound antibody. Therefore, data from negatively selected pan monocytes more accurately reflect in vivo features of ALS monocytes than positively selected CD14+ monocytes.
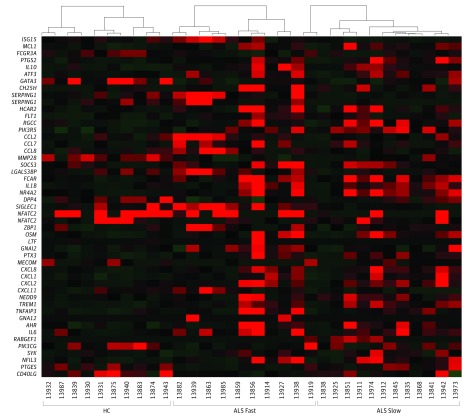
Next, we separated patients with ALS into slowly and rapidly progressing groups, using the AALS score of 1.5 points/month as a cutoff. This cutoff was defined by accessing the progression rates of more than 4000 patients’ records in our database and statistically determining the median progression rate. Using this cutoff, we previously separated 54 patients with ALS surviving less than 2 years after diagnosis from those that survive up to 6 years or more. With our past experience with increased sensitivity of the AALS in early disease progression, AALS was used to stratify slow and fast progression rates. Moreover, using the statistically determined score of 1.5 points/month for AALS and 1.0 points/month for the revised ALS functional rating, the revised ALS functional rating scores gave relatively comparable designations as slow or fast rates as the AALS scores in 18 of 22 patients (82%, eTable 2 in the Supplement). After comparing RNA-seq data of slow or fast groups with the control group, more DEGs were identified than what was observed with the combined ALS group vs controls. This is because several genes were differentially expressed in opposite directions in slow and fast ALS groups; some genes changed in one ALS group but not in another ALS group when compared with the control group. These analyses demonstrated that 65 DEGs were specifically expressed in monocytes from patients with slowly progressing ALS and 237 DEGs were solely found in monocytes from patients with rapidly progressing ALS; 43 DEGs were expressed in both the slow and fast groups (eFigure 2 and eTables 3-5 in the Supplement). Functional analysis of these DEGs revealed that monocytes from patients with rapidly progressing ALS contained more DEGs that are tightly associated with inflammatory disease or inflammatory functions than monocytes from slowly progressing patients (Table 2; the P values indicate the possibility that these genes are not involved in the corresponding disease or function category). Eighty-four DEGs participated in cell migration and chemotaxis in monocytes of patients with rapidly progressing ALS, while 30 DEGs were involved in migration and chemotaxis in monocytes from patients with slowly progressing ALS. Forty-seven genes encoded molecules acting on the activation of monocytes from rapidly progressing patients, while 22 DEGs were associated with cell activation in monocytes of slowly progressing patients. In addition, 44 monocytic DEGs were closely related with inflammatory responses in rapidly progressing patients, while 19 genes were associated with inflammatory responses in monocytes from slowly progressing patients. These DEGs associated with inflammatory responses were also illustrated in a heat map (Figure 3). These data indicate that ALS monocytes more actively participate in immune responses during fast progression than during slow progression and may contribute to the accelerating progression of ALS.
Table 2. Monocyte DEGs Involved in Activation and Inflammatory Diseases and Functions.
| Diseases or Functions Annotation | Patients With Slow ALS | Patients With Fast ALS | ||||
|---|---|---|---|---|---|---|
| P Value | DEGs | No. of DEGs | P Value | DEGs | No. of DEGs | |
| Cell migration and chemotaxis | 6.93E-05 | CORO1C, CXCL1, CXCL2, IL8, EREG, FCAR, FOSB, FOSL1, GATA3, GPR183, HBEGF, IFITM3, IL10, IL1B, LTF, NAMPT, NFATC2, NFIL3, NGFR, NR4A2, OSM, PHLDA1, PIK3R5, PTGS2, RABGEF1, SEPT9, SERPING1, SYK, TP53INP2, TREM1 | 30 | 7.60E-13 | ADGRG1, ADGRG3, AHR, ALCAM, ANXA3, AQP3, AREG, ARID5B, ATF3, CCL2, CCL7, CCL8, CD40LG, CRMP1, CXCL1, CXCL11, CXCL2, IL8, DPP4, DUSP1, EREG, ETV5, FCAR, FCGR3A/FCGR3B, FLT1, FOSB, FOSL1, GATA3, GBA, GBP1, GNA12, GNAI2, GPR183, GUCY1A3, HBEGF, HCAR2, HLX, HMMR, IFIT2, IFITM1, IFITM3, IGFBP6, IL10, IL1B, IL21R, IL6, IL9R, ITK, LGALS3BP, LIMK2, MMP28, MX1, MYO6, NAMPT, NEDD9, NFATC2, NFIL3, NKX3-1, NR4A2, OSM, PER1, PIK3CG, PIK3R5, PIM1, PLA2G16, PTGES, PTGS2, PTPRH, PTX3, RERE, RGCC, SDC2, SERPING1, SLC16A1, SLC1A3, SLC8A1, SOCS3, ST13, ST8SIA4, TIAM2, TNFAIP3, TP53INP2, TREM1, TRIB1 | 84 |
| Differentiation of cells | 7.61E-06 | ADAP1, CD83, CDH23, CXCL1, IL8, EGR3, EREG, FOSL1, GATA3, H2AFY, HBEGF, HOXB4, IL10, IL1B, IL7R, LTF, MAFF, MCL1, NAMPT, NFATC2, NFIL3, NGFR, NR4A2, OSM, PBX1, PTGS2, RCHY1, SYK, TP53INP2, TREM1, TTC7A, XRCC4, ZNF385A | 33 | 2.85E-10 | ADGRG3, AGRN, AHR, ALCAM, AQP3, AREG, ARID5B, ARL4A, ATF3, BAIAP2, BSN, CCL2, CD27, CD3G, CD40LG, CD83, CDH23, CIT, CRMP1, CXCL1, IL8, DPP4, DUSP1, EMP1, EREG, ETV5, ETV7, FFAR2, FLT1, FOSL1, GATA3, GNA12, GPC4, HBEGF, HLX, HPS4, IFITM1, IGFBP6, IL10, IL1B, IL21R, IL6, ISG15, ITK, KNDC1, LIMK2, LRG1, LY6E, MECOM, MYO6, NAMPT, NFATC2, NFIL3, NR4A2, OSM, OSR2, PEX11A, PIK3CG, PIM1, PTGS2, RCHY1, RGS6, RSAD2, SDC2, SLC1A3, SLC2A5, SOCS3, SPRY1, ST8SIA4, THOC5, TNFAIP3, TP53INP2, TREM1, TRIB1, UBE2D3, USP18, XRCC4, ZBTB7C, ZNF385A | 79 |
| Inflammation of organ | 7.90E-05 | CHRNA2, CXCL1, CXCL2, IL8, EREG, GATA3, HBEGF, IKBKE, IL10, IL1B, LTF, MCL1, NFATC2, NFIL3, NGFR, NR4A2, OSM, PTGS2, RABGEF1, SYK, TREM1 | 21 | 2.77E-09 | ABCB1, AHR, ALCAM, ANXA3, AREG, ATF3, BSN, CCL2, CCL7, CCL8, CD40LG, CH25H, CKB, CXCL1, CXCL2, IL8, DPP4, DUSP1, EREG, FCGR1B, FCGR3A/FCGR3B, FLT1, GATA3, GNAI2, GUCY1A3, HBEGF, IFI27, IL10, IL1B, IL21R, IL6, IL9R, ITK, MR1, MX1, NAV3, NEDD4L, NFATC2, NFIL3, NR4A2, OSM, PER1, PIK3CG, PIM1, PSMD2, PTGES, PTGS2, SOCS3, TIGIT, TNFAIP3, TREM1, USP18 | 52 |
| Activation of cells | 6.72E-07 | CD83, CXCL1, CXCL2, IL8, EGR3, FCAR, GATA3, HBEGF, HOXB4, IL10, IL1B, LTF, NFATC2, NFIL3, OSM, PBX1, PTGS2, RABGEF1, SERPING1, SYK, TREM1, XRCC4 | 22 | 3.61E-10 | AGRN, AHR, AREG, ATF3, CCL2, CCL7, CCL8, CD27, CD3G, CD40LG, CD83, CXCL1, CXCL2, IL8, DPP4, DUSP1, FCAR, FCGR3A/FCGR3B, FFAR2, FFAR3, GATA3, GNAI2, HBEGF, IL10, IL1B, IL21R, IL6, ITK, LGALS3BP, LYNX1, MR1, NFATC2, NFIL3, OSM, PIK3CG, PTGES, PTGS2, RGCC, SDC2, SERPING1, SIGLEC1, TIGIT, TNFAIP3, TREM1, UBE2C, XRCC4, ZBP1 | 47 |
| Inflammatory response | 9.43E-07 | CXCL1, CXCL2, IL8, FCAR, GATA3, IL10, IL1B, LTF, MCL1, NFATC2, NFIL3, NR4A2, OSM, PIK3R5, PTGS2, RABGEF1, SERPING1, SYK, TREM1 | 19 | 2.35E-09 | AHR, ATF3, CCL2, CCL7, CCL8, CD40LG, CH25H, CXCL1, CXCL11, CXCL2, IL8, DPP4, FCAR, FCGR3A/FCGR3B, FLT1, GATA3, GNA12, GNAI2, HCAR2, IL10, IL1B, IL6, ISG15, LGALS3BP, MECOM, MMP28, NEDD9, NFATC2, NFIL3, NR4A2, OSM, PIK3CG, PIK3R5, PTGES, PTGS2, PTX3, RGCC, SIGLEC1, SERPING1, SOCS3, TNFAIP3, TREM1, ZBP1 | 44 |
| Chronic inflammatory disorder | 8.92E-07 | CD83, CXCL1, CXCL2, IL8, EREG, FOSB, G0S2, HBEGF, IL10, IL1B, IL7R, LTF, MCL1, NAMPT, NGFR, NR4A2, OSM, PTGS2, SYK, TREM1, VSIG4 | 1.12E-07 | ABCB1, AHR, AREG, BSN, CCL2, CCL7, CCL8, CD83, CKB, CXCL1, CXCL2, IL8, DPP4, DUSP1, EREG, FCGR1B, FCGR3A/FCGR3B, FKBP5, FOSB, GPC4, HBEGF, IL10, IL1B, IL6, LAP3, MCTP2, NAMPT, NR4A2, OSM, PIK3CG, PSMD2, PTGS2, RASGRP3, RGCC, RGS6, SDC2, SOCS3, TAGAP, TNFAIP3, TREM1 | 40 | |
| Quantity of phagocytes | 5.08E-05 | CD83, IL8, GPR183, HBEGF, IL10, IL1B, IL7R, MCL1, NGFR, RABGEF1, SORBS1 | 11 | 9.00E-07 | ABCB1, AHR, ATF3, CCL2, CCL7, CD40LG, CD83, IL8, GBA, GNAI2, GPR183, HBEGF, IL10, IL1B, IL6, IL9R, NEDD4L, PIK3CG, PTGES, SOCS3, TNFAIP3, TRIB1 | 22 |
| Quantity of antigen presenting cells | 1.09E-03 | CD83, GPR183, HBEGF, IL10, IL1B, IL7R, MCL1 | 7 | 4.06E-06 | AHR, CCL2, CCL7, CD40LG, CD83, GBA, GPR183, HBEGF, IL10, IL1B, IL6, PIK3CG, PTGES, SOCS3, TNFAIP3, TRIB1 | 16 |
| Adhesion of phagocytes | 1.81E-04 | CXCL1, CXCL2, IL8, IL10, IL1B, PTGS2 | 6 | 1.01E-05 | ALCAM, CCL2, CD40LG, CXCL1, CXCL2, IL8, IL10, IL1B, IL6, PIK3CG, PTGS2 | 11 |
| Respiratory burst of myeloid cells | 3.89E-06 | CXCL1, IL8, FCAR, SYK, TREM1 | 5 | 5.69E-06 | CCL2, CD40LG, CXCL1, IL8, FCAR, PIK3CG, TREM1 | 7 |
Abbreviations: ALS, amyotrophic lateral sclerosis; DEGs, differentially expressed genes.
Figure 3. Heat Map of Differentially Expressed Genes Associated With Inflammatory Responses.
More proinflammatory differentially expressed genes were upregulated in monocytes isolated from patients with rapidly progressing amyotrophic lateral sclerosis (ALS fast, n = 10) than from patients with slowly progressing ALS (ALS slow, n = 12) or control groups (HC, n = 10).
Peripheral Cytokine Levels
Serum IL-1β, IL-6, TNF-α, and IFN-γ were assayed by enzyme-linked immunosorbent assays, but all were at background levels in ALS and control serum samples and were neither reproducible nor reliable. Nevertheless, serum IL-8 protein levels were measurable in patients with ALS and control individuals. Our data demonstrate that IL-8 protein levels trended higher in serum samples of patients with ALS (n = 66; mean [SD], 35.6 [1.00] pg/mL) than healthy control individuals (n = 32; mean [SD], 15.7 [3.13] pg/mL), although levels did not reach significance (eFigure 3A in the Supplement; P = .06). However, when patients with ALS were separated into slowly progressing and rapidly progressing groups, serum samples from patients with rapidly progressing ALS (n = 36; mean [SD], 51.3 [17.8] pg/mL) had increased IL-8 protein compared with serum samples from patients with slowly progressing ALS (n = 30; mean [SD], 16.9 [3.41] pg/mL) or healthy control individuals (mean [SD], 15.7 [3.13]; P = .05; eFigure 3B in the Supplement).
Discussion
Our deep RNA-seq and qRT-PCR data demonstrated that monocytes isolated from patients with ALS expressed a unique gene profile associated with proinflammatory immune responses. Nine of 10 top upregulated DEGs shown in Figure 1A are involved in inflammation. Seven of these 9 genes are directly involved in proinflammatory pathways, while 2 are involved in the negative feedback resolution of the proinflammatory response. Interleukin 1β is a major inflammatory cytokine produced by activated monocytes/macrophages. Blockade of the IL-1β receptor or an induced IL-1β deficiency resulted in less microglia activation and less motor neuron loss and prolonged survival in ALS mice, indicating that IL-1β participates in ALS progression. Nicotinamide phosphoribosyltransferase (NAMPT), also known as a pre–B-cell colony-enhancing factor, has multiple functions. In a study, silencing NAMPT gene expression in monocytes reduced IL-6 production by these cells, leading to fewer Th17 cells and reduced autoantibody titers as well as decreased infiltration of monocytes/macrophages and neutrophils in arthritic joints, suggesting the role of monocytic NAMPT in inflammation. FOSB is a key subunit of transcription factor AP-1. Stimulation of adenosine triphosphate receptor P2RX7 led to the induction of FOSB and activation of AP-1 in monocytes; subsequent upregulation of COX-2 indicated FOSB was transcriptionally active in monocytic activation. CD83 has functions of regulating antigen presentation. Upregulation of CD83 and more antigen presentation–associated DEGs (as shown in Table 2) in monocytes of patients with rapidly progressing ALS implies that monocytes may engage in presenting antigen during disease progression. SOCS3 and TNFAIP3 are 2 negative regulators of inflammation and immunity. They are induced in activated immune cells and exert their inhibitory function by downregulating overactivated NF-κB and IRF3 pathways and avoid too much proinflammatory gene expression in a negative feedback loop. Their induction in ALS monocytes implicate the activation of these cells during disease process. Interleukin 8, CXCL1, and CXCL2 are key chemokines responsible for attracting other immune cells to inflamed sites. We were also able to verify that IL8, CXCL1, and CXCL2 genes were uniquely overexpressed in ALS monocytes, similar to the demonstration that CD14+/CD16– monocytes isolated by FACS sorting exhibited increased IL-8, CXCL1, and CXCL2 expression in patients with ALS compared with healthy control individuals. All of these data suggest that ALS monocytes are involved in proinflammatory responses and may promote active inflammation.
The monocytes isolated by negative selection in our study included both CD16+ and CD16− monocytes. Including populations of both CD16+ and CD16− cells could represent a potential limitation. By our flow cytometry analyses, greater than 80% of the monocytes were CD14+/CD16−; the remainder were CD14+/CD16+ or CD14low/CD16+ 36. This major population of CD16− classic monocytes have been reported as proinflammatory and functionally altered in ALS. Nonclassic CD14low/CD16+ monocytes have been reported to be increased in patients with acute sepsis and chronic systemic lupus erythematosus, indicating their inflammatory features. Thus, both phenotypes have proinflammatory signatures, and it remains to be determined whether the gene profiles of classic and nonclassic ALS monocyte subsets are the same or different.
Proinflammatory cytokines, such as TNF-α, IL-6, and IFN-γ, have been reported to be elevated in plasma or serum samples of patients with ALS, with levels increasing as disease progresses. However, in our studies, IL-6, TNF-α, and IFN-γ protein levels measured by enzyme-linked immunosorbent assay were at background levels in ALS and control serum samples and were neither reproducible nor reliable. Furthermore, IL-1β serum protein levels were not readily detectable, despite definite evidence of IL1B RNA expression in both ALS and control monocytes, with increased IL1B mRNA expression in ALS monocytes. This result could be explained either as an extremely short cytokine half-life within plasma or serum samples, following rapid consumption in an autocrine or paracrine manner, or as enhanced secretion only following migration to tissues, including the CNS. Our inability to reproduce inflammatory cytokine serum levels published by others was a major impetus for examining the cell-based RNA expression of such inflammatory cytokines. On repetitive testing, only IL-8 protein levels were increased in serum samples of patients with rapidly progressing disease, but such levels were close to background and were not as definitively increased as IL8 gene expression. Nevertheless, a 2015 study did show that serum IL-8 protein levels increased during disease progression. Additional studies have also reported elevated IL-8 protein cerebrospinal levels in patients with ALS, especially in those patients with lower levels of physical function.
In mouse models, it has been suggested that peripheral monocytes/macrophages infiltrated spinal cords during the course of disease using Ly6C and CD169 as markers for monocyte-derived macrophages. However, parabiosis experiments indicated that peripheral monocyte/macrophage populations were not recruited to the CNS parenchymal microglia pool in transgenic ALS mice without irradiation. Furthermore, it is reported that mSOD1 microglia are not derived from infiltrating monocytes. Nevertheless, in the transgenic ALS mouse model, splenic Ly6Chigh monocytes exhibited a proinflammatory phenotype 2 months prior to disease onset and during disease progression in transgenic mice, and the comparable monocyte subpopulation (CD14+/CD16−) in human ALS exhibited a similar immune gene signature. Even if increased proinflammatory monocytes do not enter the CNS to modulate the course of disease directly, enhanced peripheral inflammation could indirectly influence CNS disease progression. The proinflammatory monocyte/macrophages might not have to migrate to the CNS to contribute to the CNS neuroinflammatory milieu; they could indirectly influence CNS neuroinflammation by modulating the proinflammatory status of T cells, dendritic, or natural killer cells in lymph nodes, spleen, or the peripheral circulation. An example of how cytokines released from activated peripheral monocyte/macrophages could influence neuroinflammation is the ability of IL-6 to enhance the conversion of protective regulatory T lymphocytes to proinflammatory Th17 lymphocytes. Following such interactions, subsequently activated immune cells could infiltrate the CNS throughout the course of disease and promote neuroinflammation.
Our results indicate that monocytes of patients with ALS may participate in the inflammatory response associated with disease progression; more inflammation-related DEGs are observed in rapid phase than slow phase. Thus, manipulating monocytes at an early stage in disease may directly promote the development of therapy for patients with ALS. In mSOD1 mice, splenic Ly6Chigh monocytes exhibited a proinflammatory phenotype. Treatment with an anti-Ly6C antibody downregulated inflammatory splenic Ly6Chigh monocytes and exerted a beneficial effect on mSOD1 mice by delaying disease onset and prolonging survival. Expression of miRNA155, which promotes macrophage inflammatory response and proinflammatory cytokine secretion, was increased in spinal cord of ALS mice; anti-miRNA155 treatment attenuated disease and prolonged survival in these ALS mice. However, limited evidence validates this therapeutic approach in patients with ALS. Nonetheless, a compound, NP001, has been suggested to reduce inflammatory markers of monocytes in patients with ALS and was possibly beneficial in the 25% of responders with greater inflammation before treatment indicated by elevated levels of IL-18 cytokine and wide-range C-reactive protein. Therefore, information as to the extent of ALS monocyte inflammatory gene expression may be useful in stratifying patients for trials of immune modulators.
Limitations
A potential limitation of our study was that the acquired gene profile was of the collective total monocyte RNA. The total monocyte can be separated into 3 subpopulations, ie, classic, intermediate, and nonclassic monocytes. Subjecting these subpopulations to deep RNA-seq may have provided us with a better understanding of which cell phenotype contributed to the observed upregulated proinflammatory response. Further experiments to examine gene expression of monocyte subpopulations would be helpful to determine their differential functions in ALS.
Conclusions
Circulating monocytes from patients with ALS have a proinflammatory gene expression profile. This proinflammatory innate immune state suggests a possible role for monocyte/macrophages in the pathogenesis of ALS. We have previously reported that decreased FOXP3 expression, the functional marker of regulatory T lymphocytes, is associated with rapid disease progression in ALS. Therefore, modulation of abnormal immune cells, including monocytes and regulatory T lymphocytes, represents a potentially important therapeutic intervention for patients with ALS.
eFigure 1. Principle Component Analysis (PAC) Indicates That Positive Selection Failed to Distinguish ALS and Control Monocyte Samples While Negative Selected ALS Monocytes Were Distinguishable From Control Monocytes.
eFigure 2. Venn Diagram of Differentially Expressed Genes (DEGs) in Monocytes From ALS Slow and Fast Patients
eFigure 3. IL-8 Protein Levels in Sera of ALS Patients and Healthy Controls.
eTable 1. DEGs of Monocytes Isolated From Total ALS Patients (vs Control)
eTable 2. Disease Progression of ALS Patients for RNA-seq Study Based on a Cutoff of 1.5 AALS Points/Month or a Cutoff of 1.0 ALS FRS Points/Month
eTable 3. 65 DEGs Solely Found in Monocytes From Slowly Progressing ALS Patients (vs Control)
eTable 4. 237 DEGs Solely Found in Monocytes From Rapidly Progressing ALS Patients (vs Control)
eTable 5. 43 DEGs Expressed in Monocytes of Both Slow and Fast Groups (vs Control)
References
- 1.Zhao W, Beers DR, Appel SH. Immune-mediated mechanisms in the pathoprogression of amyotrophic lateral sclerosis. J Neuroimmune Pharmacol. 2013;8(4):888-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner MR, Cagnin A, Turkheimer FE, et al. . Evidence of widespread cerebral microglial activation in amyotrophic lateral sclerosis: an [11C](R)-PK11195 positron emission tomography study. Neurobiol Dis. 2004;15(3):601-609. [DOI] [PubMed] [Google Scholar]
- 3.Corcia P, Tauber C, Vercoullie J, et al. . Molecular imaging of microglial activation in amyotrophic lateral sclerosis. PLoS One. 2012;7(12):e52941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henkel JS, Beers DR, Siklós L, Appel SH. The chemokine MCP-1 and the dendritic and myeloid cells it attracts are increased in the mSOD1 mouse model of ALS. Mol Cell Neurosci. 2006;31(3):427-437. [DOI] [PubMed] [Google Scholar]
- 5.Alexianu ME, Kozovska M, Appel SH. Immune reactivity in a mouse model of familial ALS correlates with disease progression. Neurology. 2001;57(7):1282-1289. [DOI] [PubMed] [Google Scholar]
- 6.Beers DR, Henkel JS, Xiao Q, et al. . Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103(43):16021-16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boillée S, Yamanaka K, Lobsiger CS, et al. . Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312(5778):1389-1392. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Sharma K, Grisotti G, Roos RP. The effect of mutant SOD1 dismutase activity on non-cell autonomous degeneration in familial amyotrophic lateral sclerosis. Neurobiol Dis. 2009;35(2):234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haverkamp LJ, Appel V, Appel SH. Natural history of amyotrophic lateral sclerosis in a database population: validation of a scoring system and a model for survival prediction. Brain. 1995;118(pt 3):707-719. [DOI] [PubMed] [Google Scholar]
- 10.Voustianiouk A, Seidel G, Panchal J, et al. . ALSFRS and appel ALS scores: discordance with disease progression. Muscle Nerve. 2008;37(5):668-672. [DOI] [PubMed] [Google Scholar]
- 11.Henkel JS, Beers DR, Wen S, et al. . Regulatory T-lymphocytes mediate amyotrophic lateral sclerosis progression and survival. EMBO Mol Med. 2013;5(1):64-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillies MA, Rau A, Aubert J, et al. ; French StatOmique Consortium . A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14(6):671-683. [DOI] [PubMed] [Google Scholar]
- 14.Lin Y, Golovnina K, Chen ZX, et al. . Comparison of normalization and differential expression analyses using RNA-Seq data from 726 individual Drosophila melanogaster. BMC Genomics. 2016;17:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meissner F, Molawi K, Zychlinsky A. Mutant superoxide dismutase 1-induced IL-1beta accelerates ALS pathogenesis. Proc Natl Acad Sci U S A. 2010;107(29):13046-13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Présumey J, Courties G, Louis-Plence P, et al. . Nicotinamide phosphoribosyltransferase/visfatin expression by inflammatory monocytes mediates arthritis pathogenesis. Ann Rheum Dis. 2013;72(10):1717-1724. [DOI] [PubMed] [Google Scholar]
- 17.Gavala ML, Hill LM, Lenertz LY, Karta MR, Bertics PJ. Activation of the transcription factor FosB/activating protein-1 (AP-1) is a prominent downstream signal of the extracellular nucleotide receptor P2RX7 in monocytic and osteoblastic cells. J Biol Chem. 2010;285(44):34288-34298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zondler L, Müller K, Khalaji S, et al. . Peripheral monocytes are functionally altered and invade the CNS in ALS patients. Acta Neuropathol. 2016;132(3):391-411. [DOI] [PubMed] [Google Scholar]
- 19.Butovsky O, Siddiqui S, Gabriely G, et al. . Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J Clin Invest. 2012;122(9):3063-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee R, Kanti Barman P, Kumar Thatoi P, Tripathy R, Kumar Das B, Ravindran B. Non-classical monocytes display inflammatory features: validation in sepsis and systemic lupus erythematous. Sci Rep. 2015;5:13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babu GN, Kumar A, Chandra R, Puri SK, Kalita J, Misra UK. Elevated inflammatory markers in a group of amyotrophic lateral sclerosis patients from northern India. Neurochem Res. 2008;33(6):1145-1149. [DOI] [PubMed] [Google Scholar]
- 22.Cereda C, Baiocchi C, Bongioanni P, et al. . TNF and sTNFR1/2 plasma levels in ALS patients. J Neuroimmunol. 2008;194(1-2):123-131. [DOI] [PubMed] [Google Scholar]
- 23.Ono S, Hu J, Shimizu N, Imai T, Nakagawa H. Increased interleukin-6 of skin and serum in amyotrophic lateral sclerosis. J Neurol Sci. 2001;187(1-2):27-34. [DOI] [PubMed] [Google Scholar]
- 24.Ehrhart J, Smith AJ, Kuzmin-Nichols N, et al. . Humoral factors in ALS patients during disease progression. J Neuroinflammation. 2015;12:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell RM, Freeman WM, Randazzo WT, et al. . A CSF biomarker panel for identification of patients with amyotrophic lateral sclerosis. Neurology. 2009;72(1):14-19. [DOI] [PubMed] [Google Scholar]
- 26.Kuhle J, Lindberg RL, Regeniter A, et al. . Increased levels of inflammatory chemokines in amyotrophic lateral sclerosis. Eur J Neurol. 2009;16(6):771-774. [DOI] [PubMed] [Google Scholar]
- 27.Philips T, Robberecht W. Neuroinflammation in amyotrophic lateral sclerosis: role of glial activation in motor neuron disease. Lancet Neurol. 2011;10(3):253-263. [DOI] [PubMed] [Google Scholar]
- 28.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10(12):1538-1543. [DOI] [PubMed] [Google Scholar]
- 29.Mildner A, Schmidt H, Nitsche M, et al. . Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10(12):1544-1553. [DOI] [PubMed] [Google Scholar]
- 30.Chiu IM, Morimoto ET, Goodarzi H, et al. . A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4(2):385-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bettelli E, Carrier Y, Gao W, et al. . Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441(7090):235-238. [DOI] [PubMed] [Google Scholar]
- 32.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24(2):179-189. [DOI] [PubMed] [Google Scholar]
- 33.Butovsky O, Jedrychowski MP, Cialic R, et al. . Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann Neurol. 2015;77(1):75-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller RG, Zhang R, Block G, et al. . NP001 regulation of macrophage activation markers in ALS: a phase I clinical and biomarker study. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(7-8):601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller RG, Block G, Katz JS, et al. ; Phase 2 Trial NP001 Investigators . Randomized phase 2 trial of NP001-a novel immune regulator: safety and early efficacy in ALS. Neurol Neuroimmunol Neuroinflamm. 2015;2(3):e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Principle Component Analysis (PAC) Indicates That Positive Selection Failed to Distinguish ALS and Control Monocyte Samples While Negative Selected ALS Monocytes Were Distinguishable From Control Monocytes.
eFigure 2. Venn Diagram of Differentially Expressed Genes (DEGs) in Monocytes From ALS Slow and Fast Patients
eFigure 3. IL-8 Protein Levels in Sera of ALS Patients and Healthy Controls.
eTable 1. DEGs of Monocytes Isolated From Total ALS Patients (vs Control)
eTable 2. Disease Progression of ALS Patients for RNA-seq Study Based on a Cutoff of 1.5 AALS Points/Month or a Cutoff of 1.0 ALS FRS Points/Month
eTable 3. 65 DEGs Solely Found in Monocytes From Slowly Progressing ALS Patients (vs Control)
eTable 4. 237 DEGs Solely Found in Monocytes From Rapidly Progressing ALS Patients (vs Control)
eTable 5. 43 DEGs Expressed in Monocytes of Both Slow and Fast Groups (vs Control)