Abstract
Formalin-fixed paraffin-embedded (FFPE) tissues are rarely used for screening DNA adducts of carcinogens because the harsh conditions required to reverse the formaldehyde-mediated DNA cross-links can destroy DNA adducts. We recently adapted a commercial silica-based column kit used in genomics to manually isolate DNA under mild conditions from FFPE tissues of rodents and humans and successfully measured DNA adducts of several carcinogens including aristolochic acid I (AA-I), 4-aminobiphenyl (4-ABP), and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) (Yun et al., (2013) Anal. Chem. 85:4251–8, and Guo et al. (2016) Anal. Chem. 88:4780–7). The DNA retrieval methodology is robust; however, the procedure is time-consuming and labor intensive, and not amenable to rapid throughput processing. In this study, we have employed the Promega Maxwell® 16 MDx system, which is commonly used in large scale genomics studies, for the rapid throughput extraction of DNA. This system streamlines the DNA isolation procedure and increases the sample processing rate by about eight-fold over the manual method (32 samples versus 4 samples processed per hour). High purity DNA is obtained in satisfactory yield for the measurements of DNA adducts by ultra performance liquid chromatography-electrospray-ion trap-multistage scan mass spectrometry (UPLC/ESI-IT-MSn). The measurements show that the levels of DNA adducts of AA-I, 4-ABP, and PhIP in FFPE rodent and human tissues are comparable to those levels measured in DNA from matching tissues isolated by the commercial silica-based column kits, and in DNA from fresh frozen tissues isolated by the conventional phenol-chloroform extraction method. The isolation of DNA from tissues is one major bottleneck in the analysis of DNA adducts. This rapid throughput methodology greatly decreases the time required to process DNA and can be employed large-scale epidemiology studies designed to assess the role of chemical exposures and DNA adducts in cancer risk.
Graphical Abstract
INTRODUCTION
Covalent modification of DNA by chemicals can result in mutations or other genetic changes and initiate chemical carcinogenesis.1,2 DNA adducts serve as biomarkers for interspecies extrapolation of toxicity data of chemicals and for human risk assessment.3,4 Identification and quantitation of DNA adducts often are the first steps in elucidating the potential role of a genotoxic chemical in the etiology of human cancer.1,4,5 However, freshly frozen human tissue samples are often not available to measure DNA adducts. In contrast, formalin-fixed samples from patients diagnosed with cancer are readily accessible. Formalin fixation, followed by paraffin embedding (FFPE) has been used as the standard storage technique for more than a century in laboratories worldwide.6–8
The screening of carcinogen DNA adducts in human FFPE tissues has been largely restricted to immunohistochemical (IHC) techniques.9–14 DNA adducts can be detected by IHC, in specific cell types within a tissue. However, an important drawback of IHC is that the specificity of many antibodies, even monoclonal antibodies, for DNA adducts is uncertain as they may cross-react with other DNA lesions or cellular components, leading to errors in identification and quantification. Additionally, the production of antibodies is limited to selected classes of carcinogen DNA adducts and thus, restricts the number of adducts that can be screened for in human cohorts. Thus, there is a critical need to develop robust methods to retrieve DNA in high yield from FFPE tissues and measure DNA adducts by specific mass spectrometry methods. However, FFPE tissues have rarely been employed as a biospecimen for the screening of DNA adducts, by mass spectrometry methods, because of the technical difficulties in recovery of DNA that is free of cross-links.8,15 Quantitative measurements require high quality DNA that is fully digestible by nucleases to obtain the chemically modified mononucleoside adducts. Formaldehyde reacts with DNA and protein to form inter- and intramolecular cross-links during the fixation process,16,17 and DNA that still contains cross-links will impede the digestion efficacy of nucleases, resulting in the recovery oligomeric adducts and an underestimation of the DNA adduct levels. The development of robust conditions to recover DNA without cross-links formed with formaldehyde while preserving the structures of DNA adducts has been challenging. The common methods of reversal of the formaldehyde-mediated DNA cross-links require incubation of FFPE tissues at elevated temperature (up to 100 °C) and strong alkaline pH.18,19 These harsh conditions reduce the quality of DNA, induce depurination, strand cleavage, and/or oxidation of nucleobases and DNA adducts.20,21
We have evaluated various conditions to isolate DNA from FFPE tissues,22,23 including phenol-chloroform extraction, and several commercial silica-based column kits used for genomic applications, which employ mild retrieval conditions to recover DNA free of cross-links.22 In our studies, the commercial, ZR FFPE DNA Miniprep™ kit (ZM) from Zymo Research was optimal. The DNA was isolated under mild temperature and neutral pH conditions and fully digestible with nucleases, demonstrating that the cross-links in the recovered DNA were completely reversed. The methodology was successfully applied to measure DNA adducts of aristolochic acids I (AA-I, 8-methoxy-6-nitrophenanthro-[3,4-d]-1,3, dioxole-5-carboxylic acid), components found naturally in Aristolochia plant species, some of which are still found in some traditional Chinese medicines in the United States and worldwide.24,25 AA-I is a potent human renal toxicant and upper urinary tract carcinogen.24,26 The major DNA adduct of AA-I, 7-(2′-deoxyadenosine-N6-yl)aristolactam (dA-AL-I) is responsible for the signature A → T transversion mutation in multiple sites of the TP53 tumor suppressor gene and leads to AA-induced upper urothelial tract carcinoma.27,28 The levels of AL-DNA adduct retrieved, by the ZM kit, from FFPE tissue blocks stored at ambient temperature for up to nine years were at comparable levels to those levels measured in the matching freshly frozen samples.22,29
Thereafter, the DNA retrieval method was successfully applied to measure other classes of carcinogen-modified DNA from FFPE tissues of rodents including the polycyclic aromatic hydrocarbon benzo[a]pyrene (B[a]P), a suspected human lung carcinogen, and its major DNA adduct 10-(2′-deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydro-benzo[a]pyrene; the aromatic amine 4-aminobiphenyl (4-ABP), a human bladder carcinogen, and its DNA adduct, N-(2′-deoxyguanosin-8-yl)-4-ABP (dG-C8-4-ABP); and the N-nitroso compound, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a lung carcinogen and two of its DNA adducts, O6-Methyl-2′-deoxyguanosine and O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine;30,31 and the heterocyclic aromatic amine (HAA), 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a rodent and potential human colorectal and prostate carcinogen,32,33 and its adduct N-(2′-deoxyguanosin-8-yl)-PhIP (dG-C8-PhIP). These chemical carcinogens are present in the environment and/or arise in tobacco smoke; some HAA also form during the high-temperature cooking of meat.30,31,34,35
The methodology of DNA retrieval and recovery of DNA adducts are robust; however, the procedure is time-consuming and not amenable for processing a large number of samples. In this investigation, we report that a commercial DNA isolation system, the Promega Maxwell® 16 MDx system, which is commonly used for high throughput genomic studies, can rapidly process DNA from archived FFPE tissues of rodents dosed with AA-I; 4-ABP, and PhIP. The high purity of the DNA and the complete reversal of the DNA cross-links permits quantitative measurements of these DNA adducts by UPLC/ESI-IT-MS3. Thereafter, we successfully applied this technology to measure dA-AL-I and dG-C8-PhIP, respectively, in human FFPE kidney and prostate specimens. The levels of adducts were comparable to those levels measured in DNA from matching fresh frozen tissue specimens isolated by the traditional phenol-chloroform method.
METHODS
Caution: AA-I, 4-ABP, and PhIP are carcinogens. These chemicals should be handled with caution in a well-ventilated fume hood with appropriate protective clothing. Human tissue specimens were processed in biohazard hood, and all tissue material was treated with bleach prior to discarding the material in biohazard waste receptacles.
Materials
AA-I was provided by Dr. H. Priestap, Department of Biological Sciences, Florida International University. PhIP was purchased from Toronto Research Chemicals (Toronto, Canada). 4-ABP, calf thymus (CT) DNA, Proteinase K, DNase I, alkaline phosphatase, nuclease P1, RNase A, and RNase T1 were purchased from Sigma-Aldrich (St. Louis, MO). Phosphodiesterase I was purchased from Worthington Biochemical Corp. (Newark, NJ). Neutral buffered formalin (NBF, 10%) was purchased from Fisher Chemical Co. (Pittsburgh, PA). DNA adducts including dA-AL-I and [15N5]-dA-AL-I,29 N-(2′-deoxyguanosin-8-yl)-4-aminobiphenyl (dG-C8-4-ABP), [13C10]-dG-C8-4-ABP, N-(2′-deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (dG-C8-PhIP), and [13C10]-dG-C8-PhIP were synthesized as described.36,37 ZR FFPE DNA Miniprep™ kit (ZM) was purchased from Zymo Research (Irvine, CA). Maxwell® 16 LEV Blood DNA kit (MXB), which is used to isolate DNA for frozen tissue, and Maxwell® 16 FFPE plus LEV DNA Purification kit (MXF), which is used to isolate DNA from formalin fixed tissues, were purchased from Promega Co. (Madison, WI). Microliter CapLC vials with silanized inserts were purchased from Wheaton (Millville, NJ). The CT DNA samples modified with 4-ABP and PhIP were kindly provided by Dr. Frederick A. Beland from the National Center for Toxicology Research, U.S. Food and Drug Administration.
Animal dosing
All protocols were reviewed and approved by the Stony Brook and University of Minnesota Institutional Animal Care and Use Committees, and followed institutional guidelines established by the National Institutes of Health Office of Laboratory Animal Welfare. Three groups of rodents were dosed by i.p. injection as following: Male C57BL/6J mice dosed with AA-I (0.1 mg/kg BW in 0.20 mL phosphate buffered saline); Male B6C3F1/J mice dosed with 4-ABP (40 mg/kg BW in 0.1 mL 80% DMSO); Male Fischer 344 rats dosed with PhIP (50 mg/kg BW in 0.9 mL DMSO). All animals were euthanized, by asphyxiation with CO2, 24 h after the dose treatment. Whole livers and kidneys were rinsed with chilled PBS, snap frozen on dry ice, and stored at −80 °C. Organs for FFPE tissues were rinsed with chilled PBS, cut into 6 mm thick pieces (approximately 1.5 cm2), and fixed in NBF (10%, 20 mL) for 24 h at room temperature. Thereafter, the tissues were processed with Sakura Tissue Tek VIP5 tissue processor at Histology Core Facility at Stony Brook University (for AA-I dosed animals)22 or at University of Minnesota (for 4-ABP- and PhIP-dosed animals).38 The FFPE section blocks were housed for at least six months prior to chemical analyses.
Human prostate and kidney specimens
The research protocol was approved by the Institutional Review Boards at the University of Minnesota and Stony Brook University. De-identified prostate specimens from the peripheral zone of the prostate were obtained from patients scheduled for radical prostatectomy at the University of Minnesota.39 The normal tumor-adjacent prostate tissues were snap frozen in liquid nitrogen and stored at −80 °C. Matching prostate tissues were fixed in 10% NBF for 24 h at room temperature, and then tissues were processed by a Sakura Tissue Tek VIP5 tissue processor at University of Minnesota.39 H&E-stained slides of prostate tissue specimens were examined by the pathologist (Dr. P. Murugan) and were confirmed as largely tumor-free. The FFPE section blocks were housed for at least six months prior to chemical analyses. Tissues were De-identified renal cortex specimens were from subjects exposed to AA-I with renal failure and provided following nephrectomy, by Dr. Bojan Jelaković, School of Medicine, University of Zagreb, Croatia.22
DNA retrieval from freshly frozen rodent and human tissues
Frozen tissue (20 or 40 mg) was thawed on ice and homogenized in 1.5 mL chilled TE lysis buffer (50 mM Tris-HCl, 10 mM EDTA, pH 8.0) containing 10 mM β-mercaptoethanol (βME) with Potter-Elvehjem homogenizer (Corning Inc. NY). The homogenate was centrifuged at 3,000 × g at 4 °C for 10 min to obtain the nuclear pellet.
Phenol-chloroform extraction: the nuclear pellet derived from 40 mg of tissue was reconstituted in 300 μL TE lysis buffer containing RNase A (150 μg) and RNase T1 (0.1 μg) and incubated at 37 °C for 1.5 h, followed by incubation with Proteinase K (400 μg) and 1% (w/v) sodium dodecyl sulfate (SDS) at 37 °C for 2 h. DNA was retrieved from the tissue lysate by phenol-chloroform extraction as previously reported.22
Rapid throughput DNA isolation from frozen rat liver tissues with the Maxwell 16 Blood kit (MXB): The nuclear pellet derived from 20 mg of tissue was reconstituted in 200 μL TE lysis buffer containing 10 mM βME, RNase A (100 μg), and RNase T1 (0.1 μg) and incubated at 37 °C for 1.5 h. Thereafter, the manufacturer’s proprietary lysis buffer (300 μL) containing Proteinase K (400 μg) was added to the mixture and tissue lysis was performed at 37 °C for 3 h. The lysate was transferred to the cartridge to extract DNA following manufacturer’s protocol. The genomic DNA was eluted in 60 μL of nuclease-free water and stored at −80 °C until further analysis.
DNA retrieval from rodent and human FFPE tissues
The detailed protocols for FFPE tissue processing, deparaffinization, rehydration, and homogenization were reported previously.22,23 Rodent FFPE tissues were removed from the paraffin block and submerged in p-xylene to remove the residual paraffin. The dry weight of FFPE tissue was measured following deparaffinization in p-xylene, and it was about 50% of the original wet weight of frozen tissue. Then, the tissues were washed with serial dilutions of ethanol to remove the infiltrated p-xylene and to rehydrate the tissue. The rehydrated FFPE tissues (~100 mg) were homogenized in 4 mL cold TE lysis buffer containing 10 mM βME using a Potter-Elvehjem homogenizer. An equivalent of 20 mg (dry weight, in triplicate) of FFPE tissue homogenate was centrifuged at 3,000 g for 10 min. The pellet was then processed with ZR FFPE DNA Miniprep™ kit (ZM) or by the Maxwell® 16 FFPE plus LEV DNA Purification kit (MXF), following the manufacturer’s protocols with s minor modifications.22,39 The FFPE tissue homogenate (20 mg) processed with the ZM kit was incubated at 50 °C overnight in 100 μL of the proprietary digestion buffer containing 10 mM βME and Proteinase K (200 μg). Then, the lysate was incubated with RNase A (150 μg) for 10 min at room temperature to eliminate RNA contamination in final DNA samples. Thereafter, the mixture of tissue lysate was processed to isolate DNA using the silica spin column, by following Zymo Research’s protocol with minor modifications.22,39 The FFPE tissue homogenate (20 mg) processed with the MXF kit, was incubated at 50 °C overnight in 200 μL of the manufacturer’s incubation buffer containing 10 mM βME and Proteinase K (400 μg). Thereafter, the lysate was incubated with RNase A (150 μg) for 10 min at room temperature. The mixture was diluted with 400 μL lysis buffer and transferred to the cartridge to process with Maxwell® 16 system. The overnight incubation of the nuclear pellets of the FFPE tissues with Proteinase K at 50 °C completely reversed the cross-links between DNA and protein.38 The concentration of DNA was determined by Agilent 8453 UV/Vis spectrometer using Traycell (Hellma, USA Inc. NY).
The FFPE sections of human renal cortex were prepared in 10 μm thickness (~ 1.5 cm2) using a microtome. Two 10 μm sections of human FFPE kidney were deparaffinized and rehydrated in 1 mL p-xylene and 95% ethanol, respectively. Then the rehydrated sections were processed with ZM or MXF kit following same protocol as FFPE rodent tissue homogenate.
Enzymatic digestion of carcinogen-modified DNA
The detailed protocol of DNA digestion was reported previously22,23,29 DNA (5 μg) was spiked with isotopically labeled internal standards ([15N5]-dA-AL-I and [13C10]-dG-C8-PhIP at a level 5 adducts per 108 nucleotides; [13C10]-dG-C8-4-ABP at a level of 10 adducts per 108 nucleotides) and digested with DNase I, nuclease P1, alkaline phosphatase, and Phosphodiesterase I overnight at 37 °C. The DNA digest was concentrated to dryness by vacuum centrifugation and reconstituted in 1:1 DMSO:H2O (25 μL), and centrifuged at 21,000 × g for 10 min. The supernatant was transferred to silanized vial insert for LC-MS analysis. The genomic or CT DNA with a known level of corresponding DNA adducts were used as the positive controls.38 The efficacy of DNA digestion and purity of DNA were assessed by HPLC analysis of unmodified 2′-deoxynucleosides as previously reported.40
Measurement of carcinogen-DNA adducts by UPLC/ESI-IT-MS3 measurements
Analyses were performed with a Waters nanoAcquity UPLC system interfaced with an Advance Captive Spray source (Michrom Bioresources Inc., Auburn, CA), and linear quadrupole ion trap (LTQ Velos Pro, Thermo Fisher Scientific, San Jose, CA). A Waters Symmetry C18 trap column (180 μm × 20 mm, 5 μm, Waters Corp., Milford, MA) was used for online sample enrichment of DNA adducts. The analytical column was a Magic C18 AQ column (300 μm × 150 mm, 3 μm, 100 Å) from Michrom Bioresources Inc. The solvents and chromatographic conditions were previously reported.39
The DNA adducts were measured in positive ion mode at MS3 scan stage. Fragmentation of precursor ions was done by collision induced dissociation (CID) with a collision energy of 28% at MS2 and 40% at the MS3 scan stage. The following transitions were employed: dA-AL-I at m/z 543.3 → 427.2 → 292.1, 293.1, and 412.1; [15N5]-dA-AL-I at m/z 548.3 → 432.2 → 292.1, 293.1, and 417.1; dG-C8-4-ABP at m/z 435.2 → 319.1 → 277.1 and 302.1; [13C10]-dG-C8-4-ABP at m/z 445.2 → 324.1 → 281.1 and 307.1; dG-C8-PhIP at m/z 490.2 → 374.1 → 329.1 and 357.1; [13C10]-dG-C8-PhIP at m/z 500.2 →379.1 → 333.1 and 362.1.
The analysis of PhIP-DNA adduct in human prostate tissues was performed with the Orbitrap Fusion Tribrid MS (Thermo Fisher Scientific, San Jose, CA) interfaced with Dionex UltiMate RSLCnano UHPLC System and a Thermo Nanospray Flex ion source. The chromatographic and mass spectra acquisition parameters were described previously.39
Method validation of carcinogen-modified DNA analysis by UPLC/ESI-IT-MSn and calibration curves
The accuracy of the method was previously validated with genomic DNA or CT DNA containing known levels of dA-AL-I, dG-C8-4-ABP and dG-C8-PhIP.29,38,39 The calibration curves were constructed as previously reported.29,38,39 The limit of quantification (LOQ) value for each carcinogen-DNA adduct approached 3 – 5 adducts per 109 nucleotides with 5 μg DNA assayed.29,38,39
Statistical methods
The unpaired t-test or one way analysis of variance (ANOVA) with unpaired Tukey’s multiple comparison test was performed using GraphPad Prism version 6.0 for Windows (GraphPad Software, La Jolla, CA). Data were expressed as mean ± SD. A p-value < 0.05 was considered statistically significant.
RESULTS
Recovery of DNA from fresh frozen tissues by manual and rapid throughput methods
The chemical structures of the carcinogens and their DNA adducts examined in this study are shown in Figure 1. The mean yields of DNA from frozen rodent tissues are expressed as μg DNA per mg of tissue. The purities of DNA isolated by two different DNA extraction methods, phenol-chloroform and Maxwell® 16 LEV Blood DNA kit (MXB) for rapid throughput method, are shown in Table 1. The MXB has been developed for rapid throughput DNA isolation from whole blood or buccal swab samples, using a low elution volume. We tested other DNA isolation kits from the Maxwell® 16 system; however, the yield of DNA obtained from MXB was consistently the highest. Therefore, we employed the MXB kit for all rapid throughput DNA isolation from fresh tissue experiments.
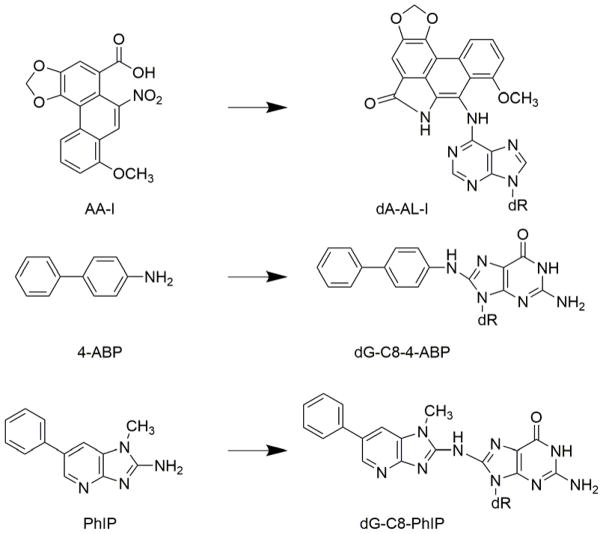
Figure 1.
Chemical structures of carcinogens and their major DNA adducts. (dR= 2′-deoxyribose).
Table 1.
The mean DNA yields recovered from fresh frozen and FFPE tissues by different DNA extraction methods.
| Method of DNA extraction | Tissue type | Tissue digestion | Number of samples (N) | DNA yield (μg/mg tissue)a | Purity (A260/280 ratio) |
|---|---|---|---|---|---|
|
Phenol-chloroform EXTRACTION (FRESH FROZEN) |
Mouse liver | 37 °C, 2 h | 27 | 2.18 ± 0.56 | 1.87 ± 0.02 |
| Mouse kidney | 15 | 2.85 ± 0.96 | 1.87 ± 0.03 | ||
| Rat liver | 9 | 1.94 ± 0.17 | 1.84 ± 0.05 | ||
|
| |||||
|
Maxwell® 16 LEV BLOOD DNA KIT (MXB, FRESH FROZEN) |
Rat liver | 37 °C, 2 h | 9 | 1.17 ± 0.17 | 1.86 ± 0.00 |
|
| |||||
|
ZR FFPE DNA MINIPREPTM KIT (ZM, FFPE) |
Mouse liver | 50 °C, 18 h | 27 | 0.63 ± 0.18 | 1.89 ± 0.07 |
| Mouse kidney | 15 | 0.52 ± 0.15 | 1.86 ± 0.01 | ||
| Rat liver | 12 | 0.46 ± 0.15 | 1.88 ± 0.03 | ||
|
| |||||
|
Maxwell® 16 FFPE PLUS LEV DNA PURIFICATION KIT (MXF, FFPE) |
Mouse Liver | 50 °C, 18 h | 27 | 0.64 ± 0.13 | 1.86 ± 0.03 |
| Mouse Kidney | 15 | 0.47 ± 0.09 | 1.85 ± 0.01 | ||
| Rat liver | 12 | 0.77 ± 0.06 | 1.81 ± 0.01 | ||
DNA yield is reported as μg per mg of tissue. DNA yields from FFPE tissues are calculated using the adjusted wet-weight (two times of dry-weight) of FFPE tissues. The dry-weight of FFPE tissue is determined after the deparaffinization process in p-xylene. The dry-weight is ~ 50% of the wet weight of frozen tissue in the three tissue types.
The purity of DNA was further confirmed by quantitation of 2′-deoxyguanosine and 2′-deoxyadenosine after enzymatic digestion using HPLC (Figure S1); FFPE, formalin-fixed paraffin-embedded tissue.
The mean DNA yields, μg DNA per mg tissue, from fresh frozen tissues by phenol-chloroform extraction were 2.18 ± 0.56 from mouse liver; 2.85 ± 0.96 from mouse kidney; and 1.94 ± 0.17 from rat liver. The ratio of absorbance at 260 nm to 280 nm (A260/280) was used to assess the purity of DNA. A ratio of ~ 1.85 at neutral pH is considered as pure for DNA.41 The mean DNA yield from frozen rat liver using MXB was 1.17 ± 0.17 μg/mg tissue, a value that was ~60% of the DNA yield by the phenol-chloroform method. The maximum DNA binding capacity of the magnetic beads in each cartridge is about 25 μg of DNA when 20 mg of tissue is used for processing. Accordingly, the reason for the lower recovery of DNA from frozen rat liver tissues extracted by the MXB than by the phenol-chloroform extraction method is likely attributed to the limit of the DNA binding capacity of the magnetic beads that carries the DNA during the purification process. Both methods recovered reproducible quantities of high purity DNA from frozen tissues.
Recovery of DNA from FFPE tissues by manual and rapid throughput methods
Our previous study revealed that even prolonged proteolysis of nuclear pellets from FFPE tissues at elevated temperature (50 °C), by the conditions of the phenol-chloroform extraction method, resulted in an incomplete reversal of DNA cross-links, and a low recovery of DNA with a concomitant underestimation of DNA adducts of AA-I.22 We successfully employed the commercial silica-based ZR FFPE DNA Miniprep™ kit (ZM), which uses mild retrieval conditions to recover DNA free of cross-links from FFPE tissues,22 to measure DNA adducts of dA-AL-I and several other important environmental and dietary carcinogens in FFPE tissues.38 The methodology, however, is a manual, labor-intensive procedure and not amenable to workup of many samples concurrently. In this study, we have advanced our method to measure DNA adducts in FFPE tissues by adapting a technology from Promega that is commonly used for the high-throughput isolation of DNA for cancer genomic studies.42,43 Starting from the nuclear pellets, approximately four samples can be manually processed by the manual phenol-chloroform extraction or the ZM kit compared to 32 samples per hour with the Promega Maxwell® 16 MDx system. There are proprietary component(s) in commercial lysis buffer (Zymo Research) which are critical for the complete reversal of formaldehyde-mediated cross-links between DNA and proteins in FFPE tissues.22,23 Here, we show that the yield of DNA and the extent of reversal of cross-links of FFPE DNA recovered with Promega’s incubation buffer and Zymo Research’s digestion buffer are comparable. The dry-weight of FFPE tissue is determined after the deparaffinization process with p-xylene, and the weight is about 50% of the wet weight of frozen tissue in the three tissue types. The DNA yield of FFPE tissues is normalized by the adjusted wet weight (~ two times the dry-weight), to directly compare the efficacy of DNA extraction method between fresh frozen and FFPE tissues. The normalized mean yields of DNA recovered from FFPE tissues of rodents using ZR FFPE DNA Miniprep™ (ZM) and Maxwell® 16 FFPE plus LEV DNA (MXF) kits are reported in Table 1. The normalized mean yields, μg DNA per mg tissue, of DNA from FFPE tissues using ZM were 0.63 ± 0.18 for mouse liver; 0.52 ± 0.15 for mouse kidney; and 0.46 ± 0.15 for rat liver. The MXF provided comparable yields of DNA to that of manual spin column method; 0.64 ± 0.13 for mouse liver, 0.47 ± 0.09 for mouse kidney, and 0.77 ± 0.06 for rat liver. The ratio A260/280 of extracted DNAs from both DNA isolation methods was close to 1.85 at neutral pH for all tissue types. The yield of DNA recovered from FFPE tissue specimens ranged from 20 to 40% of the amounts obtained from fresh frozen tissues. Similar declines in the recovery of DNA from FFPE tissue, using other methods of DNA retrieval, have been reported.23,44–46 Possible causes of lower DNA recovery from FFPE tissues include the oxidation and/or fragmentation of intact DNA during fixation process,21 incomplete reversal of DNA-protein crosslinks,45 and lower yield of DNA recovery using silica based methods compared to phenol-chloroform extraction.47
The level of RNA contamination in DNA samples processed from fresh frozen tissues, using phenol-chloroform extraction, or FFPE tissues processed by ZM and MXF kits was assessed, by HPLC and UV detection, following nuclease digestion of DNA. The chromatograms of the DNA digests showed that RNA contamination was ~ 1 to 2%, a purity comparable to that of the purity of DNA retrieved from freshly frozen tissues by phenol-chloroform extraction.22,23,38 Moreover, only the four canonical deoxynucleosides were detected in the chromatograms, demonstrating that the procedures completely removed the formaldehyde mediated crosslinks from the deoxynucleosides; there was no evidence for other residual modified deoxynucleosides in the digestion mixture.(Figure S1) The amounts of deoxynucleosides measured by HPLC-UV closely matched the concentration of DNA estimates by UV absorbance at 260 nm, demonstrating that the DNA recovered by phenol-chloroform, ZR FFPE DNA Miniprep™ and Maxwell® 16 FFPE plus LEV DNA purification kits were of high purity.
Recovery and quantification of DNA adducts in fresh frozen and FFPE rodent tissues
In our previous reports, we demonstrated that dA-AL-I29, dG-C8-4-ABP, and dG-C8-PhIP (Figure 1) were relatively stable toward formalin fixation and DNA retrieval process using ZM kit.22,38 In this study, we compared the levels of DNA adducts retrieved from fresh frozen tissue by phenol-chloroform and from FFPE tissues processed by the manual ZR FFPE DNA Miniprep™ kit and the rapid throughput method of the Maxwell® 16 system. Each set of experiments was performed on three different days with tissues from five animals assayed in triplicate. However, the amount of tissue for FFPE mouse kidney treated with AA-I was only sufficient to run triplicate assays for two days, and only two tissue specimens were assayed in triplicate for day 3.
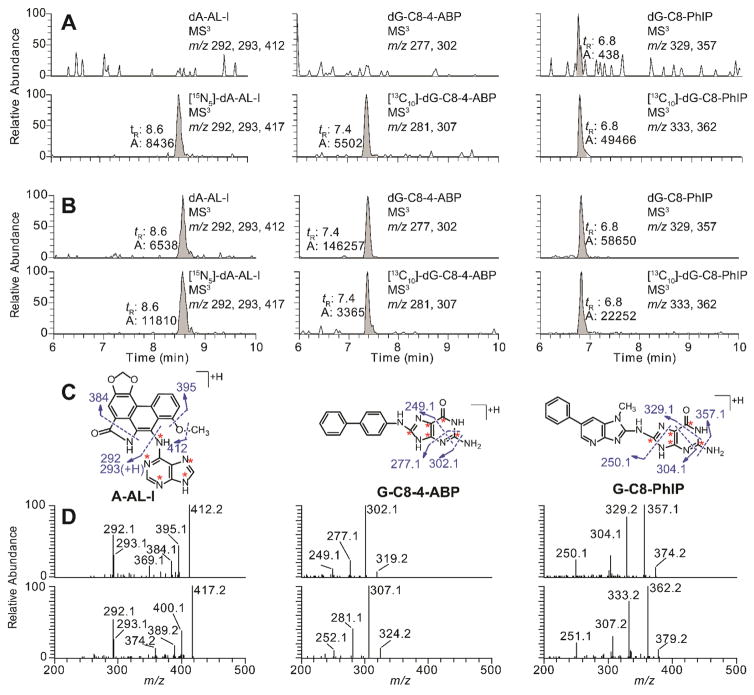
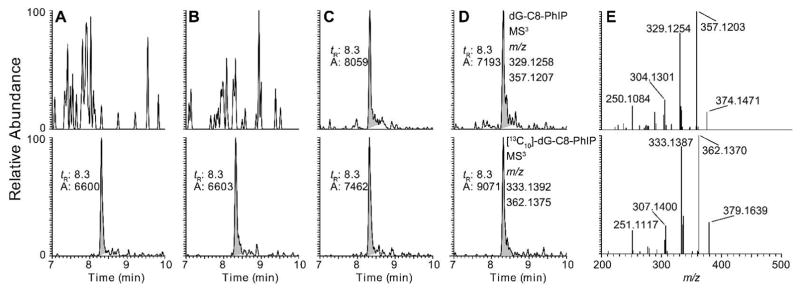
The extracted ion chromatograms (EICs) at MS3 scan stage and the product ion spectra of dA-AL-I, dG-C8-4-ABP, and dG-C8-PhIP recovered from FFPE rodent liver tissues are depicted in Figure 2. The EICs of negative control DNA samples and the adducts were from FFPE DNA samples processed by the rapid throughput DNA isolation method. Calf thymus DNA, unmodified and modified with known levels of carcinogens, and respective internal standards were used as control samples to test the efficacy of DNA digestion and to verify the absence of cross-contamination. The product ion spectra of the adducts at the MS3 scan stage, provide rich structure features about each adduct and corroborate the identities of the adduct structures.29,48,49
Figure 2.
EICs at the MS3 scan stage of rodent FFPE tissues samples targeting dA-AL-I, dG-C8-4-ABP, and dG-C8-PhIP. DNA samples were isolated from FFPE liver tissues of rodents employing the rapid throughput DNA isolation method. (A) EICs of negative control samples of calf thymus DNA spiked with isotope labeled internal standards; [15N5]-dA-AL-I and [13C10]-dG-C8-PhIP at a level of 5 adducts per 108 nucleotides, [13C10]-dG-C8-4-ABP at a level of 10 adducts per 108 nucleotides. (B) Representative EICs of FFPE liver samples from rodents dosed with AA-I, 4-ABP, and PhIP, respectively; (C) The structures of aglycone adducts of dA-AL-I, dG-C8-4-ABP, and dG-C8-PhIP, and proposed mechanism of fragmentation are present. The isotopically labeled 15N and 13C atoms of the internal standards are marked with red asterisks. (D) The product ion spectra of isotope labeled and unlabeled adduct of dA-AL-I, dG-C8-4-ABP, and dG-C8-PhIP.
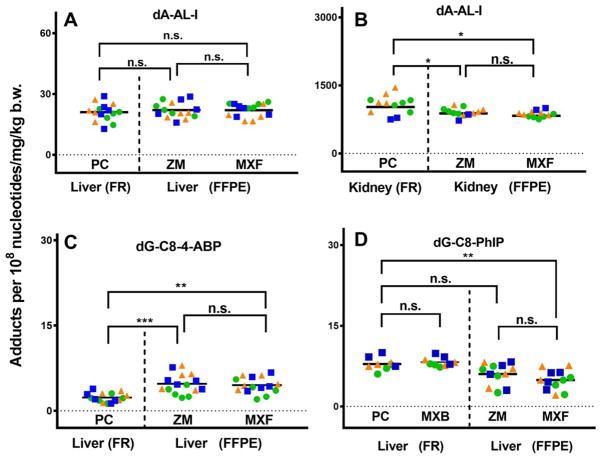
The mean levels of each DNA adduct are shown as scatter plots in Figure 3. DNA adduct levels are expressed as adducts per 108 nucleotides and normalized by dose per kilogram body weight. The % coefficient of variation (% CV) of the mean adduct level for each DNA isolation method was within 20% (or less). The mean level of dA-AL-I in DNA from fresh frozen by phenol-chloroform and FFPE mouse liver processed by manual (ZM) and rapid throughput (MXF) kits are not significantly different. However, the mean level of dA-AL-I in fresh frozen kidney processed by phenol-chloroform is a modest 1.2-fold greater than the mean levels observed in FFPE kidney processed by ZM and MXF kits (p < 0.05). The level of dG-C8-PhIP present in DNA of fresh frozen liver tissues is also slightly higher (1.3-fold) than the levels measured in FFPE tissue. In the case of dG-C8-4-ABP, we previously reported a ~1.6-fold higher level of dG-C8-4-ABP in liver from the set of rodent tissues processed by FFPE compared to the set of fresh frozen tissues.38 These levels of adduct were measured in livers of differently dosed animals and not with matching tissues within each animal. We surmise that these differences in adduct levels formed are largely attributed to differences in bioavailability of the test compound or in carcinogen metabolism among the animals since DNA adduct formation in rodents dosed with the same amounts of test article can vary by up to several-fold.50–52 Additionally, the method of processing DNA from fresh frozen and FFPE tissues is different. Frozen tissues are treated with RNase A and RNase T1, followed by the incubation with Proteinase K, whereas, the FFPE procedure requires proteolysis first, followed by RNase treatment. Therefore, we reversed the incubation sequences of Proteinase K and ribonucleases, employing frozen liver tissue from 4-ABP treated mice. However, the level of dG-C8-4-ABP measured in the fresh frozen liver of the reversed enzyme incubation was not increased, implying a possible repair of dG-C8-4-ABP did not occur during the tissue lysis process (unpublished data, BH Yun). In rats treated with PhIP, the mean level of dG-C8-PhIP in frozen tissues was slightly higher than that of FFPE tissues, but the difference was not statistically significant.
Figure 3.
Levels of carcinogen DNA adduct in rodent tissues. One way analysis of variation with Tukey’s multiple comparison test (Prism 6, San Diego, CA); ns, not significant; *p < 0.05, **p < 0.005, and ***p < 0.0005., Fresh frozen tissues were processed with phenol-chloroform extraction or Maxwell® 16 LEV Blood DNA kit (MXB, for dG-C8-PhIP only). FFPE tissues were processed with ZR FFPE DNA Miniprep™ kit (ZM) or Maxwell® 16 FFPE plus LEV DNA Purification kit (MXF). FR, freshly frozen tissue; FFPE, FFPE tissue. Data from different groups are depicted by different colors and shapes: day 1, green circle; day 2, orange triangle; and day 3, blue square. The lowest adduct level of dG-C8-PhIP in FFPE tissues processed by ZM or MXF on day 1, 2, and 3 is the same animal.
The mean levels of dA-AL-I, dG-C8-4-ABP, and dG-C8-PhIP are not significantly different between FFPE DNA processed by the manual and rapid throughput methods, and adduct levels are similar to those levels measured in DNA of fresh frozen tissues processed by phenol-chloroform, signifying that the adducts are relatively stable towards FFPE processing and during the retrieval processes for both FFPE DNA isolation methods. We also applied the rapid throughput DNA isolation method to retrieve dG-C8-PhIP from fresh frozen rat liver tissue. The levels of dG-C8-PhIP measured from frozen tissues processed by phenol-chloroform and Maxwell® 16 LEV Blood DNA kit (MXB) were not statistically different.
Measurement of dA-AL-I and dG-C8-PhIP adducts in fresh frozen and FFPE human kidney cortex and prostate tissues
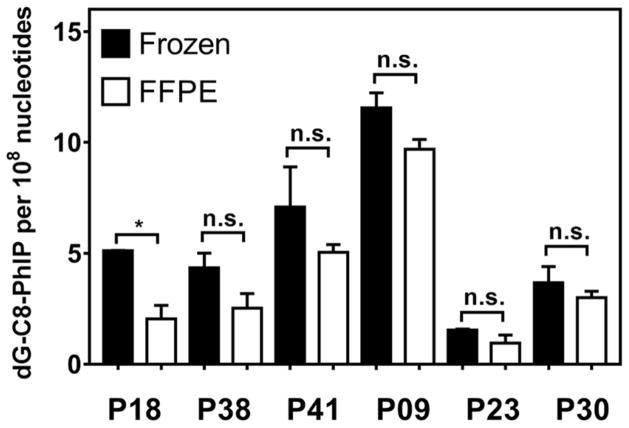
We compared the levels of PhIP-DNA adducts in freshly frozen human prostate by the phenol-chloroform extraction of DNA, and matching FFPE tissues blocks processed by Maxwell® 16 FFPE plus LEV DNA purification kit (MXF) from six patients who were positive for dG-C8-PhIP.39 The FFPE blocks were stored at room temperature at least 6 months prior to analysis. DNA extraction and measurement of adducts were performed in parallel on freshly frozen and FFPE tissues, and the results are shown in Figures 4 and 5. There was a tendency of lower levels of dG-C8-PhIP measured in FFPE specimens compared to fresh frozen specimens, but the differences in adduct levels were not statistically significant except for subject of P18, where the level of dG-C8-PhIP in FFPE tissue was 60% lower. The mean level of dG-C8-PhIP measured in the other five FFPE prostate tissues ranged from 60 to 84% of the adduct level found in fresh frozen tissues. Because of the limited amount of DNA, only two independent replicates could be performed on each sample. A larger number of sample replicate sets are required to improve the precision and to determine if there are significant differences between the levels of dG-C8-PhIP measured in fresh frozen and FFPE human tissues.
Figure 4. Levels of PhIP-DNA adduct in paired fresh frozen prostate and FFPE blocks of six patients.
The levels of adducts are reported as adducts per 108 nucleotides. Two independent analyses were done per subject; *p < 0.02, n.s.: statistically not significant. DNA isolation from frozen and FFPE prostate tissues of six patients were performed using conventional phenol-chloroform extraction and MXF kit, respectively. All measurements were performed in duplicate and reported as the mean ± SD.
Figure 5.
EICs of unlabeled dG-C8-PhIP (upper panel) and 13C-labeled dG-C8-PhIP (lower panel) of DNA from fresh frozen and FFPE human prostate tissues at the MS3 scan stage. (A) fresh frozen prostate and (B) paired FFPE block of a patient (P46) who was negative for dG-C8-PhIP; (C) fresh frozen prostate and (D) paired FFPE block of a patient (P41) who was positive for dG-C8-PhIP; and (E) product ion spectra of unlabeled and 13C-labeled dG-C8-PhIP at MS3 scan stage. The structure and proposed fragmentation mechanism of aglycone of dG-C8-PhIP are depicted in Figure 2.
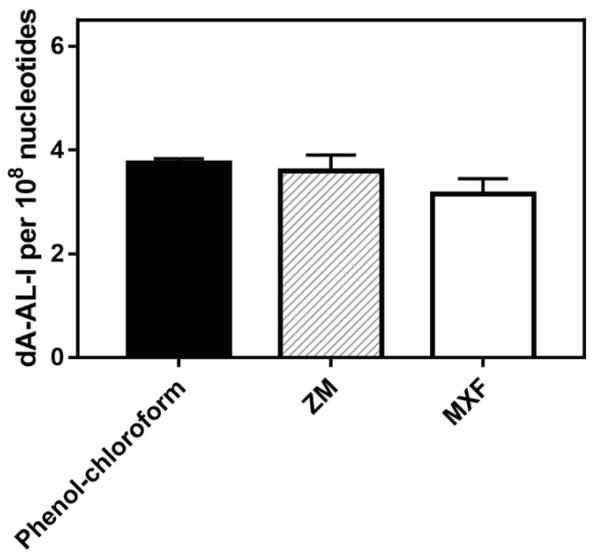
In the case of AA-I, we successfully retrieved DNA from human FFPE kidney block that was prepared in 2008 and stored at room temperature for nine years. The mean levels of dA-AL-I in DNA retrieved from human FFPE kidney using the ZM and MXF kits were 3.1 and 3.6 adducts per 108 nucleotides, respectively. (Figure 6). These of dA-AL-I measured are similar to the level of dA-AL-I measured in frozen tissues, 3.8 adducts per 108 nucleotides, that was processed by phenol-chloroform extraction.22
Figure 6.
Levels of dA-AL-I in DNA extracted from frozen and the matching FFPE tissues of human kidney cortex. DNA from frozen kidney was retrieved using conventional phenol-chloroform extraction. DNA from FFPE human kidney was isolated using ZR FFPE DNA MiniprepTM kit (ZM) or Maxwell® 16 FFPE plus LEV DNA Purification kit (MXF) kit. Each DNA (5 μg) was spiked with 15N5-dA-AL-I at a level of 5 adducts per 108 nucleotides. The FFPE tissues were acquired in 2008 and were stored at room temperature for 9 years. All measurements were performed in duplicate and reported as the mean ± SD.
DISCUSSION
Our results from previous studies and data reported here signify that the major DNA adducts of AA-I, 4-ABP, PhIP, B[a]P and NNK are relatively stable to the FFPE tissue process and that a high portion of the adducts are recovered from rodent tissues or FFPE human kidney and prostate specimens.22,38,39 The processing of high purity DNA from tissues is one major bottleneck in the analysis of DNA adducts. The rapid throughput method employing the Promega Maxwell® 16 MDx system and its DNA purification kit substantially increases the number of samples that can be processed over the manual ZR FFPE DNA Miniprep™ kit method. Thirty-two samples can be processed concurrently with the Promega Maxwell® 16 MDx system compared to about four samples per hour employing the manual ZR FFPE DNA Miniprep™ kit. Both methods use the nuclear pellets for processing of the DNA, and the cost of consumable reagents is similar for both DNA retrieval methods.. Our rapid throughput methodology paves the way for the usage of archived FFPE specimens in human epidemiology studies for which environmental exposures to hazardous chemicals and their DNA adducts may contribute to the etiology of cancer.
Supplementary Material
Acknowledgments
FUNDING INFORMATION
This research was supported by Grant R01ES019564 (RJT) from the National Institute of Environmental Health Sciences, R01CA122320 (RJT), and R01 CA220367 (RJT and TAR), and R33CA186795 (RJT) from the National Cancer Institute of the National Institutes of Health, and from Henry and Marsha Laufer (APG, KGD, and TAR). Mass spectrometry was carried out in Analytical Biochemistry Shared Resources of the Masonic Cancer Center, University of Minnesota, funded in part by Cancer Center Support Grant CA-077598.
We thank Dr. Frederick A. Beland from the National Center for Toxicology Research/US FDA for providing PhIP and 4-ABP treated CT-DNA. We thank Dr. Badrinath Konety, MD, Department of Urology, University of Minnesota, for his interest and support of this project. Dr. Suprita Krishna and Ms. Resha Tejpaul, from the Department of Urology, for administrative assistance. Drew Sciacca, Department of Laboratory Medicine and Pathology, who handled the prostatectomy specimens and Beth Fenske and Carla Heinke, from BioNet Tissue Procurement, for the collection of the prostate biospecimens. The de-identified renal cortex specimens were kindly provided by Dr. Bojan Jelaković, School of Medicine, University of Zagreb, Croatia. Technical support was provided by the Stony Brook University Research Histology Core and is gratefully acknowledged.
ABBREVIATION LIST
- AA-I
aristolochic acid I
- dA-AL-I
7-(2′-deoxyadenosine-N6-yl)aristolactam
- 4-ABP
4-aminobiphenyl
- dG-C8-4-ABP
N-(2′-deoxyguanosin-8-yl)-4-ABP
- B[a]P
benzo[a]pyrene
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- dG-C8-PhIP
N-(2′-deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- FR
freshly frozen tissue
- FFPE
formalin-fixed paraffin-embedded tissue
- NBF
neutral buffered formalin
- βME
β-mercaptoethanol
- HAAs
heterocyclic aromatic amines
- LOQ
limit of quantitation
- PC
phenol-chloroform extraction
- UPLC/ESI-IT-MSn
ultra performance liquid chromatography-electrospray-ionization trap-multistage scan mass spectrometry
Footnotes
The Supporting Information is available free of charge on the ACS Publications website.
Analyses of DNA digest from fresh frozen and FFPE tissues by HPLC-UV detection are shown in supporting information Figure S1.
References
- 1.Miller EC. Some Current Perspectives on Chemical Carcinogenesis in Humans and Experimental-Animals - Presidential-Address. Cancer Res. 1978;38:1479–1496. [PubMed] [Google Scholar]
- 2.Essigmann JM, Wood ML. The Relationship between the Chemical Structures and Mutagenic Specificities of the DNA Lesions Formed by Chemical and Physical Mutagens. Toxicol Lett. 1993;67:29–39. doi: 10.1016/0378-4274(93)90044-x. [DOI] [PubMed] [Google Scholar]
- 3.Poirier MC, Beland FA. Aromatic amine DNA adduct formation in chronically-exposed mice: considerations for human comparison. Mutat Res. 1997;376:177–184. doi: 10.1016/s0027-5107(97)00041-9. [DOI] [PubMed] [Google Scholar]
- 4.Jarabek AM, Pottenger LH, Andrews LS, Casciano D, Embry MR, Kim JH, Preston RJ, Reddy MV, Schoeny R, Shuker D, Skare J, Swenberg J, Williams GM, Zeiger E. Creating context for the use of DNA adduct data in cancer risk assessment: I. Data organization. Crit Rev Toxicol. 2009;39:659–678. doi: 10.1080/10408440903164155. [DOI] [PubMed] [Google Scholar]
- 5.Hemminki K. DNA adducts, mutations and cancer. Carcinogenesis. 1993;14:2007–2012. doi: 10.1093/carcin/14.10.2007. [DOI] [PubMed] [Google Scholar]
- 6.Fox CH, Johnson FB, Whiting J, Roller PP. Formaldehyde fixation. J Histochem Cytochem. 1985;33:845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- 7.Leong A-Y, James CL, Thomas AC. Handbook of Surgical Pathology. Churchill Livingstone; New York: 1996. [Google Scholar]
- 8.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirier MC, Beland FA. DNA adduct measurements and tumor incidence during chronic carcinogen exposure in animal models: implications for DNA adduct-based human cancer risk assessment. Chem Res Toxicol. 1992;5:749–755. doi: 10.1021/tx00030a003. [DOI] [PubMed] [Google Scholar]
- 10.Curigliano G, Zhang YJ, Wang LY, Flamini G, Alcini A, Ratto C, Giustacchini M, Alcini E, Cittadini A, Santella RM. Immunohistochemical quantitation of 4-aminobiphenyl-DNA adducts and p53 nuclear overexpression in T1 bladder cancer of smokers and nonsmokers. Carcinogenesis. 1996;17:911–916. doi: 10.1093/carcin/17.5.911. [DOI] [PubMed] [Google Scholar]
- 11.Santella RM. Immunological methods for detection of carcinogen-DNA damage in humans. Cancer Epidemiol Biomarkers Prev. 1999;8:733–739. [PubMed] [Google Scholar]
- 12.Poirier MC, Santella RM, Weston A. Carcinogen macromolecular adducts and their measurement. Carcinogenesis. 2000;21:353–359. doi: 10.1093/carcin/21.3.353. [DOI] [PubMed] [Google Scholar]
- 13.Faraglia B, Chen SY, Gammon MD, Zhang Y, Teitelbaum SL, Neugut AI, Ahsan H, Garbowski GC, Hibshoosh H, Lin D, Kadlubar FF, Santella RM. Evaluation of 4-aminobiphenyl-DNA adducts in human breast cancer: the influence of tobacco smoke. Carcinogenesis. 2003;24:719–725. doi: 10.1093/carcin/bgg013. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, Chang P, Bondy ML, Sahin AA, Singletary SE, Takahashi S, Shirai T, Li D. Detection of 2-amino-1-methyl-6-phenylimidazo[4,5-b]-pyridine-DNA adducts in normal breast tissues and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:830–837. [PubMed] [Google Scholar]
- 15.Douglas MP, Rogers SO. DNA damage caused by common cytological fixatives. Mutat Res. 1998;401:77–88. doi: 10.1016/s0027-5107(97)00314-x. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel GR, Mehrabian M, Martinson HG. Contact-site cross-linking agents. Mol Cell Biochem. 1981;34:3–13. doi: 10.1007/BF02354846. [DOI] [PubMed] [Google Scholar]
- 17.Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, Ball LM, Swenberg JA. Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J Am Chem Soc. 2010;132:3388–3399. doi: 10.1021/ja908282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L, Patten N, Yamashiro CT, Chui B. Extraction and amplification of DNA from formalin-fixed, paraffin-embedded tissues. Appl Immunohistochem Mol Morphol. 2002;10:269–274. doi: 10.1097/00129039-200209000-00015. [DOI] [PubMed] [Google Scholar]
- 19.Shi SR, Datar R, Liu C, Wu L, Zhang Z, Cote RJ, Taylor CR. DNA extraction from archival formalin-fixed, paraffin-embedded tissues: heat-induced retrieval in alkaline solution. Histochem Cell Biol. 2004;122:211–218. doi: 10.1007/s00418-004-0693-x. [DOI] [PubMed] [Google Scholar]
- 20.Shibutani S, Gentles RG, Iden CR, Johnson F. Facile Aerial Oxidation of the DNA-Base Adduct N-(2′-Deoxyguanosin-8-Yl)-2-Aminofluorene [Dg(C8)Af] J Am Chem Soc. 1990;112:5667–5668. [Google Scholar]
- 21.Peluso MEM, Munnia A, Tarocchi M, Giese RW, Annaratone L, Bussolati G, Bono R. Oxidative DNA damage and formalin-fixation procedures. Toxicol Res. 2014;3:341–349. [Google Scholar]
- 22.Yun BH, Rosenquist TA, Nikolic J, Dragicevic D, Tomic K, Jelakovic B, Dickman KG, Grollman AP, Turesky RJ. Human formalin-fixed paraffin-embedded tissues: an untapped specimen for biomonitoring of carcinogen DNA adducts by mass spectrometry. Anal Chem. 2013;85:4251–4258. doi: 10.1021/ac400612x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun BH, Yao L, Jelakovic B, Nikolic J, Dickman KG, Grollman AP, Rosenquist TA, Turesky RJ. Formalin-fixed paraffin-embedded tissue as a source for quantitation of carcinogen DNA adducts: aristolochic acid as a prototype carcinogen. Carcinogenesis. 2014;35:2055–2061. doi: 10.1093/carcin/bgu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grollman AP. Aristolochic acid nephropathy: Harbinger of a global iatrogenic disease. Environ Mol Mutagen. 2013;54:1–7. doi: 10.1002/em.21756. [DOI] [PubMed] [Google Scholar]
- 25.Vaclavik L, Krynitsky AJ, Rader JI. Quantification of aristolochic acids I and II in herbal dietary supplements by ultra-high-performance liquid chromatography-multistage fragmentation mass spectrometry. Food additives & contaminants Part A, Chemistry, analysis, control, exposure & risk assessment. 2014;31:784–791. doi: 10.1080/19440049.2014.892215. [DOI] [PubMed] [Google Scholar]
- 26.IARC. Monographs on the Evaluation of Carcinogenic Risks to Humans. Some Traditional Herbal Medicines, Some Mycotoxins, Naphthalene and Styrene. Vol. 82. IARCPress; Lyon, France: 2002. [PMC free article] [PubMed] [Google Scholar]
- 27.Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, Jakovina K, Brdar B, Slade N, Turesky RJ, Goodenough AK, Rieger R, Vukelic M, Jelakovic B. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci U S A. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CH, Dickman KG, Moriya M, Zavadil J, Sidorenko VS, Edwards KL, Gnatenko DV, Wu L, Turesky RJ, Wu XR, Pu YS, Grollman AP. Aristolochic acid-associated urothelial cancer in Taiwan. Proc Natl Acad Sci U S A. 2012;109:8241–8246. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yun BH, Rosenquist TA, Sidorenko V, Iden CR, Chen CH, Pu YS, Bonala R, Johnson F, Dickman KG, Grollman AP, Turesky RJ. Biomonitoring of aristolactam-DNA adducts in human tissues using ultra-performance liquid chromatography/ion-trap mass spectrometry. Chem Res Toxicol. 2012;25:1119–1131. doi: 10.1021/tx3000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.IARC. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Tobacco smoking. Vol. 38. IARC Press; Lyon, France: 1986. [Google Scholar]
- 31.IARC. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Smokeless Tobacco and Some Tobacco-specific N-Nitrosamines. Vol. 89. IARC Press; Lyon, France: 2007. [PMC free article] [PubMed] [Google Scholar]
- 32.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouvard V, Loomis D, Guyton KZ, Grosse Y, Ghissassi FE, Benbrahim-Tallaa L, Guha N, Mattock H, Straif K International Agency for Research on Cancer Monograph Working G. Carcinogenicity of consumption of red and processed meat. Lancet Oncol. 2015;16:1599–1600. doi: 10.1016/S1470-2045(15)00444-1. [DOI] [PubMed] [Google Scholar]
- 34.Phillips DH. Polycyclic aromatic hydrocarbons in the diet. Mutat Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 35.Felton JS, Jagerstad IM, knize MG, Skog K, Wakabayashi K. In Food Borne Carcinogens; Heterocylclic Amines. John Wiley & Sons Ltd; Chichester, England: 2000. [Google Scholar]
- 36.Jones CR, Sabbioni G. Identification of DNA adducts using HPLC/MS/MS following in vitro and in vivo experiments with arylamines and nitroarenes. Chem Res Toxicol. 2003;16:1251–1263. doi: 10.1021/tx020064i. [DOI] [PubMed] [Google Scholar]
- 37.Bessette EE, Goodenough AK, Langouet S, Yasa I, Kozekov ID, Spivack SD, Turesky RJ. Screening for DNA adducts by data-dependent constant neutral loss-triple stage mass spectrometry with a linear quadrupole ion trap mass spectrometer. Anal Chem. 2009;81:809–819. doi: 10.1021/ac802096p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo J, Yun BH, Upadhyaya P, Yao L, Krishnamachari S, Rosenquist TA, Grollman AP, Turesky RJ. Multiclass Carcinogenic DNA Adduct Quantification in Formalin-Fixed Paraffin-Embedded Tissues by Ultraperformance Liquid Chromatography-Tandem Mass Spectrometry. Anal Chem. 2016;88:4780–4787. doi: 10.1021/acs.analchem.6b00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao S, Guo J, Yun BH, Villalta PW, Krishna S, Tejpaul R, Murugan P, Weight CJ, Turesky RJ. Biomonitoring DNA Adducts of Cooked Meat Carcinogens in Human Prostate by Nano Liquid Chromatography-High Resolution Tandem Mass Spectrometry: Identification of 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine DNA Adduct. Anal Chem. 2016;88:12508–12515. doi: 10.1021/acs.analchem.6b04157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu D, Turesky RJ, Tao Y, Langouet SA, Nauwelaers GC, Yuan JM, Yee D, Yu MC. DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 4-aminobiphenyl are infrequently detected in human mammary tissue by liquid chromatography/tandem mass spectrometry. Carcinogenesis. 2012;33:124–130. doi: 10.1093/carcin/bgr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilfinger WW, Mackey K, Chomczynski P. Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques. 1997;22:474–476. 478–481. doi: 10.2144/97223st01. [DOI] [PubMed] [Google Scholar]
- 42.Kephart D, Krueger S, Grunst T, Shenoi H. Introducing the Maxwell® 16 Instrument: A Simple, Robust and Flexible Tool for DNA Purification. Promega Notes. 2006;92:20–23. [Google Scholar]
- 43.Schagat T, Koller S, Leone P, Cremonesi P, Bosetti A, Wieczorek D, Kephart D, Mann R, Storts D. The Versatility of the Maxwell® 16 System for Genomic DNA Extraction. Promega Notes. 2007;97:12–14. [Google Scholar]
- 44.Dubeau L, Chandler LA, Gralow JR, Nichols PW, Jones PA. Southern blot analysis of DNA extracted from formalin-fixed pathology specimens. Cancer Res. 1986;46:2964–2969. [PubMed] [Google Scholar]
- 45.Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, Van Marck E, Worobey M. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PloS one. 2007;2:e537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turashvili G, Yang W, McKinney S, Kalloger S, Gale N, Ng Y, Chow K, Bell L, Lorette J, Carrier M, Luk M, Aparicio S, Huntsman D, Yip S. Nucleic acid quantity and quality from paraffin blocks: defining optimal fixation, processing and DNA/RNA extraction techniques. Exp Mol Pathol. 2012;92:33–43. doi: 10.1016/j.yexmp.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 47.Bonin S, Hlubek F, Benhattar J, Denkert C, Dietel M, Fernandez PL, Hofler G, Kothmaier H, Kruslin B, Mazzanti CM, Perren A, Popper H, Scarpa A, Soares P, Stanta G, Groenen PJ. Multicentre validation study of nucleic acids extraction from FFPE tissues. Virchows Arch. 2010;457:309–317. doi: 10.1007/s00428-010-0917-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodenough AK, Schut HA, Turesky RJ. Novel LC-ESI/MS/MS(n) method for the characterization and quantification of 2′-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem Res Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bessette EE, Spivack SD, Goodenough AK, Wang T, Pinto S, Kadlubar FF, Turesky RJ. Identification of carcinogen DNA adducts in human saliva by linear quadrupole ion trap/multistage tandem mass spectrometry. Chem Res Toxicol. 2010;23:1234–1244. doi: 10.1021/tx100098f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friesen MD, Kaderlik K, Lin D, Garren L, Bartsch H, Lang NP, Kadlubar FF. Analysis of DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5- b ]pyridine in rat and human tissues by alkaline hydrolysis and gas chromatography/electron capture mass spectrometry: validation by comparison with 32 P-postlabeling. Chem Res Toxicol. 1994;7:733–739. doi: 10.1021/tx00042a004. [DOI] [PubMed] [Google Scholar]
- 51.Singh R, Gaskell M, Le Pla RC, Kaur B, Azim-Araghi A, Roach J, Koukouves G, Souliotis VL, Kyrtopoulos SA, Farmer PB. Detection and quantitation of benzo[a]pyrene-derived DNA adducts in mouse liver by liquid chromatography-tandem mass spectrometry: comparison with 32P-postlabeling. Chem Res Toxicol. 2006;19:868–878. doi: 10.1021/tx060011r. [DOI] [PubMed] [Google Scholar]
- 52.Peterson LA, Hecht SS. A Study of Chemical Carcinogenesis. 141 O6-Methylguanine Is a Critical Determinant of 4-(Methylnitrosamino)-1-(3-Pyridyl)-1-Butanone Tumorigenesis in a/J Mouse Lung. Cancer Res. 1991;51:5557–5564. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.