Abstract
Obesity is associated with development of diverse diseases, including cardiovascular diseases and dyslipidemia. MiRNA-22 (miR-22) is a critical regulator of cardiac function and targets genes involved in metabolic processes. Previously, we generated miR-22 null mice and we showed that loss of miR-22 blunted cardiac hypertrophy induced by mechanohormornal stress. In the present study, we examined the role of miR-22 in the cardiac and metabolic alterations promoted by high-fat (HF) diet. We found that loss of miR-22 attenuated the gain of fat mass and prevented dyslipidemia induced by HF diet, although the body weight gain, or glucose intolerance and insulin resistance did not seem to be affected. Mechanistically, loss of miR-22 attenuated the increased expression of genes involved in lipogenesis and inflammation mediated by HF diet. Similarly, we found that miR-22 mediates metabolic alterations and inflammation induced by obesity in the liver. However, loss of miR-22 did not appear to alter HF diet induced cardiac hypertrophy or fibrosis in the heart. Our study therefore establishes miR-22 as an important regulator of dyslipidemia and suggests it may serve as a potential candidate in the treatment of dyslipidemia associated with obesity.
Introduction
The incidence of obesity has increased intensely over the past decades and has contributed to an increase in the risk of numerous chronic disorders, including cardiovascular disease. High-fat (HF) diet-induced obesity is directly associated with development of dyslipidemia, Type 2 diabetes, and other metabolic complications associated with obesity [1,2]. Recent studies have documented adipose tissue inflammation as a critical component in glucose intolerance and insulin resistance evidenced in obesity [3,4], suggesting that reduction in adipose tissue inflammation probably leads to improvements in metabolic disorders associated with obesity.
MicroRNAs (miRNAs) are endogenous, short noncoding RNAs that post-transcriptionally regulate gene expression by inhibiting their translation and/or degrading their target mRNAs. Studies have revealed the important role of miRNAs in control of cardiac morphology and myocardial function [5,6]. Additionally, various reports have documented the critical role of miRNAs in the development of cardiovascular disorders [7–9] as well as in metabolic disorders [10,11].
Studies developed in past years revealed that miR-22 is a key regulator of cardiac remodeling and myocardial function [12–14]. Although inhibition of miR-22 have attenuated cardiomyocyte hypertrophy in vitro [12], studies in vivo showed that loss of function of miR-22 resulted in severe dilated cardiomyopathy and heart failure in response to pressure overload [13,14], suggesting miR-22 as a key regulator of cardiac remodeling in response to stress. Multiple mRNAs have been identified as targets of miR-22, including phosphatase and tensin homolog (Pten), purine rich element binding protein B (Purb), caveolin 3 (Cav3), and histone deacetylase 4 (Hdac4). In addition, miR-22 directly targets peroxisome proliferative activated receptor, γ, coactivator 1α (Pgc-1α), peroxisome proliferator-activated receptor α (Pparα), and Sirtuin 1 (Sirt1) [12–15], which are important factors involved in fatty acid metabolism, mitochondrial biogenesis, and cardiac hypertrophy [16,17].
We have previously generated miR-22 mutant mice and showed that miR-22 plays a central role in the regulation of cardiac remodeling in response to pathological stress [14]. Most recently, we have reported that cardiac hypertrophy induced by HF feeding is accompanied by altered miRNA expression pattern and that miR-22 is among the most increased miRNAs in response to HF diet [18]. However, the potential role of miR-22 in the obesity-induced cardiac hypertrophy and remodeling remains unknown. Additionally, given that miR-22 directly targets genes, such as Pgc-1α, Pparα, and Sirt1 that are involved in the control of metabolism and obesity [19], it would be extremely interesting to test whether miR-22 may contribute to metabolic alterations induced by obesity. Most recently, a report showed that miR-22 is required for normal body weight gain in male mice [20], raising the possibility that miR-22 may play a role in metabolic and cardiovascular disorders associated with obesity. In the present study, we investigated the function of miR-22 in the development of cardiac hypertrophy and metabolic disorders induced by HF diet. Surprisingly, we found that genetic deletion of miR-22 has little impact on cardiac hypertrophy and fibrosis in response to HF diet. Nevertheless, loss of miR-22 decreased the gain of fat mass and prevented dyslipidemia induced by HF feeding. Likewise, we found that loss of miR-22 attenuated the increase in expression of genes involved in inflammation and lipogenesis mediated by HF diet in the liver and adipose tissue.
Research design and methods
Animal experimentation
All animal experiments were approved by the IACUC of the Boston Children’s Hospital. The systemic (global) miR-22 knockout (miR22-KO) and miR-22 wild-type (miR22-WT) mice used in the present study were characterized previously [14] and were generated from heterozygous intercrosses. Mice were genotyped at 2 weeks of age using PCR. Five-week-old male miR22-KO and miR22-WT mice were treated with normal chow diet, with 10% kcal fat, and high-fat diet, with 60% kcal fat for 12 weeks. The diets were purchased from Teklad (Envigo, Madison, WI). Mice were housed at 22± 1°C in a 12-h light–dark cycle and had ad libitum access to water and food. Body weight gain was monitored weekly. After 12 weeks of treatment, mice were killed using carbon dioxide chamber and tissue samples were collected.
Intraperitoneal glucose tolerance test (iGTT)
Intraperitoneal glucose tolerance test (iGTT) was performed after 11 weeks of treatment with diets. Mice were fasted for 10 h and were given an intraperitoneal injection of glucose (2 g/kg body weight). Glucose levels were measured by a glucometer, following the protocol described previously [18].
Determination of glycemia, insulin, cholesterol, and triglycerides levels
Fasting glycemia was determined before the killing using the glucometer. Insulin serum levels were determined using an Elisa kit (EZRMI-13K, EMD Millipore Corporation). Total cholesterol and triglycerides serum levels were determined using commercial kits (439-17501, 461-08992; Wako Diagnostics, CA, U.S.A.).
Echocardiographic measurements
Cardiac morphology and function were determined by echocardiographic measurements, following the protocol described previously [14]. Echocardiography was performed using a Visual Sonics Vevo 2100 Imaging System (Visual Sonics) with a 40-MHz MicroScan transducer (model MS-550D). Left ventricle (LV) dimensions, such as systolic and diastolic wall thicknesses, LV end-diastolic and end-systolic chamber dimensions, and heart rate were determined using 2D short-axis views under M-mode at the level of papillary muscle. Functional parameters, including ejection fraction (EF) and percentage of fractional shortening (FS) were calculated.
Metabolic chambers
Metabolic phenotyping of miR22-WT and miR22-KO was performed using metabolic cages after 11 weeks of treatment with control and HF diets (Comprehensive Lab Animal Monitoring System; Columbus Instruments). Briefly, both miR22-WT and miR22-KO mice were adapted to the cages for 3 days, followed by the analysis of O2 consumption (VO2), CO2 production (VCO2), respiratory exchange ratio (RER; VCO2/VO2), and locomotor activity (through infrared beam sensors) during four consecutive days.
Biometric measurements and histological analyses
After killing, liver, heart, and epididymal fat were removed and weighted. Heart weight (HW) was normalized to tibia length (HW/TL) ratio. Some hearts were fixed in 4% paraformaldehyde overnight. After dehydration, samples were embedded in paraffin and serial sections of 5 μm were stained with hematoxylin and eosin for analysis of heart or Sirius Red/Fast Green for collagen quantification. Some transverse heart sections were stained with wheat germ agglutinin for quantification of cardiomyocyte area.
Quantitative real time-PCR (qPCR)
Total RNA from heart, epididymal fat, and liver was isolated using Trizol Reagent (Invitrogen). For quantification of the expression of protein-coding gene, 1 μg of total RNA was reverse-transcribed to cDNA using MMLV Reverse Transcriptase (Invitrogen), according to the manufacturer’s instruction. PCR was performed with cDNA and specific primers for the mRNA of interest using Sybr Select Master Mix (Applied Biosystems). For quantification of the expression of miRNAs, 10 ng of total RNA was reverse-transcribed to cDNA using the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems). PCR analyses were performed using specific TaqMan® Assays (Applied Biosystems) and TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (Gapdh), Actin β (Actb), and ribosomal protein L19 (Rpl19) were used to normalize mRNA expression of protein-coding genes in heart, liver, and adipose tissue respectively, since their expression were not affected by HF diet and loss of miR-22. U6 shRNA was used to normalize miRNA expression in all tissues evaluated.
Luciferase reporter assays
The wild-type 3′UTR fragments of mouse Kruppel-like factor 6 (Klf6), Suppressor of cytokine signaling 2 (Socs2), and Interleukin 13 receptor α 1 (Il13rα1) genes containing predicted miR-22 binding sites were cloned into a luciferase reporter plasmid (pGL3-Control vector, Promega), resulting in the constructs Socs2, Klf6, and Il13rα1. HEK293T cells cultured in DMEM supplemented with 10% FBS were used in the luciferase reporter assays. Cells were transfected with 50 ng of reporter plasmid, 50 ng of LacZ plasmid, and 10 nM of miR-22 mimic (Dharmacon) or duplex control (Dharmacon) using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instruction. After 24 h, cells were harvested and luciferase activity was determined. LacZ was used to normalize the luciferase activity in the cells.
Primary cardiomyocyte cultures
Neonatal rat cardiomyocyte cultures were prepared as previously described [14]. Briefly, cardiomyocytes were isolated by enzymatic dissociation of one day old neonatal rat hearts using the Neonatal Cardiomyocyte Isolation kit (Cellutron, Life Technologies). Dissociated cells were plated for 2 h to remove fibroblasts. The unattached cells were transferred to precoated plates containing medium supplemented with serum. Twenty-four hours after plating, cells were submitted to serum-free medium overnight. Cardiomyocyte cultures were transfected with 50 nM of miR-22 mimic duplex, miR-22 hairpin inhibitor, and control oligonucleotide (Life Technologies) using Lipofectamine RNAiMAX transfection reagent. Six hours later, cells were treated with 50 ng/ml of leptin (Sigma), which is a concentration sufficient to induce cardiomyocyte hypertrophy. Cells were harvested 24 h after leptin treatment for RNA isolation or 48 h later for immunochemistry analyses.
Statistical analyses
Data are expressed as mean ± standard deviations (SD). Statistical significance was calculated using one-way ANOVA or two-way ANOVA using Prism GraphPad® software. Values of P<0.05 were considered statistically significant.
Results
Loss of miR-22 attenuates fat mass gain induced by HF diet and enhances energy expenditure
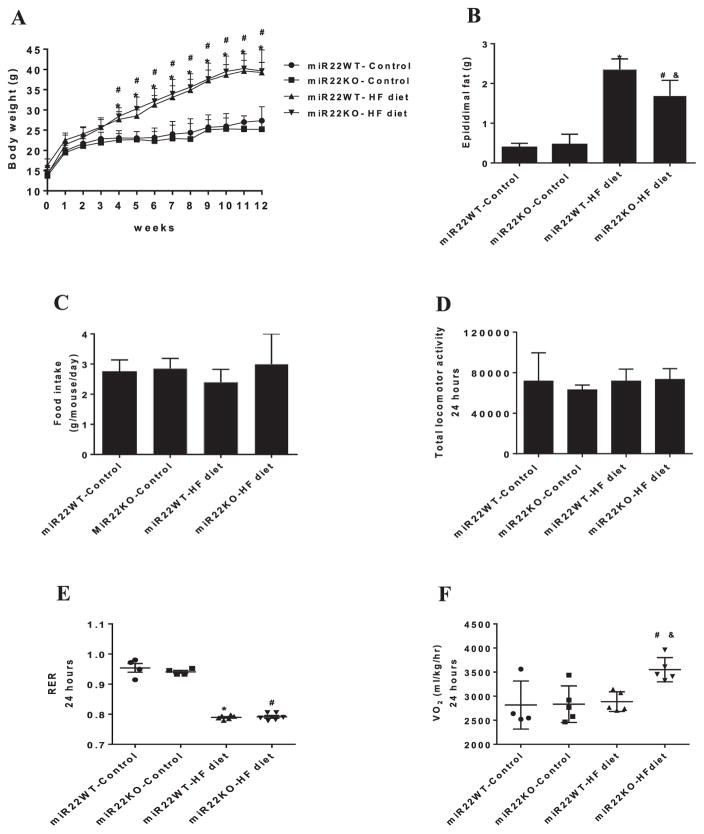
To evaluate whether loss of miR-22 could affect body weight gain in response to high-fat (HF) diet, miR22-KO and control miR22-WT mice were fed with either normal (control) or HF diet. As expected, miR22-WT mice fed HF diet gained more body weight compared with those fed control diet (Figure 1A). However, loss of miR-22 did not affect the body weight gain induced by HF diet. Supplementary Table S1 documented the morphometric data of miR22-KO and miR22-WT mice fed control or HF diet.
Figure 1. Loss of miR-22 reduces fat mass gain induced by HF diet and enhanced energy expenditure.
(A) Analysis of body weight gain in miR22-WT and miR22-KO mice fed control diet and HF diet for 12 weeks (n = 8–9). (B) Epididymal fat weight in the miR22-WT and miR22-KO mice fed control diet and HF diet (n = 7–9). (C) Food intake; (D) Total locomotor activity; (E) Respiratory exchange ratio (RER); (F) O2 consumption (VO2). * vs miR22WT-control (P<0.05); # vs miR22KO-control (P<0.05); & vs miR22WT-HF diet (n = 4–5).
Next, we determined whether loss of miR-22 could affect glucose intolerance and insulin resistance induced by HF diet. We first evaluated glucose homeostasis in miR22-WT and miR22-KO mice, before HF feeding. The miR22-WT and miR22-KO mice showed similar glucose levels after glucose challenge at 5 weeks old (Supplementary Figure S1A). Following 12 weeks of HF diet, miR22-WT and miR22-KO mice displayed higher blood glucose levels during the iGTT, compared with those fed control diet (Supplementary Figure S1B). The area under the curve was increased in miR22-WT and miR22-KO mice fed HF diet, indicating glucose intolerance (Supplementary Figure S1C). Indeed, both miR22-WT and miR22-KO mice fed HF diet exhibited higher fasting blood glucose (Supplementary Figure S1D) and insulin levels (Supplementary Figure S1E), compared with their respective controls. Similarly, HOMA-IR was increased in both miR22-WT and miR22-KO mice fed HF diet (Supplementary Figure S1F), suggesting that loss of miR-22 did not change insulin resistance induced by HF feeding. Together, these results suggest that loss of miR-22 does not change the development of obesity, glucose intolerance, and insulin resistance promoted by HF feeding.
Next, we tested whether miR-22 could regulate the fat mass gain induced by HF diet. As expected, mice fed HF diet displayed increased epididymal fat mass when compared with control diet-fed mice (Figure 1B). Interestingly, loss of miR-22 attenuated the increase in epididymal fat mass induced by HF diet (Figure 1B). This finding suggests that miR-22 may contribute to increased epididymal fat mass induced by HF diet.
Given that loss of miR-22 affected fat mass gain induced by HF diet, we asked whether miR-22 might regulate metabolic phenotype in response to HF diet. After 12 weeks of normal or HF feeding, miR22-KO and miR22-WT mice were evaluated in metabolic chambers. miR22-KO mice showed no difference in food intake when compared with miR22-WT mice (Figure 1C). We found that total locomotor activity and respiratory exchange ratio (RER) were unchanged in miR22-KO mice when compared with miR22-WT controls, either fed with normal or HF diet (Figure 1D,E). Next, we tested whether loss of miR-22 could affect energy expenditure. Interestingly, O2 consumption (VO2) was increased in miR22-KO mice fed with HF diet, when compared with miR22-WT mice fed with HF diet (Figure 1F). These results suggest that loss of miR-22 enhanced energy expenditure when the animals were under HF diet condition.
Loss of miR-22 attenuates the inflammation of adipose tissue induced by HF diet
The above results, in which miR-22 modulated the fat mass gain induced by HF feeding and increased the energy expenditure, prompt us to test whether HF diet could affect the expression levels of miR-22 in epididymal fat. qPCR analysis indicated that miR-22 expression was increased in epididymal fat of mice fed HF diet compared with those fed control diet (Figure 2A).
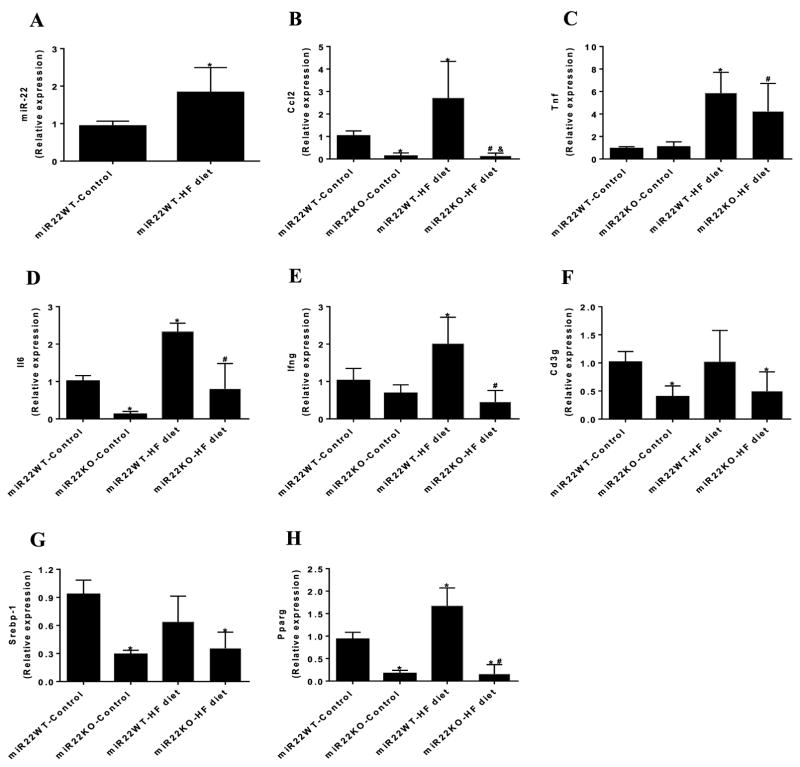
Figure 2. Loss of miR-22 is involved in adipose tissue inflammation induced by HF diet.
(A) miR-22 expression in the epididymal fat of miR22-WT mice fed control diet and HF diet evaluated by qPCR (n=4). Analysis of gene expression of (B) Ccl2, (C) Tnf, (D) Il6, (E) Ifng, (F) Cd3g, (G) Srebp-1, and (H) Pparg in the adipose tissue of miR22-WT and miR22-KO mice fed control diet and HF diet, evaluated by qPCR (n = 4–6). * vs miR22WT-control (P<0.05); # vs miR22KO-control (P<0.05); & vs miR22WT-HF diet (P<0.05).
It is well known that obesity is accompanied by the release of proinflammatory cytokines and recruitment of proinflammatory immune cells in the adipose tissue [4]. Next, we asked whether miR-22 could affect the expression of proinflammatory genes in the adipose tissue in response to HF feeding. HF diet increased gene expression of C–C motif chemokine ligand 2 (Ccl2), which is a macrophage marker (Figure 2B) in the adipose tissue of miR22-WT mice. Similarly, miR22-WT mice fed HF diet displayed elevated expression levels of proinflammatory cytokines, including tumor necrosis factor (Tnf), interleukin-6 (Il6), and interferon γ (Ifng) in the adipose tissue (Figure 2C–E). Interestingly, loss of miR-22 prevented the increase in gene expression of Ccl2, Il6, and Ifng induced by HF feeding (Figure 2B,D,E). Additionally, loss of miR-22 reduced the recruitment of T cells in the adipose tissue, as evidenced by reduced gene expression of CD3 antigen γ polypeptide (Cd3g) in miR22-KO mice (Figure 2F). These data suggest that loss of function of miR-22 attenuates adipose tissue inflammation induced by HF diet.
Attenuated increase in adipogenesis gene expression in miR-22 null mice in response to HF diet
We also evaluated the effect of miR-22 in the expression levels of factors involved in adipogenesis. Sterol regulatory element binding protein-1 (Srebp-1) is an important regulator of adipogenesis and controls fatty acid synthesis in the adipose tissue and liver [21,22]. Although HF diet did not affect gene expression of Srebp-1, loss of miR-22 was sufficient to reduce expression levels of Srebp-1 in the adipose tissue (Figure 2G). Peroxisome proliferator activated receptor γ (Pparg), which is a key regulator of adipocyte differentiation [23,24], was increased in the adipose tissue by HF diet. Interestingly, loss of miR-22 reduced the expression levels of Pparg in the adipose tissue and prevented HF diet induced increase (Figure 2H). These results suggest that loss of miR-22 attenuated the increased expression of genes involved in adipogenesis, which is induced by HF feeding.
Loss of miR-22 prevents dyslipidemia induced by HF diet
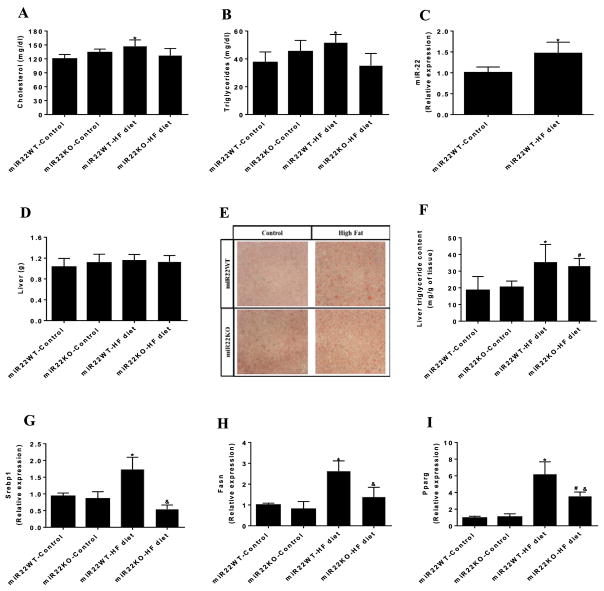
Next, we investigated whether miR-22 could mediate dyslipidemia induced by HF diet. Consistent with previous reports [18,25,26], cholesterol (Figure 3A) and triglycerides (Figure 3B) serum levels were increased in miR22-WT mice fed HF diet. However, loss of miR-22 prevented the increase in cholesterol and triglycerides serum levels in miR22-KO mice in response to HF feeding, suggesting that miR-22 contributes to dyslipidemia induced by HF diet.
Figure 3. Loss of miR-22 prevents dyslipidemia induced by HF diet.
(A) Serum levels of cholesterol and (B) triglycerides in miR22-WT and miR22-KO mice fed control diet and HF diet (n = 6–9). (C) miR-22 expression in the liver of miR22-WT mice fed control diet and HF diet evaluated by qPCR (n=4). (D) Liver weight in miR22-WT and miR22-KO mice fed control diet and HF diet (n = 8–9). (E) Analysis of fat content in the liver of miR22-WT and miR22-KO mice fed control diet and HF diet using Oil Red O staining (n=3). (F) Measurement of triglyceride content in the liver of miR22-WT and miR22-KO mice fed control diet and HF diet (n=4). Analysis of gene expression of (G) Srebp-1, (H) Fasn, and (I) Pparg in the liver of miR22-WT and miR22-KO mice fed control diet and HF diet, evaluated by qPCR (n = 4–6). * vs miR22WT-control (P<0.05); # vs miR22KO-control (P<0.05); & vs miR22WT-HF diet (P<0.05).
Loss of miR-22 attenuates the increase in expression of genes involved in fatty acid biosynthesis in the liver
Given that loss of miR-22 prevented dyslipidemia induced by HF diet, we next examined whether miR-22 could affect fatty acid biosynthesis in the liver. qPCR assays showed that the expression of miR-22 was increased in the liver of mice fed HF diet (Figure 3C). However, HF feeding did not change the liver mass both in miR22-WT and miR22-KO mice (Figure 3D). Moreover, analysis of fat content using Oil Red O staining revealed that HF diet increased the lipid droplets in the liver of miR22-WT mice (Figure 3E). Similarly, triglyceride content was increased in the liver of miR22-WT mice fed HF diet compared with those fed control diet (Figure 3F). Loss of miR-22 did not seem to affect the increase in lipid droplets and triglyceride content in the liver induced by HF feeding (Figure 3E,F). We examined the expression of genes involved in fatty acids synthesis in the liver. HF diet increased the expression levels of Srebp-1 in the liver; however, loss of miR-22 blocked the increase in the expression of Srebp-1 promoted by HF feeding (Figure 3G). We also verified the expression levels of other lipogenic genes, including fatty acid synthase (Fasn) and Pparg in the liver. miR22-WT mice fed HF diet showed elevated gene expression of Fasn and Pparg, which was attenuated by loss of miR-22 (Figure 3H,I). These data suggest that loss of miR-22 attenuates the increase in lipogenic gene expression induced by HF diet in the liver.
HF diet increased mitochondrial content in the liver, as evaluated by the ratio of mitochondrial DNA to genomic DNA, consistent with previous reports [27,28]. Interestingly, loss of miR-22 avoided the increase in mitochondrial content induced by HF diet in the liver, further suggesting that miR-22 is required for mitochondria biogenesis in response to HF feeding (Supplementary Figure S2).
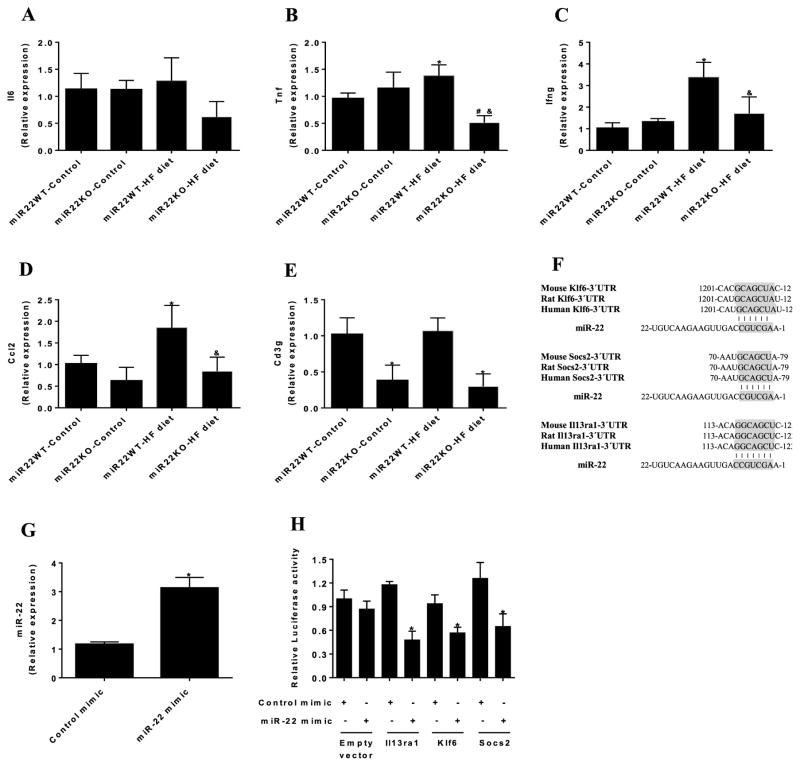
Loss of miR-22 prevents the overexpression of HF diet induced proinflammatory genes in the liver
Next, we examined whether miR-22 could affect the expression of proinflammatory genes in the liver. Analysis of qPCR showed that gene expression of Il6 in the liver was similar between the groups (Figure 4A). On the other hand, HF diet induced the expression levels of Tnfα and Ifng in the liver of miR22-WT mice. However, loss of miR-22 prevented the increase in gene expression of Tnfα and Ifng in the liver in response to HF diet (Figure 4B,C). Additionally, HF diet increased Ccl2 gene expression in the liver, which was prevented by loss of miR-22 (Figure 4D). Moreover, miR22-KO mice displayed reduced gene expression of Cd3g in the liver under both normal and HF diet feeding (Figure 4E). These data suggest that loss of miR-22 prevents the overexpression of proinflammatory genes in the liver induced by HF feeding.
Figure 4. Loss of miR-22 prevents the increased expression of proinflammatory genes in the liver induced by HF diet.
qPCR analysis of gene expression of (A) Il6, (B) Tnf, (C) Ifng, (D) Ccl2, and (E) Cd3g in the liver of miR22-WT and miR22-KO mice fed control diet and HF diet (n = 4–6). * vs miR22WT-control (P<0.05); # vs miR22KO-control (P<0.05); & vs miR22WT-HF diet (P<0.05). (F) Alignment between miR-22 and the 3′ untranslated regions (UTRs) of Klf6, Socs2, and Il13rα1 of mouse, rat, and human. Seed sequences are highlighted in gray. (G) HEK293T cells were transfected with miR-22 mimic or control mimic and miR-22 expression levels were determined by qPCR after 24 h (n=3). * vs Control mimic (P<0.05). (H) Luciferase reporters with wild type Il13rα1–3′UTR (Il13ra1), wild-type Klf6–3′UTR (Klf6) and wild-type Socs2–3′UTR (Socs2) were cotransfected with miR-22 mimic or control mimic in HEK293T cells, and luciferase activity was determined after 24 h. Values are presented as relative luciferase activity ± SD relative to the luciferase activity of reporters cotransfected with control mimic (n=4). * vs control mimic (P<0.05).
To determine the potential mechanisms involved in the metabolic effects induced by loss of miR-22, we performed in silico analysis to identify miR-22 targets. Among the predicted targets for miR-22, we found Socs2, Il13rα1, and Klf6, which are conserved cross the species (Figure 4F). These genes were selected for validation since previous reports have demonstrated their involvement in inflammation and obesity. We built luciferase reporters containing the 3′ untranslated regions (3′UTRs) of Socs2, Il13rα1 and Klf6, and tested their repression by miR-22. We verified miR-22 overexpression in cells transfected with miR-22 mimic (Figure 4G). As expected, overexpression of miR-22 decreased the luciferase activity in cells transfected with luciferase reporters containing the 3′UTR sequences of Socs2, Klf6, and Il13rα1 compared with cells transfected with control mimic (Figure 4H), confirming that they are direct targets of miR-22.
Loss of miR-22 does not affect cardiac hypertrophy or fibrosis in response to HF feeding
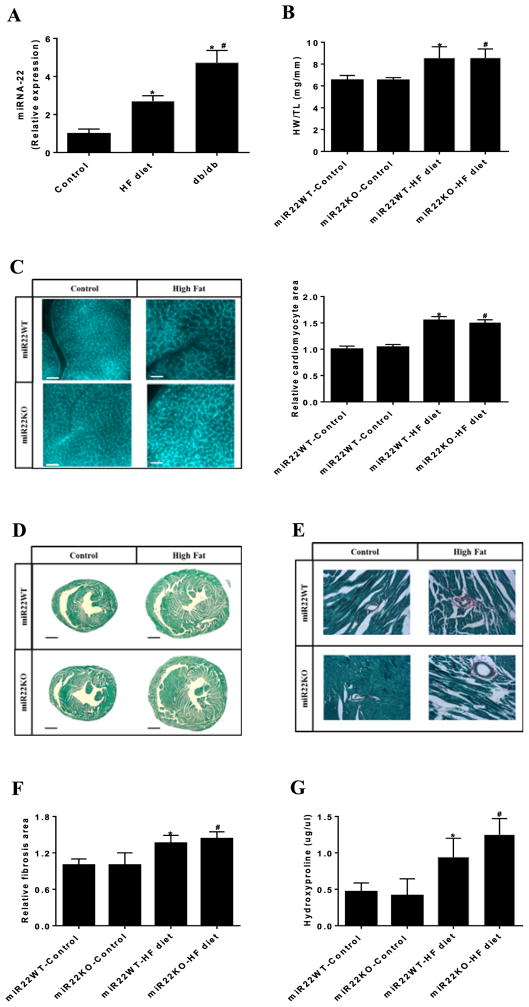
Given that miR-22 is a key factor in the control of cardiac remodeling and myocardial function in response to stress [13–15], we next evaluated whether miR-22 expression might be affected in HF diet induced hypertrophic hearts. qPCR analysis showed that the expression level of miR-22 is increased in cardiac hypertrophy induced by HF diet in C57/Bl6 mice (Figure 5A), consistent with the RNA sequencing data published recently [18]. Similarly, we found elevated miR-22 expression in the hearts of db/db mice, which is a genetic model for obesity, when compared with C57/Bl6 mice fed control diet (Figure 5A).
Figure 5. No change in cardiac hypertrophy in miR22-KO mice in response to HF feeding.
(A) miR-22 expression in the heart of C57Bl/6 mice fed control diet and HF diet and db/db mice evaluated by qPCR (n = 4–5). (B) Analysis of cardiac hypertrophy in miR22-WT and miR22-KO mice fed control diet and HF diet, evaluated by heart weight normalized by tibia length (HW/TL). (C) Wheat germ agglutinin (WGA) staining of heart transverse sections. Quanti3cation of relative cardiomyocyte area is displayed at the right panel; bars, 40 μm (n=3). (D) Fibrosis area of heart transverse sections evaluated by Sirius Red/Fast Green collagen staining; bars, 1 mm (n = 3–4). (E) Higher magni3cations of 3brosis area of heart transverse sections evaluated by Sirius Red/Fast Green collagen staining. (F) Quanti3cation of relative 3brosis area of heart transverse sections. (G) Measurement of hydroxyproline content in the heart (n=3). * vs miR22WT-control (P<0.05); # vs miR22KO-control (P<0.05); vs miR22WT-HF diet (P<0.05).
Next, we tested whether loss of miR-22 could affect cardiac hypertrophy and remodeling induced by HF diet. The miR22-WT mice fed a HF diet displayed elevated expression of miR-22 in the heart compared with those fed a control diet (Supplementary Figure S3). In addition, HF diet induced cardiac hypertrophy both in miR22-WT and miR22-KO mice, as evaluated by an increased HW/TL (heart weight/tibia length) ratio compared with their respective controls fed control diet (Figure 5B). Hematoxylin and eosin staining of heart transverse sections showed that HF diet-fed resulted in increased left ventricle (LV) thickness compared with those fed a control diet (Supplementary Figure S4). Similarly, wheat germ agglutinin (WGA) staining showed that cardiomyocyte area was increased in miR22-WT mice in response to HF feeding (Figure 5C). Unexpectedly, loss of miR-22 did not affect cardiomyocyte hypertrophy induced by HF diet. Likewise, analysis of gene expression of cardiac hypertrophy markers by qPCR indicated that miR22-WT mice fed HF diet displayed higher gene expression of Anp, Bnp, and Myh7 (Supplementary Figure S5A). Once again, loss of miR-22 did not appear to affect the increase in gene expression of the cardiac hypertrophy markers in response to HF diet.
We then investigated the potential involvement of miR-22 in the development of cardiac fibrosis induced by HF diet. Histological analysis using Sirius Red/Fast Green staining revealed that both miR22-WT and miR22-KO mice fed HF diet developed cardiac fibrosis compared with their respective controls fed control diet (Figure 5D–F). Consistent with this observation, hydroxyproline content was higher in the heart of HF-diet fed mice compared with those fed control diet (Figure 5G). Likewise, qPCR analysis showed that gene expression of Col1a, Col3a, and Fn1 was increased in heart of miR22-WT mice fed HF diet (Supplementary Figure S5B). However, loss of miR-22 did not affect the increased gene expression of fibrosis markers induced by HF diet.
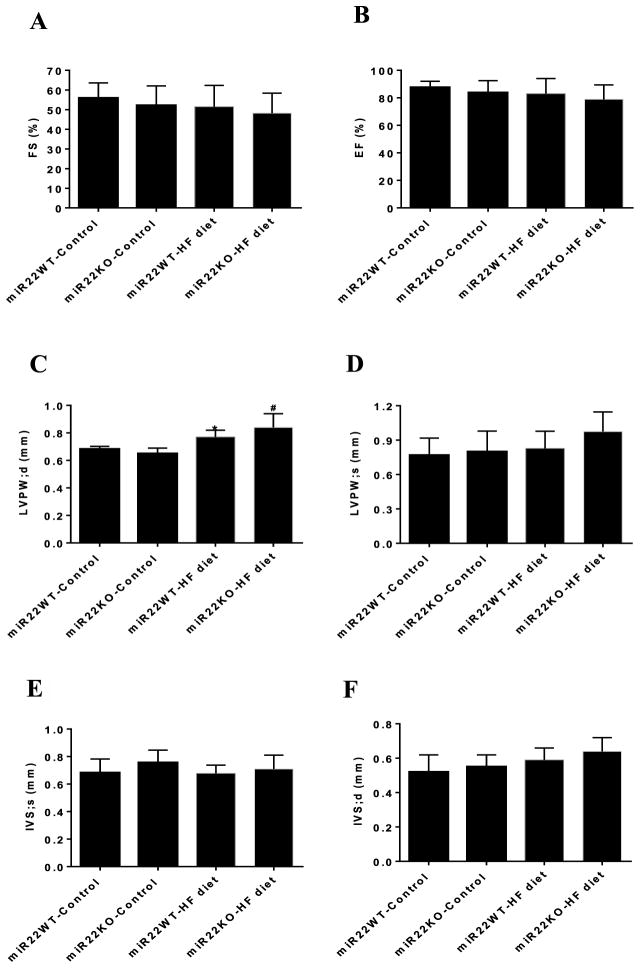
In order to determine the effect of miR-22 on cardiac function under HF diet, we performed echocardiographic analyses. Fractional shortening (FS%) (Figure 6A) and ejection fraction (EF%) (Figure 6B) were similar between groups, suggesting that left ventricle (LV) contractile function was unchanged by either HF diet or loss of miR-22 (Supplementary Table S2). Moreover, we found that HF feeding increased LV wall thickness, as evaluated by higher diastolic LV posterior wall (Figure 6C). However, the hypertrophic effect of the HF diet did not differ between miR22-WT and miR22-KO mice, suggesting that miR-22 does not directly contribute to HF diet induced cardiac hypertrophy. In addition, systolic LV posterior wall (Figure 6D) and interventricular septal thickness (Figure 6E,F) were unchanged by either HF diet or loss of miR-22.
Figure 6. Loss of miR-22 did not affect cardiac function in response to HF diet.
Echocardiographic analyses in the miR22-WT and miR22-KO mice fed control diet and HF diet. (A) Fractional shortening (FS), (B) ejection fraction (EF), (C) LV end-diastolic posterior wall thickness (LVPW; d), (D) LV end-systolic posterior wall thickness (LVPW; s), (E) end-systolic interventricular septum (IVS; s), (F) end-diastolic interventricular septum (IVS; d) (n = 5–7). * vs miR22WT-control (P<0.05); # vs miR22KO-control (P<0.05).
Finally, we tested whether miR-22 might affect cardiomyocyte hypertrophy in response to leptin in vitro. Leptin increased the expression levels of Myh7, Anp, and Acta as well as cardiomyocyte size (Supplementary Figure S6). Nonetheless, gain and loss of miR-22 function studies showed that miR-22 did not influence leptin-induced cardiomyocyte hypertrophy in vitro. These results are consistent with what we have observed in vivo in mouse hearts.
Collectively, these results suggest that although miR-22 expression is increased in HF diet induced cardiac hypertrophy, loss of function of miR-22 does not affect cardiac hypertrophy and fibrosis induced by HF diet.
miR-22 mediates the expression of genes involved in mitochondrial biogenesis and metabolism in the heart in response to HF feeding
We evaluated whether miR-22 could mediate the effect of HF diet in the expression of genes involved in mitochondrial biogenesis and metabolism. qPCR assays revealed that HF diet induced gene expression of Ppara, Pparg, and Pgc1-α in the heart of miR22-WT mice. Conversely, loss of miR-22 blocked the increased expression of these genes in the heart induced by HF feeding (Supplementary Figure 7A–E). These results are consistent with what we have observed in the liver (Supplementary Figure S2), and suggest that miR-22 is required for the expression of Ppara, Pparg, and Pgc1-α in the heart in response to HF diet.
Discussion
In the present study, we examined the impact of loss of miR-22 on cardiac and metabolic alterations associated with obesity. Several studies support a link between obesity and cardiovascular diseases [1]. Consistent with data from RNA sequencing [18], in the present study we confirmed that expression of miR-22 is increased in cardiac hypertrophy induced by HF diet. Additionally, we showed that db/db mice exhibited higher miR-22 expression in the heart. Unexpectedly, we found that loss of miR-22 did not affect HF feeding-induced cardiac hypertrophy and remodeling, suggesting that miR-22 does not directly contribute to the development of cardiac hypertrophy and fibrosis induced by obesity. Additionally, gain- and loss-of-function studies in cardiomyocyte cultures indicated that miR-22 does not contribute to cardiomyocyte hypertrophy induced by leptin. The reason for the discrepancy between our study and others previously published, which demonstrated that miR-22 is critical for cardiomyocyte hypertrophy in response to several stimuli is currently unknown. It is important of note that we did not find altered cardiac function in response to HF diet. Given that miR-22 plays a critical role in the control of cardiac remodeling and myocardial function under stress condition, we speculate that the time of dietary fat feeding used in the present study might not be sufficient to act as a pathological factor in the heart. In addition, our findings reinforce the importance of elucidating the role of miR-22 in different models of cardiovascular disorders.
Obesity is a major risk factor for cardiovascular disorders. Several studies have revealed that obesity leads to a chronic inflammatory state, which mediates many of the deleterious effects on cardiovascular system [29,30]. Although we have observed that loss of miR-22 attenuated the expression of proinflammatory genes and lipogenic genes induced by HF feeding, we did not observe the involvement of miR-22 on HF-feeding induced cardiomyopathy under our experimental conditions. It would be interesting to test whether increased exposition to HF diet could affect cardiac function. Similarly, it would be important to evaluate whether the loss of miR-22 is sufficient to influence the metabolic and cardiovascular disorders induced by long-term exposition to HF diet.
Recent studies have revealed the critical role of several miRNAs in the control of metabolism by affecting the expression of genes and proteins involved in metabolic processes [10,11]. Diverse miRNAs have been implicated in the control of body weight gain, glucose homeostasis, insulin resistance, and cholesterol and lipid metabolism [31,32], suggesting that miRNAs may be used as therapeutic targets for the treatment of metabolic disorders associated with obesity. A recent report demonstrated that miR-22 controls lipid metabolism of skeletal muscle, which influences the amount of white adipose tissue and body weight in male mice [20]. Our results showed that although loss of miR-22 did not affect HF diet induced body weight gain, glucose intolerance and insulin resistance, loss of miR-22 did prevent the increase on cholesterol and triglycerides levels promoted by HF feeding (Figure 7). Given that abnormalities in circulating lipid profiles represent a critical risk factor for cardiovascular disease [10], future investigations will be important to determine whether miR-22 may serve as a suitable candidate for therapeutic intervention in the treatment of some metabolic disorders observed in obesity, such as dyslipidemia, as well as the potential side effects. Our previous studies did not detect gender-dependent cardiac phenotype in miR-22 mutant mice [14] and our present study has only used male mice as an experimental model of obesity. Intriguingly, a recent study revealed that miR-22 promotes gender-specific regulation in muscle lipid metabolism by repressing estrogen receptor α [20]. Therefore, it would be interesting to test whether the contribution of miR-22 to metabolic disorders in response to HF diet may be affected by gender differences.
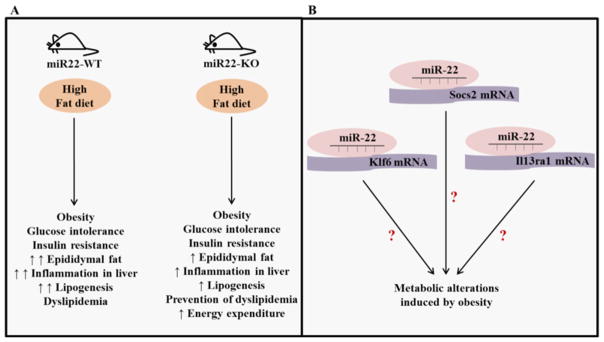
Figure 7. Schematic representation of the role of miR-22 in metabolic disorders induced by HF diet.
(A) HF diet increased body weight and epididymal fat mass as well as induced glucose intolerance, insulin resistance, and dyslipidemia. In addition, HF diet increased the expression of genes involved in inflammation and adipogenesis. Loss of miR-22 attenuated the gain of fat mass and prevented dyslipidemia. Moreover, loss of miR-22 attenuated the increased expression of genes involved in inflammation and adipogenesis mediated by HF diet, and increased the energy expenditure. (B) Socs2, Il13rα1, and Klf6, which have been implicated in inflammation and obesity, are direct targets of miR-22.
Adipose tissue plays a key role in the pathophysiology of obesity. A recent study revealed that reduced Sirt1 mRNA level in adipose tissue of obese patients was correlated negatively with the level of miR-22 [33]. Interestingly, activation of Sirt1 induces the browning of white adipose tissue and promotes resistance to obesity [34], suggesting that factors that regulate Sirt1 might attenuate the metabolic disorders associated with obesity. Intriguingly, our study revealed that the expression of miR-22 was elevated in the adipose tissue of obese mice, and that loss of miR-22 attenuated the increase in epididymal fat mass induced by HF diet. Given that miR-22 direct targets Sirt1, we speculate that Sirt1 may represent one of the mechanisms by which miR-22 mediates partial increase in epididymal fat mass induced by HF feeding. Future studies are required to determine whether loss of miR-22 can modulate Sirt1 expression in the adipose tissue or the browning of white adipose tissue.
It is well known that obesity is associated with inflammation in the adipose tissue [35]. We found that the increased expression levels of proinflammatory genes in response to HF diet, including Ccl2, Il6, and Ifng, were blunted in the adipose tissue of miR22-KO mice, suggesting that miR-22 contributes at least in part to adipose tissue inflammation induced by HF feeding. Moreover, we observed that loss of miR-22 reduced expression of genes involved in adipogenesis, such as Pparg and Srebp-1 in the epididymal fat, further suggesting that miR-22 may participate in the regulation of adipogenesis. In the future, gain- and loss-of-function studies will be important to characterize the role of miR-22 targets in adipogenesis.
The liver plays a critical role in metabolic homeostasis, including control of cholesterol synthesis, gluconeogenesis, and lipogenesis [36]. We observed that although loss of miR-22 had not affected the increased triglyceride content in the liver induced by HF diet, loss of miR-22 did repress the higher expression of genes involved in fatty acid synthesis, including Srebp-1, Fasn, and Pparg. These results indicate that miR-22 by itself is not sufficient to govern the triglyceride content, additional factors are probably needed. Similar to the data obtained from adipose tissue, loss of miR-22 blocked the overexpression of proinflammatory genes in the liver, underscoring the involvement of this miRNA in distinct biological processes. It will be interesting to determine whether miR-22 regulates similar targets and molecular pathways in adipogenesis and inflammation.
Prediction analyses identified Socs2, Klf6, and Il13rα1 as potential targets of miR-22. Socs2 is a master regulator of hepatic steatosis and inflammation in response to HF feeding [37]. Moreover, macrophage activation in the adipose tissue is regulated by IL-13 and overexpression of IL-13 prevented the expression of inflammatory genes induced by HF diet [38]. In addition, Klf6 contributes to adipogenesis [39] and has been shown to affect glucose and lipid metabolism in response to HF diet [40]. Here, we experimentally verified that miR-22 directly targets Socs2, Il13rα1, and Klf6. Given that Socs2 regulates the hepatic metabolic disorder induced by HF diet [37], it would be interesting to test whether the role of miR-22 in the metabolic alterations induced by HF feeding may be influenced, at least in part, by Socs2. In addition, considering that IL-13 attenuates the expression of genes involved in inflammation in response to HF diet [38], it is possible that part of the miR-22 effects in the expression of inflammatory genes may be influenced by Il13rα1. However, future investigations are needed to determine whether Il13rα1 mediates part of the miR-22 effect in the expression of inflammatory genes. Moreover, given that Klf6 controls adipogenesis [39], and that miR-22 targets Klf6, it would be interesting to test whether the influence of miR-22 in the expression of lipogenic genes may be mediated, at least in part, by Klf6. Clearly, future studies will be important to determine whether regulation of the expression of Socs2, Il13rα1, and Klf6 mediated by miR-22 may play a critical role in the control of metabolic dysregulation associated with obesity. Additionally, future investigations are critical to clarify the potential use of miR-22 as a therapeutic approach for the metabolic disorders associated with obesity.
Supplementary Material
Clinical perspectives.
The incidence of obesity has increased significantly over recent decades and has contributed to an increase in the risk of numerous chronic disorders. However, the molecular nature of such disorders remains unclear.
The goal of our study is to uncover the role of a small noncoding RNA, miR-22, in metabolism. We used a mouse loss-of-function genetic model to provide in vivo evidence that miR-22 regulates energy expenditure. We found that loss of miR-22 reduces the formation of epididymal fat and miR-22 mutant mice exhibit a reduction in lipogenesis in response to a high-fat diet.
Our results suggest that miR-22 could be a novel therapeutic target for metabolic disorders.
Acknowledgments
Funding
This work was supported by American Heart Association, Muscular Dystrophy Association and NIH grants [HL085635 and HL116919 (to D.Z.W.)]. G.P.D. was supported by CAPES grant [6733-14-0] and FAPESP (Sao Paulo Research Foundation) grant [2015/21859-5]. J.D. Jr was supported by FAPESP grant [2014/50140-6]. Z.P.H. is supported by American Heart Association Scientist Development Grant; J.L. and J.D. are supported by NIH [5T32HL007572].
Abbreviations
- FS
fractional shortening
- iGTT
intraperitoneal glucose tolerance test
- LV
Left ventricle
- Ppar
peroxisome proliferator-activated receptor
- RER
respiratory exchange ratio
- Sirt1
Sirtuin 1
- WGA
wheat germ agglutinin
Footnotes
Author Contribution
G.P.D. and J.L. designed the study and the protocols. G.P.D. and D.Z.W wrote the manuscript. Z.P.H. and J.D. constructed luciferase reporters. Z.P.H. generated miR-22 null mice. J.C. analyzed echocardiographic data. R.I.F. and J.D.J. performed metabolic cage analyses. X.H. performed echocardiography and histology experiments. M.L.B.C. contributed with protocols and critical revision of the paper. D.Z.W is the guarantor of this study.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Ortega FB, Lavie CJ, Blair SN. Obesity and cardiovascular disease. Circ Res. 2016;118:1752–1770. doi: 10.1161/CIRCRESAHA.115.306883. https://doi.org/10.1161/CIRCRESAHA.115.306883. [DOI] [PubMed] [Google Scholar]
- 2.Malik S, Huang CF, Schmidt J. The role of the CANNTG promoter element (E box) and the myocyte-enhancer-binding-factor-2 (MEF-2) site in the transcriptional regulation of the chick myogenin gene. Eur J Biochem. 1995;230:88–96. doi: 10.1111/j.1432-1033.1995.tb20537.x. https://doi.org/10.1111/j.1432-1033.1995.0088i.x. [DOI] [PubMed] [Google Scholar]
- 3.Perry RJ, Camporez JP, Kursawe R, Titchenell PM, Zhang D, Perry CJ, et al. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell. 2015;160:745–758. doi: 10.1016/j.cell.2015.01.012. https://doi.org/10.1016/j.cell.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lancaster GI, Kammoun HL, Kraakman MJ, Kowalski GM, Bruce CR, Febbraio MA. PKR is not obligatory for high-fat diet-induced obesity and its associated metabolic and inflammatory complications. Nat Commun. 2016;7:10626. doi: 10.1038/ncomms10626. https://doi.org/10.1038/ncomms10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Wang DZ. microRNAs in cardiovascular development. J Mol Cell Cardiol. 2012;52:949–957. doi: 10.1016/j.yjmcc.2012.01.012. https://doi.org/10.1016/j.yjmcc.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espinoza-Lewis RA, Wang DZ. MicroRNAs in heart development. Curr Top Dev Biol. 2012;100:279–317. doi: 10.1016/B978-0-12-387786-4.00009-9. https://doi.org/10.1016/B978-0-12-387786-4.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11:860–872. doi: 10.1038/nrd3864. https://doi.org/10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duygu B, de Windt LJ, da Costa Martins PA. Targeting microRNAs in heart failure. Trends Cardiovasc Med. 2016;26:99–110. doi: 10.1016/j.tcm.2015.05.008. https://doi.org/10.1016/j.tcm.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. https://doi.org/10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. https://doi.org/10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deiuliis JA. MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. Int J Obes (Lond) 2016;40:88–101. doi: 10.1038/ijo.2015.170. https://doi.org/10.1038/ijo.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu XD, Song XW, Li Q, Wang GK, Jing Q, Qin YW. Attenuation of microRNA-22 derepressed PTEN to effectively protect rat cardiomyocytes from hypertrophy. J Cell Physiol. 2012;227:1391–1398. doi: 10.1002/jcp.22852. https://doi.org/10.1002/jcp.22852. [DOI] [PubMed] [Google Scholar]
- 13.Gurha P, Abreu-Goodger C, Wang T, Ramirez MO, Drumond AL, van Dongen S, et al. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation. 2012;125:2751–2761. doi: 10.1161/CIRCULATIONAHA.111.044354. https://doi.org/10.1161/CIRCULATIONAHA.111.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang ZP, Chen J, Seok HY, Zhang Z, Kataoka M, Hu X, et al. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res. 2013;112:1234–1243. doi: 10.1161/CIRCRESAHA.112.300682. https://doi.org/10.1161/CIRCRESAHA.112.300682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang ZP, Wang DZ. miR-22 in cardiac remodeling and disease. Trends Cardiovasc Med. 2014;24:267–272. doi: 10.1016/j.tcm.2014.07.005. https://doi.org/10.1016/j.tcm.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oka S, Alcendor R, Zhai P, Park JY, Shao D, Cho J, et al. PPARalpha-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 2011;14:598–611. doi: 10.1016/j.cmet.2011.10.001. https://doi.org/10.1016/j.cmet.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. https://doi.org/10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 18.Guedes EC, Franca GS, Lino CA, Koyama FC, do Moreira LN, Alexandre JG, et al. MicroRNA expression signature is altered in the cardiac remodeling induced by high fat diets. J Cell Physiol. 2016;231:1771–1783. doi: 10.1002/jcp.25280. https://doi.org/10.1002/jcp.25280. [DOI] [PubMed] [Google Scholar]
- 19.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. https://doi.org/10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 20.Schweisgut J, Schutt C, Wust S, Wietelmann A, Ghesquiere B, Carmeliet P, et al. Sex-specific, reciprocal regulation of ERalpha and miR-22 controls muscle lipid metabolism in male mice. EMBO J. 2017;36:1199–1214. doi: 10.15252/embj.201695988. https://doi.org/10.15252/embj.201695988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 22.Shimano H. SREBPs: physiology and pathophysiology of the SREBP family. FEBS J. 2009;276:616–621. doi: 10.1111/j.1742-4658.2008.06806.x. https://doi.org/10.1111/j.1742-4658.2008.06806.x. [DOI] [PubMed] [Google Scholar]
- 23.Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20:107–114. doi: 10.1016/j.tem.2008.11.005. https://doi.org/10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci USA. 2012;109:15330–15335. doi: 10.1073/pnas.1207605109. https://doi.org/10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceylan-Isik AF, Kandadi MR, Xu X, Hua Y, Chicco AJ, Ren J, et al. Apelin administration ameliorates high fat diet-induced cardiac hypertrophy and contractile dysfunction. J Mol Cell Cardiol. 2013;63:4–13. doi: 10.1016/j.yjmcc.2013.07.002. https://doi.org/10.1016/j.yjmcc.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Carabelli J, Burgueno AL, Rosselli MS, Gianotti TF, Lago NR, Pirola CJ, et al. High fat diet-induced liver steatosis promotes an increase in liver mitochondrial biogenesis in response to hypoxia. J Cell Mol Med. 2011;15:1329–1338. doi: 10.1111/j.1582-4934.2010.01128.x. https://doi.org/10.1111/j.1582-4934.2010.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Martinez I, Santoro N, Chen Y, Hoque R, Ouyang X, Caprio S, et al. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest. 2016;126:859–864. doi: 10.1172/JCI83885. https://doi.org/10.1172/JCI83885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taube A, Schlich R, Sell H, Eckardt K, Eckel J. Inflammation and metabolic dysfunction: links to cardiovascular diseases. Am J Physiol Heart Circ Physiol. 2012;302:H2148–H2165. doi: 10.1152/ajpheart.00907.2011. https://doi.org/10.1152/ajpheart.00907.2011. [DOI] [PubMed] [Google Scholar]
- 30.Fuster JJ, Ouchi N, Gokce N, Walsh K. Obesity-induced changes in adipose tissue microenvironment and their impact on cardiovascular disease. Circ Res. 2016;118:1786–1807. doi: 10.1161/CIRCRESAHA.115.306885. https://doi.org/10.1161/CIRCRESAHA.115.306885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. https://doi.org/10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu X, Dong B, Tian Y, Lefebvre P, Meng Z, Wang X, et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest. 2015;125:2497–2509. doi: 10.1172/JCI75438. https://doi.org/10.1172/JCI75438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurylowicz A, Owczarz M, Polosak J, Jonas MI, Lisik W, Jonas M, et al. SIRT1 and SIRT7 expression in adipose tissues of obese and normal-weight individuals is regulated by microRNAs but not by methylation status. Int J Obes (Lond) 2016;40:1635–1642. doi: 10.1038/ijo.2016.131. https://doi.org/10.1038/ijo.2016.131. [DOI] [PubMed] [Google Scholar]
- 34.Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. https://doi.org/10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121:2094–2101. doi: 10.1172/JCI45887. https://doi.org/10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyons CL, Kennedy EB, Roche HM. Metabolic inflammation-differential modulation by dietary constituents. Nutrients. 2016:8. doi: 10.3390/nu8050247. https://doi.org/10.3390/nu8050247. [DOI] [PMC free article] [PubMed]
- 37.Zadjali F, Santana-Farre R, Vesterlund M, Carow B, Mirecki-Garrido M, Hernandez-Hernandez I, et al. SOCS2 deletion protects against hepatic steatosis but worsens insulin resistance in high-fat-diet-fed mice. FASEB J. 2012;26:3282–3291. doi: 10.1096/fj.12-205583. https://doi.org/10.1096/fj.12-205583. [DOI] [PubMed] [Google Scholar]
- 38.Darkhal P, Gao M, Ma Y, Liu D. Blocking high-fat diet-induced obesity, insulin resistance and fatty liver by overexpression of Il-13 gene in mice. Int J Obes (Lond) 2015;39:1292–1299. doi: 10.1038/ijo.2015.52. https://doi.org/10.1038/ijo.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li D, Yea S, Li S, Chen Z, Narla G, Banck M, et al. Kruppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J Biol Chem. 2005;280:26941–26952. doi: 10.1074/jbc.M500463200. https://doi.org/10.1074/jbc.M500463200. [DOI] [PubMed] [Google Scholar]
- 40.Bechmann LP, Vetter D, Ishida J, Hannivoort RA, Lang UE, Kocabayoglu P, et al. Post-transcriptional activation of PPAR alpha by KLF6 in hepatic steatosis. J Hepatol. 2013;58:1000–1006. doi: 10.1016/j.jhep.2013.01.020. https://doi.org/10.1016/j.jhep.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.