Abstract
Human mutations of the GRID1 gene encoding the orphan delta1 glutamate receptor-channel (GluD1) are associated with schizophrenia but the explicit role of GluD1 in brain circuits is unknown. Based on the known function of its paralog GluD2 in cerebellum, we searched for a role of GluD1 in slow glutamatergic transmission mediated by metabotropic receptor mGlu1 in midbrain dopamine neurons, whose dysfunction is a hallmark of schizophrenia. We found that an mGlu1 agonist elicits a slow depolarizing current in HEK cells co-expressing mGlu1 and GluD1, but not in cells expressing mGlu1 or GluD1 alone. This current is abolished by additional co-expression of a dominant-negative GluD1 dead pore mutant. We then characterized mGlu1-dependent currents in dopamine neurons from midbrain slices. Both the agonist-evoked and the slow postsynaptic currents are abolished by expression of the dominant-negative GluD1 mutant, pointing to the involvement of native GluD1 channels in these currents. Likewise, both mGlu1-dependent currents are suppressed in GRID1 knockout mice, which reportedly display endophenotypes relevant for schizophrenia. It is known that mGlu1 activation triggers the transition from tonic to burst firing of dopamine neurons, which signals salient stimuli and encodes reward prediction. In vivo recordings of dopamine neurons showed that their spontaneous burst firing is abolished in GRID1 knockout mice or upon targeted expression of the dominant-negative GluD1 mutant in wild-type mice. Our results de-orphanize GluD1, unravel its key role in slow glutamatergic transmission and provide insights into how GRID1 gene alterations can lead to dopaminergic dysfunctions in schizophrenia.
Introduction
Genome wide association studies of several distinct human cohorts have pointed out variations in the GRID1 gene linked with schizophrenia and bipolar disorder.1, 2, 3, 4 Likewise, GRID1 knockout mice exhibit endophenotypes relevant for these pathologies and have been proposed as a model of schizophrenia and depression co-morbidity.5, 6, 7, 8 GRID1 encodes GluD1, which forms with GluD2 the delta family of ionotropic glutamate receptors.9 GluDs have remained orphan of a pore-opening ligand since their cloning, raising doubts on their function as ion channels,9, 10, 11 until the recent demonstration that GluD2 channel opening is triggered by mGlu1 activation through the canonical Gq/phospholipase C/protein kinase C pathway, and plays a key role in slow glutamatergic transmission in the cerebellum.12, 13 In support of this function, native GluDs physically interact with mGlu1 and mGlu5 metabotropic glutamate receptors of the group I (mGlu1/5).14, 15 GluD1 is widely expressed in the brain and largely predominates over GluD2 outside the cerebellum.16, 17 GluD1 is localized at the postsynaptic density of excitatory synapses and its expression increases during postnatal development,16, 17 consistent with a role in glutamatergic transmission in the adult.
Alterations of dopamine (DA) and glutamate neurotransmission are critical factors in the physiopathology of schizophrenia.18 The main source of DA in the brain originates in midbrain neurons of the substantia nigra (SN) and ventral tegmental area (VTA). The firing of these neurons consists in single spikes, but also in bursts of action potentials19 that enhance DA release and have important behavioral correlates.20, 21 Accordingly, an imbalance between these two firing modes is thought to be a key factor in the physiopathology of schizophrenia22, 23 and is linked to alterations of brain circuits and behaviors in a mouse model of schizophrenia.24 Interestingly, DA neurons of both the SN and the VTA express mGlu1/5 receptors,25 whose activation promotes their burst firing,26, 27, 28 and exhibit a prominent slow excitatory postsynaptic current (EPSC), which is blocked by mGlu1/5 antagonists.29, 30 These observations suggest that GluD1 may play a role in slow glutamatergic transmission onto DA neurons, and in the firing pattern of these neurons.
The present study examines the functional coupling between mGlu1/5 and GluD1 and its role in DA neurons using a combination of electrophysiology, molecular inactivation and histochemistry. We first tested the ability of mGlu1 to gate GluD1 channels in a heterologous expression system. We then examined the expression of GluD1 mRNA and protein in DA neurons. Finally, we probed the role of GluD1 in the slow EPSC and the firing pattern of DA neurons in midbrain slices of wild type (WT) and GRID1 knockout mice and in vivo. The results indicate that mGlu1/5 activation triggers the opening of GluD1 channels, which are key determinants of the slow EPSC and the burst firing of DA neurons.
Materials and methods
Detailed materials and methods are provided as Supplementary Information
Plasmids and viruses
The plasmids encoding rat mGlu1a-YFP,31 mGlu5a-Venus,32 mouse GluD1 (Source Bioscience, Nottingham, UK) or human 5-HT2C33 under the control of a cytomegalovirus promoter were transfected in human embryonic kidney cells (HEK293T line) using the calcium phosphate precipitation method. Sindbis viruses (or lentiviruses) expressing both the GluD1V617R mutant and green fluorescent protein (GFP), or GFP alone, were constructed from pSinEGdsp34 (or pLV-PGK-β2-ires-GFP-WPRE,35 pLV-PDGF-lox-mCherry-lox-β236), or already described (pLV-PGK-GFP,35 pLV-PDGF-lox-mCherry-lox-GFP37). Recombinant pseudo-virions were produced as described35, 38 and stereotaxically injected in the SN/VTA of WT or DAT-Cre39 mice 14–24 h (Sindbis) or 4–6 weeks (lentivirus) before electrophysiological recordings.
Mice and electrophysiological recordings
Animal breeding and killing were performed in accordance to European Commission guidelines and French legislation (2010/63/UE) and procedures were approved by the French Ministry of Research (Agreement # 01479.01). GluD1 knockout,5 GluD2 Hotfoot (HO)40 and DAT-Cre39 mice were genotyped as described.16, 39 Homozygous GluD1-knockout mice were obtained from breeding heterozygous parents. WT mice were purchased from Janvier Labs (Le Genest-Saint-Isle, France). All mice had C57BL/6 background. Whole-cell electrophysiological recordings in midbrain slices from P14–P18 mice, and in vivo juxtacellular recordings in 6–12 week-old anesthetized mice were performed using standard procedures.
Single-cell real-time-PCR and immunochemistry
Single-cell real-time PCR (RT-PCR) after patch-clamp was performed as described41, 42 using primer pairs given in Supplementary Table S1. We used standard procedures for immunocyto-, immunohisto-chemistry and immunoprecipitation. Immunohistochemical reactions at the electron microscopic level were carried out using the post-embedding immunogold method as described.43
Statistical analyses
Results are given as mean±s.e.m. In vitro data were analyzed using Mann–Whitney or Wilcoxon rank signed nonparametric tests. Differences were considered significant if P<0.05. In the in vivo section, three-group comparisons were performed using a Student t-test with Bonferroni correction, and two group comparisons were performed using Student t-test (if n>30) or Wilcoxon rank signed test (if n<30) with significance set at P<0.05.
Results
The activation of mGlu1/5 triggers the opening of GluD1 channels in a heterologous expression system
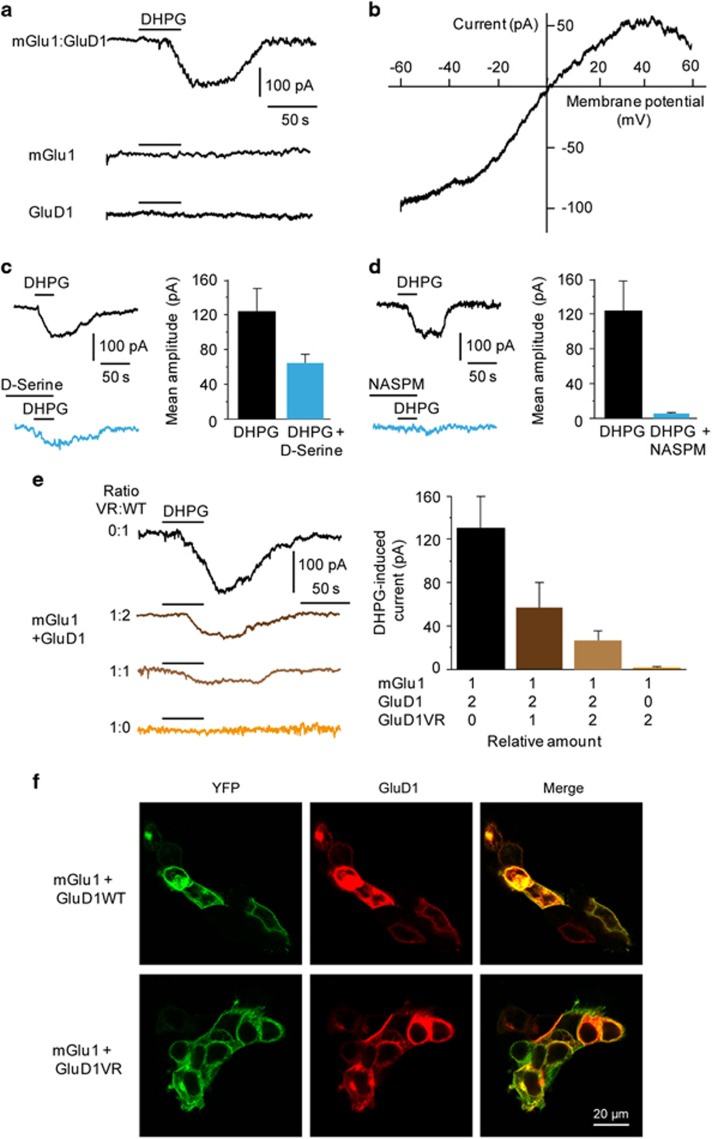
We tested the existence of a functional coupling between mGlu1 and GluD1 in HEK 293 cells transfected with plasmids encoding GluD1 and an mGlu1-YFP fusion protein. Expression of these proteins, separately or together, resulted in their correct targeting to the plasma membrane (Figure 1f, Supplementary Figures S1 and S2). Local application of the mGlu1/5 agonist 3,5-dihydroxyphenylglycine (DHPG, 100 μM) on cells expressing both mGlu1 and GluD1 induced a slow and reversible inward current (IDHPG, 131±31 pA, n=13; Figure 1a). Similar results were obtained on HEK cells co-transfected with plasmids encoding mGlu5-Venus and GluD1 (IDHPG, 78±7 pA, n=9; not shown). The IDHPG observed in mGlu1+GluD1-expressing cells was almost abolished in the presence of the mGlu1/5 antagonist 1-aminoindan-1,5-dicarboxylic acid (AIDA, 150 μM; 7±5 pA, n=5, P<0.03; not shown), indicating that this current was triggered by activation of mGlu1. No DHPG-induced current was observed in cells expressing either mGlu1 (n=15) or GluD1 (n=6) alone (Figure 1a), attesting to the critical dependence of the IDHPG on both proteins. The mutual requirement of mGlu1/5 and GluD1 to generate currents was further tested by stimulating other Gq-coupled receptors in GluD1-expressing HEK cells. No response was observed upon stimulation of the endogenous muscarinic M3 receptor44, 45, 46 by carbachol (100 μM) in DHPG-responsive HEK cells co-expressing mGlu1 and GluD1 (n=5, IDHPG: 80±19 pA; Supplementary Figure S1). Similarly, application of the 5-HT2C serotonin receptor agonist 2,5-dimethoxy-4-iodoamphetamine (50 μM) failed to induce appreciable response in HEK cells co-expressing 5-HT2C and GluD1 (1.7±2.6 pA, n=10; Supplementary Figure S1). These results are in line with the observation that PKC activation is not sufficient to elicit GluD2 currents,13 and suggest that mGlu1/5-GluD coupling in HEK cells may rely on their physical interaction, which is documented for native receptors in the brain.14, 15 Indeed, we found that antibodies against either GluD1, mGlu1 or mGlu5 co-immunoprecipitated recombinant mGlu1/5 and GluD1 expressed in HEK cells (Supplementary Figure S3). These results indicate that the current caused by the activation of mGlu1/5 in HEK cells relies on the physical interaction of mGlu1/5 receptors with GluD1.
Figure 1.
Activation of mGlu1 triggers the opening of GluD1 channels in HEK cells. (a) Bath application of DHPG (100 μM) induced a slow inward current in cells co-expressing mGlu1 and GluD1, but not in cells expressing either mGlu1 or GluD1 alone. (b) Example of I/V curve of the DHPG-induced current exhibiting a reversal potential around 0 mV and inward rectification at positive potentials. (c,d) The DHPG-induced current was reduced by D-serine (10 mM) and almost abolished by NASPM (100 μM). (e) Co-expression of the dominant-negative GluD1VR dead pore mutant with mGlu1 and WT GluD1 dose-dependently reduced the DHPG-induced current. (f) Correct targeting of recombinant proteins at the plasma membrane. DHPG, 3,5-dihydroxyphenylglycine; NASPM, naphtyl-acetyl spermine; WT, wild type.
We next examined the involvement of the GluD1 channel pore in the mGlu1-dependent current by characterizing its biophysical and pharmacological properties. The current–voltage (I–V) relationship of the IDHPG exhibited a reversal potential around 0 mV and a marked inward rectification at positive potentials (n=8, see example in Figure 1b). This voltage-dependence is similar to that reported for GluD2-mediated currents.12, 47, 48 We also tested the effect of two inhibitors of GluD1 and GluD2, the calcium-permeable AMPA receptor-channel blocker naphtyl-acetyl spermine (NASPM) and D-serine.12, 47, 49, 50, 51 NASPM (100 μM) and D-serine (10 mM) reduced the IDHPG by 86±4% (n=5, P<0.001) and 42±8% (n=5, P<0.05), respectively (Figures 1c and d), consistent with the involvement of GluD1 channels. To assess permeation of the IDHPG through GluD1 channels, we used a dominant-negative GluD1 dead pore mutant obtained by a valine to arginine substitution at position 617 (GluD1VR), as described for GluD2 and AMPA/kainate receptor subunits.12, 52, 53 We verified the correct targeting of GluD1VR to the plasma membrane and its co-immunoprecipitation with mGlu1/5 (Supplementary Figure S1 and Supplementary Figure S3). No IDHPG was observed in cells co-expressing mGlu1 and GluD1VR (n=8). Moreover, increasing the amount of GluD1VR plasmid in co-transfection with mGlu1 and WT GluD1 gradually decreased IDHPG amplitude (n⩾4 for each GluD1VR/GluD1WT ratio tested, Figure 1e). These results indicate that mGlu1/5 activation triggers the opening of the GluD1 channel in HEK cells.
GluD1 is co-expressed with mGlu1/5 in DA neurons
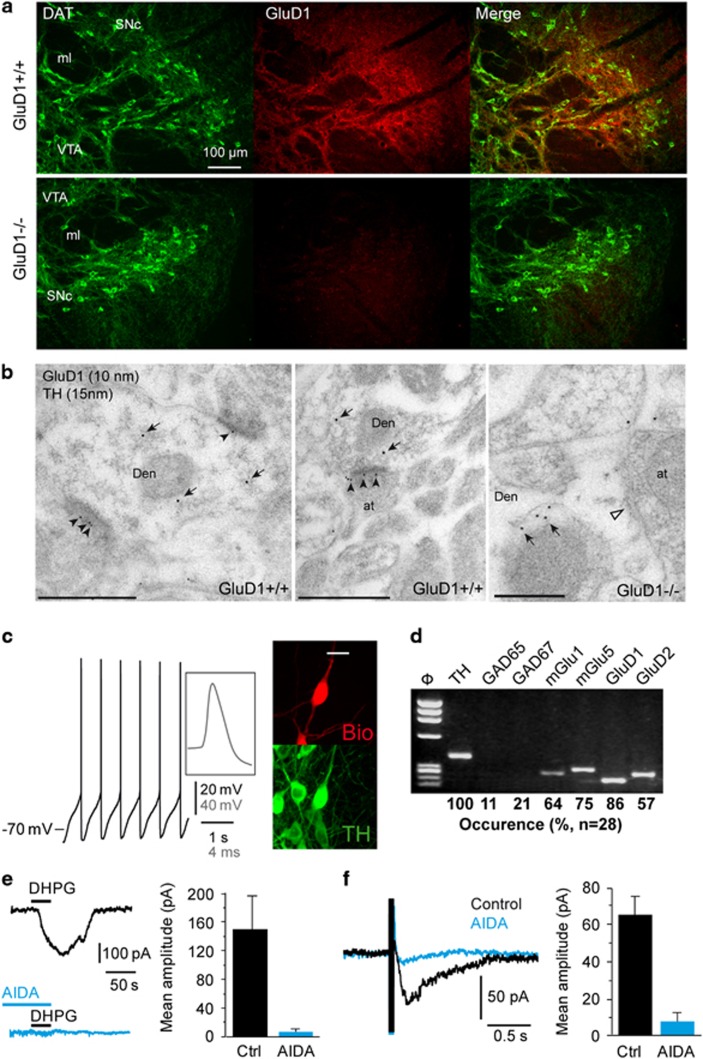
Currents evoked by activation of mGlu1/5 in midbrain DA neurons of the SN pars compacta (SNc) and the VTA share biophysical properties with mGlu1/5-activated GluD currents in HEK cells and cerebellar Purkinje cells (see above and references 12, 29, 30). We thus probed the expression of GluD1 in DA neurons using anti-GluD1 together with anti-DA transporter (DAT) or anti-tyrosine hydroxylase (TH, the DA-synthesizing enzyme) immunohistochemistry. At the light microscopic level, we observed a diffuse expression of GluD1 in somata and processes of DAT-positive neurons in the SNc and VTA (Figure 2a). At higher magnification, GluD1 immunopositive puncta were observed at intracellular and putative plasma membrane locations of DAT-positive somata and dendritic shafts (Supplementary Figure S4), as described in the cerebellum.17 Further examination at the electron microscopic level in the SNc revealed that immunogold particles for GluD1 were localized along the postsynaptic density of excitatory synapses in TH-positive neurons (Figure 2b). Quantitative analysis showed that most immunoparticles were found at postsynaptic sites (98.3%, n=339) and only few (1.7%, n=6) at presynaptic sites. Of the immunoparticles located at postsynaptic sites, 81.4% (n=276) were found in the plasma membrane of dendritic shafts, and 18.6% (n=63) at cytoplasmic site associated in intracellular membranes. Of the immunoparticles located in the plasma membrane, 80.1% (n=221) were found in the postsynaptic density of excitatory synapses, and 19.9% (n=55) along the extrasynaptic plasma membrane. GluD1 immunolabeling was absent in GluD1−/− mice (Figures 2a and b), consistent with earlier observations.16 These results show that GluD1 is expressed in DA neurons at excitatory postsynaptic sites, as found in other brain regions.16, 17
Figure 2.
GluD1 is co-expressed with mGlu1/5 in DA neurons. (a) Fluorescence micrographs of the SNc and VTA showing diffuse GluD1 immunostaining of somata and processes of DAT-positive DA neurons. Immunostaining almost disappeared in GluD1−/− mice. Scale bar: 100 μm. (b) Electron micrographs of the SNc showing immunoparticles for GluD1 (arrowheads) and TH (arrows), as detected using a double-labeling post-embedding immunogold method. GluD1 immunoparticles were detected along the postsynaptic density of dendritic shafts (Den) of TH-labeled DA neurons, establishing asymmetrical synapses with axon terminal (at). No GluD1 immunoparticles were detected in GluD1−/− mice, as highlighted by open arrowhead pointing to postsynaptic density of a TH-labeled neuron. Scale bars: 0.5 μm. (c) Characterization of a DA neuron from the SNc typically exhibiting long duration action potentials and spontaneous pacemaker firing at low rate. This biocytin-filled neuron (Bio) was TH-immunopositive (scale bar: 20 μm). (d) RT-PCR analysis of a single DA neuron detected expression of TH, mGlu1, mGlu5, GluD1 and GluD2 upon agarose gel electrophoresis of PCR products (ΦX HaeIII molecular weight marker). Summary of single-cell RT-PCR data obtained in SNc DA neurons is given below the agarose gel picture. (e,f) The mGlu1/5 antagonist AIDA (150 μM) prevented the DHPG-induced current and the sEPSC triggered by local electrical stimulation in DA neurons. AIDA, 1-aminoindan-1,5-dicarboxylic acid; DA, dopamine; RT-PCR, real-time PCR; sEPCS, slow excitatory postsynaptic current; SN, substantia nigra; SNc, SN pars compacta; VTA, ventral tegmental area.
We next examined GluD1 expression in DA neurons of the SNc using patch-clamp recordings and single-cell RT-PCR in acute midbrain slices. We selected DA neurons based on their long duration action potentials (above 1.5 ms at half-width) and their spontaneous pacemaker firing at low rate (below 2 Hz at resting membrane potential, Figure 2c), which discriminate them from local GABAergic neurons.54, 55 The identity of recorded DA neurons was confirmed by post hoc TH immunostaining (n=122 TH-positive neurons out of 126 tested throughout this study, Figure 2c). We also verified the responsiveness of DA neurons to mGlu1/5 activation in our experimental conditions. Indeed, in the presence of ionotropic glutamate and GABA receptor antagonists (CNQX 10 μM, APV 50 μM, gabazine 10 μM), we observed robust inward currents in response to DHPG application (100 μM, 102±13 pA, n=33), and slow EPSCs (sEPSC) in response to bursts of local electrical stimulation (97±10 pA, n=27), as earlier described.29, 30 Both currents were almost abolished in the presence of AIDA (150 μM; IDHPG: n=5, P<0.05; sEPSC: n=7, P<0.01; Figures 2e and f), consistent with the reported involvement of mGlu1/5 in these responses.29, 30 DA neurons identified by their electrophysiological properties were all TH-positive upon analysis by single-cell RT-PCR (n=28, Figure 2d), whereas mRNAs for the GABA synthesizing enzymes were only detected in11% (GAD65) and 21% (GAD67) of these cells. A majority of these DA neurons (93%) were positive for mGlu1 (64%) and/or mGlu5 (75%), in line with functional and immunohistochemical observations.25 GluD1 expression was also detected with a high occurrence (86%) in this sample, in agreement with our above immunochemical data, whereas GluD2 occurrence was lower (57%), consistent with its low abundance in brain regions outside the cerebellum.16 These results show that GluD1 is co-expressed with mGlu1/5 in DA neurons.
GluD1 channels mediate mGlu1/5-dependent currents in DA neurons
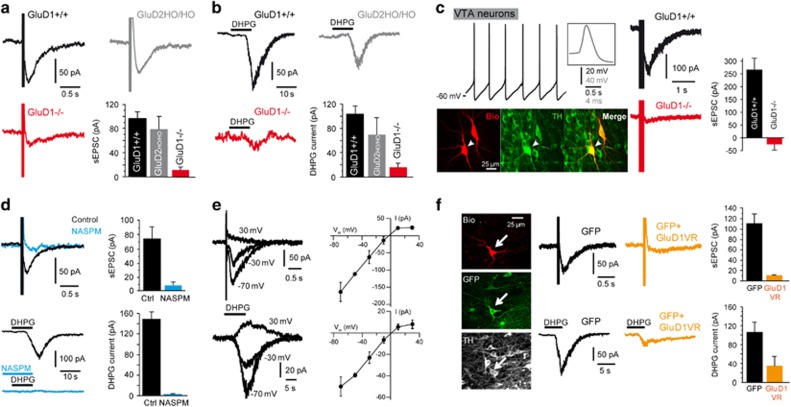
To test the involvement of GluD1 in mGlu1/5-gated currents of DA neurons, we investigated the effects of GluD1 deletion on the sEPSC and IDHPG recorded in acute midbrain slices. We first compared fast EPSCs in DA neurons from WT and GluD1−/− mice to rule out major alteration of excitatory synaptic inputs onto GluD1−/− DA neurons. Fast EPSC amplitudes did not significantly differ between WT and GluD1−/− cells (SNc: 91±12 pA (n=27) and 123±28 pA (n=17), respectively, P=0.5; VTA: 154±34 pA (n=12) and 280±71 pA (n=10), respectively, P=0.14). Moreover, AMPA and NMDA receptor-mediated components of fast EPSCs in SNc DA neurons were not significantly different in WT and GluD1−/− mice (NMDA amplitude: 82±10 pA and 106±13 pA, AMPA/NMDA ratio: 3.1±0.6 and 2.6±0.3, NMDA decay time-constant: 69±7 ms and 77±6 ms, n=16 and 19, respectively, P⩾0.24; see Supplementary Figure S5). In contrast, we found that the sEPSC recorded in SNc DA neurons was strongly reduced in GluD1−/− mice (13±5 pA, n=20) relatively to WT animals (97±12 pA, n=27; P<0.001; Figure 3a). The IDHPG of SNc DA neurons was also largely diminished in GluD1−/− vs WT mice (16±7 pA, n=14 vs 102±13 pA, n=33; P<0.001; Figure 3b). Conversely, mGlu1/5-gated currents in SNc DA neurons were not significantly altered in Hotfoot-Nancy mutant mice40 (GluD2HO/HO) that bear a deletion of the GluD2 gene (sEPSC: 80±20 pA, n=7, P=0.46; IDHPG: 69±28 pA, n=8, P=0.55; Figures 3a and b), as expected from the low abundance of GluD2 in DA neurons. We further examined the involvement of GluD1 in the mGlu1/5-gated currents of DA neurons by recording their sEPSC in the VTA. Similar to our findings in the SNc, the sEPSC of VTA DA neurons recorded in WT mice (265±45 pA, n=12) was almost abolished in GluD1−/− mice (Figure 3c), unmasking an outward current in half of the recorded cells (mean of GluD1−/− cells: 26±22 pA outward, n=12; P<0.001). These results show that mGlu1/5-gated currents in DA neurons critically rely on GluD1.
Figure 3.
GluD1 channels mediate mGlu1/5-dependent currents in DA neurons. (a,b) The sEPSC and the IDHPG of SNc DA neurons were strongly reduced in GluD1−/− mice, but were not significantly altered in mice bearing a deletion of the GluD2 gene (GluD2HO/HO). (c) In the VTA, GluD1 deletion also hampered the sEPSC (right panels) of DA neurons identified from their long duration action potentials, spontaneous pacemaker firing at low rate and TH immunoreactivity (left panels). Traces above fluorescence pictures stem from recording of the biocytin-filled cell indicated by an arrow. (d) Application of the GluD channel blocker NASPM (100 μM) almost abolished the sEPSC and the IDHPG in DA neurons of the SNc. (e) I/V curves of the sEPSC and the IDHPG recorded at different preset potentials in DA neurons of the SNc, as exemplified by superimposed traces in left panels. (f) Fluorescence pictures show histological analysis of a GFP-positive DA neuron recorded 20 h after injection of a Sindbis virus co-expressing GluD1VR and GFP in the SNc of a WT mouse and labeled with biocytin (Bio). Note the sparse transduction achieved using sindbis viral transfer in the SNc. The sEPSC and the IDHPG were largely reduced in SNc DA neurons co-expressing GluD1VR and GFP compared to DA neurons transduced with a control Sindbis virus expressing GFP alone. DA, dopamine; NASPM, naphtyl-acetyl spermine; sEPCS, slow excitatory postsynaptic current; SNc, SN pars compacta; SNR, substantia nigra pars reticulata; VTA, ventral tegmental area; WT, wild type.
To evaluate the contribution of GluD1 channels to mGlu1/5-gated currents, we next characterized their pharmacological and biophysical properties in SNc DA neurons. The application of NASPM (100 μM) strongly reduced both the sEPSC (by 82±5%, n=7, P<0.001) and the IDHPG (by 98±1%, n=9, P<0.001, Figure 3d) of DA neurons, similar to NASPM effects on the GluD1 current in HEK cells. The I–V curves of mGlu1/5-gated currents in DA neurons have been earlier described29, 30 and resemble that of the GluD1 current in HEK cells. Indeed, we observed that the I–V curves of the sEPSC (n=8) and the IDHPG (n=4) were both roughly linear with a reversal potential around 0 mV and exhibited inward rectification at positive potentials (Figure 3e), in agreement with previous reports.29, 30 Hence, the pharmacological and biophysical properties of mGlu1/5-gated currents in DA neurons point to the key contribution of GluD1 channels.
We assessed the dependence of mGlu1/5-gated currents on the GluD1 channel pore by co-expressing the dominant-negative GluD1VR pore mutant and GFP in DA neurons through viral transfer with a Sindbis vector in the SNc of WT mice. We found that the sEPSC was largely reduced in DA neurons co-expressing GluD1VR and GFP (9±2 pA, n=7) compared to DA neurons expressing GFP alone (110±20 pA, n=6, P<0.001, Figure 3f). The IDHPG was also hampered by GluD1VR and GFP co-expression in DA neurons (34±21 pA, n=6) compared to DA neurons expressing GFP alone (103±22 pA, n=7, P<0.03, Figure 3f). The sparse transduction achieved using sindbis viral transfer in the SNc (Figure 3f) suggests that GluD1VR expression suppresses mGlu1/5-gated currents in DA neurons via a cell-autonomous effect. These results indicate that mGlu1/5-gated currents in DA neurons critically depend on ion flow through GluD1 channels.
Hence, our pharmacological, biophysical and genetic disruption analyses show that native GluD1 channels are opened by activation of mGlu1/5 and are a key determinant of the sEPSC and the IDHPG in DA neurons.
GluD1 controls the burst firing of DA neurons in vivo
The firing of DA neurons in vivo consists in either single spikes or bursts of action potentials.56 It is known that activation of mGlu1/5 promotes bursting of DA neurons in brain slices and increases their bursting frequency in vivo.26, 27, 28 We thus explored the consequences of genetic disruption of the GluD1 channel on the firing of DA neurons in vivo using juxtacellular recordings in anesthetized mice. DA neurons were identified from their electrophysiological hallmarks56 (Supplementary Methods). Furthermore, some recorded neurons were electroporated with neurobiotin for post hoc immunohistochemistry. Among all recorded neurons, those successfully labeled for neurobiotin in the SNc (n=27/124) and the VTA (n=43/230) were all TH-positive (Figures 4b and d, Figure 5c and Supplementary Figure S6A), assessing a low misidentification rate of DA neurons.
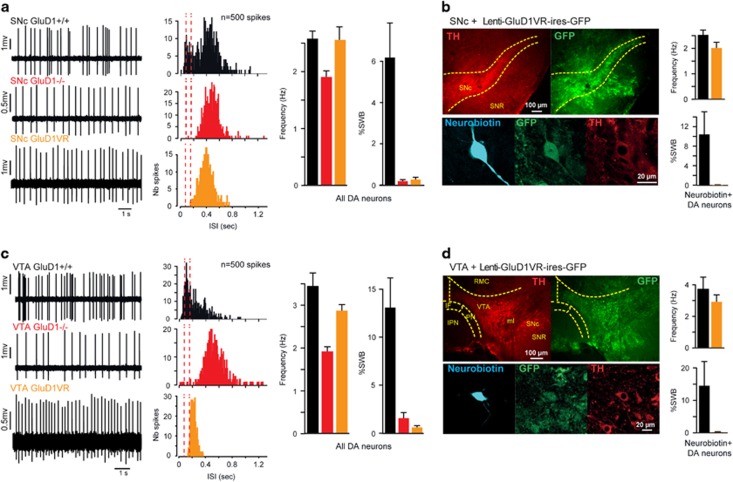
Figure 4.
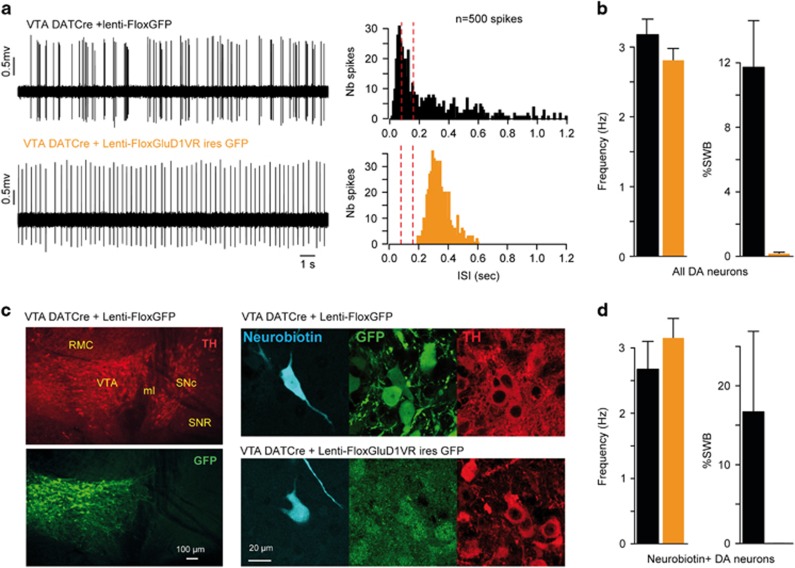
GluD1 controls the burst firing of DA neurons in vivo. (a,c) Examples of spontaneous spike firing patterns (left panels) and corresponding histograms of ISI (middle panels, red dotted lines indicate the limits of burst onset <80 ms and termination >160 ms) obtained from DA neurons recorded in GluD1+/+ and GluD1−/− mice and in WT mice injected with a lentivirus co-expressing GluD1VR and GFP in the SNc or VTA. Right panels show the mean spike frequency and %SWB of recorded DA neurons. Note that burst firing of DA neurons is virtually abolished by global (GluD1−/−) or local (GluD1VR) genetic disruption of GluD1. (b,d) Left panels: Histological analysis of virally transduced DA neurons recorded 4–6 weeks after injection of lentivirus LV-PGK-GluD1V617R-ires-GFP and filled with neurobiotin. DA, dopamine; IF, interfascicular nucleus; IPN, interpeduncular nucleus; ISI, interspike intervals; ml, medial lemniscus; PN, paranigral nucleus; RMC, red nucleus; SNc, SN pars compacta; SNR, substantia nigra pars reticulata; VTA, ventral tegmental area; WT, wild type; %SWB, percentage of spikes within burst. Right panels: same analyses as in (a,c) restricted to neurobiotin+/TH+ (GluD1+/+) and neurobiotin+/GFP+/TH+ (GluD1VR) recorded neurons.
Figure 5.
Selective expression of GluD1VR mutant in DA neurons suppresses burst firing. (a) Examples of spontaneous spike firing patterns (left panels) and corresponding histograms of ISI (middle panels) obtained from VTA DA neurons recorded in DAT-Cre mice after injection of a Cre-dependent lentivirus expressing either GFP, or GluD1VR and GFP, in the VTA. (b) Mean spike frequency and percentage of spikes within burst of recorded DA neurons. Note that selective expression of GluD1VR in DA neurons abolishes burst firing without significant effect on firing frequency. (c) Left: Whole field fluorescence micrographs showing selective expression of GFP in VTA TH+ neurons after viral transduction. Right: histological analysis of virally transduced DA neurons recorded 6 weeks after injection of indicated lentivirus and filled with neurobiotin. The oblique neurobiotin-labeled dendrite in lower left picture belongs to another GFP+/TH+ recorded neuron whose soma is out of field. (d) Same analyses as in (b) restricted to neurobiotin+/GFP+/TH+ recorded neurons. DA, dopamine; ISI, interspike intervals; VTA, ventral tegmental area;.
We first compared the firing pattern, measured as spike frequency and percentage of within burst (%SWB), of DA neurons from both the SNc and the VTA in GluD1−/− spikes and WT mice (Figures 4a and c and Supplementary Figure S7A). The spiking frequency of DA neurons recorded in WT mice (SNc: 2.6±0.1 Hz, n=45; VTA: 3.4±0.3 Hz, n=45) exhibited a modest but significant decrease in GluD1−/− mice (SNc: 1.9±0.1 Hz, n=39, P<0.001; VTA: 1.9±0.1 Hz, n=45, P<0.001; Figures 4a and c). Strikingly, the typical burst firing of DA neurons observed in WT mice (SNc: 6.2±1.7%SWB, n=45; VTA: 13.1±3.1%SWB, n=45) was abolished in GluD1−/− mice (SNc: 0.2±0.1%SWB, n=39, P<0.001; VTA: 1.6±0.6%SWB, n=45, P<0,001; Figures 4a and c). Hence, GluD1 deletion hampers burst firing of DA neurons, consistent with above observations on the critical role of GluD1 in mGlu1/5-dependent currents and with the involvement of mGlu1/5 in the burst firing of these neurons.26, 27, 28
We next tested the dependence of burst firing on the GluD1 channel pore by co-expressing the dominant-negative GluD1VR mutant and GFP in DA neurons via injection of a lentiviral vector in the SNc or VTA of WT mice (Figures 4b and d). All of the recorded neurons successfully labeled for neurobiotin in the SNc (n=14/40) and the VTA (n=10/71) of injected mice were both GFP- and TH-positive (Figures 4b and d). We found that expression of GluD1VR had no significant effect on the firing rate of DA neurons (SNc: 2.6±0.2 Hz, n=40, P=0.93; VTA: 2.9±0.1 Hz, n=71, P=0.09; Figures 4a and c), but abolished the burst firing of DA neurons in both the SNc (0.3±0.1%SWB, n=40, P<0,002) and the VTA (0.6±0.2%SWB, n=71, P<0,001; Figures 4a and b and Supplementary Figure S7A). Similar results were obtained in our subsets of neurobiotin-labeled control and GluD1VR-expressing DA neurons (SNc: n=13 and 14, VTA: n=15 and 10, respectively; Figures 4b and d) for spiking frequency (SNc control vs GluD1VR: 2.5±0.2 vs 2.0±0.2 Hz, P=0.07; VTA control vs GluD1VR: 3.7±0.8 vs 2.9±0.4 Hz, P=0.07) and burst firing (SNc control vs GluD1VR: 10.4±4.6 vs 0.04±0.03%SWB, P<0,01; VTA control vs GluD1VR: 14.5±7.5 vs 0.2±0.1%SWB, P<0,05). Conversely, transduction of VTA DA neurons with a control lentivirus expressing GFP alone did not significantly alter their firing rate (3.0±0.3 Hz, n=23, P=0.6) or burst firing (13.8±5.4%SWB, P=0.9; Supplementary Figure S6B). Hence, local GluD1VR expression in WT mice and global GluD1 deletion in GluD1−/− mice similarly hamper DA neuron burst firing, pointing to a cell-autonomous effect of GluD1 genetic disruption on DA neuron functions.
We assessed the cell-autonomous nature of GluD1VR effects on DA neurons using injection of Cre-dependent lentiviral vectors encoding both GluD1VR and GFP, or GFP alone (control), in the VTA of DAT-Cre mice. Selective expression of GluD1VR in VTA DA neurons did not significantly change firing frequency as compared to control (2.8±0.2 vs 3.2±0.2 Hz, n=28 vs 41, respectively; P=0.2) but abolished burst firing (0.2±0.1 vs 11.7±2.9%SWB, P<0.001; Figures 5a and b and Supplementary Figure S7B). Neurobiotin-labeled cells were all GFP- and TH-positive and yielded similar results (frequency: 2.7±0.4 vs 3.2±0.3 Hz, P=0.1; burst: 16.7 ±10.2 vs 0.01±0.01%SWB, P<0.01; n=7 and 7 for control and GluD1VR, respectively; Figures 5c and d). These observations indicate that the burst firing of DA neurons depends on ion flow through GluD1 channels expressed by these neurons, as observed above for mGlu1/5-dependent currents.
Our results show that GluD1 channels are a key determinant of the burst firing of DA neurons in vivo and suggest that GluD1 alterations can impair dopaminergic functions.
Discussion
We probed the existence of a functional coupling between mGlu1/5 and GluD1, and addressed its role in the sEPSC and the firing pattern of DA neurons. Our data indicate that the activation of mGlu1/5 triggers the opening of GluD1 channels and that these channels are essential to the sEPSC of DA neurons in midbrain slices and to the spontaneous burst firing of these neurons in vivo. These results establish a causal link between GRID1 gene alteration and dopaminergic dysfunction.
We found that the IDHPG obtained in HEK cells co-expressing mGlu1/5 and GluD1 is abolished by the mGlu1/5 antagonist AIDA and the GluD pore blocker NASPM, is prevented by the dominant-negative GluD1VR dead pore mutant, and is reduced by the GluD inhibitor D-serine. Moreover, mGlu1-5 co-immunoprecipitate with GluD1, and activation of other Gq-coupled receptors fail to induce GluD1-dependent currents in HEK cells. These observations show that mGlu1/5 activation triggers the opening of GluD1 channels in HEK cells, and suggest that this functional coupling depends on physical interaction between mGlu1/5 and GluD1. The lower potency of the above GluD antagonists on constitutively open GluD1 channel mutants51 suggests that such mutations alter the pharmacological profile of GluD1. Conversely, the voltage-dependence and pharmacological properties of the mGlu1-dependent GluD1 current closely resemble those described for GluD2,12 indicating that current reversal at ~0 mV, inward rectification at positive potential, as well as inhibition by NASPM and D-serine are fingerprints of the GluD channel family. Hence, our results de-orphanize GluD1 and indicate that both members of the GluD channels family are gated by mGlu1/5 activation and share common biophysical and pharmacological properties.
GluD1 is widely expressed in the brain16, 17 but its cellular distribution in the midbrain is unknown. We found that the GluD1 mRNA is expressed in single DA neurons together with the TH and the mGlu1/5 mRNAs. The GluD2 mRNA is also detected in single DA neurons, albeit at a lower frequency. Expression of GluD1 protein in DA neurons is assessed by our immunohistochemical experiments, and earlier reports established the presence of both mGlu1 and mGlu5 proteins in these neurons.25 We also found that GluD1 is localized at the postsynaptic density of excitatory synapses in DA neurons, as observed at hippocampal and cerebellar synapses.16, 17 These results show that GluD1 is present at excitatory postsynaptic sites in midbrain DA neurons and is co-expressed with mGlu1/5, consistent with a role of GluD1 in mGlu1-5-dependent synaptic excitation of these neurons.
The mGlu1/5-dependent currents in DA neurons are closely similar to those carried by GluD channels in HEK cells. Indeed, both the IDHPG and the slow EPSC reverse at ~0 mV, inwardly rectify at positive potentials, and are suppressed by AIDA, NASPM or expression of GluD1VR. The prominent involvement of GluD1 in both currents is confirmed by their strong reduction in GluD1−/− mice. The non-significant decrease of mGlu1/5-dependent currents in GluD2HO/HO mice may indicate that GluD2 homomers or GluD1-2 heteromers have a minor contribution to these currents, consistent with detection of the GluD2 mRNA in single DA neurons and its low abundance outside the cerebellum.16 Earlier reports provided pharmacological evidence for the involvement of transient receptor potential canonical channels in mGlu1/5-dependent currents of DA neurons.29, 57 This is reminiscent of the slow EPSC in cerebellar Purkinje cells that reportedly depends on both transient receptor potential canonicals and GluD2, which both belong to a complex also comprising mGlu1.12, 14, 58, 59 In the present case, the close resemblance of mGlu1/5-dependent currents in DA neurons and in HEK cells indicates that GluD1 is the major ion channel underlying these currents, but does not rule out a contribution of transient receptor potential canonical channels. GluD2 acts as a trans-synaptic scaffold through the interaction of its N-terminal domain with the presynaptic terminal.60, 61 Several observations indicate that disruption of such trans-synaptic scaffolding, which has not been reported for GluD1, is unlikely to be the cause of slow EPSC suppression upon GluD1 genetic inactivation. Indeed, (i) AMPA and NMDA components of fast EPSCs are not significantly altered in GluD1−/− mice, (ii) in our Sindbis viral transduction experiments, mGlu1-5 currents are hampered by the rapid, sparse expression of GluD1VR whose N-terminal domain is intact, and (iii) the IDHPG, which does not rely solely on synaptic GluD1, is similarly hampered by genetic inactivation. Hence, our results demonstrate that GluD1 is a critical determinant of slow glutamatergic transmission onto DA neurons and point to the direct involvement of the GluD1 pore in the postsynaptic current.
We found that the spontaneous burst firing of DA neurons in vivo is virtually abolished in anesthetized animals upon genetic inactivation of GluD1. The overall firing frequency of these neurons is almost unchanged upon local expression of GluD1VR in the SN/VTA or selective expression of GluD1VR in VTA DA neurons, but is significantly reduced by approximately one third in GluD1−/− mice, beyond the decrease expected from burst suppression (6–13%SWB in WT mice). This suggests that GluD1 controls the transition to burst firing via a cell-autonomous effect in DA neurons, whereas changes of the overall firing frequency in GluD1−/− mice may reflect larger scale circuit dysfunctions involving other brain structures where molecular alterations reportedly occur in GluD1−/− mice.7, 8, 16, 17 Our data further suggest a causal link between the GluD1-dependent slow EPSC and the burst firing of DA neurons, consistent with the involvement of mGlu1-5 in both phenomena.26, 27, 28, 30 Although the transition to burst firing relies on multiple inputs and intrinsic conductances,20, 21 the slow EPSC appears well-suited as a trigger of bursting that converts high frequency excitatory inputs into a long-lasting depolarization susceptible to enhancement or decrease upon integration of other inputs. Our results thus show that GluD1 critically controls the burst firing of DA neurons in vivo and indicate that this control results from the key contribution of GluD1 channels to the slow EPSC of these neurons.
Schizophrenia is characterized by episodic ‘positive’ symptoms like delusions and psychosis, and/or persistent ‘negative’ symptoms, such as social withdrawal and cognitive impairments. The ‘dopamine hypothesis’ of schizophrenia postulates that DA neuron firing and neurotransmission dysfunction22, 23 underlies these behavioral abnormalities. This hypothesis is based on the treatments given to patients but also on neurophysiological evidence. Indeed, patients with schizophrenia show reduced midbrain activation and mesocortical connectivity during reward processing,62, 63 consistent with alterations of DA neuron firing pattern. Likewise, an imbalance between tonic and burst firing of DA neurons has been linked to alterations of brain circuits and behaviors in a mouse model of schizophrenia.24 However, an alternative explanation of the pathology is the ‘glutamate dysfunction’ hypothesis, originally based on the observation that intoxication with the glutamate receptor antagonist phencyclidine mimics schizophrenic symptoms.18 Our work on the role of GluD1 in glutamate transmission onto DA neurons underlines the strong interconnection between glutamate and DA transmission abnormalities. By showing that genetic disruption of GluD1 hampers the burst firing of DA neurons, we provide an explanation of how genetic alterations of the GRID1 gene can impair glutamate transmission and lead to DA dysfunctions that are relevant for schizophrenia.
Acknowledgments
We thank Martine Soudan and the IBPS Cell Imaging Facility for their valuable help. We thank Jian Zuo, Carole Levenes and Carine Becamel for sharing GRID1 knockout, GRID2 hotfoot mouse lines and the 5-HT2C plasmid, respectively. This work was supported by grants from Agence Nationale de la Recherche (ANR# BLAN-SVSE4-LS-110624 to BL and ANR-16-CE16-0014-01 to LT), Brain and Behavior Research foundation (2014 and 2016 NARSAD Young Investigator grant to LT), Fondation pour la Recherche Médicale to PF, Fondation Bettencourt Schueller to PF, Fondation pour la Recherche sur le Cerveau/Rotary Club de France to BL and PF, Ecole des Neurosciences de Paris (‘Network for Viral Transfer’), and by grants from the Spanish Ministry of Education and Science (BFU2015-63769-R) and Junta de Comunidades de Castilla-La Mancha (PPII-2014-005-P) to RL. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
The authors declare no conflict of interest.
Supplementary Material
References
- Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet 2005; 77: 918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ et al. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry 2011; 168: 930–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SZ, Huang K, Shi YY, Tang W, Zhou J, Feng GY et al. A case-control association study between the GRID1 gene and schizophrenia in the Chinese Northern Han population. Schizophr Res 2007; 93: 385–390. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Muhleisen TW, Frank J, Mattheisen M, Herms S, Ludwig KU et al. Dissection of phenotype reveals possible association between schizophrenia and Glutamate Receptor Delta 1 (GRID1) gene promoter. Schizophr Res 2009; 111: 123–130. [DOI] [PubMed] [Google Scholar]
- Gao J, Maison SF, Wu X, Hirose K, Jones SM, Bayazitov I et al. Orphan glutamate receptor delta1 subunit required for high-frequency hearing. Mol Cell Biol 2007; 27: 4500–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsom JN, Wong AH. Schizophrenia and depression co-morbidity: what we have learned from animal models. Front Psychiatry 2015; 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Gupta SC, Hillman BG, Bhatt JM, Stairs DJ, Dravid SM. Deletion of glutamate delta-1 receptor in mouse leads to aberrant emotional and social behaviors. PLoS ONE 2012; 7: e32969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Hillman BG, Gupta SC, Suryavanshi P, Bhatt JM, Pavuluri R et al. Deletion of glutamate delta-1 receptor in mouse leads to enhanced working memory and deficit in fear conditioning. PLoS ONE 2013; 8: e60785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SM, Hollmann M. To gate or not to gate: are the delta subunits in the glutamate receptor family functional ion channels? Mol Neurobiol 2008; 37: 126–141. [DOI] [PubMed] [Google Scholar]
- Araki K, Meguro H, Kushiya E, Takayama C, Inoue Y, Mishina M. Selective expression of the glutamate receptor channel delta 2 subunit in cerebellar Purkinje cells. Biochem Biophys Res Commun 1993; 197: 1267–1276. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Sprengel R, Laurie DJ, Kohr G, Herb A, Seeburg PH et al. The rat delta-1 and delta-2 subunits extend the excitatory amino acid receptor family. FEBS Lett 1993; 315: 318–322. [DOI] [PubMed] [Google Scholar]
- Ady V, Perroy J, Tricoire L, Piochon C, Dadak S, Chen X et al. Type 1 metabotropic glutamate receptors (mGlu1) trigger the gating of GluD2 delta glutamate receptors. EMBO Rep 2014; 15: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadak S, Bouquier N, Goyet E, Fagni L, Levenes C, Perroy J. mGlu1 receptor canonical signaling pathway contributes to the opening of the orphan GluD2 receptor. Neuropharmacology 2016; 115: 92–99. [DOI] [PubMed] [Google Scholar]
- Kato AS, Knierman MD, Siuda ER, Isaac JT, Nisenbaum ES, Bredt DS. Glutamate receptor delta2 associates with metabotropic glutamate receptor 1 (mGluR1), protein kinase Cgamma, and canonical transient receptor potential 3 and regulates mGluR1-mediated synaptic transmission in cerebellar Purkinje neurons. J Neurosci 2012; 32: 15296–15308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi PS, Gupta SC, Yadav R, Kesherwani V, Liu J, Dravid SM. Glutamate delta-1 receptor regulates metabotropic glutamate receptor 5 signaling in the hippocampus. Mol Pharmacol 2016; 90: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepp R, Hay YA, Aguado C, Lujan R, Dauphinot L, Potier MC et al. Glutamate receptors of the delta family are widely expressed in the adult brain. Brain Struct Funct 2015; 220: 2797–2815. [DOI] [PubMed] [Google Scholar]
- Konno K, Matsuda K, Nakamoto C, Uchigashima M, Miyazaki T, Yamasaki M et al. Enriched expression of GluD1 in higher brain regions and its involvement in parallel fiber-interneuron synapse formation in the cerebellum. J Neurosci 2014; 34: 7412–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laruelle M, Kegeles LS, Abi-Dargham A. Glutamate, dopamine, and schizophrenia: from pathophysiology to treatment. Ann N Y Acad Sci 2003; 1003: 138–158. [DOI] [PubMed] [Google Scholar]
- Grace AA, Bunney BS. Intracellular and extracellular electrophysiology of nigral dopaminergic neurons—1. Identification and characterization. Neuroscience 1983; 10: 301–315. [DOI] [PubMed] [Google Scholar]
- Faure P, Tolu S, Valverde S, Naude J. Role of nicotinic acetylcholine receptors in regulating dopamine neuron activity. Neuroscience 2014; 282C: 86–100. [DOI] [PubMed] [Google Scholar]
- Paladini CA, Roeper J. Generating bursts (and pauses) in the dopamine midbrain neurons. Neuroscience 2014; 282C: 109–121. [DOI] [PubMed] [Google Scholar]
- Grace AA. Phasic versus tonic dopamine release and the modulation of dopamine system responsivity: a hypothesis for the etiology of schizophrenia. Neuroscience 1991; 41: 1–24. [DOI] [PubMed] [Google Scholar]
- Stopper CM, Floresco SB. Dopaminergic circuitry and risk/reward decision making: implications for schizophrenia. Schizophr Bull 2015; 41: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krabbe S, Duda J, Schiemann J, Poetschke C, Schneider G, Kandel ER et al. Increased dopamine D2 receptor activity in the striatum alters the firing pattern of dopamine neurons in the ventral tegmental area. Proc Natl Acad Sci USA 2015; 112: E1498–E1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert GW, Paquet M, Smith Y. Differential subcellular localization of mGluR1a and mGluR5 in the rat and monkey Substantia nigra. J Neurosci 2001; 21: 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer LT, Serpa KA, Christoffersen CL. Metabotropic glutamate receptor-mediated inhibition and excitation of substantia nigra dopamine neurons. Synapse 1997; 26: 184–193. [DOI] [PubMed] [Google Scholar]
- Prisco S, Natoli S, Bernardi G, Mercuri NB. Group I metabotropic glutamate receptors activate burst firing in rat midbrain dopaminergic neurons. Neuropharmacology 2002; 42: 289–296. [DOI] [PubMed] [Google Scholar]
- Zheng F, Johnson SW. Group I metabotropic glutamate receptor-mediated enhancement of dopamine cell burst firing in rat ventral tegmental area In vitro. Brain Res 2002; 948: 171–174. [DOI] [PubMed] [Google Scholar]
- Bengtson CP, Tozzi A, Bernardi G, Mercuri NB. Transient receptor potential-like channels mediate metabotropic glutamate receptor EPSCs in rat dopamine neurones. J Physiol 2004; 555: 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen KZ, Johnson SW. A slow excitatory postsynaptic current mediated by G-protein-coupled metabotropic glutamate receptors in rat ventral tegmental dopamine neurons. Eur J Neurosci 1997; 9: 48–54. [DOI] [PubMed] [Google Scholar]
- Perroy J, Raynaud F, Homburger V, Rousset MC, Telley L, Bockaert J et al. Direct interaction enables cross-talk between ionotropic and group I metabotropic glutamate receptors. J Biol Chem 2008; 283: 6799–6805. [DOI] [PubMed] [Google Scholar]
- Ango F, Albani-Torregrossa S, Joly C, Robbe D, Michel JM, Pin JP et al. A simple method to transfer plasmid DNA into neuronal primary cultures: functional expression of the mGlu5 receptor in cerebellar granule cells. Neuropharmacology 1999; 38: 793–803. [DOI] [PubMed] [Google Scholar]
- Labasque M, Reiter E, Becamel C, Bockaert J, Marin P. Physical interaction of calmodulin with the 5-Hydroxytryptamine(2C) Receptor C-Terminus Is essential for G protein-independent, arrestin-dependent receptor signaling. Mol Biol Cell 2008; 19: 4640–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Yamada N, Kakegawa W, Tsuzuki K, Kawamura M, Nawa H et al. Sindbis viral-mediated expression of Ca2+-permeable AMPA receptors at hippocampal CA1 synapses and induction of NMDA receptor-independent long-term potentiation. Eur J Neurosci 2001; 13: 1635–1643. [DOI] [PubMed] [Google Scholar]
- Maskos U, Molles BE, Pons S, Besson M, Guiard BP, Guilloux JP et al. Nicotine reinforcement and cognition restored by targeted expression of nicotinic receptors. Nature 2005; 436: 103–107. [DOI] [PubMed] [Google Scholar]
- Tolu S, Eddine R, Marti F, David V, Graupner M, Pons S et al. Co-activation of VTA DA and GABA neurons mediates nicotine reinforcement. Mol Psychiatry 2013; 18: 382–393. [DOI] [PubMed] [Google Scholar]
- Tolu S, Avale ME, Nakatani H, Pons S, Parnaudeau S, Tronche F et al. A versatile system for the neuronal subtype specific expression of lentiviral vectors. FASEB J 2010; 24: 723–730. [DOI] [PubMed] [Google Scholar]
- Drobac E, Tricoire L, Chaffotte AF, Guiot E, Lambolez B. Calcium imaging in single neurons from brain slices using bioluminescent reporters. J Neurosci Res 2010; 88: 695–711. [DOI] [PubMed] [Google Scholar]
- Turiault M, Parnaudeau S, Milet A, Parlato R, Rouzeau JD, Lazar M et al. Analysis of dopamine transporter gene expression pattern - generation of DAT-iCre transgenic mice. FEBS J 2007; 274: 3568–3577. [DOI] [PubMed] [Google Scholar]
- Lalouette A, Lohof A, Sotelo C, Guenet J, Mariani J. Neurobiological effects of a null mutation depend on genetic context: comparison between two hotfoot alleles of the delta-2 ionotropic glutamate receptor. Neuroscience 2001; 105: 443–455. [DOI] [PubMed] [Google Scholar]
- Cauli B, Audinat E, Lambolez B, Angulo MC, Ropert N, Tsuzuki K et al. Molecular and physiological diversity of cortical nonpyramidal cells. J Neurosci 1997; 17: 3894–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambolez B, Ropert N, Perrais D, Rossier J, Hestrin S. Correlation between kinetics and RNA splicing of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors in neocortical neurons. Proc Natl Acad Sci USA 1996; 93: 1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan R, Nusser Z, Roberts JD, Shigemoto R, Somogyi P. Perisynaptic location of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur J Neurosci 1996; 8: 1488–1500. [DOI] [PubMed] [Google Scholar]
- Bonnot A, Guiot E, Hepp R, Cavellini L, Tricoire L, Lambolez B. Single-fluorophore biosensors based on conformation-sensitive GFP variants. FASEB J 2014; 28: 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann GP, Yasuda RP, Xiao YX, Wolfe BB, Kellar KJ. Endogenously expressed muscarinic receptors in HEK293 cells augment up-regulation of stably expressed alpha 4 beta 2 nicotinic receptors. J Biol Chem 2011; 286: 39726–39737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancellin N, Preisser L, Le Maout S, Barbado M, Creminon C, Corman B et al. Homologous and heterologous phosphorylation of the vasopressin V1a receptor. Cell Signal 1999; 11: 743–751. [DOI] [PubMed] [Google Scholar]
- Schmid SM, Kott S, Sager C, Huelsken T, Hollmann M. The glutamate receptor subunit delta2 is capable of gating its intrinsic ion channel as revealed by ligand binding domain transplantation. Proc Natl Acad Sci USA 2009; 106: 10320–10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, Kuner T, Jatzke C, Seeburg PH, Heintz N, Zuo J. The Lurcher mutation identifies delta 2 as an AMPA/kainate receptor-like channel that is potentiated by Ca(2+). J Neurosci 2000; 20: 5973–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda K, Wang Y, Yuzaki M. Mutation of a glutamate receptor motif reveals its role in gating and delta2 receptor channel properties. Nat Neurosci 2000; 3: 315–322. [DOI] [PubMed] [Google Scholar]
- Naur P, Hansen KB, Kristensen AS, Dravid SM, Pickering DS, Olsen L et al. Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci USA 2007; 104: 14116–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Rimerman R, Scofield MA, Dravid SM. Mutations in the transmembrane domain M3 generate spontaneously open orphan glutamate delta1 receptor. Brain Res 2011; 1382: 1–8. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Hume RI, Heinemann SF. Structural determinants of barium permeation and rectification in non-NMDA glutamate receptor channels. J Neurosci 1992; 12: 4080–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa W, Kohda K, Yuzaki M. The delta2 'ionotropic' glutamate receptor functions as a non-ionotropic receptor to control cerebellar synaptic plasticity. J Physiol 2007; 584: 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CD, Shiroyama T, Kitai ST. Electrophysiological and immunocytochemical characterization of GABA and dopamine neurons in the substantia nigra of the rat. Neuroscience 1997; 80: 545–557. [DOI] [PubMed] [Google Scholar]
- Wolfart J, Neuhoff H, Franz O, Roeper J. Differential expression of the small-conductance, calcium-activated potassium channel SK3 is critical for pacemaker control in dopaminergic midbrain neurons. J Neurosci 2001; 21: 3443–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace AA, Bunney BS. Intracellular and extracellular electrophysiology of nigral dopaminergic neurons—2. Action potential generating mechanisms and morphological correlates. Neuroscience 1983; 10: 317–331. [DOI] [PubMed] [Google Scholar]
- Tozzi A, Bengtson CP, Longone P, Carignani C, Fusco FR, Bernardi G et al. Involvement of transient receptor potential-like channels in responses to mGluR-I activation in midbrain dopamine neurons. Eur J Neurosci 2003; 18: 2133–2145. [DOI] [PubMed] [Google Scholar]
- Hartmann J, Dragicevic E, Adelsberger H, Henning HA, Sumser M, Abramowitz J et al. TRPC3 channels are required for synaptic transmission and motor coordination. Neuron 2008; 59: 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Kim YS, Yuan JP, Petralia RS, Worley PF, Linden DJ. Activation of the TRPC1 cation channel by metabotropic glutamate receptor mGluR1. Nature 2003; 426: 285–291. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science 2010; 328: 363–368. [DOI] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 2010; 141: 1068–1079. [DOI] [PubMed] [Google Scholar]
- Gradin VB, Waiter G, O'Connor A, Romaniuk L, Stickle C, Matthews K et al. Salience network-midbrain dysconnectivity and blunted reward signals in schizophrenia. Psychiatry Res 2013; 211: 104–111. [DOI] [PubMed] [Google Scholar]
- Murray GK, Corlett PR, Clark L, Pessiglione M, Blackwell AD, Honey G et al. Substantia nigra/ventral tegmental reward prediction error disruption in psychosis. Mol Psychiatry 2008; 13: 239–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.