Abstract
The development of the cerebral cortex is a tightly regulated process that relies on exquisitely coordinated actions of intrinsic and extrinsic cues. Here, we show that the communication between forebrain meninges and apical neural progenitor cells (aNPC) is essential to cortical development, and that the basal compartment of aNPC is key to this communication process. We found that Celsr1, a cadherin of the adhesion G protein coupled receptor family, controls branching of aNPC basal processes abutting the meninges and thereby regulates retinoic acid (RA)-dependent neurogenesis. Loss-of-function of Celsr1 results in a decreased number of endfeet, modifies RA-dependent transcriptional activity and biases aNPC commitment toward self-renewal at the expense of basal progenitor and neuron production. The mutant cortex has a reduced number of neurons, and Celsr1 mutant mice exhibit microcephaly and behavioral abnormalities. Our results uncover an important role for Celsr1 protein and for the basal compartment of neural progenitor cells in fate decision during the development of the cerebral cortex.
Introduction
The cerebral cortex is the seat of higher brain functions and its formation requires the production and positioning of a right number and diversity of neurons for intricate circuits assembly. During early cortical development, neural stem cells (NSC) divide symmetrically in the ventricular zone (VZ) and both daughter cells re-enter the cell cycle, leading to an increased number of NSC. At the onset of neurogenesis, radial or apical (aNPC), the progenitors of excitatory cortical neurons, which derive from NSC, shift from a symmetric/proliferative to an asymmetric/differentiative mode of division. Asymmetric divisions produce aNPC as well as neurons, either directly or indirectly through basal/intermediate progenitors (BP).1 Each BP divides symmetrically to give rise to postmitotic neurons, thus increasing the final output of neurons.1, 2 Hence, a delicate balance between the proliferation and differentiation of aNPC must be maintained during neurogenesis. This balance is regulated by intrinsic and extrinsic factors at the level of individual aNPC, which undergo a rigorous sequence of fate choices. Errors in fate decisions often lead to devastating disorders that vary from severe and lethal malformations, to cognitive, social and motor disabilities.3
The Celsr1 gene codes for an atypical cadherin usually located at epithelial cells adherens junctions where it coordinates polarity in the plane of the epithelium, in cooperation with a set of conserved ‘core’ planar cell polarity (PCP) genes/proteins such as Vangl2 or Fzd3 and 6.4 Mice carrying mutations in Celsr1 have defects in neural tube closure,5, 6, 7 inner ear hair cells,6 cilia polarity,8, 9 as well as in cell movement and rearrangement.10, 11 In humans, mutations of CELSR1 are associated with craniorachischisis,12 the most severe form of neural tube closure defects. Despite the robust and maintained expression of Celsr1 in the developing forebrain germinal zones,13 its function, after neural tube closure, has not been assessed.
Here, we report that at the onset of neurogenesis, the Celsr1 protein localizes to the endfeet of aNPC and has an important role in shaping their basal compartment. Compared with those of control mice, Celsr1-deficient aNPC endfeet have fewer contact with the basal lamina and are less accessible to retinoic acid (RA) delivered by meningeal cells. They undergo more proliferative divisions, thereby expanding the pool of aNPC at the expense of BP and neuron production, and leading to marked morphological and behavioral defects. Finally, we provide evidence that RA signaling controls fate decision of neural progenitors.
Materials and methods
Mutant mice and in vivo treatment
All procedures were carried out in accordance with European guidelines and approved by the animal ethics committee of the Université catholique de Louvain. Mouse lines used in this study are: Celsr1−/− and Celsr1f/f,7 Nestin-CreERT (C57BL/6-Tg(Nes-cre/ERT2)KEisc/J; Jackson Lab, Bar Harbor, ME, USA), Rosa-dTomato (B6.CgGt(ROSA)26Sortm9(CAG-dTomato)Hze/J; Jackson Lab), Dlx5/6-Cre-IRES-eGFP (Tg(mI56i-cre,EGFP)1Kc/J; Jackson Lab) and Emx1-Cre (B6.Cg-Emx1tm1(cre)Krj/J; Jackson Lab). For in utero RA supplementation, pregnant females were subjected to a daily intraperitoneal injection of 20 mg kg−1 of RA. Cell fate experiments in Celsr1−/−;Nestin-CreERT2; lox-STOP-lox dTomato mice were carried out at e14.5 after a single 4-Hydroxytamoxifen injection (1.25 mg) in pregnant female at e13.5. Clonal analyses in Celsr1f/f;Nestin-CreERT2;lox-STOP-lox dTomato embryos were performed at e14.5 after a single 4-Hydroxytamoxifen injection at e10.5 (0.3 mg per pregnant female).
Behavioral tests
All the mice were tested at 3 months of age. The open-field test was used to assess a non-forced ambulation as mice can move freely without any influence of the examiner. Mice were placed in a square arena (60 × 60 cm) and video tracked (Ethovision 6.1, Noldus; Wageningen, The Netherlands) for 20 min. The total distance covered by animals and the time spent in the center versus periphery were measured.
The elevated plus maze test was used to assess anxiety. Mice were placed in an elevated plus maze consisting of two opposing open arm (exposed place) and two opposing closed arm (safer place). Time spent in each arm and distance covered were recorded by a video tracking system for 5 min.
Morris water maze was used to assess spatial learning and memory. Water maze was made of a round pool with a diameter of 113 cm virtually divided into four quadrants (North, South, West and East) and filled with water (26 C). Several visual cues were placed around the pool. Mice were tested during 4 days with three consecutive trials per day. During the trials, animals were placed in the pool facing the sidewall and allowed to swim freely to the platform visible the first day and submerged 1 cm under the water surface for the 3 next days. The platform was placed at the center of the North-East quadrant of the pool and maintained in this position throughout the 4 days. The initial position in which the animal was left in the pool varied among trials. If the animal did not find the platform during a period of 60 s, it was gently guided to it. Then, it was allowed to remain on the platform for 5 s and removed from the pool before being placed in the next initial starting position in the pool. On the 5th day, the animals were submitted to a probe test that consisted in allowing the animals to swim freely for 1 min in the pool without the escape platform. During the trial and the probe test, mice were video-tracked. The time latency to reach the platform, the swim speed, and the time spent in each quadrant were measured.
The three-chamber test was used to assess sociability. The test box was divided into three equal compartments (20 cm each), and dividing walls had retractable doorways allowing access into each chamber. First, for habituation, test mice were placed in the middle chamber for 5 min with the doorways closed. Then, to evaluate sociability, the test mice were placed in the central compartment and an unfamiliar mouse, called stranger, was restricted in a wire cage placed in one of the side compartments, whereas the other side compartment contains an empty wire cage. The test mice were video tracked for 10 min. The total distance covered by the animals and the time spent in each chamber were measured.
Immunohistochemistry
We used 12 μm-thick cryosections from embryos, fixed 30–45 min in 4% paraformaldehyde and 50 μm-thick slices (sliding microtome) from postnatal brains fixed with 4% paraformaldehyde. Sections were blocked in PBS supplemented with 0.5% Triton X-100 and 3% Bovine serum albumin, and incubated with the following primary antibodies: Celsr1 (homemade, 1/500), Laminin (Sigma-Aldrich, Overijse, Belgium, Cat no. L9393, 1/50), Nestin (Chemicon/Millipore, Overijse, Belgium, Cat no. MAB353, 1/1000), Pax6 (Covance, Cat no. MAB353, 1/500), Tbr2 (Abcam, Brussels, Belgium, Cat no. ab23345, 1/500), Tuj1 (Covance, Cat no. MMS-435P-0250, 1/500), Crabp2 (Abcam, Cat no. ab74365, 1/100), Sox2 (Millipore, Cat no. AB5603, 1/500), Mash1 (BD Pharmigen, Erembodegem, Belgium, Cat no. 556604, 1/200), phospho-Vimentin (Abcam, Cambdrige, UK, Cat no. ab22651, 1/100), Satb2 (Abcam, Cat no. ab51502, 1/1000), Cux1 (Santa Cruz, Santa Cruz, CA, USA, at no. SC-13024, 1/500), Foxp2 (Abcam, Cat no. ab16046, 1/1000) and Ctip2 (Abcam, Cat no. ab18465, 1/500).
Whole-mount preparation of the ventricular wall
Lateral ventricular walls were dissected and immediately immersed in PBS as described previously.8 Whole-mount preparations were fixed in 4% paraformaldehyde and 0.1% Triton X-100 for 12 min, washed three times in PBS-0.1% Triton X-100, blocked in PBS supplemented with 0.5% Triton X-100 and 3% BSA, and incubated with β-Catenin antibody (Cell Signaling, Danvers, MA, USA, Cat no. 9581, 1/500), ZO-1 (Invitrogen, Brussels, Belgium, Cat no. 61-7300, 1/200), γ-Tubulin (Abcam, Cat no. ab11317, 1/500) and GFAP (Millipore, Cat no. AB5804, 1/1000).
In utero electroporation
E14.5 embryos were electroporated with 1 μg pCAG-Gfp and collected at e15.5. Injection needles were pulled from Wiretrol II glass capillaries (Drummond Scientific, Broomall, PA, USA) and calibrated for 1 μl injections. Plasmid DNAs were resuspended in 10 mM Tris, pH 8.0, with 0.01% Fast Green. Forceps-type electrodes (Nepagene, Sonidel, Dublin, Ireland) with 5 mm pads were used for electroporation (five 50 ms pulses of 25 V at E12.5 or five 50 ms pulses of 40 V at E14.5). Newborn (P0) mice were electroporated with a pcx-Gfp plasmid as previously described.8
Real-time PCR
Total mRNA was isolated from e14.5 telencephalon using RNeasy mini kit according to the supplier’s instructions. Reverse transcription was carried out using a RT cDNA synthesis Kit, 1 μg of total RNA in a total reaction volume of 20 μl (Qiagen, Antwerp, Belgium), SYBR green super mix, and an iCycler real-time PCR detection system (Bio-Rad, Gent, Belgium).
FACS
For aNPC sorting experiments, cortices of e14.5 control and mutant were processed in parallel. Single cell suspensions were produced using the MACS Neural Tissue Dissociation kit containing papain (MiltenyiBiotec, Paris, France) following the manufacturer’s instructions. Cell viability (typically >95%) was assessed with Trypan blue staining. Cell surface staining of CD133 (prominin1) was performed on cell suspensions with rat anti-13A4-APC conjugated (1:100, eBioscience, Brussels, Belgium, Clone 13A4). Cells were incubated with CD133 antibody on ice for 60 min in PBS (1000 μl final volume per brain). Rat IgG-APC (1:100, eBioscience) was used as isotype controls. Cells were sorted according to their APC staining. FACS were performed on a BD FACSAria III sorter (BD Bioscience), and analyzed using the FACS Diva software (BD Bioscience).
Stranded RNA-Seq on purified aNPC
Total RNA from FACS purified e14.5 CD133-positive aNPC was extracted using Reliaprep RNA miniprep system (Promega, Leiden, Netherlands). Two brains of each genotype were pooled to get one biological replicate. The sequencing was performed by Beckman Coulter Genomics. Total RNA was further purified by ribodepletion and cDNA synthesis and library construction was performed on an automated Illumina TruSEQ stranded RNA-Seq library construction with a Biomek liquid handling platform. Detection of sequencing fragments was performed using the Illumina HiSeq 2500 platform. Alignment was made with Bowtie/Tophat, gene counts with Cufflinks and expression values/differential expression with edgeR. The sequencing output was on average 70.106 reads per sample.
Quantification
For quantification on coronal sections, pictures were acquired so as to cover the entire VZ/SVZ (subventricular zone) or all cortical layers. Both hemispheres were analyzed. ImageJ was used for counts and measurements.
Statistics
For pairwise analysis of treatment conditions or genotypes, Student’s t-tests were used. For three chamber test, one-way analysis of variance (ANOVA) was used. For cell population quantification during development, effect of treatment on different genotypes and water maze, two-way ANOVA and Bonferoni post-test were used. Data are represented as mean±s.e.m. Chi contingency test was used for distribution analyses.
Results
Loss of Celsr1 promotes self-renewal of progenitors at the expense of neurogenesis
Celsr1 mRNA is specifically expressed by neural progenitors in mouse and human germinal zones13 (Supplementary Figure 1a, b; adapted from http://brainspan.org14). We used a tamoxifen inducible system to label aNPC and trace their progeny. Tamoxifen was injected to pregnant females at e13.5, and Celsr1−/−;NestinCreERT2;loxP-stop-loxP-dTomato embryos were collected at e14.5 and compared to control littermates (Celsr1+/+;NestinCreERT2;loxP-stop-loxP-dTomato). Mutant aNPC generated more Tomato-positive cells in the VZ and less cells in the SVZ than controls (Supplementary Figure 2a–c; percentage of Tomato+ cells in VZ: Wild type=53.6%, Celsr1−/−=70.2%, n=3, P=0.0008, Student’s t-test). This observation was corroborated by in utero electroporation of a Gfp coding plasmid at e14.5, which disclosed an accumulation of Gfp- and Pax6-positive cells in the mutant versus control VZ at e15.5 (Figures 1a–c, percentage of Pax6+ cells in Gfp cell population: Wild type=66.3%, Celsr1−/−=72.7%, n=4, P=0.0064, Student’s t-test). To further investigate the effect of Celsr1 loss-of-function on fate decision, we injected low concentrations of tamoxifen to mice carrying a floxed allele (Celsr1f) at e10.5, with the aim to inactivate the gene in sparse aNPC and enable a clonal analysis. Examination of the ventricular surface at e14.5 detected larger clones in conditional mutants (Celsr1f/f;NestinCreERT2;loxP-stop-loxP-dTomato) than in controls (Celsr1+/+;NestinCreERT2;loxP-stop-loxP-dTomato) (Figures 1d–f).
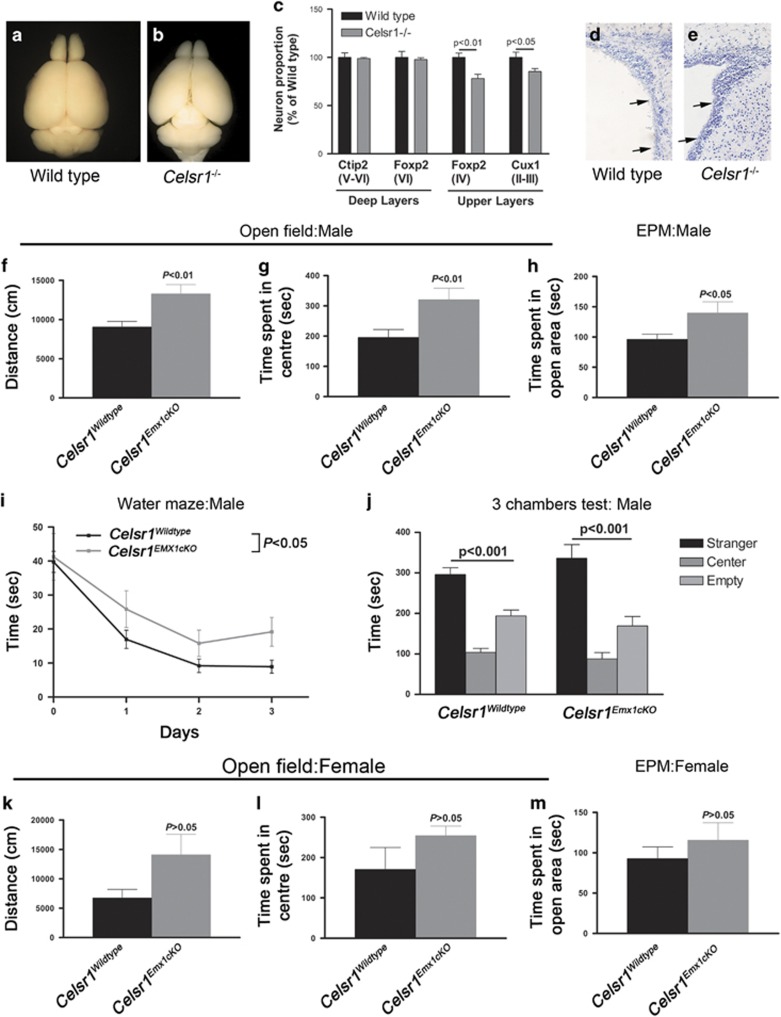
Figure 1.
Cortical phenotype of Celsr1 mutant mice. (a–c) Pax6 (red) and Gfp (green) staining of coronal brain sections from wild type (a) and Celsr1−/− (b) embryos, electroporated with a Gfp- coding plasmid at e14.5 and collected at e15.5. Scale bars 50 μm. (c) Quantification of Gfp-positive cells residing in the VZ, 24 h after electroporation. n=4 brains for each genotype, error bars are s.e.m., Student’s t-test, P<0.01. (d–f) clonal analysis of aNPC progeny in whole-mounts of the ventricular wall from Celsr1+/+;NestinCreERT2;dTomato (d) and Celsr1f/f;NestinCreERT2;dTomato (e) Scale bars 50 μm. (f) Quantification of the number of cells per clone. 18 % of mutant clones have more than 17 cell per clone (0 % in controls), n=6 whole-mounts for each genotype. (g–l) Immunochemistry for aNPC (Pax6) and BPs (Tbr2) at e12.5 (g, h), e14.5 (i, j) and e16.5 (k, l). Scale bars 50 μm. (m) Quantification of Tbr2-positive cells in lateral pallium at indicated stages. n=3 brains per stage and per genotype, bars represent s.e.m., two-way ANOVA, P<0.001.
These results suggest that Celsr1-deficient aNPC undergo more symmetric, proliferative divisions than their control counterparts, leading to accumulation of aNPC (Pax6+ cells) in the VZ. Reasoning that this could disrupt the radial versus tangential expansion of the developing cerebral cortex, we measured the length of the VZ (which reflects the expansion of the pool of progenitors) and found that it was similar in mutant and control embryos at e10.5, but increased significantly in mutant samples from e12.5 onwards (Supplementary Figure 2d, e, n=3 brains per genotype and per stage, P<0.001, two-way ANOVA). This increase resulted in a local distortion of the VZ at e16.5 (Supplementary Figure 2f, g), and correlated with a higher number of Pax6-positive aNPC in the VZ (Wild type=410.7±11.3, Celsr1−/−=452±14.5, n=3 brains per stage and per genotype, P<0.01, two-way ANOVA; Supplementary Figure 2h). During corticogenesis, aNPC produce Tbr2+ BP, which divide in the SVZ to scale up neuron production. Concomitant to the increased number of aNPC progenitors, the number of Tbr2-positive BP decreased in mutants (Figures 1g–m, Supplementary Figure 3a–n). The reduction of BPs was significant starting from e12.5 in the lateral pallium (Figure 1m, Tbr2+ cells in VZ/SVZ, wild type=366±72.6, Celsr1−/−=293.8±49.3, n=3, P<0.001, two-way ANOVA), and from e14.5 in the dorsal pallium (Supplementary Figure 3n, Tbr2+ cells in VZ/SVZ, Wild type=351.8±67.8, Celsr1−/−=307.9±26.5, n=3, P<0.05, two-way ANOVA). This was further confirmed by the observation of a decreased number of mitotic, Phospho-Vimentin+ cells in the SVZ at e14.5 (Supplementary Figure 4a–f, dorsal pallium: wild type=4.2±0.5, Celsr1−/−=2.3±0.4, n=3, P=0.0185, Student’s t-test; lateral pallium: Wild type=3.2±0.5, Celsr1−/−=1.5±0.3, n=3, P=0.0046, Student’s t-test).
Celsr1-deficient aNPC produce more adult NSCs
According to their localization in the developing telencephalon, aNPC cells produce cortical or striatal neurons. To test whether the effect of Celsr1 on fate decision is region specific, we characterized other neurogenic areas in the developing and adult brain. In absence of Celsr1, aNPC in ganglionic eminences, which generate striatal neurons and cortical interneurons, underwent more proliferative divisions than control ones, as illustrated by: (i) Increased number of Sox2-positive cells (Supplementary Figure 5a–e, Sox2+ cells in VZ/SVZ at e12.5: wild type=207±5.9, Celsr1−/−=271.3±11.7, n=3; at e16.5: Wild type=96.2±5.6, Celsr1−/−=161.9±5.6, n=3, P<0.0001, two-way ANOVA); (ii) Decreased number of Mash1+ BPs (Supplementary Figure 5a–d, f, Mash1+ cells in VZ/SVZ at e12.5: wild type=340.7±7.2, Celsr1−/−=312±9.6, n=3; at e16.5: wild type=128±5.8, Celsr1−/−=110.3±5.9, n=3, P<0.002, two-way ANOVA); and (iii) Narrowing of the intermediate zone containing Dlx5/6+ postmitotic neurons (Supplementary Figure 5g–i, width of Dlx5/6-Gfp+ area at e16.5, wild type=283.3±3.3 μm, Celsr1−/−=234±4.9 μm, n=3, P=0.0005, Student’s t-test).
In addition to postmitotic neurons, B1 cells, a pool of neural progenitors that remain quiescent until reactivation in the adult brain, are mainly derived from aNPC during early neurogenesis.15 In the adult VZ/SVZ, B1 cells display an epithelial apicobasal organization reminiscent of aNPC cells. The morphology of these cells was similar in Celsr1 mutants and controls. Following electroporation at early postnatal stages, Gfp-filled B1 cells extended from the ventricle apically (Supplementary Figure 6a, b) to the VZ/SVZ blood vessel plexuses basally (Supplementary Figure 6c, d). Similarly, B1 cells exhibited apical processes abutting the ventricle (Supplementary Figure 6e, f) and basal endfeet lining blood vessels (Supplementary Figure 6g, h) at P90. Whole-mount staining of the ventricular lateral wall showed that B1 cells expressing GFAP were surrounded by ependymal GFAP-negative cells (Supplementary Figure 6e, f), forming the so called pinwheel structure.16 Contrary to ependymal multiciliated cells, B1 cells are decorated by a single cilium. We used the basal body marker γ-tubulin to visualize the pinwheels and found that these structures were more abundant in mutant than in control mice at P30 and P90. (mean number of pinwheels per field (0.044 mm2) in the dorso-lateral wall at P30: Wild type=16.6±1.3, Celsr1−/−=28.2±1.5, n=6 whole mounts, P=0.0004, Student’s t-test, Supplementary Figure 7a–e; at P90: wild type=7.5±0.8, Celsr1−/−=12.8±1.5, n=6 whole mounts, P=0.0016, Student’s t-test, Supplementary Figure 7h–q). In addition, the mutant lateral wall contained larger clusters of B1 cells than control, both at P30 (Supplementary Figure 7f) and P90 (Supplementary Figure 7r); leading to a dramatic increase of the density of adult NSC (number of NSC per field in the dorso-lateral wall at P30: wild type=39.6±3.9, Celsr1−/−=104.4±7, n=6 whole mounts, P<0.0001; Student’s t-test, Supplementary Figure 7g; at P90: Wild type=16.4±1.7, Celsr1−/−=47.1±5, n=6 whole mounts, P<0.0001, Student’s t-test, Supplementary Figure 7s). Thus, in Celsr1 mutants, aNPC produce more adult NSCs (B1 cells), and less neurons than in controls.
Celsr1 mutant mice have cortical hypoplasia and disturbed behavior
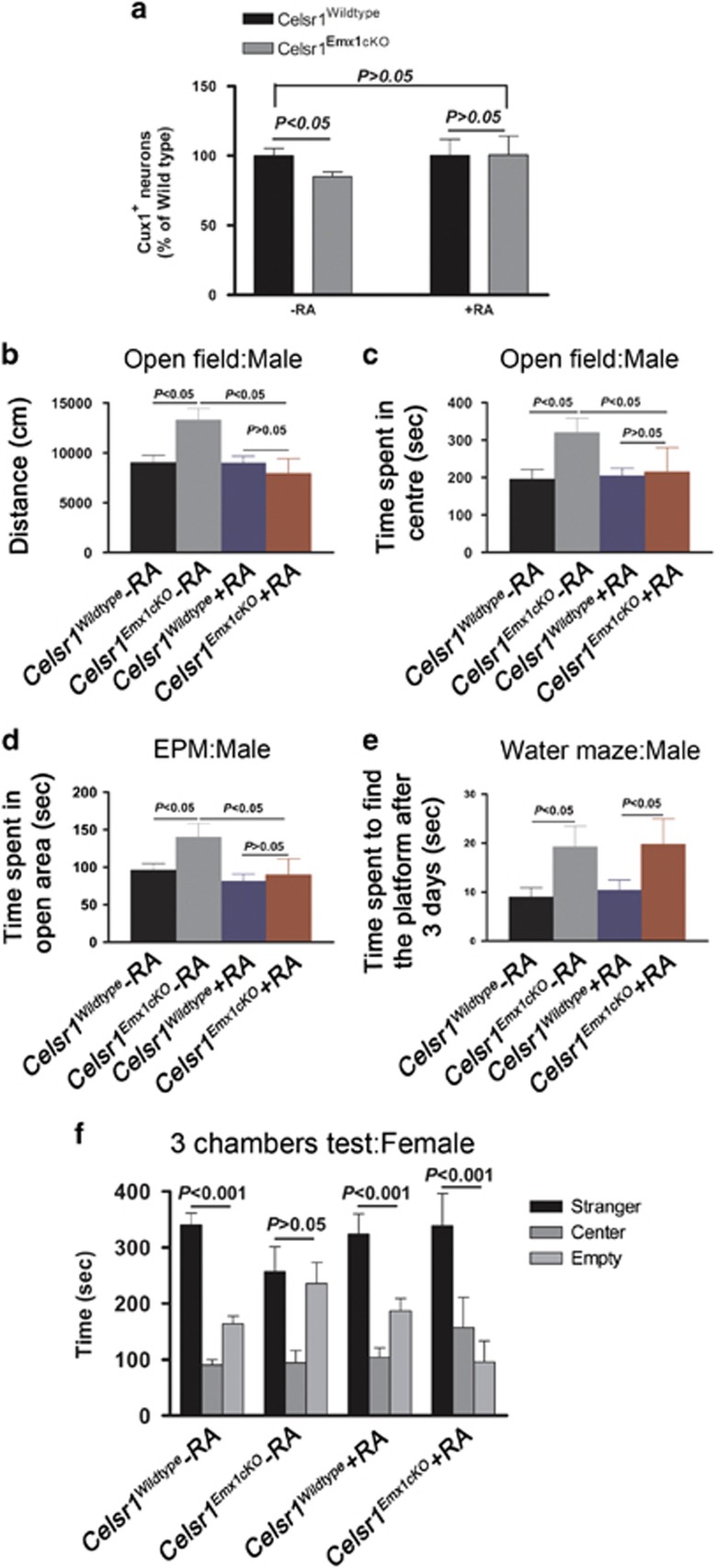
We measured brain weight at P0, P4, P8, P10 and P12, and found significant differences between Celsr1−/− and controls starting from P4 (Supplementary Figure 8a; wild type=171±7.6 mg, Celsr1−/−=138.6±2.5 mg, P=0.0002, n=15, Student’s t-test). At P90, the hypoplasia was particularly apparent in cerebral cortex and cerebellum, with unmasking of the superior and inferior colliculi (Figures 2a, b). Immunolabeling using layer specific markers showed a reduction in the number of Foxp2+ and Cux1+ neurons (Figure 2c, number of Foxp2+ cells (wild type=341±15.3, Celsr1−/−=266±15.1, n=3, P=0.0083; number of Cux1+ cells: wild type=644.4±34.8, Celsr1−/−=550±20, n=3, P=0.0467, Student’s t-test). In addition, histological examination of brain sections revealed that Celsr1−/− mice displayed a thicker VZ/SVZ (Figures 2d, e). Celsr1−/− adult mice exhibited turning behavior, a distinctive feature of inner ear/vestibular system dysfunction (Supplementary Movie 1). To investigate further the impact of loss of Celsr1 on cortical development, we selectively inactivated Celsr1 in neocortical areas by crossing mice carrying the Celsr1f allele with Emx1-Cre mice. Like the constitutive mutants, Celsr1f/f;Emx1-Cre conditional knockout mice (hereafter referred to as Celsr1Emx1cKO) displayed smaller cortices than controls (Celsr1+/+;Emx1-Cre) (Supplementary Figure 8b–e, weight of the Emx1-Cre-Tomato+ area: control males=87±3.3 mg, Celsr1Emx1cKO males=75±9.9 mg, n=3, P=0.0256; control females= 75.6±4.2 mg, Celsr1Emx1cKO females=59.1±2.4 mg, n=4, P=0.0138, Student’s t-test). We assessed the impact of these morphological changes on behavior. Celsr1Emx1cKO mice exhibited gender-specific defects in activity, attention, anxiety, spacial learning and social interactions (Figures 2f–o). Males displayed more spontaneous motor activity and spent more time than controls in exposed areas in open field and elevated plus maze (distance covered in open field: controls=8220±726.2 cm, Celsr1Emx1cKO=13670±1747 cm, n=14 controls and 10 Celsr1Emx1cKO, P=0.0040, Student’s t-test, Figure 2f; time spent in exposed areas in open field: controls=186.7±24.7 s, Celsr1Emx1cKO=287.5±23.7 s, P=0.0096, Student’s t-test Figure 2g; time spent in open area in elevated plus maze, controls=94.95±7.2 s, Celsr1Emx1cKO=127.6±14 s, P=0.0352, Student’s t-test, Figure 2h). The hiding behavior defect suggests attention and or anxiety deficit in addition to hyperactivity. Males also exhibited a spatial learning deficit in the water maze setup (time spent to find the platform controls at day 3=8.9±1.9 sec, Celsr1Emx1cKO=19.2±4.2 sec, n=5–9 animals per genotype, P<0.05, two-way ANOVA, Figure 2i), but no social behavior defect in the ‘three-chamber’ test (time spent in empty chamber controls=193.9±14.7 sec, with stranger=296.2±16.5 sec, Celsr1Emx1cKO in empty chamber=169.1±23.3 sec, with stranger=336.3±33.5 sec, n=5–9 animals per genotype, P<0.001 for both genotype, one-way ANOVA, Figure 2j).
Figure 2.
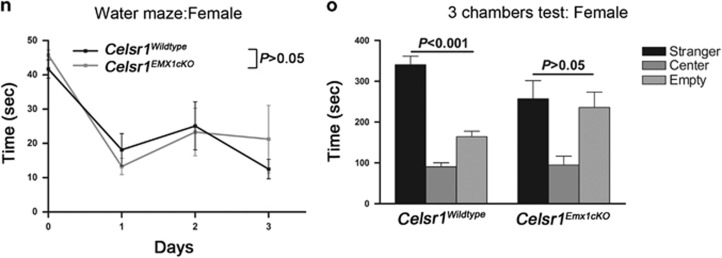
Cortical hypoplasia and abnormal behavior in Celsr1-deficient mice. (a,b) Mature brains from Wild type (a) and Celsr1−/− (b) mice. Note the reduced size in the mutant. (c) Quantification of neurons of different cortical layers. n=3 brains per genotype, bars represent s.e.m., Student’s t-test, P<0.05 for Cux1-positive cells (layers II and III); and P<0.01 for Foxp2-positive cells (layer IV). The number of cells in the Wild type is set to 100%. (d, e) Nissl staining of the adult VZ/SVZ in wild type (d) and Celsr1−/− (e). (f, g) Activity of males in open field. Bars represent s.e.m., Student’s t-test, P<0.01. (h) Time spent by males in open areas of elevated plus maze (EPM). Bars represent s.e.m., Student’s t-test, P<0.05; (i) Learning ability of males in the Water maze. Bars represent s.e.m., Student’s t-test, P<0.05. (j) Social behavior in the ‘3 chamber’ test for males. Bars represent s.e.m., one-way ANOVA, P<0.001 for both genotypes (stranger versus empty chamber). (k, l) Activity of females in open field. Bars represent s.e.m., Student’s t-test, P>0.05. (m) Time spent by females in open areas of elevated plus maze. Bars represent s.e.m., Student’s t-test, P>0.05. (n) Learning ability of females in the Water maze. Bars represent s.e.m., Student’s t-test, P>0.05 (o) Social behavior in the ‘3 chamber’ test for females. Bars represent s.e.m., one-way ANOVA, P<0.001 control females, P>0.05 for mutant females (stranger versus empty chamber). n=14 controls (Celsr1+/+;Emx1-Cre: nine males, five females), n=10 Celsr1Emx1cKO (five males, five females).
Although mutant females tended to be more active and less anxious than controls, the differences were not statistically significant (distance covered in open field: controls =6735±1461 cm, Celsr1Emx1cKO=14070±3505 cm, n=5 per genotype, P=0.0894, Student’s t-test, time spent in exposed areas in open field: controls=170.6±54.5 s, Celsr1Emx1cKO=254.4±23.7 s, P=0.1963, Student’s t-test, Figures 2k), elevated plus maze (time spent in open area in elevated plus maze, controls=92.74±14.6 s, Celsr1Emx1cKO=115.6±21.6 s, P=0.4063, Student’s t-test, Figure 2m), and water maze (Day 1–3, P>0.05, two-way ANOVA, Figure 2n); Yet, they exhibited a social interaction defect, spending similar time periods in the empty chamber as with the stranger mouse (controls in empty chamber=164.3±13.4 s, with stranger=340.6s±20.8 s, n=5, P<0.001; Celsr1Emx1cKO in empty chamber=235.8±37.8 s, with stranger=257.1 s±44.5 s, n=5 animals per genotype, P>0.05, one-way ANOVA, Figure 2o). This combination of traits has been described in rodent models of attention deficit/hyperactivity disorder (ADHD),17 and autism spectrum disorder (ASD),18, 19 two neurodevelopmental conditions with high co-morbidity.20 Taken together, these results suggest that the loss of Celsr1 promotes proliferation of aNPC at the expense of neurogenesis, decreases the number of neurons, leading to cortical hypoplasia and disrupted behavior.
Celsr1 inactivation modifies the transcriptome of aNPC
To investigate further how Celsr1 controls the balance between proliferation and differentiation of aNPC, we purified control and mutant Prominin-positive cells by FACS at e14.5, extracted RNA and compared their transcriptome using RNA-Seq (Supplementary Figure 9). We found that 276 genes displayed an altered expression in Celsr1-deficient aNPC (Supplementary Table 1, False discovery rate P<0.01, fold changes >1.5). Compared with controls, 20% of misregulated genes were upregulated, and 80% were downregulated (Supplementary Figure 9a). Hierarchically clustered gene ontology terms for biological processes emphasized genes involved in Wnt/PCP signaling, cilia assembly and movement, brain development and generation of neurons, cell proliferation and differentiation, positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway, and regulation of cellular response to growth factor stimulus (Supplementary Table 2). Changes in Wnt/PCP, cilia movement and assembly, and microtubule cytoskeleton were predictable given the established role of Celsr1 in PCP and cilia biogenesis, and the relationship between Wnt and PCP.5, 8, 9 On the other hand, several genes involved in Wnt signaling such as Sulf1, Sulf2, GPC3, Rspo1-3 are also implicated in transmembrane receptor protein serine/threonine kinase signaling.
Consistent with the increased proliferation of aNPC in Celsr1 mutants, we found that 19% of the upregulated genes promote proliferation (Supplementary Figure 9b). Those include Tmsb4x,21 Rpl34,22 Nsg2,23 Tnc,24 Snhg6,25 Pttg1 (Supplementary Figure 9d),26 and Dbi (Supplementary Figure 9e).27 Of note, in humans, the extracellular matrix protein TNC (Tenascin C) is highly expressed in the outer SVZ, a region which, in gyrencephalic brains, contains highly proliferative outer radial glia.24 Moreover, Thymosin beta4 (Tmsb4x) strongly promotes proliferation of neural progenitors.21 On the other hand, 13% of the downregulated genes are known tumor suppressors (Supplementary Figure 9c). They include Sulf1,28 Sostdc1,29 Ccdc67,30 Ptprt,31 Igfbp5,32 Rbm47,33 Sepp134 (Supplementary Figure 9f), Mamdc2,35 Mecom,36 Zmat4,37 Dab2,38 Gpx3,39 Ptprg,40 Cds141 or Spred242 (Supplementary Figure 9g). In addition, Ptpn6,43 Kank1,44 Lrp1b,45 Lrig3,46 Nkd1,47 Crabp2,48 Igfbp349 and Cbx750 have been implicated in glioblastoma progression, confirming that invasiveness of glioblastoma and proliferation of RG progenitors use common molecules and signaling pathways.24
The most enriched terms in gene ontology for cellular component pointed to cilia, proteinaceous ECM (extracellular matrix) and intrinsic component of plasma membrane (Supplementary Table 3, Supplementary Figure 9h), suggesting that Celsr1 may be instrumental to the communication between aNPC and their environment. Indeed, many differentially expressed genes encode extracellular proteins (Sulf1, Metallothionein-3, (Supplementary Figure 9i), Mmp2, Adamts2, Adamts18 (Supplementary Figure 9j), Galnt10 and Galnt12), membrane-bound proteins (Ptpn6, Ptprt, Sgms2, Tmsb4x (Supplementary Figure 9k) and Mamdc2 (Supplementary Figure 9l), ECM components (Tnc and Col1a2), or growth factor-binding proteins (Igfbp3 and Igfbp5). All these molecules can promote and regulate interactions between cell surface receptors, ligands and ECM.
Finally, it is worth mentioning that 10% (26/276) of misregulated genes are associated with ASD (Supplementary Table 4), and 2.5% (7/276) with ADHD (Supplementary Table 5).
Celsr1 is required in aNPC endfeet to mediate RA signaling
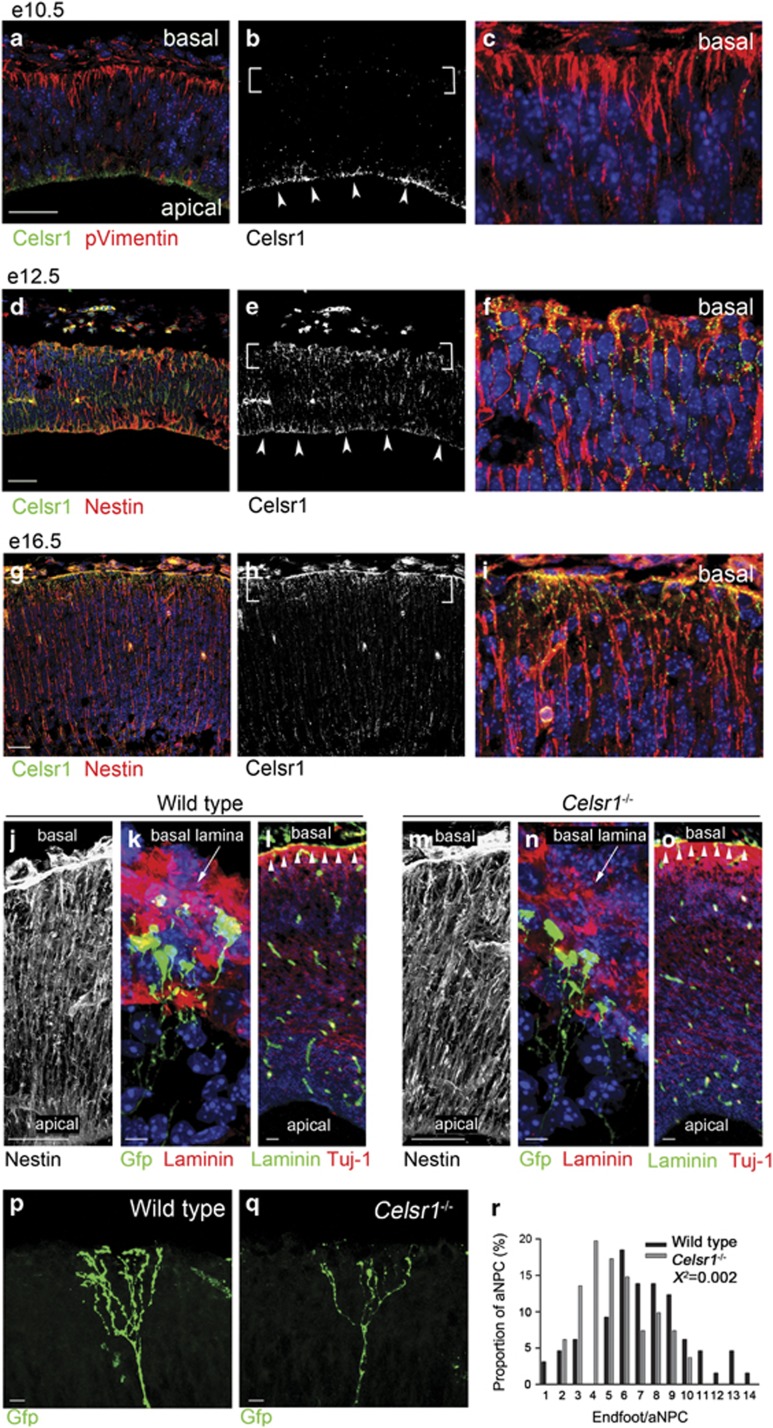
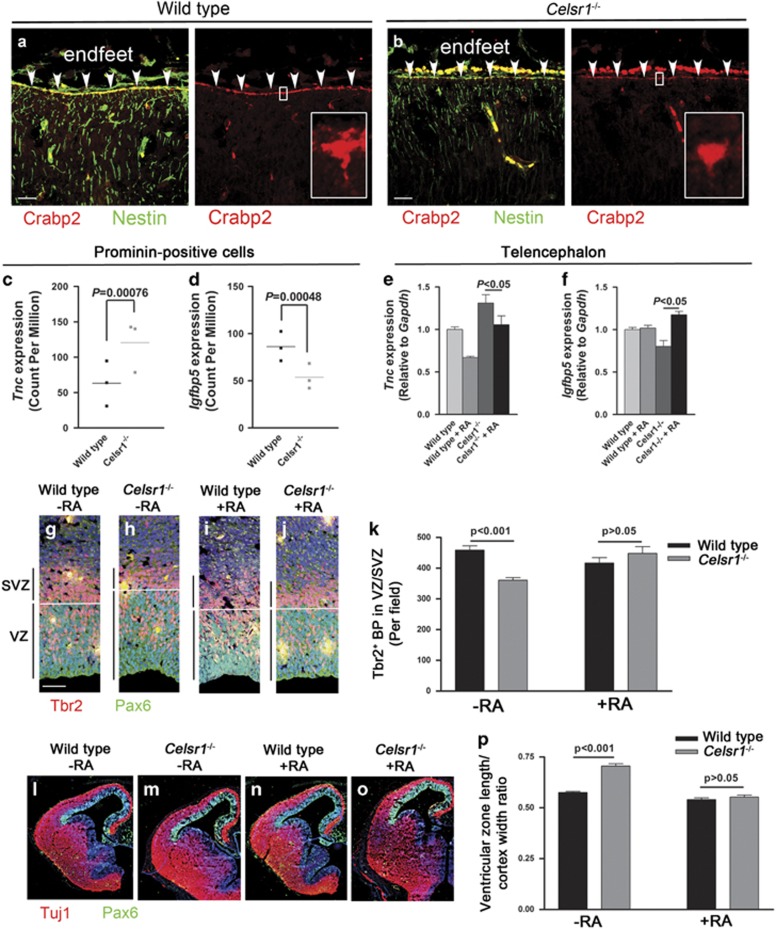
The phenotype and transcriptional landscape of Celsr1−/− mutant aNPC indicate that they fail to perceive critical neurogenic signals. To examine the relationship between the loss of Celsr1 and the cortical phenotype, we studied the distribution of Celsr1 protein in the embryonic mouse telencephalon and found that, prior to the onset of neurogenesis, Celsr1 was confined to the apical junctions of NSC (Figures 3a–c). From e12.5 onwards, Celsr1 accumulated progressively in basal processes and endfeet of RG progenitors (Figures 3d–i), as was shown in the brainstem and spinal cord.11, 51 Because Celsr1 is a putative adhesion protein, we investigated the attachment of RG processes to the pia in Celsr1−/− mice. Immunohistochemistry with antibodies to the intermediate filament-associated protein Nestin showed that mutant aNPC were correctly oriented (Figures 3j, m) and reached the basal lamina (Figures 3k, n). Furthermore, we did not observe discontinuities in the basal lamina or neuronal heterotopia in meningeal spaces (Figures 3l, o). However, close scrutiny of the aNPC basal compartment after electroporation of a Gfp encoding plasmid revealed that the number of basal processes was dramatically reduced in Celsr1−/− mice (Figures 3p–r). Hence, Celsr1 redistributes to basal processes of aNPC at the onset of neurogenesis, and its deficiency affects their complexity. As mutant aNPC endfeet were drastically affected, we considered signals emanating from meninges, which were previously proposed to regulate brain development through secreted molecules.52, 53 Chief among meningeal signals is RA, which is believed to trigger the switch from proliferative to neurogenic divisions at the onset of neurogenesis.52 Meningeal cells, the main source of RA in the developing cortex, appear progressively around the telencephalon in a lateral to dorsal gradient between e12.5 and e14.5,52 which correlates perfectly with the defect in BP production seen in Celsr1−/− cortex (Figure 1m, Supplementary Figure 3n). Furthermore, we found that Crabp2, a cytosol-to-nuclear shuttling protein that facilitates the binding of RA to its cognate nuclear receptor, was concentrated in RG endfeet (Figures 4a, b) where it could mediate RA uptake and its relocation to the nucleus. In agreement with this hypothesis, the expression of Crabp248 as well as other RA target genes involved in neural differentiation such as Tnc,24, 54 and Igfbp555, 56 was misregulated in Celsr1−/− mice (Supplementary Table 1, Figures 4c, d).
Figure 3.
Celsr1 expression during cortical development. (a–i) Coronal sections stained for Celsr1 (green, white), and Phospho-Vimentin (pVimentin, a–c) or Nestin (d–i) (red). At e10.5 (a–c), Celsr1 protein localizes at the apical side (arrowheads) but not in endfeet (brackets). Scale bars 50 μm. At e12.5 (d–f) and e16.5 (g–i), Celsr1 is distributed both in apical and basal compartments. Scale bars 50 μm. (j–o) Coronal sections in forebrain of Wild type (j–l) and Celsr1−/− mice (m–o) immunostained for Nestin (j, m), Gfp (green) and Laminin (red) (k, n), Laminin (green) and Tuj–1 (red) (l, o). Note the intact basal lamina (arrowheads). Scale bars 50 μm (j, m), 10 μm (k, n), 100 μm (l, o). (p, r) Gfp-filled basal processes of aNPC from Wild type (p) and Celsr1−/− mice (q). Note the simplified morphology in the mutant. Scale bars 20 μm. (r) Quantification of endfeet in Wild type and Celsr1−/− mice at P1. n=65 aNPC for Wild type, and 81 for Celsr1−/−; Chi square test, X2=0.002.
Figure 4.
RG endfeet modulate the RA neurogenic signal. (a, b) The RA shuttle protein Crabp2 is enriched in aNPC endfeet. Scale bars 50 μm. (c–f) Modification of transcription of RA target genes Tnc and Igfbp5 revealed by RNA-Seq on isolated aNPC (c, d) and confirmed by qRT-PCR on e14.5 telencephalon (e, f). n=3 brains for RNA-Seq and four brains for qRT-PCR. Bars represent s.e.m. Two-way ANOVA for qRT-PCR. (g–p) RA supplementation from e11.5 to e14.5 rescues the production of BPs (g–k) and lateral expansion of the VZ (l–p). n=4 embryos per condition. Bars represent s.e.m. Two-way ANOVA, P<0.001 without RA and P>0.05 with RA.
To test further whether RA is involved in the altered neurogenesis observed in mutant mice, we subjected Celsr1−/− embryos to RA supplementation in utero between e11.5 and e14.5. This treatment restored the expression of Tnc and Igfbp5 (Figures 4e, f). It also rescued the number of Tbr2-positive BP cells (Figures 4g and k, Tbr2+ cells in VZ/SVZ at e14.5, wild type=458.8±13.8, Celsr1−/− =360.9±8.6, n=4, P<0.001, two-way ANOVA; wild type+RA=416.8±17.6, Celsr1−/−+RA=448.3±21.9, n=4, P>0.05, two-way ANOVA); and the length of the VZ (Figures 4l–p, wild type=0.57±0.005, Celsr1−/−=0.70±0.011, n=4, P<0.001, two-way ANOVA; wild type+RA=0.54±0.008, Celsr1−/−+RA=0.55±0.010, n=4, P>0.05, two-way ANOVA). Long-term RA supplementation (from e11.5 to e17.5) had profound consequences on the mature brain, and on behavior of adult mice (Figure 5). The treatment rescued the number of cortical neurons (number of Cux1+ cells: wild type-RA=100%, wild type+RA=100.2%, Celsr1−/−-RA=85.1%, Celsr1−/−+RA=100.6%, n=5, P>0.05, Figure 5a;), and the hyperactivity and hiding behaviors (distance covered in open field: controls-RA=9045±708.4 cm, Celsr1Emx1cKO-RA=13279±1175 cm, controls+RA=8994±678.9 cm, Celsr1Emx1cKO+RA=7942±1493 cm, n=9, P>0.05 for RA treatment, one-way ANOVA, Figure 5b; time spent in exposed areas in open field: controls-RA=195.7±26.3 cm, Celsr1Emx1cKO-RA=320.5±37.8 cm, controls+RA=204±21.1 cm, Celsr1Emx1cKO+RA=215.5±64.9 cm, n=9, P>0.05 for RA treatment, one-way ANOVA, Figure 5c; time spent in open area in elevated plus maze: controls-RA=1193±61.1 cm, Celsr1Emx1cKO-RA=1625±176.8 cm, controls+RA=1381±85.6 cm, Celsr1Emx1cKO+RA=1279±136.2, n=9, P>0.05 for RA treatment, one-way ANOVA, Figure 5d), but it did not restore the learning abilities of the mutant males (time spent to find the platform after 3 days, controls-RA =8.9±1.9 s, Celsr1Emx1cKO-RA=19.2±4.2 s, controls+RA=10.3±2.1 s, Celsr1Emx1cKO+RA=19.8±5.2 s, n=9, P<0.05 for day 1–3, two-way ANOVA, Figure 5e). Long-term repetitive treatment with RA also rescued social interaction defects in Celsr1Emx1cKO females (time spent in empty chamber Celsr1Emx1cKO-RA=235.8±37.8 s, with stranger=257.1±44.5 s, P>0.05, Celsr1Emx1cKO+RA in empty chamber=96±37 s, with stranger=339±37 s, P<0.001, n=9 animals per genotype, Student’s t-test; Figure 5f). These results support a link between RA and the Celsr1−/− phenotype.
Figure 5.
Effect of RA supplementation on cortical development and behavior in Celsr1 Emx1cKO mice. (a) Quantification of Cux1-positive neurons (layers II and III) in Celsr1wildtype, Celsr1Emx1cKO-RA and Celsr1Emx1cKO+RA. n=5 brains per genotype; bars represent s.e.m.; Student’s t-test; Celsr1Wildtype versus Celsr1Emx1cKO-RA, P<0.05; Celsr1Wildtype versus Celsr1Emx1cKO+RA, P>0.05. The number of cells in the Wild type is set to 100%. (b) Locomotor activity of males in open field. Bars represent s.e.m.; Student’s t-test; Celsr1Wildtype versus Celsr1Emx1cKO-RA, P<0.05; Celsr1Wildtype versus Celsr1Emx1cKO+RA, P>0.05. (c, d) Time spent by males in exposed areas of open field (c) and open arms of elevated plus maze (d). Bars represent s.e.m., Student’s t-test, Celsr1Wildtype versus Celsr1Emx1cKO-RA, P<0.05; Celsr1Wildtype versus Celsr1Emx1cKO+RA, P>0.05; (e) Learning ability of males in the water maze. Bars represent s.e.m., Student’s t-test, Celsr1Wildtype versus Celsr1Emx1cKO-RA, P<0.05; Celsr1Wildtype versus Celsr1Emx1cKO+RA, P<0.05. (f) Social behavior in the ‘3 chamber’ test for females. Bars represent s.e.m., one-way ANOVA, P>0.05 for Celsr1Emx1cKO-RA (stranger versus empty chamber); P<0.001 for Celsr1Wildtype and Celsr1Emx1cKO+RA (stranger versus empty chamber). For the RA rescue experiments, n=9 animals per genotype and per condition.
Discussion
The control of neurogenesis during embryonic development is crucial to regulate the number, diversity and position of neurons, and to normalize cortical size and architecture. At the onset of neurogenesis, aNPC integrate intrinsic and extrinsic signals that determine their fate choices. Here, we report that Celsr1, which localizes to the basal compartment of aNPC,51 contributes to such signals and regulates the type of division of progenitors. We propose that the basal compartment of aNPC interprets signals secreted from the meninges, such as that of RA, and thereby regulates expression of genes involved in the neurogenic switch.
Celsr1 mutant mice exhibit errors in neural progenitor fate decision that lead to a reduced number of cortical neurons, abnormal brain architecture (that is, thicker VZ/SVZ, and thinner upper layers of the neocortex), microcephaly and behavioral impairment. Celsr1Emx1cKO mice are hyperactive and display abnormal exploratory behavior, spending more time than controls in exposed regions, and in unprotected arms of the elevated plus maze. The fact that mutant mice explore uncomfortable, risky, or threatening environments could be linked to anxiety-related responses or reflect a diminished attention to their environment, a conclusion supported by their reduced spacial learning abilities. In addition to the increased locomotor and reduced anxiety and/or attention in males, Celsr1Emx1cKO females spend significantly less time than controls interacting with a stranger mouse, a sign of social withdrawal. This combination of phenotypes is evocative of neurodevelopmental human disorders where: (i) ASD and ADHD co-occur with high frequency (up to 50% of children with ADHD meet criteria for ASD, and up to 80% of ASD children meet criteria for ADHD;20 and (ii) Brains of patients with both ASD and hyperactivity displayed thickening of the VZ/SVZ and altered neurogenesis,57 in addition to microcephaly.58, 59 In humans, neural tube closure defects and fetal lethality may have prevented the identification of causative links between CELSR1 and neurodevelopmental disorders in large cohorts. Nevertheless, CELSR1 mutations have been associated with autism, hyperactivity, delayed speech and intellectual disability (https://decipher.sanger.ac.uk/).60 Further studies using appropriately designed genome editing strategies and brain organoids should help assess the role of CELSR1 in human cortical development.
Mechanistically, we find that the Celsr1 protein is confined to apical junctions in NSC but distributes to the basal compartment in aNPC, and this localization is essential to shape the basal processes. Accumulation of proteins that regulate proliferation versus differentiation of neural progenitors in specific cell compartments and especially in aNPC endfeet has been reported. For instance, Cyclin D2, a crucial cell cycle regulator, accumulates in aNPC endfeet and, among cells that inherit basal processes after division, may promote re-entry into the cell cycle.61 Likewise, the Fragile X mental retardation protein FMRP involved in the transition from aNPC to BP is also distributed in aNPC basal processes,62 where it controls transport and localization of kinesin Kif26a.63 Accumulation of the Celsr1 protein in aNPC endfeet may promote the branching and/or stabilization of the basal compartment of aNPC. Both processes require a coordinated reorganization of the cytoskeleton and involve actin and microtubules dynamics, a typical feature of PCP signaling.64 Celsr1 is a seven-pass transmembrane protein of the adhesion G protein-coupled receptor family with an ectodomain containing cadherin repeats, EGF, laminin and hormone receptor motifs, thus making it a good candidate to bridge the ECM and the cytoskeleton. Such an interaction is instrumental to aNPC morphogenesis and communication with meninges since the simplified shape of basal processes in Celsr1-deficient mice, correlates with alterations in gene expression, and with a defective neurogenic switch. Of note, the Celsr1 paralog Celsr3 is required at later embryonic stages for the gliogenic switch.65
The meninges are a source of developmental cues. The pia produces the chemoattractant Cxcl12 (Sdf1), which regulates the positioning of cortical interneurons and Cajal-Retzius cells.53 The dura is involved in skull development by producing TGFβ and FGF2 to induce bone formation.66, 67 RA derived from meninges may play a role in the neurogenic induction during corticogenesis,52 even though this is debated.68 Our RNA-Seq data revealed that receptors and binding proteins involved in RA signaling (for example, Rarα, Rarβ, Rxrα, Rxrβ, Nr2f1, Crabp2, Ppard and Fabp5) are expressed in aNPC. Furthermore, removing RA-producing meninges by Foxc1 inactivation produces phenotypic features rather similar to those in Celsr1−/− mice (this study), and in RARα/γ double knockout,69 namely an increased number of self-renewing divisions of aNPC with lateral expansion of the VZ. In addition, neurogenesis defects found in Celsr1−/− and Foxc1−/− mutants are both rescued by RA treatment. RA is a strong morphogen whose activity must be tightly regulated. Restricting its production to the meninges and its uptake by Crabp2, a shuttle protein which accumulates in aNPC endfeet, may provide an efficient way to deliver the appropriate amount of RA to the distant nucleus.
Transcriptional dysregulations found in Celsr1−/− point to common pathways and mechanisms by which Celsr1, through the basal processes of aNPC, controls the switch from self-renewal to differentiation. Many genes and proteins that are differentially expressed in Celsr1 mutant aNPC modulate growth factor signaling at the membrane or via extracellular space. Tyrosine phosphatases (Ptpn6, Ptprt, Ptprg) which blocks the activation of tyrosine kinase receptors with known proliferation inducing activity, such as Egfr70 or Pdgfrb, are reduced. Likewise, Sulf1, which decreases receptor affinity to ligands by modifying sulfation of their co-receptor heparin sulfate proteoglycans,71 is diminished. At the plasma membrane level, Lrig3, Myof and Spred2 (through receptor degradation and through interaction with PI3K) also negatively regulate growth factors (FGF or EGF) signaling.72
Acknowledgments
We thank Dr Xavier Morin for providing the pcx-Gfp plasmid, Catherine Lombard for her help with the UCL-IREC FACS platform and Valérie Bonte, R El Kaddouri, I Lambermont and E Paitre for technical support. This study made use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust. This work was supported by the following grants: FNRS PDR T0002.13, FNRS PDR T00075.15, Interuniversity Poles of Attraction (SSTC, PAI p6/20 and PAI7/20), Fondation Médicale Reine Elisabeth and Fondation JED-Belgique. FT is a Research Director of the Belgian Fund for Scientific Research (FNRS).
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
The authors declare no conflict of interest.
Supplementary Material
References
- Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development 2014; 141: 2182–2194. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Contribution of intermediate progenitor cells to cortical histogenesis. Arch Neurol 2007; 64: 639–642. [DOI] [PubMed] [Google Scholar]
- Fernandez V, Llinares-Benadero C, Borrell V. Cerebral cortex expansion and folding: what have we learned? EMBO J 2016; 35: 1021–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nathans J. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development 2007; 134: 647–658. [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. Shaping the nervous system: role of the core planar cell polarity genes. Nat Rev Neurosci 2013; 14: 525–535. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, Copp AJ et al. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr Biol 2003; 13: 1129–1133. [DOI] [PubMed] [Google Scholar]
- Ravni A, Qu Y, Goffinet AM, Tissir F. Planar cell polarity cadherin Celsr1 regulates skin hair patterning in the mouse. J Invest Dermatol 2009; 129: 2507–2509. [DOI] [PubMed] [Google Scholar]
- Boutin C, Labedan P, Dimidschstein J, Richard F, Cremer H, Andre P et al. A dual role for planar cell polarity genes in ciliated cells. Proc Natl Acad Sci USA 2014; 111: E3129–E3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Komatsu K, Hirao M, Toyooka Y, Koyama H, Tissir F et al. Celsr1 is required for the generation of polarity at multiple levels of the mouse oviduct. Development 2014; 141: 4558–4568. [DOI] [PubMed] [Google Scholar]
- Tatin F, Taddei A, Weston A, Fuchs E, Devenport D, Tissir F et al. Planar cell polarity protein Celsr1 regulates endothelial adherens junctions and directed cell rearrangements during valve morphogenesis. Dev Cell 2013; 26: 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Glasco DM, Zhou L, Sawant A, Ravni A, Fritzsch B et al. Atypical cadherins Celsr1-3 differentially regulate migration of facial branchiomotor neurons in mice. J Neurosci 2010; 30: 9392–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A, Escuin S, Doudney K, Vekemans M, Stevenson RE, Greene ND et al. Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum Mutat 2012; 33: 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin C, Goffinet AM, Tissir F. Celsr1-3 cadherins in PCP and brain development. Curr Top Dev Biol 2012; 101: 161–183. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A et al. Transcriptional landscape of the prenatal human brain. Nature 2014; 508: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S, Miya H, Watanabe T, Kawai H, Yamasaki N, Harada Y et al. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat Neurosci 2015; 18: 657–665. [DOI] [PubMed] [Google Scholar]
- Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell 2008; 3: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell VA. Neurobiology of animal models of attention-deficit hyperactivity disorder. J Neurosci Methods 2007; 161: 185–198. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Young NB, Nonneman RJ, Grossman AW, Murphy DL et al. Social approach in genetically engineered mouse lines relevant to autism. Genes Brain Behav 2009; 8: 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Revi Neuroscience. 2010; 11: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelse NN, Franke B, Geurts HM, Hartman CA, Buitelaar JK. Shared heritability of attention-deficit/hyperactivity disorder and autism spectrum disorder. Eur Child Adolesc Psychiatry 2010; 19: 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsching HG, Kretz O, Morosan-Puopolo G, Chernogorova P, Theiss C, Brand-Saberi B. Thymosin beta4 induces folding of the developing optic tectum in the chicken (Gallus domesticus). J Compar Neurol 2012; 520: 1650–1662. [DOI] [PubMed] [Google Scholar]
- Yang S, Cui J, Yang Y, Liu Z, Yan H, Tang C et al. Over-expressed RPL34 promotes malignant proliferation of non-small cell lung cancer cells. Gene 2016; 576: 421–428. [DOI] [PubMed] [Google Scholar]
- Ueda T, Nagamachi A, Takubo K, Yamasaki N, Matsui H, Kanai A et al. Fbxl10 overexpression in murine hematopoietic stem cells induces leukemia involving metabolic activation and upregulation of Nsg2. Blood 2015; 125: 3437–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR et al. Molecular identity of human outer radial glia during cortical development. Cell 2015; 163: 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Fang N, Gu J, Zhang Y, Li X, Zu L et al. Noncoding RNA small nucleolar RNA host gene 1 promote cell proliferation in nonsmall cell lung cancer. Indian J Cancer 2014; 51: e99–e102. [DOI] [PubMed] [Google Scholar]
- Noll JE, Vandyke K, Hewett DR, Mrozik KM, Bala RJ, Williams SA et al. PTTG1 expression is associated with hyperproliferative disease and poor prognosis in multiple myeloma. J Hematol Oncol 2015; 8: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vied CM, Freudenberg F, Wang Y, Raposo AA, Feng D, Nowakowski RS. A multi-resource data integration approach: identification of candidate genes regulating cell proliferation during neocortical development. Fronti Neurosci 2014; 8: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CT, Zhu ST, Li P, Wang YJ, Zhang H, Zhang ST. SULF1 inhibits proliferation and invasion of esophageal squamous cell carcinoma cells by decreasing heparin-binding growth factor signaling. Digest Dis Sci 2013; 58: 1256–1263. [DOI] [PubMed] [Google Scholar]
- Liang W, Guan H, He X, Ke W, Xu L, Liu L et al. Down-regulation of SOSTDC1 promotes thyroid cancer cell proliferation via regulating cyclin A2 and cyclin E2. Oncotarget 2015; 6: 31780–31791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Jang HR, Kim M, Kim JH, Kwon OH, Park JL et al. Epigenetic alteration of CCDC67 and its tumor suppressor function in gastric cancer. Carcinogenesis 2012; 33: 1494–1501. [DOI] [PubMed] [Google Scholar]
- Scott A, Wang Z. Tumour suppressor function of protein tyrosine phosphatase receptor-T. Biosci Rep 2011; 31: 303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ding N, Li Y, Cheng H, Wang D, Yang Q et al. Insulin-like growth factor binding protein 5 (IGFBP5) functions as a tumor suppressor in human melanoma cells. Oncotarget 2015; 6: 20636–20649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A et al. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. eLife 2014; 3, doi: 10.7554/eLife.02734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CW, Reddy VK, Short SP, Motley AK, Lintel MK, Bradley AM et al. Selenoprotein P influences colitis-induced tumorigenesis by mediating stemness and oxidative damage. J Clin Invest 2015; 125: 2646–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darda L, Hakami F, Morgan R, Murdoch C, Lambert DW, Hunter KD. The role of HOXB9 and miR-196a in head and neck squamous cell carcinoma. PloS ONE 2015; 10: e0122285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou A, Zhao L, Zhao F, Wang W, Niu J, Li B et al. Expression of MECOM is associated with unfavorable prognosis in glioblastoma multiforme. OncoTargets Ther 2016; 9: 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Gao Y, Zhao X, Wu Q, Fu X, Shao Y et al. The association between the copy-number variations of ZMAT4 and hematological malignancy. Hematology 2011; 16: 20–23. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chen Y, Xie X, Tang J. The expression of disabled-2 is common reduced in meningiomas. Neurol India 2014; 62: 57–61. [DOI] [PubMed] [Google Scholar]
- Barrett CW, Ning W, Chen X, Smith JJ, Washington MK, Hill KE et al. Tumor suppressor function of the plasma glutathione peroxidase gpx3 in colitis-associated carcinoma. Cancer Res 2013; 73: 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AK, Ip JC, Chu AC, Cheng Y, Leong MM, Ko JM et al. PTPRG suppresses tumor growth and invasion via inhibition of Akt signaling in nasopharyngeal carcinoma. Oncotarget 2015; 6: 13434–13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan CH. Checking in on Cds1 (Chk2): a checkpoint kinase and tumor suppressor. BioEssays 2002; 24: 502–511. [DOI] [PubMed] [Google Scholar]
- Kachroo N, Valencia T, Warren AY, Gnanapragasam VJ. Evidence for downregulation of the negative regulator SPRED2 in clinical prostate cancer. Br J Cancer 2013; 108: 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sooman L, Ekman S, Tsakonas G, Jaiswal A, Navani S, Edqvist PH et al. PTPN6 expression is epigenetically regulated and influences survival and response to chemotherapy in high-grade gliomas. Tumour Biol 2014; 35: 4479–4488. [DOI] [PubMed] [Google Scholar]
- Guo X, Fan W, Bian X, Ma D. Upregulation of the Kank1 gene-induced brain glioma apoptosis and blockade of the cell cycle in G0/G1 phase. Int J Oncol 2014; 44: 797–804. [DOI] [PubMed] [Google Scholar]
- Tabouret E, Labussiere M, Alentorn A, Schmitt Y, Marie Y, Sanson M. LRP1B deletion is associated with poor outcome for glioblastoma patients. J Neurol Sci 2015; 358: 440–443. [DOI] [PubMed] [Google Scholar]
- Guo D, Yang H, Guo Y, Xiao Q, Mao F, Tan Y et al. LRIG3 modulates proliferation, apoptosis and invasion of glioblastoma cells as a potent tumor suppressor. J Neurol Sci 2015; 350: 61–68. [DOI] [PubMed] [Google Scholar]
- Gotze S, Wolter M, Reifenberger G, Muller O, Sievers S. Frequent promoter hypermethylation of Wnt pathway inhibitor genes in malignant astrocytic gliomas. Int J Cancer 2010; 126: 2584–2593. [DOI] [PubMed] [Google Scholar]
- Campos B, Warta R, Chaisaingmongkol J, Geiselhart L, Popanda O, Hartmann C et al. Epigenetically mediated downregulation of the differentiation-promoting chaperon protein CRABP2 in astrocytic gliomas. Int J Cancer 2012; 131: 1963–1968. [DOI] [PubMed] [Google Scholar]
- Yang CH, Yue J, Pfeffer SR, Fan M, Paulus E, Hosni-Ahmed A et al. MicroRNA-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3). J Biol Chem 2014; 289: 25079–25087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Warden C, Zou Z, Neman J, Krueger JS, Jain A et al. Altered expression of polycomb group genes in glioblastoma multiforme. PloS ONE 2013; 8: e80970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstone CJ, Moxon C, Murdoch J, Little P, Mason I. Basal enrichment within neuroepithelia suggests novel function(s) for Celsr1 protein. Mol Cell Neurosci 2010; 44: 210–222. [DOI] [PubMed] [Google Scholar]
- Siegenthaler JA, Ashique AM, Zarbalis K, Patterson KP, Hecht JH, Kane MA et al. Retinoic acid from the meninges regulates cortical neuron generation. Cell 2009; 139: 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell V, Marin O. Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat Neurosci 2006; 9: 1284–1293. [DOI] [PubMed] [Google Scholar]
- Alvarez-Dolado M, Gonzalez-Sancho JM, Navarro-Yubero C, Garcia-Fernandez LF, Munoz A. Retinoic acid and 1,25-dihydroxyvitamin D3 inhibit tenascin-C expression in rat glioma C6 cells. J Neurosci Res 1999; 58: 293–300. [DOI] [PubMed] [Google Scholar]
- Chang YS, Cho JY, Cho HA, Kim HJ, Chang J, Ahn CM et al. 9-cis retinoic acid induces insulin-like growth factor binding protein-3 through DR-8 retinoic acid responsive elements. Cancer Biol Ther 2006; 5: 586–592. [DOI] [PubMed] [Google Scholar]
- Dong Y, Canalis E. Insulin-like growth factor (IGF) I and retinoic acid induce the synthesis of IGF-binding protein 5 in rat osteoblastic cells. Endocrinology 1995; 136: 2000–2006. [DOI] [PubMed] [Google Scholar]
- Wegiel J, Kuchna I, Nowicki K, Imaki H, Wegiel J, Marchi E et al. The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol 2010; 119: 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Itzchak E, Ben-Shachar S, Zachor DA. Specific neurological phenotypes in autism spectrum disorders are associated with sex representation. Autism Res 2013; 6: 596–604. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Molloy E, Castellanos FX. Brain imaging of attention deficit/hyperactivity disorder. Ann N Y Acad Sci 2001; 931: 33–49. [DOI] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet 2009; 84: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunekawa Y, Britto JM, Takahashi M, Polleux F, Tan SS, Osumi N. Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J 2012; 31: 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffary R, Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J Neurosci 2011; 31: 1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz LJ, Lennox AL, Rouanet JP, Silver DL. Dynamic mRNA transport and local translation in radial glial progenitors of the developing brain. Curr Biol 2016; 26: 3383–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler PN. The frizzled/stan pathway and planar cell polarity in the Drosophila wing. Curr Top Dev Biol 2012; 101: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Jossin Y, Chai G, Lien WH, Tissir F, Goffinet AM. Feedback regulation of apical progenitor fate by immature neurons through Wnt7-Celsr3-Fzd3 signalling. Nat Commun 2016; 7: 10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P Jr., Nakajima A et al. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 2003; 130: 5269–5280. [DOI] [PubMed] [Google Scholar]
- Mehrara BJ, Greenwald J, Chin GS, Dudziak M, Sagrioglu J, Steinbrech DS et al. Regional differentiation of rat cranial suture-derived dural cells is dependent on association with fusing and patent cranial sutures. Plastic Reconstruct Surg 1999; 104: 1003–1013. [DOI] [PubMed] [Google Scholar]
- Chatzi C, Cunningham TJ, Duester G. Investigation of retinoic acid function during embryonic brain development using retinaldehyde-rescued Rdh10 knockout mice. Dev Dynamics 2013; 242: 1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dolle P, Dierich A, Gorry P et al. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development 1994; 120: 2723–2748. [DOI] [PubMed] [Google Scholar]
- Forbes K, Skinner L, Aplin JD, Westwood M. The tyrosine phosphatase SHP-1 negatively regulates cytotrophoblast proliferation in first-trimester human placenta by modulating EGFR activation. Cell Mol Life Sci 2012; 69: 4029–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA et al. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem 2003; 278: 23107–23117. [DOI] [PubMed] [Google Scholar]
- Mardakheh FK, Yekezare M, Machesky LM, Heath JK. Spred2 interaction with the late endosomal protein NBR1 down-regulates fibroblast growth factor receptor signaling. J Cell Biol 2009; 187: 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.