Abstract
Background:
To date, efforts for the prevention and treatment of human respiratory syncytial virus (RSV) infection have been still vain, and there is no safe and effective clinical accepted vaccine. Arisaema genus has claimed for various traditional bioactivities, but scientific assessments are quite limited.
Objective:
This encouraged us to carry out our present study on around 60 phytoconstituents of different Arisaema species as a natural inhibitor against the human RSV.
Materials and Methods:
Selected 60 phytochemical entities were evaluated on the docking behavior of human RSV receptor (PDB: 4UCC) using Maestro 9.3 (Schrödinger, LLC, Cambridge, USA). Furthermore, kinetic properties and toxicity nature of top graded ligands were analyzed through QikProp and ProTox tools.
Results:
Notably, rutin (glide score: −8.49), schaftoside (glide score: −8.18) and apigenin-6,8-di-C-β-D-galactoside (glide score − 7.29) have resulted in hopeful natural lead hits with an ideal range of kinetic descriptors values. ProTox tool (oral rodent toxicity) has resulted in likely toxicity targets of apex-graded tested ligands.
Conclusion:
Finally, the whole efforts can be explored further as a model to confirm its anti-human RSV potential with wet laboratory experiments.
SUMMARY
Rutin, schaftoside, and apigenin-6,8-di-C-β-D-galactoside showed promising top hits docking profile against human respiratory syncytial virus
Moreover, absorption, distribution, metabolism, excretion properties (QikProp) of top hits resulted within an ideal range of kinetic descriptors
ProTox tool highlighted toxicity class ranges, LD50 values, and possible toxicity targets of apex-graded tested ligands.
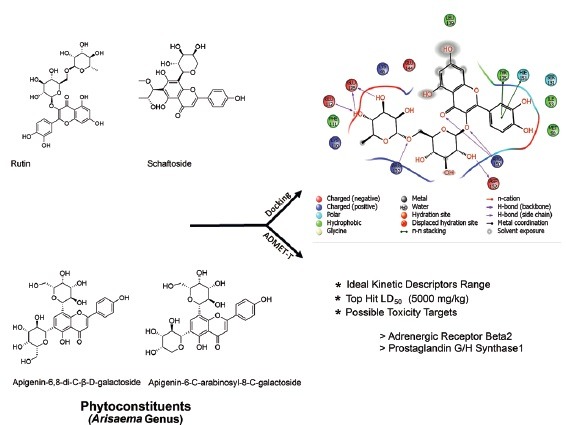
Abbreviations used: RSV: Respiratory syncytial virus, PRRSV: Porcine respiratory and reproductive syndrome virus, ADME-T: Absorption, distribution, metabolism, excretion, and toxicity.
Keywords: Apigenin-6, 8-di-C-β-D-galactoside, Arisaema, ProTox, Rutin, Schaftoside
INTRODUCTION
Human respiratory syncytial virus (RSV) represents as an important respiratory pathogens predecessor for the development of lower respiratory tract infections such as bronchiolitis and pneumonia in infants and young children worldwide. It causes diseases with an expected of 125,000 hospitalizations and 66,000–199,000 mortality rate in 2005 among children >5 years of age. To date, attempts for the prevention and treatment of RSV infection have been still unsuccessful and there is no safe and effective clinical accepted vaccine. Notably, ribavirin showed promising licensed antiviral treatment, and moreover, passive immunization with palivizumab has too resulted in 50% protection to high-risk children but due to questionable efficacy and toxicity put a big question mark among researchers.[1,2] Thus, there is an urgent need to explore and unlock safe effective therapy for RSV. RSV is an enveloped and nonsegmented with negative-strand RNA virus (family; Paramyxoviridae) belonging to the genus Pneumovirus. Previous studies have indicated that RNA-dependent polymerase complex offers promising targets for RSV-specific drugs. Remarkably, the ribonucleoprotein complex involves of genomic RNA encapsulated by the RSV nucleoprotein, N. This detection proceeds through interaction between the phosphoprotein P, which is the chief polymerase cofactor, and N.[3] Thus, we have tried to explore natural chemical entities with the nucleocapsid/phosphoprotein pocket site as an alternative to synthetic drugs to reduce or minimize the side effects. A series of phytochemical entities are accumulated as secondary metabolites during biosynthesis. The literature findings have already documented that the nature of these chemical substances varies due to the unique form of biosynthetic processes with the particular period of the plants and may worth for exceptional antiviral action.[4,5]
Arisaema genus consists of monocotyledon plant species belonging to family Araceae. Around 150 species are accessible throughout the world, out of which 140 species are found in Asia, Africa, and Arab continents.[6] Previous study has revealed that Arisaema franchetianum confirmed hopeful biological spectrum against porcine respiratory and reproductive syndrome virus.[7] Very few species of Arisaema genus have been documented for their biological actions to date.[8,9,10,11,12] Thus, herein, an attempt was designed to computationally discover the in silico natural lead hits concerning their kinetic and toxicity nature against RSV [Figure 1].
Figure 1.
Chemical structures of chief phytoconstituents reported from Arisaema species
MATERIALS AND METHODS
Molecular docking simulations were operated on Maestro 9.3 (Schrödinger, LLC, Cambridge, USA) furnished with Core i5 processor, 8 GB RAM, and 500 GB with Window 10 as the operating system. The tested ligands details were retrieved from various search engines such as SciFinder, PubMed, and Google Scholar.[7,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29]
Target protein identification and preparation
The three-dimensional crystal structure of target RSV receptor (PDB: 4UCC) with a resolution of 2.05 Å was obtained from the Research Collaboratory for Structural Bioinformatics, PDB database (Anonymous, www.rcsb.org). The protein was linked in complex with 1-[(2,4-dichlorophenyl) methyl] pyrazole-3,5-dicarboxylic acid as a reference ligand. Target receptor preparation was started in the course of protein preprocess step which agrees with the insertion of polar hydrogen and amputation of metal ions, cofactor, and water molecule outside 5 Å. In addition, ionization (pH: 6.7–7.3), optimization of hydrogen bond, and restorative energy minimization steps were also performed to attained the proper geometry of the receptor. The probable binding pocket area was indicated through grid box formation by clicking on the internal ligand.
Ligand preparation
The tested ligands were sketched in ChemDraw Ultra 10.0 (CambridgeSoft) in.mol file format, followed by exportation into Maestro software. Outstandingly, ligands preparations were done using least square OPLS_2005 force field plus conformer generations and filtration to their energy minima with probable state creation (pH 7 ± 2.0).
Docking simulation
Extra Precision (XP) Glide docking simulations were applied to the indicated receptor grid of human RSV protein receptor. Finally, the results outcome was analyzed by the way of XP Visualizer not only in the form of glide score but also reviewing various probable interactions such as H-bonding, π-π interactions, and hydrophobic interactions, correspondingly.[30]
Absorption, distribution, metabolism, excretion, and toxicity prediction
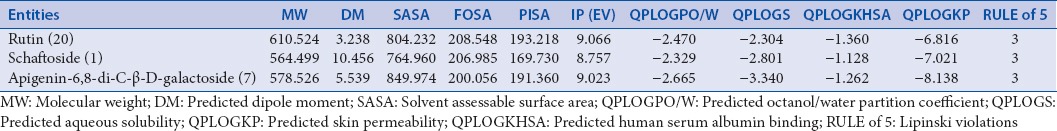
Both of absorption, distribution, metabolism, excretion, and toxicity prediction (ADME-T) were accomplished using QikProp (Maestro 9.3) and ProTox tools, respectively. Various kinetic descriptors as indicated in Table 1 were scrutinized. As for the ProTox analysis, oral toxicity in rodents (LD50 in mg/kg of body weight) descriptors with likely toxicity targets was too studied.[31]
Table 1.
Absorption, distribution, metabolism, excretion descriptors of top ranked entities from QikProp
RESULTS AND DISCUSSION
Overall, the results of tested chemical entities having particular glide score with human RSV receptors are summarized in Table 2. From the results, it has been observed that out of 60 phytoconstituents of Arisaema genus, rutin (glide score: −8.49), schaftoside (glide score: −8.18), and apigenin-6,8-di-C-β-D-galactoside (glide score −7.29) attained top hits with an ideal range of kinetic descriptors values [Table 3].[32] Furthermore, toxicity profiles of tested chemical constituents (ProTox) were also highlighted with probable toxicity targets as mentioned in Table 4.
Table 2.
Phytoconstituents from Arisaema species docked with respiratory syncytial virus receptor

Table 3.
Binding affinity of top three hits of Arisaema species with human respiratory syncytial virus receptor
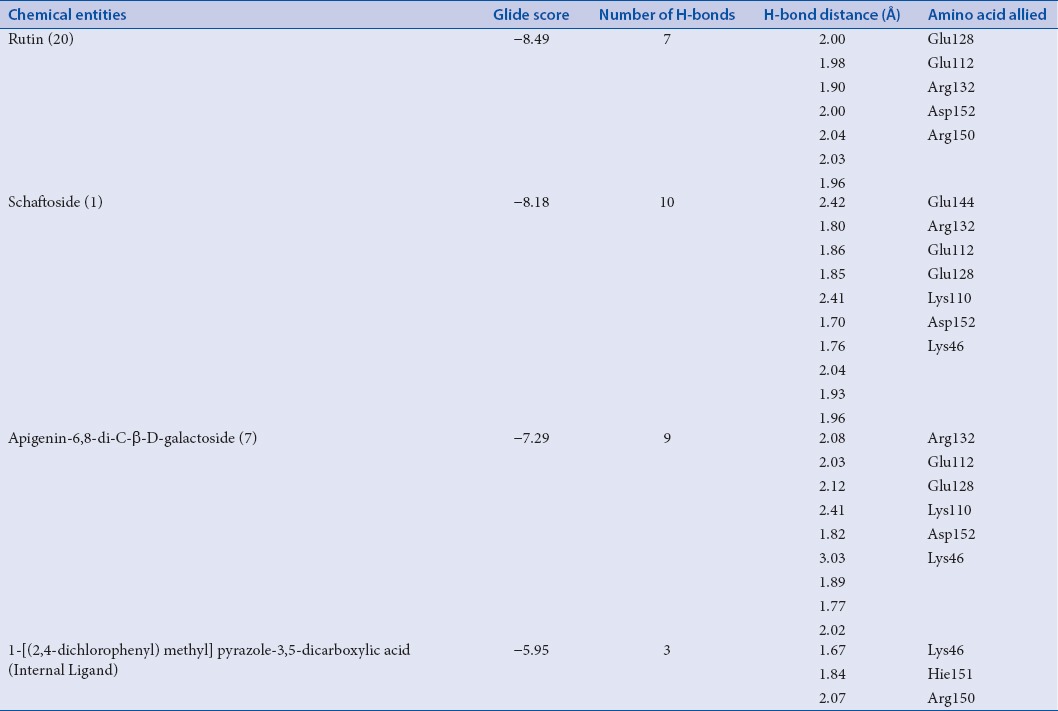
Table 4.
Toxicity profiles of top hits from ProTox database

Top hits phytoconstituents
Rutin
This compound resulted as a first rank great hit with remarkably, H-bonding interactions of Glu128, Glu112, Arg132, Asp152, and Arg150, followed by hydrophobic interactions of amino acid residues such as Phe111, Met50, Tyr135, leu139, respectively. Interestingly, π-π stacking (Tyr135 and Hie151) were too seen.
Schaftoside
Schaftoside compound confirmed as the second best hit with H-bonding interactions such as Glu144, Arg132, Glu112, Glu128, Lys110, Asp152, and Lys46. Moreover, π-π stacking (Arg150) was examined. The hydrophobic interactions (Leu139, Tyr135, and Phe111) were also analyzed.
Apigenin-6,8-di-C-β-D-galactoside
Apigenin-6,8-di-C-β-D-glucopyranoside was observed as the third-ranked promising hit with capable of H-bonding (Arg132, Glu112, Glu128, Lys110, Asp152, and Lys46) and hydrophobic interactions (Tyr135, Phe111). In addition, π-π stacking (Arg150) was observed.
Among all top hits, rutin showed possible predicted interaction (LD50: 5000 mg/kg rodents) with adrenoceptor beta 2 (ADR β2) and prostaglandin G/H synthase 1 targets, which might be a valuable tool for future researchers to understand its precise mechanistic nature [Figure 2].
Figure 2.
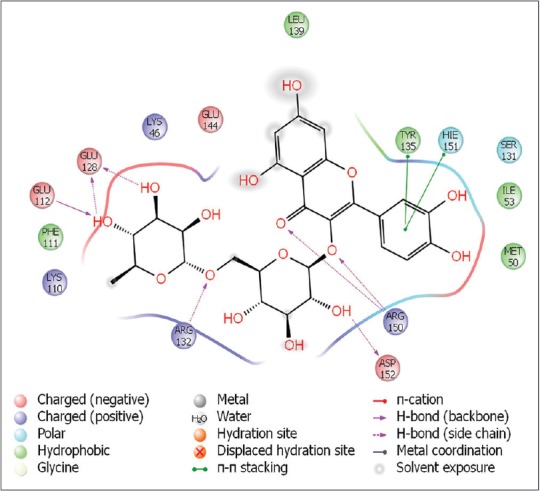
Binding interactions of rutin with human respiratory syncytial virus receptor
CONCLUSION
In the present study, glide scores of tested phytoconstituents of Arisaema genus were recorded from −1.41 to −8.49. Apex-ranked hits such as rutin, schaftoside and apigenin-6,8-di-C-β-D-glucopyranoside have found to have superior interactions and binding affinity with human RSV. Computational ADME-T studies were also emphasized to ensure the safety and effectiveness of drugs based on natural origin. Therefore, these preliminary studies might be quite useful for future researchers to validate it with wet laboratory experiments.
Financial support and sponsorship
This study financially backed by UGC-BSR, New Delhi, India, under grant agreement no. F.7-36/2007.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We would like to thank the Head, Department of Pharmaceutical Sciences and Technology, Birla Institute of Technology, Mesra, Ranchi, for his encouragement and providing basic facilities during the study.
REFERENCES
- 1.Feng S, Hong D, Wang B, Zheng X, Miao K, Wang L, et al. Discovery of imidazopyridine derivatives as highly potent respiratory syncytial virus fusion inhibitors. ACS Med Chem Lett. 2015;6:359–62. doi: 10.1021/acsmedchemlett.5b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui R, Wang Y, Wang L, Li G, Lan K, Altmeyer R, et al. Cyclopiazonic acid, an inhibitor of calcium-dependent ATPases with antiviral activity against human respiratory syncytial virus. Antiviral Res. 2016;132:38–45. doi: 10.1016/j.antiviral.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Ouizougun-Oubari M, Pereira N, Tarus B, Galloux M, Lassoued S, Fix J, et al. A druggable pocket at the nucleocapsid/phosphoprotein interaction site of human respiratory syncytial virus. J Virol. 2015;89:11129–43. doi: 10.1128/JVI.01612-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukherjee PK, Wahile A. Integrated approaches towards drug development from Ayurveda and other Indian system of medicines. J Ethnopharmacol. 2006;3(103):25–35. doi: 10.1016/j.jep.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 5.Dias DA, Urban S, Roessner U. A historical overview of natural products in drug discovery. Metabolites. 2012;2:303–36. doi: 10.3390/metabo2020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava P, Banerji BK. Gender biasing in Arisaema – A unique and rare phenomenon. Curr Sci. 2012;102:189–3. [Google Scholar]
- 7.Su Y, Xu JJ, Bi JL, Wang YH, Hu GW, Yang J, et al. Chemical constituents of Arisaema franchetianum tubers. J Asian Nat Prod Res. 2013;15:71–7. doi: 10.1080/10286020.2012.723202. [DOI] [PubMed] [Google Scholar]
- 8.Azam S, Saqib MS, Zar F, Ahmad B, Khan I, Zeb Z, et al. Antibacterial, antifungal, phytotoxic, antioxidant and hemagglutination activities of organic fractions of Arisaema tortuosum. Pak J Pharm Sci. 2016;29:991–7. [PubMed] [Google Scholar]
- 9.Singla N, Jain S, Kachroo M, Gakhar A. Review on functions of dietary agents and plants in cancer prevention and treatment. Syst Rev Pharm. 2013;4:31–9. [Google Scholar]
- 10.Zhu L, Dai J, Yang L, Qiu J. Anthelmintic activity of Arisaema franchetianum and Arisaema lobatum essential oils against Haemonchus contortus. J Ethnopharmacol. 2013;148:311–6. doi: 10.1016/j.jep.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 11.Ahn CB, Shin TS, Seo HK, Je JY. Phenolic composition and antioxidant effect of aqueous extract of Arisaema cum Bile, the oriental herb medicine, in human fibroblast cells. Immunopharmacol Immunotoxicol. 2012;34:661–6. doi: 10.3109/08923973.2011.649289. [DOI] [PubMed] [Google Scholar]
- 12.Chunxia C, Peng Z, Huifang P, Hanli R, Zehua H, Jizhou W. Extracts of Arisaema rhizomatum C.E.C. Fischer attenuate inflammatory response on collagen-induced arthritis in BALB/c mice. J Ethnopharmacol. 2011;133:573–82. doi: 10.1016/j.jep.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 13.Du SS, Lei N, Xu YC, Wei LX. Study on flavonoids of Arisaema erubescens. Chin Pharm J. 2005;40:1457–9. [Google Scholar]
- 14.Ducki S, Hadfield JA, Zhang X, Lawrence NJ, McGown AT. Isolation of aurantiamide acetate from Arisaema erubescens. Planta Med. 1996;62:277–8. doi: 10.1055/s-2006-957878. [DOI] [PubMed] [Google Scholar]
- 15.Ducki S, Hadfield JA, Lawrence NJ, Zhang X, McGown AT. Isolation of paeonol from Arisaema erubescens. Planta Med. 1995;61:586–7. doi: 10.1055/s-2006-959390. [DOI] [PubMed] [Google Scholar]
- 16.Shen J, Song DS, Ying L. A study on the constituents of ten species of the Chinese medicinal plant “tian nan xin”. J Shanghai Med Univ. 1989;16:203–8. [Google Scholar]
- 17.Li X, Yin J, Fan B, Yan J. Studies on the chemical constituents of Arisaema amurense. Zhongguo Zhongyao Zazhi. 2000;36:89–1. [Google Scholar]
- 18.Chung W, Goo YM, Na DS, Kim KJ. A phospholipase A 2 inhibitor from Arisaema amurense Max. var. serratum. Nakai Arch Pharm Res. 1995;18:293–4. [Google Scholar]
- 19.Jung JH, Lee H, Kang SS. Diacylglycerylgalactosides from Arisaema amurense. Phytochemistry. 1996;42:447–52. doi: 10.1016/0031-9422(95)00929-9. [DOI] [PubMed] [Google Scholar]
- 20.Jung JH, Lee CO, Kim YC, Kang SS. New bioactive cerebrosides from Arisaema amurense. J Nat Prod. 1996;59:319–22. doi: 10.1021/np960201+. [DOI] [PubMed] [Google Scholar]
- 21.Verma H, Lal V, Pant K, Soni NA. Ethno medicinal Review on Arisaema tortuosum. Int J Adv Pharm Biol Chem. 2012;1:176–9. [Google Scholar]
- 22.Nile SH, Park SW. HPTLC analysis, antioxidant, anti-inflammatory and antiproliferative activities of Arisaema tortuosum tuber extract. Pharm Biol. 2014;52:221–7. doi: 10.3109/13880209.2013.831110. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad VU, Hussain H, Hussain J, Ullah F. Chemical constituents of Arisaema flavum. Proc Pak Acad Sci. 2003;40:85–0. [Google Scholar]
- 24.Rehman HU, Siddiqui F, Khokhar I. Anticancer agents of Arisaema jacquemontii Blume. Pak J Sci Ind Res. 1992;35:406–8. [Google Scholar]
- 25.Tanveer M, Sims J, Choudhary MI, Hamann MT. First ever isolation of cytotoxic triterpenoid 2-hydroxydiplopterol from plant source. J Med Plants Res. 2013;7:2040–2. [Google Scholar]
- 26.Jeelani S, Khuroo MA, Razadan TK. New triterpenoids from Arisaema jacquemontii. J Asian Nat Prod Res. 2010;12:157–61. doi: 10.1080/10286020903505057. [DOI] [PubMed] [Google Scholar]
- 27.Inatomi H, Orii Y, Sugiyama T, Suyama Y. Identification of cis-ribosylzeatin from immature seeds of Arisaema negishii Makino. Bull Fac Agric Meji Univ. 1990;87:17–21. [Google Scholar]
- 28.Song ZZ, Jia ZJ. Studies on the chemical constituents of Arisaema fargesii Buchett. Zhongguo Zhong Yao Za Zhi. 1989;14:480–2. 511. [PubMed] [Google Scholar]
- 29.Zhao FW, Luo M, Wang YH, Li ML, Tang GH, Long CL. A piperidine alkaloid and limonoids from Arisaema decipiens, a traditional antitumor herb used by the Dong people. Arch Pharm Res. 2010;33:1735–9. doi: 10.1007/s12272-010-1104-6. [DOI] [PubMed] [Google Scholar]
- 30.Kumar M, Dagar V, Gupta VK, Sharma A. In silico docking studies of bioactive natural plant products as putative DHFR antagonists. Med Chem Res. 2014;23:810–7. [Google Scholar]
- 31.Drwal MN, Banerjee P, Dunkel M, Wettig MR, Preissner R. ProTox: A web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014;42:W53–8. doi: 10.1093/nar/gku401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choudhary D, Gupta GK, Khokra SL. Structure based designing and ADME-T studies of butenolide derivatives as potential agents against receptor ICAM-1: A drug target for cerebral malaria. J Comput Sci. 2015;10:156–5. [Google Scholar]


