Abstract
Background
MYCN is amplified in small cell lung cancers and several pediatric tumors, including alveolar rhabdomyosarcomas and neuroblastomas. MYCN protein is known to play a key oncogenic role in both alveolar rhabdomyosarcomas and neuroblastomas. MYCN opposite strand (MYCNOS) is a gene located on the antisense strand to MYCN that encodes alternatively spliced transcripts, two of which (MYCNOS-01 and MYCNOS-02) are known to be expressed in neuroblastoma and small cell lung cancer with reciprocal regulation between MYCNOS-02 and MYCN reported for neuroblastomas. We sought to determine a functional role for MYCNOS-01 in alveolar rhabdomyosarcoma and neuroblastoma cells and identify any associated regulatory effects between MYCN and MYCNOS-01.
Methods
MYCNOS-01, MYCNOS-02 and MYCN expression levels were assessed in alveolar rhabdomyosarcoma and neuroblastoma cell lines and tumor samples from patients using Affymetrix microarray data and quantitative RT-PCR. Following MYCNOS-01 or MYCN siRNA knockdown and MYCNOS-01 overexpression, transcript levels were assayed by quantitative RT-PCR and MYCN protein expression assessed by Western blot and immunofluorescence. Additionally, effects on cell growth, apoptosis and cell cycle profiles were determined by a metabolic assay, caspase activity and flow cytometry, respectively.
Results
MYCNOS-01 transcript levels were generally higher in NB and RMS tumor samples and cell lines with MYCN genomic amplification. RNA interference of MYCNOS-01 expression did not alter MYCN transcript levels but decreased MYCN protein levels. Conversely, MYCN reduction increased MYCNOS-01 transcript levels, creating a negative feedback loop on MYCN protein levels. Reduction of MYCNOS-01 or MYCN expression decreased cell growth in MYCN-amplified alveolar rhabdomyosarcoma and neuroblastoma cell lines. This is consistent with MYCNOS-01-mediated regulation of MYCN contributing to the phenotype observed.
Conclusions
An alternative transcript of MYCNOS, MYCNOS-01, post-transcriptionally regulates MYCN levels and affects growth in MYCN-amplified rhabdomyosarcoma and neuroblastoma cells.
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4129-8) contains supplementary material, which is available to authorized users.
Keywords: MYCNOS, MYCN, Rhabdomyosarcoma, Neuroblastoma, Long non-coding RNA
Background
Several pediatric cancers feature amplification at the chromosomal region 2p24 including alveolar rhabdomyosarcoma (ARMS), neuroblastoma (NB), medulloblastoma, Wilms’ tumor, and retinoblastoma [1–3]. The minimum common region of amplification at 2p24 in rhabdomyosarcoma (RMS) and NB has been found to consistently include the oncogene MYCN and amplification of MYCN is used clinically as a prognostic marker in NB [3–7]. Amplification or overexpression of MYCN leads to dysregulation of proliferation, differentiation and the cell cycle in NB [8] and contributes to cell growth in ARMS [9]. MYCN is also capable of positive auto-regulation as well as auto-suppression in NB, potentially fine-tuning MYCN levels [10–12]. In ARMS, MYCN transcription is driven by PAX3-FOXO1, the protein product of a fusion between the PAX3 and FOXO1 genes that has prognostic significance in these tumors [9, 13]. RMS and NB are a major cause of cancer related death in children with a five-year survival rate of around 50% for high-risk NB cases, including those with MYCN amplification, and 40% for PAX3-FOXO1 positive RMS cases [13, 14].
MYCN opposite-strand (MYCNOS, N-CYM, MYCN-AS1, NYCM, CYMN) is produced by antisense transcription across exon 1 and intron 1 of MYCN that has been shown to be highly expressed in MYCN-amplified NB and small cell lung cancer [15, 16]. Two alternative transcripts denoted MYCNOS-01 and MYCNOS-02 (Additional file 1: Figure S1A) are fully sequence-verified [15]. There is an emerging body of evidence for roles of MYCNOS-02 through an encoded protein (NCYM) that promotes NB tumorigenesis, in particular via its regulation of MYCN expression, and also its role as a long non-coding RNA (lncRNA) [17–22]. NCYM has been shown to mediate expression of MYCN protein by both direct interaction and also indirectly via inhibition of GSK3β, leading to decreased MYCN phosphorylation and a concomitant increase in MYCN protein stability [17]. This was associated with increased tumor growth and metastasis [17]. The same study also concluded that MYCN positively drives the promoter of MYCNOS-02 in an E-box-dependent manner [17]. As well as increasing MYCN protein, NCYM has been found to increase MYCN cleavage to produce the anti-apoptotic protein Myc-nick in NB [22]. NCYM has also been shown to promote aggressiveness in NB by increasing OCT4 expression via its stabilization of MYCN [20].
LncRNAs are commonly defined as transcripts of over 200 nucleotides in length that in general do not code for a protein [23]. MYCNOS-02 lncRNA is able to regulate the usage of two MYCN promoters and therefore expression of different MYCN transcripts via interaction with binding partners such as G3BP1. This in turn results in expression of different isoforms of the MYCN protein [18]. MYCNOS-02 lncRNA has also been found to recruit CTCF to the MYCN promoter to increase recruitment of activating chromatin marks and thus increase MYCN expression [19]. This positive regulation of MYCN suppressed differentiation and increased growth, invasion and metastasis in NB [19]. Additionally, a recent study has shown MYCNOS-02 lncRNA can interact with the RNA-binding protein NonO to indirectly increase MYCN transcript levels post-transcriptionally [21]. Overall, these studies show that both the MYCNOS-02 encoded protein and lncRNA play a role in growth, invasion and metastasis of NB cells [17, 19, 20].
Unlike MYCNOS-02, a functional role for the MYCNOS-01 transcript has not yet been investigated, despite original annotation of its sequence being consistent with a lncRNA [15]. In this study we therefore investigated the role of MYCNOS-01 as a lncRNA in RMS and NB. We demonstrate that MYCNOS-01 post-transcriptionally regulates MYCN protein levels without affecting MYCN mRNA levels, whilst MYCN regulates MYCNOS-01 transcription. We show that silencing of MYCNOS-01 in RMS and NB cell lines with MYCN amplification reduces cell viability, similar to the effects of MYCN reduction. Thus, we conclude that regulation of MYCN by MYCNOS-01 contributes to the reduction in cell growth in RMS and NB cell lines after MYCNOS-01 silencing.
Methods
Translation and Kozak sequence prediction tools
Translation prediction for the MYCNOS-01 transcript sequence was carried out using the ExPASy translate tool (http://web.expasy.org/translate/) [24] and Kozak sequence prediction was carried out using ATGpr (http://atgpr.dbcls.jp/) [25].
Cell culture
Human ARMS cell line RMS-01 was available directly from the authors [26] and the RH30 cell line was a gift from Peter Houghton (St Jude Children’s Research Hospital, Memphis, Tennessee). The human NB cell lines KELLY and SY5Y were obtained from ECACC (cat. No. 92110411) and ATCC (cat. No. CRL-2266) respectively. RMS-01 and RH30 were cultured in DMEM (Thermo Fisher Scientific, MA, USA) and KELLY and SY5Y were cultured in RPMI-1640 medium (Thermo Fisher Scientific, MA, USA) supplemented with 10% Foetal Bovine Serum (FBS), 2 mM L-glutamine and 1% penicillin/streptomycin. The MYCN overexpressing and matched empty vector expressing RH30 lines were generated as previously described in Tonelli et al., (2012) [9] and cultured in DMEM supplemented with 400 μg/ml geneticin. Cells were maintained at 37 °C and 5% CO2. Data from short tandem repeat testing of the cell lines using the GenePrint 10 system (Promega, WI, USA) were compared with records for these cell lines in a repository database or our own archival records. This was consistent with the origin of these cell lines.
Analyses of expression profiling data
Data uploaded to R2 Genomics Analysis and Visualisation Platform (http://r2.amc.nl) were used for analyses. These included 101 RMS samples (ITCC) [6], a set of 19 RMS cell lines (Versteeg) [27], 88 NB samples (Versteeg) and 24 NB cell lines (Versteeg) that had been previously profiled using the Affymetrix GeneChip with the HGU133 Plus2 array. Probe sets could distinguish MYCNOS-01 and MYCNOS-02 transcripts: probe set 216188_at detects MYCNOS-01, set 207028_at detects MYCNOS-02 and set 209757_s_at was used to detect MYCN.
qRT-PCR
MYCNOS-01 and MYCNOS-02 expression data was available from primary sample biopsies from RMS patients. Samples and details of RNA extraction were previously described [6, 28] with appropriate approvals for investigation. RNA was isolated from cell lines using the RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Cell line cDNA was synthesised using the High Capacity cDNA Reverse Transcriptase kit (Applied Biosystems, CA, USA) and patient sample cDNA was synthesised using SuperScript II reverse transcriptase (Invitrogen, CA, USA) following the manufacturers’ protocol. Samples were run for qRT-PCR on the ViiA™ 7 Real-Time PCR System (Applied Biosystems, CA, USA). The following Taqman® probe and primer sets were used: MYCNOS-01 Hs01032821_m1, MYCNOS-02 Hs01040745_m1, MYCN Hs_00232074_m1. Human ACTB (Beta Actin) endogenous control (Applied Biosystems, CA, USA) was used to normalise gene expression. Each sample was run in triplicate. Analysis of MYCN expression and copy number in patient samples is described in [6].
siRNA transfection
Oligonucleotides for specific silencing of MYCNOS-01, MYCNOS-02 and MYCN were transfected into cells using Lipofectamine RNAimax (Invitrogen, CA, USA) according to the manufacturer’s instructions. All siRNAs were obtained from GE Dharmacon (CO, USA). The sequence from 5′ to 3′ for the three siRNAs against MYCNOS-01 were as follows: siMYCNOS-01 1 GGGACAAGAGCACAGUUUCUU, siMYCNOS-01 2 GGUAAGUUAAGGUACAGCCUU, siMYCNOS-01 3 GGAGUAUUUGUUUAGUGCUUU. The sequences for the three siRNAs against MYCNOS-02 were GAAAGAAGGGUAGUCCGAAUU for siMYCNOS-02 1, GACCGAUGCUUCUAACCCAUU for siMYCNOS-02 2, CCGCUUUGACUGCGUGUUGUU for siMYCNOS-02 3. For knockdown of MYCN a pool of three siRNAs was used with sequences GAAGAAAUCGACGUGGUCA, CCAAGGCUGUCACCACAUU, AAUUGAACACGCUCGGACU, as previously described [9]. The control siRNA used was the ON-TARGETplus non-targeting control pool (GE Dharmacon, CO, USA). Samples were analyzed by qRT-PCR, Western blot, flow cytometry or phenotypic assays at time-points indicated in the relevant figures.
Western blotting
Protein lysates were prepared using Cell Lysis Buffer (Cell Signaling Technology, MA, USA) and their concentration measured by the Pierce™ BCA protein assay kit (Thermo Fisher Scientific, MA, USA). Protein samples were resolved by SDS-PAGE and transferred onto PVDF membranes. Blots were incubated with the following primary antibodies: MYCN SC-791 (1:200, Santa Cruz, TX, USA), PARP 9542 (1:1000, Cell Signaling Technology, MA, USA), Phospho-C-Myc (Thr58/Ser62) 04–217 (1:4000, Merck Millipore, MA, USA), GAPDH MAB374 (1:10000, Merck Millipore, MA, USA). Blots were then incubated with rabbit (sc-2313, Santa Cruz, TX, USA) or mouse (A9044, Sigma-Aldrich, MO, USA) horseradish peroxidase-conjugated secondary antibody diluted to 1:4000 depending on primary antibody species. Blots were developed using the ECL™ Prime Western Blotting System (GE Healthcare, IL, USA) on the Chemidoc Touch Imaging System (Bio-Rad, CA, USA). Densitometry was performed using Bio-Rad Image Lab 5.2.1 (Bio-Rad, CA, USA).
Immunofluorescence staining
Cells cultured in chamber slides were fixed with 2% paraformaldehyde for 15 min at room temperature and permeabilised with 0.1% Triton X-100. Samples were blocked in PBS with 10% goat serum and 1% BSA for 1 h. Samples were incubated with primary MYCN antibody SC-53993 (1:500, Santa Cruz, TX, USA) overnight at 4 °C followed by secondary antibody Alexa Fluor 555 goat anti-mouse (1:400, Invitrogen, CA, USA) for 30 min at room temperature. Cells were counterstained with DAPI. Fluorescent images were captured using a Zeiss Axioplan 2 microscope (Oberkochen, Germany) using a 16× objective and a standard exposure time optimised for control treated cells. The sum of the intensity for MYCN staining was measured using Image J software and made relative to the number of cells in that field of view, indicated by DAPI.
Plasmid production and transfection
Full-length MYCNOS-01 transcript (RefSeq NR_110230) was cloned into the pcDNA5/TO vector (Invitrogen, CA, USA) and the construct verified by Sanger sequencing (Eurofins Genomics, Ebersberg, Germany). For plasmid transfection, Lipofectamine 2000 (Invitrogen, CA, USA) was used following the manufacturer’s instructions. Samples were analyzed by qRT-PCR and Western blot at time-points indicated in the relevant figures.
Proteasome inhibition
For protein stability experiments, cells were transfected for a total of 48 h and treated for the final 4 h with either 10 μM MG132 (Sigma-Aldrich, MO, USA) in DMSO or DMSO control. Protein was then extracted from cells for analysis by Western blot.
Cell viability assay
Cells were transfected as six repeats in a 96-well plate to assess the effects of gene knockdown. Cell viability was assessed by the MTS method using the CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega, WI, USA). Fresh media plus 20 μl assay reagent were added at the indicated time-point. After 2.5 h incubation at 37 °C and 5% CO2 the absorbance of each well was measured at 492 nm on a FLUOstar Optima plate reader (BMG Labtech, Ortenberg, Germany).
Apoptosis assay
Cells were transfected in quadruplicate in a 96-well plate and apoptosis measured by evaluating the activation of caspase 3/7 at the indicated time-point by replacement of 50 μl of media with 50 μl of Caspase-Glo® 3/7 Assay (Promega, WI, USA). After 1 h incubation protected from light at room temperature, the samples were transferred to a white-walled 96-well plate and luminescence of each well was read on a FLUOstar Optima plate reader (BMG Labtech, Ortenberg, Germany). The caspase signal intensity was normalised by the absorbance measurement from the corresponding MTS assay.
Flow cytometry
Cells were fixed in 70% ethanol at -20 °C overnight, washed with PBS and resuspended in 1 ml PBS containing 100 μg/ml RNase A and 40 μg/ml Propidium Iodide. Samples were incubated for 30 min at 37 °C then analyzed on a BD™ LSRII Flow Cytometer (BD Biosciences, CA, USA).
Statistical analysis
Graphs represent means ± standard deviation from multiple independent experiments as stated in figure legends. Statistical significance was measured by unpaired two-tailed Student t-test or by one-way analysis of variance (ANOVA) with post hoc Dunnett’s test for multiple comparisons. For linear correlation studies of gene expression Pearson’s coefficient (R) was calculated between each pair of variables to indicate the strength of the linear association. p < 0.05 was considered significant and indicated by a single asterisk, p < 0.01 is indicated by a double asterisk, and p < 0.001 is indicated by a triple asterisk.
Results
MYCNOS-01 is predicted to be non-protein coding and correlations between MYCNOS and MYCN transcript levels in RMS and NB patients and cell lines
Previous data showed protein encoding potential for MYCNOS-02 transcripts [15]. In contrast, translation and Kozak sequence analysis for MYCNOS-01 predicted transcripts to be non-protein coding since the probability any start codon present in the MYCNOS-01 sequence is an initiation codon is very low (Additional file 1: Figure S1B and C). The relationship between MYCN and MYCNOS-01 transcript levels was then investigated by mining publically available expression profiling data for RMS and NB samples from patients and cell lines. A significant correlation between levels of MYCNOS-01 and MYCN expression and MYCNOS-02 and MYCN expression was identified considering all RMS samples (Fig. 1a and b) and cell lines (Fig. 1c and d). There was also a significant positive correlation between both MYCNOS transcripts and MYCN transcript expression in NB patient samples (Fig. 1e and f) and cell lines (Fig. 1g and h) that is consistent with the literature for MYCNOS-02 [17, 19]. For NB patient samples and cell lines, levels of MYCNOS-01 and 02 were higher in cases with MYCN amplification (Fig. 1e-h) versus cases without. Amplification at 2p24 in RMS is also associated with high MYCNOS-01 and MYCNOS-02 expression levels, as indicated in data mined for the cell lines (Fig. 1c and d) and qRT-PCR analyses of a limited number of cases with known MYCN amplification status as well as the cell lines (Additional file 2: Figure S2A–D). MYCN and MYCNOS-01 transcript levels in RMS and NB showed no significant correlations when MYCN amplified cases were excluded (RMS p = 0.72, R = 0.078; NB p = 0.42, R = 0.097).
Fig. 1.
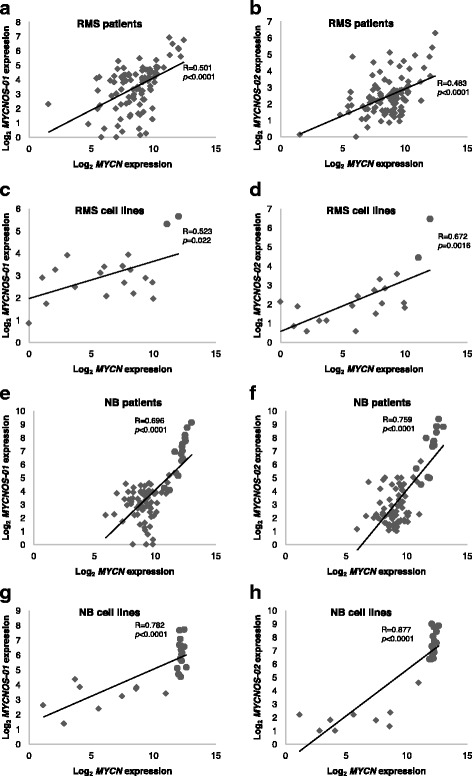
Correlation of MYCNOS transcripts and MYCN transcript expression in RMS and NB. a Correlation between MYCNOS-01 and MYCN measured by Affymetrix microarray in 101 RMS patient samples (R = 0.501, p < 0.0001) and (b) correlation between MYCNOS-02 and MYCN in these patient samples (R = 0.483, p < 0.0001). c Correlation between MYCNOS-01 and MYCN measured by Affymetrix microarray in 19 RMS cell lines (R = 0.523, p = 0.022) and (d) correlation between MYCNOS-02 and MYCN (R = 0.672, p = 0.0016). e Correlation between MYCNOS-01 and MYCN measured by Affymetrix microarray in 88 NB patient samples (R = 0.696, p < 0.0001) and (f) correlation between MYCNOS-02 and MYCN in these patient samples (R = 0.759, p < 0.0001). g Correlation between MYCNOS-01 and MYCN measured by Affymetrix microarray in 24 NB cell lines (R = 0.782, p < 0.0001) and (h) correlation between MYCNOS-02 and MYCN (R = 0.877, p < 0.0001). Data derived from R2 genomics analysis and visualisation platform. MYCN-amplified cases or cell lines are depicted with circles
MYCNOS-01 regulates MYCN protein but not transcript levels in MYCN-amplified RMS and NB cells
To determine whether MYCNOS-01 regulates MYCN transcript levels, which may be consistent with the correlations in their expression levels in NB and RMS derived samples, we performed siRNA-mediated silencing of MYCNOS-01 in cell lines. RMS cell lines tested had either high (RMS-01) or intermediate (RH30) MYCN expression levels, and NB cell lines used had high (KELLY) and low (SY5Y) levels of MYCN [7–9]. The relative MYCNOS and MYCN transcript levels for the four cell lines used are indicated in Additional file 2: Figure S2E.
In high MYCN-expressing lines, no consistent effect on MYCN transcript levels was observed after silencing of MYCNOS-01 for 72 h (Fig. 2a-d). To determine whether MYCNOS-01 is regulating MYCN post-transcriptionally, we also assessed MYCN protein levels by Western blotting. MYCN protein was decreased in cells transfected with MYCNOS-01 siRNAs compared to non-targeting control in high MYCN-expressing cell lines RMS-01 and KELLY (Fig. 2e, f). This is supported by the decrease in MYCN immunofluorescence signal after MYCNOS-01 depletion compared to control in these cell lines (Additional file 3: Figure S3A-F). Furthermore, MYCNOS-01 overexpression has no effect on MYCN transcript levels but causes a slight increase in MYCN protein; the effect observed is limited since RMS-01 and KELLY cells express very high basal levels of MYCN (Additional file 4: Figure S4). Experiments on RMS-01 cells including a proteasome inhibitor showed no increase in phosphorylated MYCN with MYCNOS-01 knockdown, indicating that MYCN protein stability was not affected (Additional file 5: Figure S5). However, MYCNOS-01 silencing had no strong effect on either MYCN transcript or protein levels in the ARMS cell line RH30 with intermediate MYCN expression (Fig. 3a-c). Due to the low expression levels of MYCN protein in the NB cell line SY5Y, no bands were visible by Western blot but there was similarly no MYCNOS-01-mediated effect on MYCN transcript expression (Fig. 3d and e).
Fig. 2.
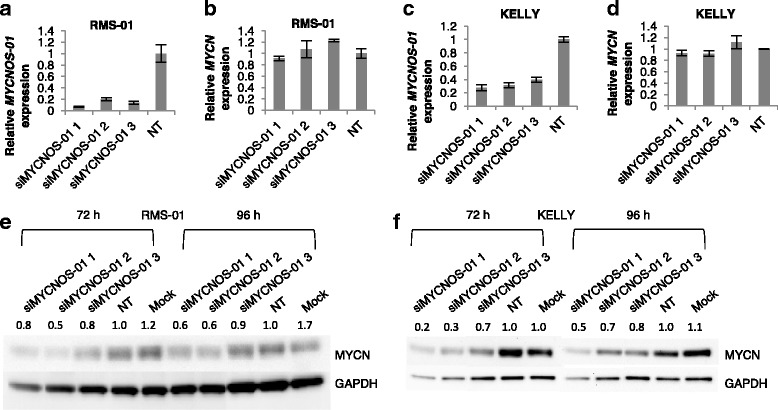
MYCN transcript and protein expression after MYCNOS-01 knockdown in high MYCN-expressing RMS and NB. qRT-PCR detecting MYCNOS-01 transcript level after MYCNOS-01 knockdown for 72 h with three siRNAs in (a) RMS-01 and (c) KELLY. Corresponding MYCN transcript level shown in (b) RMS-01 and (d) KELLY. Expression relative to NT control. (e) and (f) MYCN protein levels after MYCNOS-01 knockdown for 72 h and 96 h with three siRNAs in RMS-01 and KELLY respectively. GAPDH used as loading control. Densitometry values shown above each blot normalised to GAPDH and relative to NT control. Data representative of three repeats. NT = non-targeting control. Relative expression of transcripts in these cell lines are indicated in Additional file 2: Figure S2E
Fig. 3.
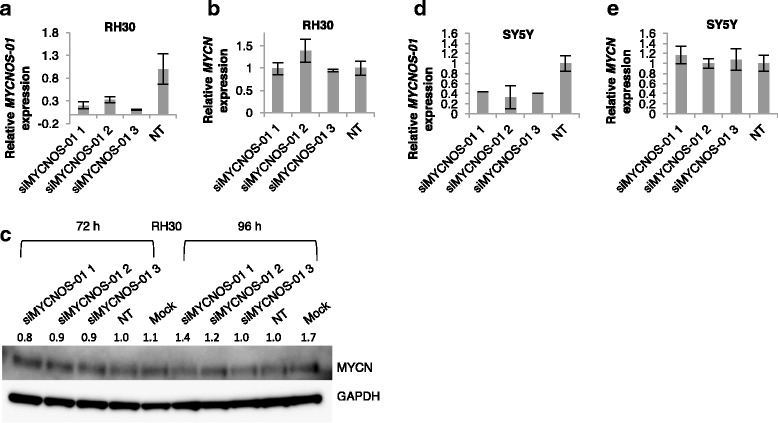
MYCN transcript and protein expression after MYCNOS-01 knockdown in intermediate and low MYCN-expressing RMS and NB. qRT-PCR detecting MYCNOS-01 transcript level after MYCNOS-01 knockdown for 72 h with three siRNAs in (a) RH30 and (d) SY5Y. Corresponding MYCN transcript level shown in (b) RH30 and (e) SY5Y. Expression relative to NT control. (c) MYCN protein levels after MYCNOS-01 knockdown for 72 h and 96 h with three siRNAs in RH30. GAPDH used as loading control. Densitometry values shown above each blot normalised to GAPDH and relative to NT control. Data representative of three repeats. NT = non-targeting control. Relative expression of transcripts in these cell lines are indicated in Additional file 2: Figure S2E
As a role for MYCNOS-02 in RMS has not been previously evaluated, we also performed siRNA-mediated silencing of MYCNOS-02 to determine the effects on MYCN mRNA and protein expression in RMS-01 and KELLY cells (Additional file 6: Figure S6). Similar levels of MYCNOS-02 silencing were observed with all 3 siRNAs, but there was no consistent effect on MYCN expression at the RNA or protein level.
Overall, these results demonstrate that MYCNOS-01 reduces MYCN protein levels in MYCN-amplified RMS and NB cell lines but not through altering MYCN phosphorylation.
MYCN regulates MYCNOS-01 transcript levels
Silencing MYCN in RMS and NB caused a significant increase in MYCNOS-01 expression in both high MYCN-expressing (RMS-01, KELLY) and intermediate MYCN-expressing (RH30) RMS and NB cell lines (Fig. 4a-i). Conversely, overexpressing MYCN in RH30 decreased MYCNOS-01 expression (Fig. 4j-l). MYCN was found to similarly regulate MYCNOS-02 in RMS and NB with MYCN amplification (Additional file 7: Figure S7). Overall, these results indicate MYCN negatively regulates MYCNOS-01 expression whilst MYCNOS-01 positively regulates MYCN protein levels.
Fig. 4.
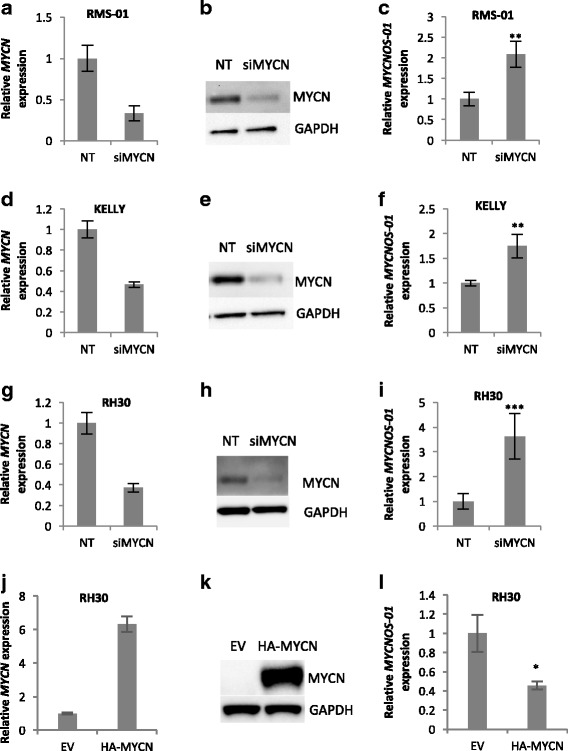
Effect of MYCN knockdown on MYCNOS-01 transcript expression in RMS and NB. a qRT-PCR and (b) Western blot for MYCN, and (c) qRT-PCR for MYCNOS-01 after treatment with three pooled MYCN siRNAs for 72 h in RMS-01. d-f in KELLY as for (a)-(c). g-i in RH30 as for (a)-(c). j qRT-PCR and (k) Western bot for MYCN, and (l) qRT-PCR for MYCNOS-01 in RH30 stably transfected with empty vector (EV) or HA-MYCN expressing vector. Graphs and Western blots representative of three repeats. NT = non-targeting control
Decreasing MYCNOS-01 levels results in decreased cell viability in MYCN-amplified RMS and NB cells
We next investigated the phenotypic effects of MYCNOS-01 and MYCNOS-02 depletion on RMS (RMS-01, RH30) and NB (KELLY, SY5Y) cells. Decreasing MYCNOS-01 expression resulted in a significant decrease in cell viability compared to negative control with all MYCNOS-01 siRNAs in MYCN-amplified RMS-01 and KELLY cells (Fig. 5a and b), similar to the reduction in cell viability that is seen with siRNA-mediated MYCN silencing (Fig. 5a and b). Silencing MYCNOS-01 did not reduce MYCN protein lower than direct MYCN silencing (Fig. 5c), but showed a similar or greater effect on RMS cell viability (Fig. 5a) raising the possibility that MYCNOS-01 may have targets in addition to MYCN in RMS. Decreasing MYCNOS-02 expression also significantly decreased cell viability in these cell lines (Additional file 8: Figure S8A and B). In contrast, silencing of MYCNOS-01 did not significantly affect cell viability in intermediate (RH30) and low (SY5Y) MYCN expressing cells overall (Fig. 6a and b). However, although the effect was less for RH30 compared to RMS-01, MYCN knockdown did significantly decrease cell growth as we have previously reported [9] (Fig. 6a). MYCN reduction in SY5Y had no effect, although levels of MYCN are very low (Fig. 6b). There was no significant increase in caspase 3/7 activation or PARP cleavage in MYCNOS-01 or MYCN siRNA treated cells compared to non-targeting control indicating apoptosis was not induced (Additional file 9: Figure S9). However, MYCNOS-02 knockdown promoted apoptosis in RMS-01 and KELLY cells (Additional file 10: Figure S10). Decreasing MYCNOS-01 expression had no effect on cell cycle progression but silencing MYCN caused a G1 arrest in both RMS-01 and KELLY (Additional file 11: Figure S11). Based on all our results, we propose a feedback model for MYCNOS-01 and MYCN regulation in RMS and NB (Fig. 7).
Fig. 5.
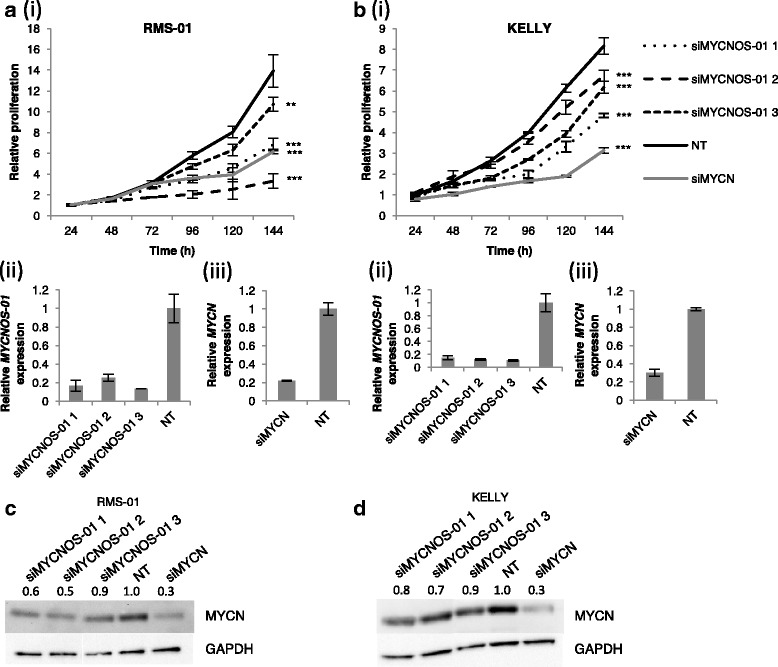
Knockdown of MYCNOS-01 inhibits cell viability in high MYCN-expressing RMS and NB. (ai) MTS assay showing cell viability of RMS-01 up to 144 h after transfection with three MYCNOS-01 siRNAs or three pooled MYCN siRNAs relative to NT siRNA. (bi) KELLY as in (ai). Corresponding qRT-PCR for MYCNOS-01 (ii) and MYCN (iii) at 24 h shown below line graph. c MYCN protein levels after MYCNOS-01 or MYCN knockdown for 72 h in RMS-01 and (d) in KELLY. Densitometry values shown above each blot normalised to GAPDH and relative to NT control. Data representative of three repeats. Statistical analysis relative to non-targeting control. NT = non-targeting
Fig. 6.
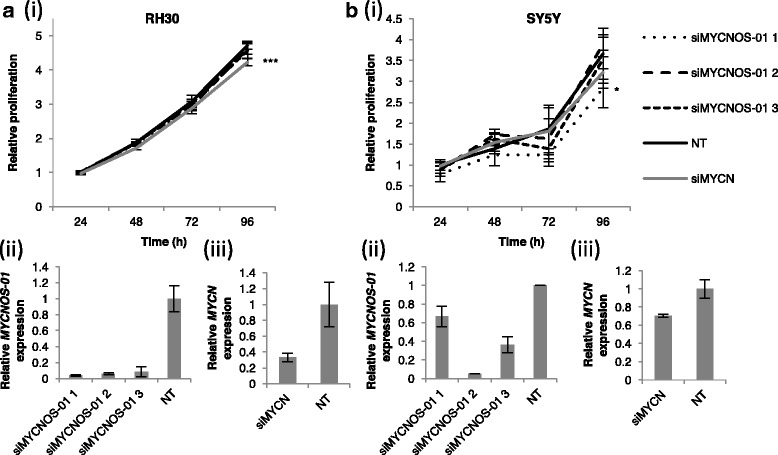
Knockdown of MYCNOS-01 does not inhibit cell viability in intermediate or low MYCN-expressing RMS and NB. (ai) MTS assay showing cell viability of RH30 up to 96 h after transfection with three MYCNOS-01 siRNAs or three pooled MYCN siRNAs relative to NT siRNA. (bi) SY5Y as in (ai). Corresponding qRT-PCR for MYCNOS-01 (ii) and MYCN (iii) at 24 h shown below line graph. Data representative of three repeats. Statistical analysis relative to non-targeting control. NT = non-targeting
Fig. 7.
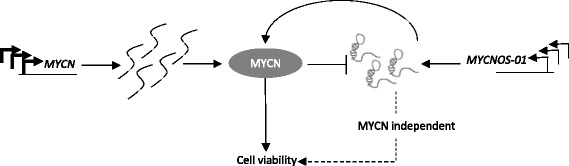
Schematic of proposed model for regulation between MYCN and MYCNOS-01. MYCN and MYCNOS-01 expression are both driven by genomic amplification, although MYCN is expressed at higher levels than MYCNOS-01. MYCNOS-01 positively regulates MYCN protein levels and MYCN negatively regulates MYCNOS-01 transcript levels as a form of negative feedback on MYCN. MYCNOS-01-mediated regulation of MYCN has effects on cell viability but there is also likely a MYCN-independent signaling pathway
Discussion
In this study, we have shown that MYCNOS-01 and MYCNOS-02 can play important roles in RMS as well as NB cell growth, at least in part via their regulation of MYCN. Roles for MYCNOS-01 in RMS and NB and MYCNOS-02 in RMS have not been previously explored whilst our data for the effect of MYCNOS-02 on growth of NB cells is consistent with previous findings [17–22]. Regulation of MYCN by MYCNOS-01 and MYCNOS-02 was readily apparent in MYCN-amplified RMS and NB, which express these transcripts at high levels, presumably as a result of their co-amplification at the genomic level. In contrast, effects of MYCNOS transcripts on MYCN protein levels in RMS and NB without high level MYCN amplification were either less marked or not seen. The positive regulation of MYCN by MYCNOS-01 and MYCNOS-02 likely contribute to the cell growth of RMS and NB. This is consistent with the phenotypic dose dependent effects and dependencies of RMS and NB cells on MYCN levels seen in this and previous studies [9, 29, 30].
In addition to the positive regulation of MYCN by MYCNOS-01, MYCN also negatively regulates MYCNOS-01 transcription, summarized in Fig. 7. As MYCN protein has been found to be recruited to its own intron 1 [10], in line with the transcription start site for MYCNOS-01 on the opposite strand, this binding activity could be involved in MYCN-mediated regulation of MYCNOS-01. Previous studies have also identified indirect MYCN negative feedback mechanisms involving trans-acting factors [12]. This is likely a mechanism that fine-tunes MYCN expression levels.
An increasing number of lncRNAs have now been characterized and many have been linked to cancer progression [31]. Here we have shown that MYCNOS-01 can act as a cis-antisense lncRNA on its sense partner MYCN. Although MYCN transcript expression was not regulated by MYCNOS-01, we have identified a post-transcriptional role for this lncRNA in regulating MYCN protein levels. This is consistent with the lack of correlation we found between the two transcripts in non-amplified lines.
There are several examples of lncRNAs that regulate protein partners post-transcriptionally without affecting transcript expression [32–35]. For example, the lncRNA PVT1 has been shown to be required for increasing MYC protein stability and high expression levels in 8q24-amplified cancers [32]. In our study, we found no evidence that MYCNOS-01 altered the stability of MYCN protein via Thr58/Ser62 phosphorylation. Another example of post-transcriptional regulation is the lncRNA treRNA, which has been shown to play a role in tumor invasion and metastasis in breast cancer, and which regulates the translation of E-cadherin mRNA in these cells via redistribution of CDH1 to low molecular weight polysomes to suppress translation [34]. Potentially MYCNOS-01 could have a similar mechanism to regulate translation efficiency of MYCN and thus its protein expression. In addition, the lncRNA BACE1-AS regulates translation of BACE1 by masking the binding site for miR-485-5p, thus preventing miRNA-induced translational repression and mRNA decay [35]. Another possibility therefore is that MYCNOS-01 interacts with an miRNA that targets MYCN for degradation, therefore increasing MYCN protein expression by sequestering away a negative regulating factor. The above examples indicate possible post-transcriptional mechanisms that could be explored for MYCNOS-01-mediated regulation of MYCN. Further investigations are required to determine molecular interactions with MYCNOS-01 and how these regulate MYCN protein levels.
Both MYCNOS-01 and MYCNOS-02 shown in this study in RMS and NB, and MYCNOS-02 shown previously in NB, regulate MYCN protein levels [17, 19, 20]. However, data on whether MYCNOS-02 is able to regulate MYCN at the transcriptional level is conflicting. One report for MYCNOS-02 indicates silencing of MYCNOS-02 does not affect MYCN expression at the transcriptional level [17]. However, other studies suggest that MYCNOS-02 can affect MYCN transcript expression in NB [18, 19, 21]. MYCNOS-02 has been found to interact with CTCF to affect chromatin remodeling at the MYCN promoter therefore MYCN transcript expression [19]. However, in silico prediction techniques suggest CTCF interacts with a region of MYCNOS-02 that does not overlap with the sequence of MYCNOS-01, supporting the possibility that the two transcripts could have different binding partners that are involved in MYCN regulation. It is possible for two overlapping lncRNAs from the same locus to have different characteristics and function. For example, the CCAT1 locus encodes CCAT1-L that is located in the nucleus and positively regulates MYC transcription and CCAT1-S that is mainly located in the cytoplasm with no effect on MYC transcript levels [36–38].
Previous studies of MYCNOS-02 have identified its role in NB tumor growth and metastasis. In vitro, MYCNOS-02 can suppress differentiation and promote metastasis, invasion and cell proliferation partially due to its indirect regulation of MYCN [19]. Our study found MYCNOS-01 also plays a role in MYCN-amplified RMS and NB cell viability, although no effect was seen on cell cycle progression. Often a decrease in proliferation occurs with concomitant cell cycle arrest, however it is possible for these two effects to be separated. For example, one study found decreasing the tumor suppressors RPL5 or RPL11 resulted in a reduction in ribosome content and translation capacity, causing cells to progress at a lower rate through all stages of the cell cycle thus resulting in decreased proliferation without cell cycle arrest [39].
MYCNOS-01 was also found to play a role in cell growth via regulation of MYCN; silencing of MYCNOS-01 resulted in a reduction in MYCN protein levels. However, MYCNOS-01 knockdown did produce a slightly different phenotype to MYCN knockdown due to differences in MYCN protein levels achieved. We have previously identified that using different molecular tools to diminish MYCN can affect the strength of phenotype detected depending on the magnitude and endurance of MYCN reduction [9]. MYCNOS-01 knockdown did not decrease MYCN protein sufficiently to produce a G1 arrest, in contrast to direct MYCN knockdown. However, MYCNOS-01 reduction affecting other protein targets and signaling pathways that contribute to the phenotype observed cannot be excluded. Further defining how MYCNOS-01 regulates MYCN, and possibly other proteins, may lead to new approaches to perturb the clinically aggressive phenotype of RMS and NB tumors.
Conclusions
MYCNOS-01 can be added to the growing list of lncRNAs involved in tumorigenesis; MYCNOS-01 positively regulates MYCN and MYCN negatively regulates MYCNOS-01, potentially to fine-tune MYCN protein levels. MYCNOS-01 affects cell growth of MYCN-amplified RMS and NB and could also play a role in other MYCN-driven cancers. Although directly targeting lncRNAs and MYCN is challenging, future therapeutic strategies could disrupt specific lncRNA-protein interactions to reduce MYCN levels.
Additional files
Figure S1. Relative genomic positions of MYCNOS-01, MYCNOS-02 and MYCN and protein-coding potential of MYCNOS-01. (A) Diagram illustrating relative positions of MYCNOS-01, MYCNOS-02 and MYCN exons. Modified from UCSC Genome Browser. (B) Predicted sequence for three forward reading frames of MYCNOS-01. (B) Table of results for ATGpr analysis of MYCNOS-01. The reliability score indicates the likelihood a start codon is in the correct context for translation initiation, with a score closer to one indicating increased likelihood. (PDF 594 kb)
Figure S2. Correlation of MYCNOS transcripts and MYCN transcript expression in RMS measured by qRT-PCR. (A) Correlation between MYCNOS-01 and MYCN measured by qRT-PCR in 80 RMS patient samples (R = 0.281, p = 0.012) and (B) correlation between MYCNOS-02 and MYCN in 53 of these patient samples (R = 0.308, p = 0.025). (C) Correlation between MYCNOS-01 and MYCN measured by qRT-PCR in 18 RMS cell lines (R = 0.821, p < 0.0001) and (D) correlation between MYCNOS-02 and MYCN in 14 of these cell lines (R = 0.980, p < 0.0001). MYCN-amplified cases or cell lines are depicted with circles. RMS-01 and RH30 are indicated specifically. (E) Expression of MYCNOS-01, MYCNOS-02 and MYCN transcripts measured by qRT-PCR in the cell lines used in experiments; RMS-01, RH30, KELLY and SY5Y. Expression relative to normal cells. (PDF 50 kb)
Figure S3. Effect of MYCNOS-01 knockdown on MYCN protein expression in RMS and NB measured by immunofluorescence. (A) Immunofluorescence staining of MYCN (red) with DAPI shown in blue after MYCNOS-01 knockdown for 72 h with three siRNAs in RMS-01. (B) Quantification of MYCN staining intensity from (A) relative to total number of cells and normalised to non-targeting control. (C) qRT-PCR detecting MYCNOS-01 transcript level matching experiment shown in (A). (D)-(F) in KELLY as for (A)-(C). Data representative of three repeats. Statistical analysis relative to non-targeting control. NT = non-targeting. (PDF 74664 kb)
Figure S4. Effect of MYCNOS-01 overexpression on MYCN transcript and protein expression. qRT-PCR detecting MYCNOS-01 transcript level after 72 h transient transfection with two MYCNOS-01 overexpressing vectors or empty vector control in (A) RMS-01 and (D) KELLY. Corresponding MYCN transcript level shown in (B) RMS-01 and (E) KELLY. Expression relative to empty vector control. MYCN protein levels after MYCNOS-01 overexpression for 72 h shown in (C) RMS-01 and (F) KELLY. GAPDH used as loading control. Densitometry values shown above each blot normalised to GAPDH and relative to empty vector control. EV = empty vector. (PDF 3518 kb)
Figure S5. Effect of MYCNOS-01 knockdown on MYCN protein stability. (A) RMS-01 cells treated with three MYCNOS-01 siRNAs for 48 h including 4 h treatment with DMSO or MG132. Densitometry values shown above each blot normalised to GAPDH and relative to NT control for each condition. Blots representative of experiments run in triplicate. (B) MYCNOS-01 transcript expression after MYCNOS-01 knockdown for 24 h measured by qRT-PCR. NT = non-targeting control. (PDF 584 kb)
Figure S6. Effect of MYCNOS-02 knockdown on MYCN transcript and protein expression in RMS and NB. qRT-PCR detecting MYCNOS-02 transcript level after MYCNOS-02 knockdown with three siRNAs in (A) RMS-01 and (D) KELLY. Corresponding MYCN transcript level shown in (B) RMS-01 and (E) KELLY. Expression relative to NT control. Western blots showing MYCN protein levels after MYCNOS-02 knockdown with three siRNAs shown in (C) RMS-01 and (F) KELLY. GAPDH used as loading control. Densitometry values shown above each blot normalised to GAPDH and relative to NT control. Data representative of 3 repeats. NT = non-targeting control. Relative expression of transcripts in these cell lines are indicated in Additional file 2: Figure S2E. (PDF 3709 kb)
Figure S7. Effect of MYCN knockdown on MYCNOS-02 transcript expression in RMS and NB. (A) qRT-PCR and (B) Western blot for MYCN, and (C) qRT-PCR for MYCNOS-02 after treatment with three pooled MYCN siRNAs for 72 h in RMS-01. (D)-(F) in KELLY as for (A)-(C). Graphs and Western blots representative of three repeats. NT = non-targeting control. (PDF 4860 kb)
Figure S8. Knockdown of MYCNOS-02 inhibits cell viability. (Ai) MTS assay showing cell viability of RMS-01 over 96 h after transfection with three MYCNOS-02 siRNAs relative to NT siRNA. (Bi) KELLY as in (Ai). Corresponding qRT-PCR of MYCNOS-02 (ii) at 24 h shown below line graph. Data representative of two repeats. Statistical analysis relative to non-targeting control. NT = non-targeting. (PDF 82 kb)
Figure S9. Knockdown of MYCNOS-01 does not induce apoptosis. (A) Caspase 3/7 signalling intensity in RMS-01 after transfection with three MYCNOS-01 siRNAs or three pooled MYCN siRNAs for 96 h relative to cell viability and normalised to NT siRNA. (B) Western blot of total and cleaved PARP in RMS-01 after transfection with three MYCNOS-01 siRNAs or three pooled MYCN siRNAs. (C, D) in KELLY, (E, F) in RH30 and (G, H) in SY5Y as for (A, B). Data representative of three repeats. NT = non-targeting, tPARP = total PARP, cPARP = cleaved PARP. (PDF 5703 kb)
Figure S10. Knockdown of MYCNOS-02 does induce apoptosis. (A) Caspase 3/7 signalling intensity in RMS-01 after transfection with three MYCNOS-02 siRNAs for 96 h relative to cell viability and normalised to NT siRNA. (B) Western blot of total and cleaved PARP in RMS-01 after transfection with three MYCNOS-02 siRNAs. (C, D) in KELLY as for (A, B). Data representative of two repeats. NT = non-targeting, tPARP = total PARP, cPARP = cleaved PARP. (PDF 3672 kb)
Figure S11. Knockdown of MYCNOS-01 does not affect cell cycle progression. (A) Flow cytometry 96 h post-transfection with three MYCNOS-01 siRNAs or three pooled MYCN siRNAs in RMS-01, (B) KELLY, (C) RH30, and (D) SY5Y. Graphs show an average of three repeats. NT = non-targeting. (PDF 111 kb)
Acknowledgements
We thank the Children’s Cancer and Leukaemia Group (CCLG) Tissue Bank for access to samples, and contributing CCLG centres, including members of the ECMC pediatric network. The CCLG Tissue Bank is funded by Cancer Research UK and CCLG. We acknowledge NHS funding to the NIHR Biomedical Research Center.
Funding
This work was supported by a studentship for EOB from the Institute of Cancer Research and Cancer Research UK grants C5066/A8815, that included support for ZSW, C5066/A9541 that supported ASM and C5066/A1099 that funded sample collection and processing.
Availability of data and materials
The public datasets supporting the conclusions of this article are available in the R2 Genomics Analysis and Visualisation Platform repository, (http://r2.amc.nl).
Abbreviations
- lncRNA
long non-coding RNA
- NB
Neuroblastoma
- RMS
Rhabdomyosarcoma
Authors’ contributions
EO carried out the majority of the experiments, interpretation of the data and drafted the manuscript. JLS helped with designing experiments and the interpretation of results. ASM was involved in the design of initial experiments. ZSW carried out qRT-PCR of RMS patient samples. ZSW and JMS conceived the study, supervised the investigation and interpretation of data as well as drafting the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study of patient materials was conducted according to the principles expressed in the Declaration of Helsinki. The study of rhabdomyosarcoma samples had both local and national ethical approval (Local Research Ethics Committee No 1836 and 2015 and Multi-Regional Research Ethics Committee 98/4/023) and complied with requirements for informed written consent from all patients or their guardians.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4129-8) contains supplementary material, which is available to authorized users.
Contributor Information
Eleanor M. O’Brien, Email: eleanor.obrien@icr.ac.uk
Joanna L. Selfe, Email: joanna.selfe@icr.ac.uk
Ana Sofia Martins, Email: anasofia_ba@hotmail.com.
Zoë S. Walters, Email: zoe.walters@icr.ac.uk
Janet M. Shipley, Phone: +44 20 8722 4273, Email: janet.shipley@icr.ac.uk
References
- 1.Schwab M. MYCN in neuronal tumours. Cancer Lett. 2004;204:179–187. doi: 10.1016/S0304-3835(03)00454-3. [DOI] [PubMed] [Google Scholar]
- 2.Gordon AT, Brinkschmidt C, Anderson J, Coleman N, Dockhorn-Dworniczak B, Pritchard-Jones K, Shipley J. A novel and consistent amplicon at 13q31 associated with alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2000;28:220–226. doi: 10.1002/(SICI)1098-2264(200006)28:2<220::AID-GCC11>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 3.Barr FG, Duan F, Smith LM, Gustafson D, Pitts M, Hammond S, Gastier-Foster JM. Genomic and clinical analyses of 2p24 and 12q13-q14 amplification in alveolar rhabdomyosarcoma: a report from the Children's oncology group. Genes Chromosomes Cancer. 2009;48:661–672. doi: 10.1002/gcc.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman LC, Castleberry RP, Cantor A, Joshi V, Cohn SL, Smith EI, Yu A, Brodeur GM, Hayes FA, Look AT. Genetic staging of unresectable or metastatic neuroblastoma in infants: a pediatric oncology group study. J Natl Cancer Inst. 1997;89:373–380. doi: 10.1093/jnci/89.5.373. [DOI] [PubMed] [Google Scholar]
- 5.Lipska BS, Koczkowska M, Wierzba J, Ploszynska A, Iliszko M, Izycka-Swieszewska E, Adamkiewicz-Drozynska E, Limon J. On the significance of germline cytogenetic rearrangements at MYCN locus in neuroblastoma. Mol Cytogenet. 2013;6:43. doi: 10.1186/1755-8166-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson D, Lu YJ, Gordon T, Sciot R, Kelsey A, Fisher C, Poremba C, Anderson J, Pritchard-Jones K, Shipley J. Relationship between MYCN copy number and expression in rhabdomyosarcomas and correlation with adverse prognosis in the alveolar subtype. J Clin Oncol. 2005;23:880–888. doi: 10.1200/JCO.2005.11.078. [DOI] [PubMed] [Google Scholar]
- 7.Weber A, Starke S, Bergmann E, Christiansen H. The coamplification pattern of the MYCN amplicon is an invariable attribute of most MYCN-amplified human neuroblastomas. Clin Cancer Res. 2006;12:7316–7321. doi: 10.1158/1078-0432.CCR-06-0837. [DOI] [PubMed] [Google Scholar]
- 8.Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29:1249–1259. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tonelli R, McIntyre A, Camerin C, Walters ZS, Di Leo K, Selfe J, Purgato S, Missiaglia E, Tortori A, Renshaw J, et al. Antitumor activity of sustained N-myc reduction in rhabdomyosarcomas and transcriptional block by antigene therapy. Clin Cancer Res. 2012;18:796–807. doi: 10.1158/1078-0432.CCR-11-1981. [DOI] [PubMed] [Google Scholar]
- 10.Suenaga Y, Kaneko Y, Matsumoto D, Hossain MS, Ozaki T, Nakagawara A. Positive auto-regulation of MYCN in human neuroblastoma. Biochem Biophys Res Commun. 2009;390:21–26. doi: 10.1016/j.bbrc.2009.09.044. [DOI] [PubMed] [Google Scholar]
- 11.Sivak LE, Tai KF, Smith RS, Dillon PA, Brodeur GM, Carroll WL. Autoregulation of the human N-myc oncogene is disrupted in amplified but not single-copy neuroblastoma cell lines. Oncogene. 1997;15:1937–1946. doi: 10.1038/sj.onc.1201363. [DOI] [PubMed] [Google Scholar]
- 12.Kim MK, Carroll WL. Autoregulation of the N-myc gene is operative in neuroblastoma and involves histone deacetylase 2. Cancer. 2004;101:2106–2115. doi: 10.1002/cncr.20626. [DOI] [PubMed] [Google Scholar]
- 13.Missiaglia E, Williamson D, Chisholm J, Wirapati P, Pierron G, Petel F, Concordet JP, Thway K, Oberlin O, Pritchard-Jones K, et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol. 2012;30:1670–1677. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 14.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F, Schleiermacher G, Park JR, et al. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;33:3008–3017. doi: 10.1200/JCO.2014.59.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armstrong BC, Krystal GW. Isolation and characterization of complementary DNA for N-cym, a gene encoded by the DNA strand opposite to N-myc. Cell Growth Differ. 1992;3:385–390. [PubMed] [Google Scholar]
- 16.Krystal GW, Armstrong BC, Battey JF. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990;10:4180–4191. doi: 10.1128/MCB.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suenaga Y, Islam SM, Alagu J, Kaneko Y, Kato M, Tanaka Y, Kawana H, Hossain S, Matsumoto D, Yamamoto M, et al. NCYM, a cis-antisense gene of MYCN, encodes a de novo evolved protein that inhibits GSK3beta resulting in the stabilization of MYCN in human neuroblastomas. PLoS Genet. 2014;10:e1003996. doi: 10.1371/journal.pgen.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vadie N, Saayman S, Lenox A, Ackley A, Clemson M, Burdach J, Hart J, Vogt PK, Morris KV. MYCNOS functions as an antisense RNA regulating MYCN. RNA Biol. 2015;12:893–899. doi: 10.1080/15476286.2015.1063773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Li D, Pu J, Mei H, Yang D, Xiang X, Qu H, Huang K, Zheng L, Tong Q. CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene. 2016;35(27):3565-76. [DOI] [PubMed]
- 20.Kaneko Y, Suenaga Y, Islam SM, Matsumoto D, Nakamura Y, Ohira M, Yokoi S, Nakagawara A. Functional interplay between MYCN, NCYM, and OCT4 promotes aggressiveness of human neuroblastomas. Cancer Sci. 2015;106:840–847. doi: 10.1111/cas.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu PY, Atmadibrata B, Mondal S, Tee AE, Liu T. NCYM is upregulated by lncUSMycN and modulates N-Myc expression. Int J Oncol. 2016;49(6):2464-70. [DOI] [PubMed]
- 22.Shoji W, Suenaga Y, Kaneko Y, Islam SM, Alagu J, Yokoi S, Nio M, Nakagawara A. NCYM promotes calpain-mediated Myc-nick production in human MYCN-amplified neuroblastoma cells. Biochem Biophys Res Commun. 2015;461:501–506. doi: 10.1016/j.bbrc.2015.04.050. [DOI] [PubMed] [Google Scholar]
- 23.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 24.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salamov AA, Nishikawa T, Swindells MB. Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics. 1998;14:384–390. doi: 10.1093/bioinformatics/14.5.384. [DOI] [PubMed] [Google Scholar]
- 26.Garvin AJ, Stanley WS, Bennett DD, Sullivan JL, Sens DA. The in vitro growth, heterotransplantation, and differentiation of a human rhabdomyosarcoma cell line. Am J Pathol. 1986;125:208–217. [PMC free article] [PubMed] [Google Scholar]
- 27.Missiaglia E, Selfe J, Hamdi M, Williamson D, Schaaf G, Fang C, Koster J, Summersgill B, Messahel B, Versteeg R, et al. Genomic imbalances in rhabdomyosarcoma cell lines affect expression of genes frequently altered in primary tumors: an approach to identify candidate genes involved in tumor development. Genes Chromosomes Cancer. 2009;48:455–467. doi: 10.1002/gcc.20655. [DOI] [PubMed] [Google Scholar]
- 28.Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G, Thway K, Orbach D, Lae M, Freneaux P, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28:2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 29.Bell E, Premkumar R, Carr J, Lu X, Lovat PE, Kees UR, Lunec J, Tweddle DA. The role of MYCN in the failure of MYCN amplified neuroblastoma cell lines to G1 arrest after DNA damage. Cell Cycle. 2006;5:2639–2647. doi: 10.4161/cc.5.22.3443. [DOI] [PubMed] [Google Scholar]
- 30.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 32.Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 34.Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA, Tchou J, Liu Q, Zhang L, Speicher DW, et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 2013;32:2672–2684. doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang JF, Yang L, Chen LL. The long noncoding RNA regulation at the MYC locus. Curr Opin Genet Dev. 2015;33:41–48. doi: 10.1016/j.gde.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim T, Cui R, Jeon YJ, Lee JH, Lee JH, Sim H, Park JK, Fadda P, Tili E, Nakanishi H, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci U S A. 2014;111:4173–4178. doi: 10.1073/pnas.1400350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng T, Mercer CA, Hexley P, Thomas G, Fumagalli S. Loss of tumor suppressor RPL5/RPL11 does not induce cell cycle arrest but impedes proliferation due to reduced ribosome content and translation capacity. Mol Cell Biol. 2013;33:4660–4671. doi: 10.1128/MCB.01174-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Relative genomic positions of MYCNOS-01, MYCNOS-02 and MYCN and protein-coding potential of MYCNOS-01. (A) Diagram illustrating relative positions of MYCNOS-01, MYCNOS-02 and MYCN exons. Modified from UCSC Genome Browser. (B) Predicted sequence for three forward reading frames of MYCNOS-01. (B) Table of results for ATGpr analysis of MYCNOS-01. The reliability score indicates the likelihood a start codon is in the correct context for translation initiation, with a score closer to one indicating increased likelihood. (PDF 594 kb)
Figure S2. Correlation of MYCNOS transcripts and MYCN transcript expression in RMS measured by qRT-PCR. (A) Correlation between MYCNOS-01 and MYCN measured by qRT-PCR in 80 RMS patient samples (R = 0.281, p = 0.012) and (B) correlation between MYCNOS-02 and MYCN in 53 of these patient samples (R = 0.308, p = 0.025). (C) Correlation between MYCNOS-01 and MYCN measured by qRT-PCR in 18 RMS cell lines (R = 0.821, p < 0.0001) and (D) correlation between MYCNOS-02 and MYCN in 14 of these cell lines (R = 0.980, p < 0.0001). MYCN-amplified cases or cell lines are depicted with circles. RMS-01 and RH30 are indicated specifically. (E) Expression of MYCNOS-01, MYCNOS-02 and MYCN transcripts measured by qRT-PCR in the cell lines used in experiments; RMS-01, RH30, KELLY and SY5Y. Expression relative to normal cells. (PDF 50 kb)
Figure S3. Effect of MYCNOS-01 knockdown on MYCN protein expression in RMS and NB measured by immunofluorescence. (A) Immunofluorescence staining of MYCN (red) with DAPI shown in blue after MYCNOS-01 knockdown for 72 h with three siRNAs in RMS-01. (B) Quantification of MYCN staining intensity from (A) relative to total number of cells and normalised to non-targeting control. (C) qRT-PCR detecting MYCNOS-01 transcript level matching experiment shown in (A). (D)-(F) in KELLY as for (A)-(C). Data representative of three repeats. Statistical analysis relative to non-targeting control. NT = non-targeting. (PDF 74664 kb)
Figure S4. Effect of MYCNOS-01 overexpression on MYCN transcript and protein expression. qRT-PCR detecting MYCNOS-01 transcript level after 72 h transient transfection with two MYCNOS-01 overexpressing vectors or empty vector control in (A) RMS-01 and (D) KELLY. Corresponding MYCN transcript level shown in (B) RMS-01 and (E) KELLY. Expression relative to empty vector control. MYCN protein levels after MYCNOS-01 overexpression for 72 h shown in (C) RMS-01 and (F) KELLY. GAPDH used as loading control. Densitometry values shown above each blot normalised to GAPDH and relative to empty vector control. EV = empty vector. (PDF 3518 kb)
Figure S5. Effect of MYCNOS-01 knockdown on MYCN protein stability. (A) RMS-01 cells treated with three MYCNOS-01 siRNAs for 48 h including 4 h treatment with DMSO or MG132. Densitometry values shown above each blot normalised to GAPDH and relative to NT control for each condition. Blots representative of experiments run in triplicate. (B) MYCNOS-01 transcript expression after MYCNOS-01 knockdown for 24 h measured by qRT-PCR. NT = non-targeting control. (PDF 584 kb)
Figure S6. Effect of MYCNOS-02 knockdown on MYCN transcript and protein expression in RMS and NB. qRT-PCR detecting MYCNOS-02 transcript level after MYCNOS-02 knockdown with three siRNAs in (A) RMS-01 and (D) KELLY. Corresponding MYCN transcript level shown in (B) RMS-01 and (E) KELLY. Expression relative to NT control. Western blots showing MYCN protein levels after MYCNOS-02 knockdown with three siRNAs shown in (C) RMS-01 and (F) KELLY. GAPDH used as loading control. Densitometry values shown above each blot normalised to GAPDH and relative to NT control. Data representative of 3 repeats. NT = non-targeting control. Relative expression of transcripts in these cell lines are indicated in Additional file 2: Figure S2E. (PDF 3709 kb)
Figure S7. Effect of MYCN knockdown on MYCNOS-02 transcript expression in RMS and NB. (A) qRT-PCR and (B) Western blot for MYCN, and (C) qRT-PCR for MYCNOS-02 after treatment with three pooled MYCN siRNAs for 72 h in RMS-01. (D)-(F) in KELLY as for (A)-(C). Graphs and Western blots representative of three repeats. NT = non-targeting control. (PDF 4860 kb)
Figure S8. Knockdown of MYCNOS-02 inhibits cell viability. (Ai) MTS assay showing cell viability of RMS-01 over 96 h after transfection with three MYCNOS-02 siRNAs relative to NT siRNA. (Bi) KELLY as in (Ai). Corresponding qRT-PCR of MYCNOS-02 (ii) at 24 h shown below line graph. Data representative of two repeats. Statistical analysis relative to non-targeting control. NT = non-targeting. (PDF 82 kb)
Figure S9. Knockdown of MYCNOS-01 does not induce apoptosis. (A) Caspase 3/7 signalling intensity in RMS-01 after transfection with three MYCNOS-01 siRNAs or three pooled MYCN siRNAs for 96 h relative to cell viability and normalised to NT siRNA. (B) Western blot of total and cleaved PARP in RMS-01 after transfection with three MYCNOS-01 siRNAs or three pooled MYCN siRNAs. (C, D) in KELLY, (E, F) in RH30 and (G, H) in SY5Y as for (A, B). Data representative of three repeats. NT = non-targeting, tPARP = total PARP, cPARP = cleaved PARP. (PDF 5703 kb)
Figure S10. Knockdown of MYCNOS-02 does induce apoptosis. (A) Caspase 3/7 signalling intensity in RMS-01 after transfection with three MYCNOS-02 siRNAs for 96 h relative to cell viability and normalised to NT siRNA. (B) Western blot of total and cleaved PARP in RMS-01 after transfection with three MYCNOS-02 siRNAs. (C, D) in KELLY as for (A, B). Data representative of two repeats. NT = non-targeting, tPARP = total PARP, cPARP = cleaved PARP. (PDF 3672 kb)
Figure S11. Knockdown of MYCNOS-01 does not affect cell cycle progression. (A) Flow cytometry 96 h post-transfection with three MYCNOS-01 siRNAs or three pooled MYCN siRNAs in RMS-01, (B) KELLY, (C) RH30, and (D) SY5Y. Graphs show an average of three repeats. NT = non-targeting. (PDF 111 kb)
Data Availability Statement
The public datasets supporting the conclusions of this article are available in the R2 Genomics Analysis and Visualisation Platform repository, (http://r2.amc.nl).


