Abstract
Objective
CD16+/CD163+ macrophages (MΦ)s and microglia accumulate in the brains of patients with HIV encephalitis (HIVE), a neuropathological correlate of the most severe form of HIV-associated neurocognitive disorders (HAND), HIV-associated dementia (HIV-D). Recently, we found that some parenchymal microglia in brain of HIV+ subjects without encephalitis (HIV/noE) but with varying degrees of neurocognitive impairment express CD16 and CD163, even in the absence of detectable virus production. To further our understanding of microglial activation in HIV, we investigated expression of specific genes by profiling parenchymal microglia from archival brain tissue of patients with HIVE, HIV/noE, and HIV− controls.
Methods
Single-population microarray analyses were performed on ~2,500 laser capture microdissected CD163+, CD16+ or CD68+ MΦs/microglia per case, using terminal continuation (TC) RNA amplification and a custom-designed array platform.
Results
Several classes of microglial transcripts in HIVE and HIV/noE, were altered, relative to HIV− subjects, including factors related to cell stress, immune activation, and apoptosis. Additionally, several neurotrophic factors are reduced in HIV infection, suggesting an additional mechanism of neuropathogenesis. The majority of transcripts altered in HIVE displayed intermediate changes in HIV/noE.
Interpretation
Our results support the notion that microglia contribute to the maintenance of brain homeostasis and their potential loss of function in the context of chronic inflammation contributes to neuropathogenesis. Furthermore, they indicate the utility of profiling MΦs/microglia to increase our understanding of microglia function, as well as ascertain alterations in specific pathways, genes, and, ostensibly, encoded proteins that may be amenable to targeted treatment modalities in diseases affecting the brain.
Introduction
Varying degrees of neurocognitive dysfunction remain a common complication of HIV infection, even in the context of successful pharmacological intervention. Collectively referred to as HIV-associated neurocognitive disorders (HAND), impairment ranges from mild to severe, and significantly impacts the quality-of-life of those living with HIV. Combination anti-retroviral therapy (cART) has decreased the incidence of the most severe form of neurocognitive impairment, HIV-associated dementia (HIV-D), which has as one of its substrates perivascular cuffs and nodular lesions of microglia and macrophages (MΦ)s. These sites serve as the major reservoir of productive virus in this neuropathological HIV-D correlate, HIV encephalitis (HIVE) 1. Yet, even in the absence of productive virus and/or the distinguishing pathological features of encephalitis, we observed marked neuroinflammation in autopsy-derived brain tissues of individuals who died with HIV infection, irrespective of pharmacological intervention 2. This is evidenced not only by expression of markers that suggest inflammatory responses by astrocytes and microglia, but also by prominent morphological changes of the two cell types indicative of an activated state 2. This expands on earlier histopathological studies performed before and after the introduction of highly active antiretroviral therapy (HAART) that demonstrate widespread microglial activation in HIV infection, even in the absence of viral products 3–5. Together, these findings provide critical insights into the mechanisms of the less severe form of HAND, minor neurocognitive disorder (MND), which is observed frequently in the cART-era, and support the contention that chronic inflammation of the brain plays a key role in the pathogenesis of all forms of HAND.
Persistent CNS inflammation is associated with, and may even predispose individuals to, many neurodegenerative diseases 6–8. Moreover, neuroinflammation is recognized as a major component of several neurocognitive and psychological disorders, including Alzheimer’s disease, major depressive disorder, and schizophrenia 9, 10. How inflammation contributes to neurocognitive impairment is not completely clear, however, several inflammation-associated factors, including cytokines, chemokines, and reactive oxygen and nitrogen species, from activated microglia can directly and/or indirectly promote neuronal injury or death. Additionally, continuing advances in our understanding of the role of microglia in neuronal function and maintaining CNS homeostasis suggest that microglia may also contribute to neuropathogenesis through mechanisms other than those associated with immunological processes. As such, the varying degrees of neurocognitive dysfunction observed in the context of HIV infection may also be a consequence of chronic neuroinflammation that impairs microglial function.
To further understand microglial function in the context of chronic inflammation, we investigated expression of specific classes of transcripts and individual genes by parenchymal MΦs/microglia isolated from brain of patients with HIVE, HIV infection without encephalitis (HIV/noE), and seronegative (HIV−) controls. Custom-designed microarray analyses on laser capture microdissected CD163+, CD16+ or CD68+ MΦs/microglia revealed altered expression of several classes of transcripts in HIV infection with and without encephalitis, as compared to seronegative controls. The majority of transcripts significantly altered in HIVE show intermediate changes in HIV/noE but with impaired cognition, supporting the notion that the varying degrees of neurocognitive impairment described by HAND are a continuum of the same pathogenic processes 2.
Materials and Methods
Human subjects and tissue acquisition
Formalin-fixed paraffin-embedded frontal white matter (FWM) from patients with HIVE, HIV/noE, and HIV− without CNS disease were provided by the Manhattan HIV Brain Bank (MHBB), a member of the National NeuroAIDS Tissue Consortium (NNTC; U24 MH100931). All tissues were acquired under the Mount Sinai Institutional Review Board approved protocol 98-477(0003) and the institutional PPHS Federal-Wide assurance number 00005656. Written, informed consent was obtained from all subjects at the time of enrollment into the MHBB of NNTC. Both HIV groupings included patients on- and off-cART at the time of death (Table 1). HIV-D, minor cognitive motor disorder (MCMD), or normal cognition designations were determined by a licensed MHBB neuropsychologist in accordance with the DANA modification of the American Academy of Neurology (AAN) criteria 11, 12, as described 2. All HIV/noE subjects in our study demonstrated some degree of neurocognitive impairment (Table 1). Cognitive status was available for 7 of the 9 HIVE subjects in our study (Table 1). The retrospective ascertainment of encephalopathy at the time of autopsy for the two HIVE subjects that had not undergone neuropsychological testing suggests that these individuals similarly had impaired cognition in life.
Table 1.
Human Subjects. Autopsy frontal white matter (FWM) from of 9 HIVE, 10 HIV without encephalitis (HIV+/no E) and 5 seronegative (HIV−) individuals without CNS disease were investigated.
| PID | Class | Age | Sex | Race | CD4 | VL (copies/ml) | NI* | HIV in brain† | Cog status‡ |
|---|---|---|---|---|---|---|---|---|---|
| 500 | HIVE | 47 | M | W | 20 | 210000 | 4+ | 3+ | HIV-D |
| 537 | HIVE | 45 | F | B | 6 | n/a | 4+ | 2+ | n/a |
| 540 | HIVE | 47 | M | B | 1 | 683333 | 4+ | 2½+ | psychosis |
| 603 | HIVE | 49 | M | B | 2 | 493381 | 4+ | 2½+ | HIV-D |
| 629 | HIVE | 57 | F | B | 25 | 730085 | 3+ | n/d | HIV-D |
| 10017 | HIVE | 44 | M | W | 7 | 389120 | 3+ | 3½+ | npi-o |
| 10070 | HIVE | 37 | M | B | 1 | >750000 | 4+ | n/d | ms change |
| 10133 | HIVE | 48 | M | W | 3 | 173921 | 4+ | 4+ | MCMD po |
| 10231 | HIVE | 47 | M | H | 98 | 208516 | 4+ | 1+ | n/a |
| 10001 | HIV+/no E | 64 | F | B | 72 | 359 | 3+ | neg | HIV-D po |
| 10011 | HIV+/no E | 44 | M | H | 16 | 162642 | 4+ | neg | MCMD po |
| 10013 | HIV+/no E | 33 | M | W | 9 | >750000 | 2½+ | neg | MCMD pr |
| 10016 | HIV+/no E | 58 | F | B | 98 | 576000 | 2+ | neg | MCMD pr |
| 10066 | HIV+/no E | 54 | F | B | 8 | 469163 | 3+ | neg | MCMD po |
| 10067 | HIV+/no E | 28 | M | A | 5 | 2097 | 1+ | neg | MCMD pr |
| 10119 | HIV+/no E | 33 | M | B | 1 | 312240 | 4+ | neg | HIV-D po |
| 10127 | HIV+/no E | 39 | F | H | 402 | 1013 | ½+ | n/d | npi-o |
| 10203 | HIV+/no E | 44 | F | H | 86 | 36035 | 3½+ | neg | npi-o |
| 30024 | HIV+/no E | 43 | M | B | 104 | <50 | 1½+ | neg | MCMD po |
| 551 | HIV− | 30 | F | H | - | - | ½+ | n/a | n/a |
| 567 | HIV− | 63 | M | H | - | - | <½+ | n/a | n/a |
| 588 | HIV− | 21 | M | H | - | - | 1+ | n/a | n/a |
| 594 | HIV− | 57 | F | W | - | - | <½+ | n/a | n/a |
| 601 | HIV− | 58 | F | H | - | - | <½+ | n/a | n/a |
NI (neuroinflammation index) is an assigned value (0+ to 4+) based on the overall severity of neuroinflammation, as determined by our previous histopathological evaluation that investigated specific MΦ/microglial (CD16, CD163, and HLA-DR) and astrocytic (GFAP and vimentin) markers of inflammation 2. The majority of seronegative individuals had very mild indications of neuroinflammation. Morphological alterations suggestive of inflammation/activation of microglia and astrocytes, such as thickened/retracted cell processes and loss of individual cellular domains, were also used to assign the NI value for each case.
HIV in brain – Individuals were assigned a degree of positivity (+) or listed as negative (neg) for evidence of productive HIV infection in FWM. These values were derived from our previous immunohistochemical studies investigating HIVp24 protein expression, as well as in situ hybridization 2.
Cog status (cognitive status) reflects the neurocognitive diagnosis assigned to individuals who underwent neuropsychological testing by a licensed neuropsychologist at the MHBB. Assignments were made in accordance with the DANA modification of the American Academy of Neurology (AAN) criteria, which includes two degrees of HIV-related neurocognitive impairment, HIV-associated dementia (HIV-D) and minor cognitive motor disorder (MCMD).
Abbreviations: HIV-D = HIV-associated dementia; MCMD = minor cognitive motor disorder; ms change = mental status change: patient has altered cognitive capacity, however, the individual observing this is unsure if this constitutes dementia or a transient abnormality; pr = probable: no confounds; po = possible: confounds exist, but observer believes cognitive impairment is most likely HIV-associated; npi-o = neuropsychological impairment-other: patient is cognitively impaired and the observer believes there are enough ancillary factors/co-morbid processes to account for the dementia.
Immunohistochemistry (IHC)
FWM was cut at 4 μm thickness from a total of 9 HIVE, 10 HIV/noE, and 5 HIV- cases. Instruments used for acquiring tissue sections were decontaminated of RNase (RNaseZap, ThermoFisher, Waltham, MA), including the microtome and blades. RNase-free techniques, including cleaning all work surfaces with RNaseZap and using new, sterile RNase-free pipette tips and microcentrifuge tubes, disposable gloves and new, certified nucleic acid-free plasticware are employed during all RNA work. IHC was performed as described previously by our laboratory 1, 2, 13. Briefly, brain tissue sections were deparaffinized and rehydrated and underwent heat induced epitope retrieval (HIER) horizontally, rather than vertically, to prevent loss of tissue. Additional xylene washes were performed to ensure complete paraffin removal. All solutions were prepared using DEPC-treated water to remove RNase contamination. HIER was performed through three 10-minute applications of boiling citrate buffer, pH 6.0 (Dako). Tissues were allowed to incubate with primary mouse monoclonal CD163 (1:100; Vector Laboratories, clone 10D6), CD16 (1:40; Vector, clone 2H7), or CD68 (1:50; Dako, clone KP1). Antigen-specific staining was detected with horse-α-mouse biotinylated antibodies (Vector), followed by Vectastain ABC Alkaline Phosphatase and Vector Red Alkaline Phosphatase Substrate Kit (Vector), according to the manufacturer’s instructions. Slides were rinsed for 5 minutes in 100% ethanol to increase the intensity of Vector Red fluorescence. Following dehydration, tissues were allowed to air dry in a dark, protected area at least 3 days prior to laser capture microdissection (LCM). Tissue sections on plus slides (ThermoFisher) were immunostained in tandem with tissues on PEN membrane slides, coverslipped with Permount (ThermoFisher) and viewed by light microscopy to verify specificity of staining.
Laser capture microdissection (LCM) and RNA extraction
LCM and terminal continuation (TC) RNA amplification procedures have been described in detail previously by our group 14–17. Approximately 2,500 microglia from FWM of each study subject were microaspirated via LCM (Arcturus PixCell IIe, ThermoFisher, South San Francisco, CA). A rhodamine filter cube was used to identify Vector Red positive CD163+, CD16+, or CD68+ microglia in the brain parenchyma, with care taken to avoid perivascular MΦs, which may have an expression profile distinct from that of resident microglia. Caps with captured cells were immediately snap-secured into the opening of RNase-free 0.5 ml microcentrifuge tubes containing 200 μl TRIzol Reagent (ThermoFisher), inverted, and stored at −80 °C. Microarrays (containing 2,500 LCM-captured microglia each) were performed per human brain (range 1–6 per brain). The TC RNA amplification protocol is available at http://cdr.rfmh.org/pages/ginsberglabpage.html. This method entails synthesizing first strand cDNA complementary to the RNA template, re-annealing the primers to the cDNA, and finally in vitro transcription using the synthesized cDNA as a template. Briefly, microaspirated microglia were homogenized in Trizol reagent (ThermoFisher), chloroform extracted, and precipitated 16, 18, 19. RNAs were reverse transcribed and single-stranded cDNAs were then subjected to RNase H digestion and re-annealing of the primers to generate cDNAs with double-stranded regions at the primer interfaces. Single stranded cDNAs were digested and samples were purified by Vivaspin 500 columns (Sartorius Stedim Biotech, Goettingen, Germany). Hybridization probes were synthesized by in vitro transcription using 33P and radiolabeled TC RNA probes were hybridized to custom-designed cDNA arrays without further purification.
Custom-designed microarray platforms and hybridization
Array platforms consist of 1 μg of linearized cDNA purified from plasmid preparations adhered to high-density nitrocellulose (Hybond XL, GE Healthcare, Piscataway, NJ) using an arrayer robot (VersArray, Bio-Rad, Hercules, CA) 20, 21. Each cDNA and/or expressed sequence-tagged cDNA (EST) was verified by sequence analysis and restriction digestion. Mouse and human clones were employed on the custom-designed array. Approximately 576 cDNAs/ESTs were utilized, organized into 19 gene ontology (GO) groups. Most genes are represented by one transcript on the array platform, although the neurotrophin receptors TrkA, TrkB, and TrkC are represented by ESTs that contain the extracellular domain (ECD) as well as the tyrosine kinase domain (TK) 17, 22.
Statistical analyses
Statistical procedures for custom-designed microarray analysis using postmortem human brain tissues have been described in detail previously 17, 23 HIVE, HIV/noE, and HIV− subjects were compared with respect to the hybridization signal intensity ratio of 576 genes. For each gene, the signal intensity ratio was modeled using mixed effects models with random effect to account for the correlation between repeated assays in the same subject 24. Significance was judged at the level (α = 0.01), two-sided; false discovery rate (FDR) based on an empirical null distribution due to strong correlation between genes 25, 26 was controlled at level 0.1. Expression levels were graphed using a bioinformatics software package (GeneLinker Gold, Predictive Patterns, Kingston, ON).
Results
Significant overall expression level changes in HIVE and HIV/noE relative to HIV− microglia
Results indicated significant (p<0.02) downregulation of the majority of transcripts evaluated on the custom-designed array for HIVE (63.7%; a total of 367/576 genes significantly downregulated), and HIV/noE (22.9%; a total of 132/576 genes significantly downregulated) relative to HIV−, indicating a massive effect of HIV infection and chronic inflammation on the genetic fingerprint of microglia. In contrast, statistically significant upregulation was observed in a relatively small subset of genes in HIVE (10.4%; a total of 60/576 genes) and HIV/noE (5.2%; a total of 30/576 genes) compared to HIV−.
Alterations in stress-related and immunological transcripts in HIV infection
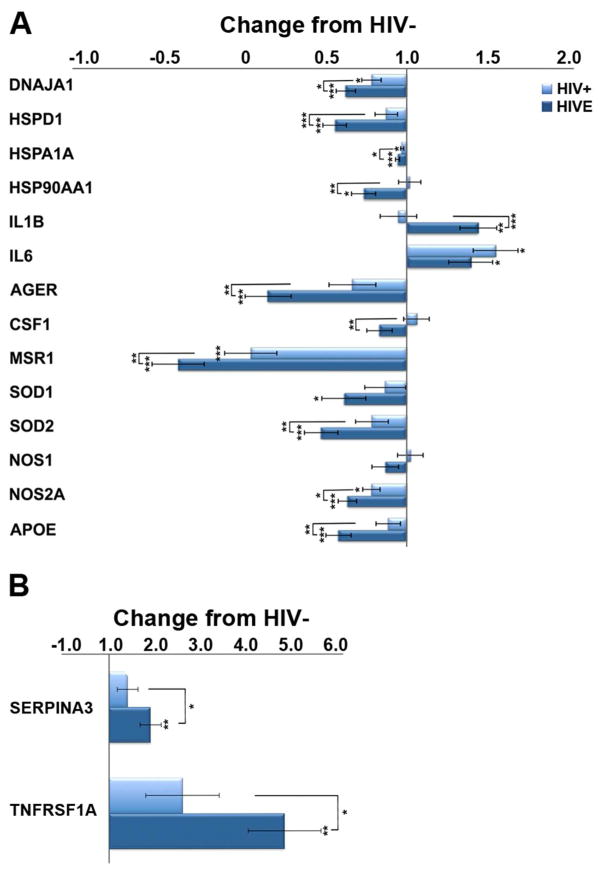
Several stress-associated factors are significantly reduced in HIVE, relative to seronegative subjects, with intermediate expression seen in HIV/noE subjects (Figure 1). Specifically, downregulation of heat-shock proteins (HSPs), Dnaja1, Hspd1, Hspa1a, and Hsp90aa1, as well as both the constitutive and inducible forms of nitric oxide synthase (NOS), Nos1 and Nos2a, and superoxide dismutase (SOD) Sod1 and Sod2 was found. In addition, factors associated with ameliorating or suppressing inflammation are decreased in HIV/noE and HIVE, including apolipoprotein E (Apoe) and colony stimulating factor 1 (Csf1), while key pro-inflammatory interleukins, IL-1β and IL-6, were significantly upregulated. Upregulation of the serpin peptidase inhibitor, Serpina3 and a primary tumor necrosis factor alpha (TNFα) receptor, Tnfrsf1a, further supports chronic pro-inflammatory stimulation and signaling in HIV infection.
Figure 1.
A. Histograms depicting results obtained from custom-designed microarray analysis of LCM-captured microglia that reveal altered expression of cellular stress and immune activation genes in HIV infection and HIVE. Compared to HIV-, heat-shock and antioxidant activity genes are significantly downregulated in HIVE, with intermediate expression seen in HIV/noE but with MND. Several anti-inflammatory transcripts are decreased in HIV and HIVE, while pro-inflammatory factors, including IL-1β and IL-6, are upregulated. B. Upregulation of pro-inflammatory markers SerpinA3 and Tnfrsf1A are observed in HIVE to a significantly greater degree than in HIV/noE. Key: * p<0.02; **p<0.001; ***p<0.0001. DnaJ (Hsp40) homolog, subfamily A, member 1 (DNAJA1); heat shock protein family D (Hsp60) member 1 (HSPD1); heat shock protein family A (Hsp70) member 1A (HSPA1A); heat shock protein 90kDa alpha (cytosolic), class A member 1 (HSP90AA1); interleukin 1 beta (IL1b); interleukin 6 (IL6); RAGE advanced glycosylation and product-specific receptor (AGER); colony stimulating factor 1 (CSF1); macrophage scavenger receptor 1 (MSR1); superoxide dismutase 1 (SOD1); superoxide dismutase 2 (SOD2); nitric oxide synthase 1 (NOS1); nitric oxide synthase 2A (NOS2A); apolipoprotein E (APOE); serpin peptidase inhibitor, clade A member 3 (SERPINA3); tumor necrosis factor receptor superfamily member 1A (TNRFSF1A).
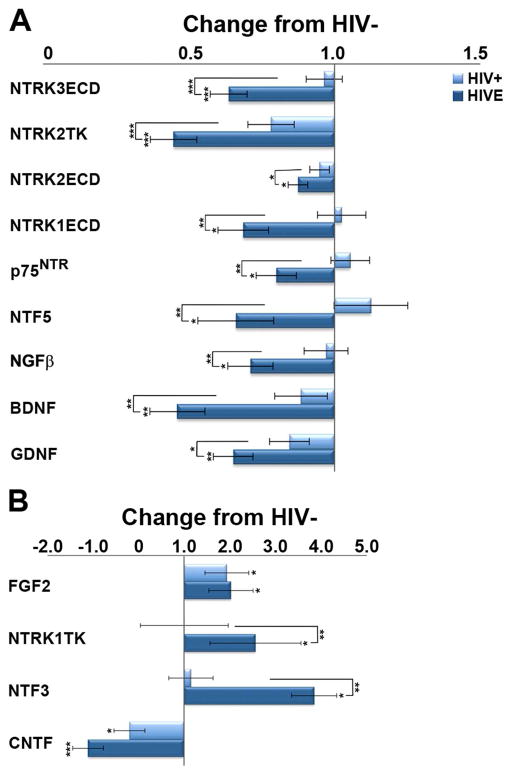
Dysregulation of microglia-derived neurotrophic factors
Relative to seronegative controls, several microglia-derived neurotrophic factor transcripts are downregulated in HIV infection, with and without encephalitis (Figure 2), including brain-derived neurotrophic factor (Bdnf), glial cell-derived neurotrophic factor (Gdnf), and ciliary neurotrophic factor (Cntf). Upregulation of neurotrophic factors is rarely observed, with statistical significance reached primarily in those with encephalitis (Figure 2B).
Figure 2.
A. Histograms demonstrating downregulation of several microglia-derived neurotrophic factor transcripts in HIV+, and to a greater degree, in HIVE. B. Upregulation of a select few neurotrophins is observed predominantly in HIVE. Key: * p<0.02; **p<0.001; ***p<0.0001. neurotrophin-3 receptor TrkC, extracellular domain (NTRK3ECD); BDNF receptor TrkB tyrosine kinase domain (NTRK2TK); BDNF receptor TrkB extracellular domain (NTRK2ECD) NGF receptor TrkA, extracellular domain (NTRK1ECD); pan-neurotrophin growth factor receptor (p75NTR); neurotrophin 4/5 (NTF5); nerve growth factor-β (NGFB); brain-derived neurotrophic factor (BDNF); glial cell-derived neurotrophic factor (GDNF); fibroblast growth factor 2 (basic; FGF2); NGF receptor TrkA, tyrosine kinase domain (NTRK1TK); neurotrophin-3 (NTF3); ciliary neurotrophic factor (CNTF).
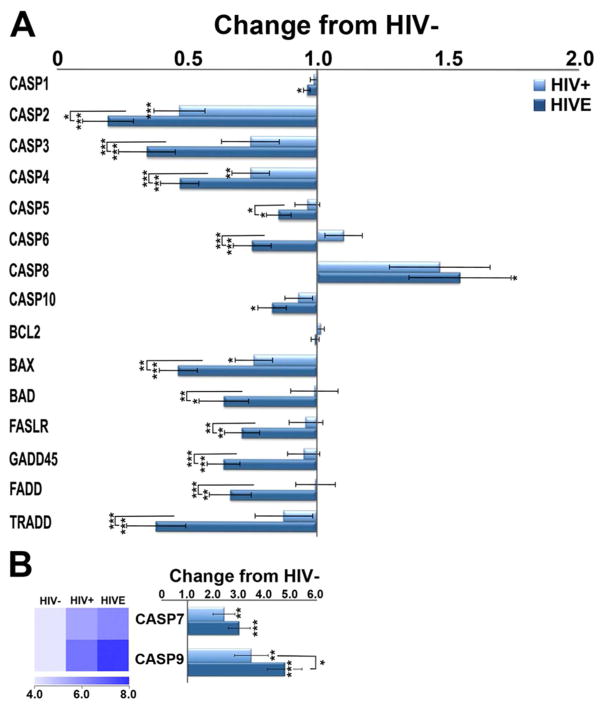
Dysregulation of select caspase mRNAs
We observed downregulation of several caspases that induce pyroptotic cell death, including caspase (Casp) Casp1, Casp4, and Casp5, despite overwhelming evidence of sustained neuroinflammation (Figure 3). Executioners of cell death, Casp3 and Casp6, are also downregulated in HIV/noE and HIVE, whereas Casp7 is significantly increased in both HIVE and HIV/noE relative to HIV− subjects. Initiators of cell death in extrinsic (Casp8) and intrinsic (Casp9) apoptotic pathways, respectively, are upregulated in HIVE and HIV/noE relative to HIV− subjects (Figure 3). Pro-apoptotic factors, bcl2-antagonist of cell death (Bad) and bcl2-associated x protein (Bax), as well related mediators of apoptotic pathways including fas-associated via death domain (Fadd), fas ligand receptor (Faslr), and TNFRSF1A-associated via death domain (Tradd) are significantly downregulated in HIVE (Figure 3A). Interestingly, B-cell lymphoma 2 (Bcl2), which can inhibit or induce cell death, did not differ significantly among the three groups investigated.
Figure 3.
A. Histograms illustrating downregulation of select caspases and cell-death-related genes are observed in HIVE to a greater degree than HIV+. Notably, the initiators of cell death, Casp8 (A) and Casp9 (B), are increased in HIV, as compared to non-infected persons, with an even greater increase seen in HIVE. Executioner Casp7 (B) is elevated in HIV infection with and without encephalitis, while Casp6 is increased in HIV+ but decreased in HIVE. Interestingly, Casp3, which cleaves and activates Casp6 and Casp7, is decreased in both HIV groups. Key: * p<0.02; **p<0.001; ***p<0.0001. Caspase 1 (CASP1); Caspase 2 (CASP2); Caspase 3 (CASP3); Caspase 4 (CASP4); Caspase 5 (CASP5); Caspase 6 (CASP6); Caspase 7 (CASP7); Caspase 8 (CASP8); Caspase 9 (CASP9); Caspase 10 (CASP10); b-cell lymphoma 2 (BCL2); bcl2-associated x protein (BAX); bcl2-antagonist of cell death (BAD); fas ligand receptor (FASLR); growth arrest and DNA-damage-inducible protein (GADD45); fas-associated death domain (FADD); TNFRSF1A-associated via death domain (TRADD).
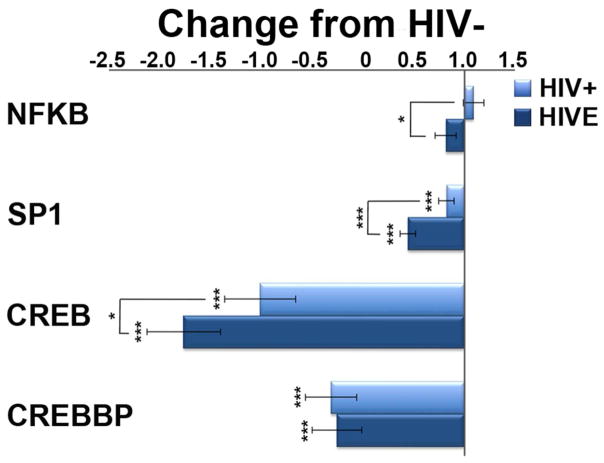
Microglial production of ubiquitously expressed transcription factors is decreased in HIVE and HIV/noE
Downregulation of key transcription factors nuclear factor kappa B (Nfkb), specific protein 1 (Sp1), and cAMP responsive element-binding protein (Creb) is observed in HIVE and HIV/noE microglia (Figure 4). The ubiquitously expressed transcriptional co-activator, CREB binding protein (Crebbp) is also significantly decreased in both HIV groups.
Figure 4.
Reduction in ubiquitously expressed transcription factors may reflect cellular senescence due to long-tern neuroinflammation in HIV infection and HIVE. Microglial transcription of Nfkb is upregulated in HIV/noE, but significantly decreased in HIVE. Sp1 and Creb are downregulated in HIV infection, with a greater reduction seen in HIVE. The transcriptional co-activator of CREB, Crebbp, is similarly reduced in both HIV groupings. Key: * p<0.02; ***p<0.0001. nuclear factor kappa B (NFκB); specific protein 1 transcription factor (SP1) cAMP responsive element-binding protein (CREB); CREB binding protein (CREBBP).
Select protein kinase and phosphatase transcripts are dysregulated in HIVE and HIV/noE
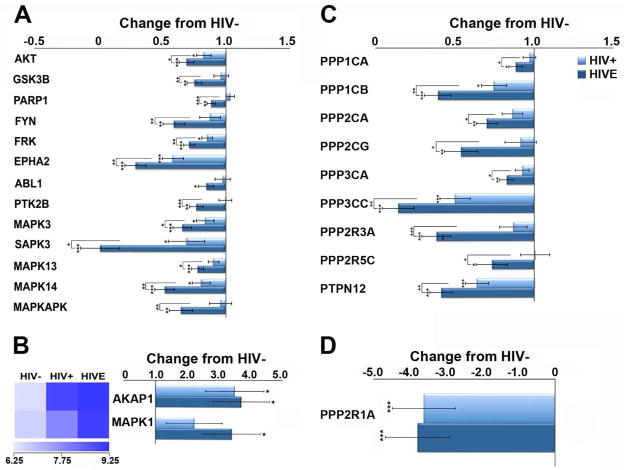
The majority of significantly altered protein kinase and phosphatase transcripts on the custom-designed array are downregulated in HIVE and HIV/noE (Figure 5). Downregulated kinases include protein kinase B (Akt), glycogen synthase kinase-3β (Gsk3β), protein tyrosine kinase 2β (focal adhesion kinase; Ptk2β) and mitogen-activated protein kinase 3 (Mapk3) among several others (Figure 5). Downregulated protein phosphatase subunits include protein phosphatase 1, catalytic subunit, α isoform (Ppp1ca), protein phosphatase 1, catalytic subunit, β isoform (Ppp1cb) protein phosphatase 2, catalytic subunit, α isoform (Ppp2ca), protein phosphatase 2, catalytic subunit, γ isoform (Ppp2cg), and protein phosphatase 3, catalytic subunit, α isoform (Ppp3ca) among several others (Figure 5). These coordinated expression level changes in protein phosphatases and kinases may indicate microglial senescence, which has been demonstrated in the context of long-term inflammation in the brain. In contrast, only the mitochondrial kinase anchoring protein A-kinase anchor protein 1 (Akap1) and mitogen-activated protein kinase 1 (Mapk1) are significantly increased in HIV infection (Figure 5B).
Figure 5.
A significant reduction in select protein kinases and phosphatases in HIV+ and HIVE may indicate reduced microglial signal transduction and associated activity/function. A. Downregulation of several protein kinase mRNAs is observed in HIVE to a greater degree that HIV+. B. Although the vast majority of altered transcripts are reduced in the context of HIV infection, upregulation of Akap1, and Mapk1 was found. C. Downregulation of select protein phosphatase subunits was found in HIVE to a greater degree than in HIV+. D. Highly significant downregulation of Ppp2r1A was found in both HIV+ and HIVE. Key: * p<0.02; **p<0.001; ***p<0.0001. protein kinase B (AKT); glycogen synthase kinase-3β (GSK3β); FYN kinase (FYN); FYN-related kinase (FRK); eph/elk epithelial cell protein tyrosine kinase (EPHA2); Abelson murine leukemia viral oncogene homolog 1 (ABL1); protein tyrosine kinase 2β (focal adhesion kinase; PTK2β); mitogen-activated protein kinase 3(MAPK3); stress-activated protein kinase 3(SAPK3); mitogen-activated protein kinase 13 (MAPK13); mitogen-activated protein kinase 14 (MAPK14); MAP kinase-activated protein kinase 2 (MAPKAPK); A kinase anchor protein 1(AKAP1); mitogen-activated protein kinase 1, ERK2 (MAPK1); protein phosphatase 1, catalytic subunit, α isoform (PPP1CA); protein phosphatase 1, catalytic subunit, β isoform (PPP1CB); protein phosphatase 2, catalytic subunit, α isoform (PPP2CA); protein phosphatase 2, catalytic subunit, γ isoform (PPP2CG); protein phosphatase 3, catalytic subunit, α isoform (PPP3CA); protein phosphatase 3, catalytic subunit, γ isoform (PPP3CC); protein phosphatase 2, regulatory subunit B, α isoform (PPP2R3A); protein phosphatase 2, regulatory subunit B, γ isoform (PPP2R5C); protein tyrosine phosphatase, non-receptor type 12 (PTPN12); protein phosphatase 2, regulatory subunit A, α isoform (PPP2R1A)
Discussion
Microglia have been implicated as key players in promoting and advancing neurodegenerative disease, largely through immune activation and associated soluble factors that are directly or indirectly neurotoxic. Pro-inflammatory cytokines, TNF-α, IL-1β, and IL-6, as well as increased reactive oxygen species (ROS) and reactive nitrogen species (RNS), can be injurious to healthy cells and are commonly seen in brain or CSF of individuals suffering CNS disease, including HIVE 27–29. In our study, both HIV groups demonstrated altered transcripts related to immune activation including the pro-inflammatory cytokines, IL-1β and IL-6, and the major TNFα receptor, Tnfrsf1a.
Increased transcription of IL-6 and Tnfrsf1a supports our earlier immunohistochemical findings within the same subjects that show substantial widespread microglial activation in HIV, including individuals on cART prior to death and without detectable virus in brain 2. We suggested that neuroinflammation is a common complication of HIV infection that may impact the severity of HAND, however, cART was not effective in the majority of patients studied, as evidenced by high plasma viral loads and low CD4+ T-cell counts. Thus, our study may not be relevant in determining whether neuroinflammation is present in aviremic patients with immunologic reconstitution. Recent positron emission tomography (PET) neuroimaging revealed microglial activation in living HIV+ subjects under successful pharmacological intervention with restored CD4+ T-cells and without cognitive impairment 30, 31. These findings support the notion that neuroinflammation is common to HIV infection and continues under effective cART.
In contrast to IL-6 and Tnfrsf1a, transcription of IL-1β was only upregulated in HIVE. Interestingly, Casp1, which cleaves IL-1β into its active form 32 was modestly downregulated in both HIV groups. As such, it is unclear if increased transcription of IL-1β in HIVE is associated with increased activity of this important pyrogen.
Downregulation of genes was far more common than upregulation in both HIV groups, as compared to seronegative donors. An unprecedented number of genes were impacted in HIVE patients, with downregulation of 63.7% of genes interrogated. By contrast, 22.9% of genes were downregulated in HIV/noE. Overall, this indicates a substantial influence of HIV infection and chronic inflammation on the genetic fingerprint of vulnerable microglia.
Decreased levels of factors associated with resolving immune responses may also perpetuate neuroinflammation. Tissue MΦs, including microglia, participate in all aspects of immunity, including promoting and resolving inflammation. To perform these diverse functions, monocyte/MΦ activation is considerably plastic and heterogeneous, broadly classified into classically activated “pro-inflammatory” (M1) and alternatively activated “anti-inflammatory” (M2) MΦs. M1-associated stimuli, IL-6 and IL-1β, are increased in HIV, while a number of transcripts associated with M2 activation are decreased, including Csf1, Sod1, and Sod2.
A key MΦ hematopoietic factor, CSF1 [macrophage colony stimulating factor (M-CSF)], promotes M2-like activation of MΦs. In brain of simian immunodeficiency virus (SIV) infected rhesus macaques, increased M-CSF expression by MΦs that accumulate perivascularly and within nodular lesions corresponds with the principal reservoir of productive virus 33, however, parenchymal microglia had less detectable M-CSF, as compared to non-infected animals 34. Here, we show Csf1 transcription by parenchymal microglia is not significantly changed in HIV/noE or HIVE, as compared to seronegative controls, although a trend towards downregulation is seen in HIVE.
How reduced CSF1 transcription impacts HIV neuropathogenesis remains unknown, but may impair microglial survival. Ligation of the translated product to its cognate receptor, Cfms, activates several signaling cascades that mediate MΦ survival through suppression of the ‘initiator caspase’, CASP9, and/or activation of Erk. Our studies show a significant increase in Casp9 transcription in both HIV groupings that is greatest in HIVE. Similarly, the “executioner caspase”, Casp7, is significantly increased in HIVE, with intermediate expression seen in HIV/noE. Additionally, M-CSF supports MΦ survival through activation of NFκB 35, which is also reduced at the transcription level in HIVE.
Activated MΦs/microglia produce high levels of ROS, which are needed to perform a variety of functions, such as phagocytosis. Disproportionate levels of ROS, however, can promote cellular oxidative stress and accumulation of ROS is often seen in the context of neuroinflammation. SODs assist in reducing ROS through enzymatic dismutation of superoxides to hydrogen peroxide. They may also help in resolving inflammation, as SOD1 has been demonstrated to directly mediate M2 polarization 36. Here, we found microglia-associated Sod1 and Sod2 are downregulated in HIV, which could profoundly impact microglial health and function, as lost or reduced SODs would, presumably, impair the ability of microglia to kill and/or degrade ingested particles.
The consequence of reduced microglial Apoe is unclear but may also contribute to chronic neuroinflammation. APOE exists in three isoforms, APOE2-4. The E4 variant is associated with increased CNS inflammation and oxidative stress and is a genetic risk factor for several CNS pathologies 37–39. Interestingly, pro-inflammatory cytokines in APOE−/− mice are elevated in plasma following lipopolysaccharide injection or when fed a high-fat diet, comparable to observations with the E4 allele 40, 41. If APOE has similar immunomodulatory functions in the CNS, loss of the more ‘favorable’ APOE genotypes may contribute to neuroinflammation seen in HIV infection.
Microglia may also contribute to HIV-associated cognitive impairment through loss of neurotrophic factors that help spare neurons from injury or death. Our studies reveal several key neurotrophic factors are downregulated in both HIV groupings but most only reach statistical significance in HIVE. Reduced or lost microglial production of neurotrophic factors suggest an additional mechanism of neuropathogenesis and underscore the role microglia potentially play in brain homeostasis by supporting neuronal function and survival through paracrine mechanisms, as well as regulating microglial activation through autocrine mechanisms.
In the GO categories of transcription factors, kinases, and phosphatases, downregulation was primarily observed, apart from Akap1 and Mapk1. The encoded product of AKAP1 binds the two regulatory subunits of the cyclic adenosine monophosphate (cAMP)-dependent protein kinase A (PKA) and anchors the enzyme to the mitochondrial membrane. This compartmentalizes cAMP-PKA signaling to the mitochondria through PKA activation of the cAMP response element-binding protein (CREB), which is bound to mitochondrial DNA 42, 43. Increased AKAP1 transcription in HIV may be in response to CREB, which is also reduced at the RNA level in both HIV groupings. This may suggest impaired mitochondrial function and metabolic stress caused by chronic immune activation. Neuroinflammation may also influence upregulation of MAPK1, which has been shown to repress transcription of many genes in the context of chronic inflammation, including those induced by interferon (IFN)γ 44.
While our findings are intriguing and provide important insights into microglial function/loss of function in healthy and diseased brain, there are study limitations. LCM was performed on formalin-fixed, paraffin-embedded brain tissue acquired from autopsy, which can impair the quality of extracted RNA. To lessen this concern, tissues were acquired from an NNTC site and collected within the same protocol and defined post-mortem interval. IHC was performed using a high-heat antigen retrieval method that interferes with formalin cross-linking of RNA, DNA, and protein and we collected many cells from each case. Extracted RNA was amplified using a TC RNA amplification to limit amplification bias and increase the reliability of comparisons between groups. Additionally, we were unable to validate our findings by qPCR, due to low RNA yields and lack of matched frozen tissues. We previously tried to validate single population microarray findings via qPCR from single- and double-labeled LCM-captured neurons from paraffin-embedded postmortem human tissue sections with equivocal results 45–47, suggesting RNA abundance is the limiting factor in fixed tissues and is not specific to HIV-infected cohorts.
Even with qPCR validation, the question remains of whether abnormal levels of transcripts result in similar changes to protein expression and/or function. For example, in our subjects, Casp1 showed significant downregulation in HIVE; yet, prior immunohistochemical analyses of HIVE patients demonstrated significant increases in microglial caspase 1 staining compared to controls 48. In our current study, we found significantly decreased NFκB transcripts in HIVE brain, as compared to HIV/noE, yet transcription of IL-1β and IL6, which is regulated by NFκB, is elevated. This may be a reflection of the complexities of NFκB signaling, where activated cytoplasmic NFκB may continue to participate in transcriptional activity even while NFκB production is reduced. TNFα signaling through TNFRSF1A, which is also upregulated in HIVE, induces NFκB activation. Although we do not know if TNFα production is increased in our patients, previous immunohistochemical studies have shown microglia-associated TNFα in brain of patients with HIVE 49 and may suggest a mechanism through which pro-inflammatory cytokine production continues, even in the context of reduced NFκB transcription.
Despite these limitations, our work has yielded unparalleled insight and new avenues of investigation into the role of microglia in health and disease, particularly within the context of chronic HIV infection. Additional studies are needed to better understand the mechanisms driving the observed differential gene regulation and their consequences, which cannot infer causality in postmortem tissue.
In summary, single-population custom-designed microarray analysis demonstrates numerous microglial genes are significantly altered in HIV infection, even in the absence of encephalitis or detectable virus in brain. Changes in microglial transcripts related to immune activation and function, kinases, phosphatases, and pro-/anti-apoptotic and neurotrophic factors suggest several possible mechanisms of neuronal injury or death. Most of the significantly altered transcripts in HIVE displayed intermediate changes in HIV/noE but with impaired cognition, supporting the notion that the varying degrees of HAND are a continuum of the same pathogenic processes. Additionally, our microarray results indicate the utility of profiling brain MΦs/microglia in HIV infection to ascertain alterations in specific pathways, genes, and, presumably, encoded proteins that may be amenable to targeted treatment modalities.
Acknowledgments
This work was supported by NIH grants R01 NS063605 (TF), P01 MH105303 (TF), R01 AG043375 (SDG), P01 AG014449 (SDG), P01 AG017617 (SDG), and U24 MH100931 (SM).
Footnotes
Author Contributions
TF and SDG were responsible for the study concept and design; TF, SDG, MJA, SMG, CS, SHL, and SM were responsible for data acquisition and analyses; TF, SDG, MJA, SMG, CS, SHL, and SM contributed to drafting the manuscript and figures.
Potential Conflicts of Interest
The authors have no ethical or financial conflicts of interest to declare.
References
- 1.Fischer-Smith T, Croul S, Sverstiuk AE, et al. CNS invasion by CD14+/CD16+ peripheral blood-derived monocytes in HIV dementia: perivascular accumulation and reservoir of HIV infection. J Neurovirol. 2001 Dec;7(6):528–41. doi: 10.1080/135502801753248114. [DOI] [PubMed] [Google Scholar]
- 2.Tavazzi E, Morrison D, Sullivan P, Morgello S, Fischer T. Brain Inflammation is a Common Feature of HIV-Infected Patients Without HIV Encephalitis or Productive Brain Infection. Curr HIV Res. 2014 May 26; doi: 10.2174/1570162x12666140526114956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelman BB. Diffuse microgliosis associated with cerebral atrophy in the acquired immunodeficiency syndrome. Ann Neurol. 1993 Jul;34(1):65–70. doi: 10.1002/ana.410340112. [DOI] [PubMed] [Google Scholar]
- 4.Weis S, Neuhaus B, Mehraein P. Activation of microglia in HIV-1 infected brains is not dependent on the presence of HIV-1 antigens. Neuroreport. 1994 Jul 21;5(12):1514–6. doi: 10.1097/00001756-199407000-00026. [DOI] [PubMed] [Google Scholar]
- 5.Xing HQ, Hayakawa H, Gelpi E, Kubota R, Budka H, Izumo S. Reduced expression of excitatory amino acid transporter 2 and diffuse microglial activation in the cerebral cortex in AIDS cases with or without HIV encephalitis. J Neuropathol Exp Neurol. 2009 Feb;68(2):199–209. doi: 10.1097/NEN.0b013e31819715df. [DOI] [PubMed] [Google Scholar]
- 6.Kempuraj D, Thangavel R, Selvakumar GP, et al. Brain and Peripheral Atypical Inflammatory Mediators Potentiate Neuroinflammation and Neurodegeneration. Front Cell Neurosci. 2017;11:216. doi: 10.3389/fncel.2017.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gironi M, Borgiani B, Farina E, et al. A global immune deficit in Alzheimer’s disease and mild cognitive impairment disclosed by a novel data mining process. J Alzheimers Dis. 2015;43(4):1199–213. doi: 10.3233/JAD-141116. [DOI] [PubMed] [Google Scholar]
- 8.De Virgilio A, Greco A, Fabbrini G, et al. Parkinson’s disease: Autoimmunity and neuroinflammation. Autoimmun Rev. 2016 Oct;15(10):1005–11. doi: 10.1016/j.autrev.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 9.Steiner J, Bielau H, Brisch R, et al. Immunological aspects in the neurobiology of suicide: elevated microglial density in schizophrenia and depression is associated with suicide. J Psychiatr Res. 2008 Jan;42(2):151–7. doi: 10.1016/j.jpsychires.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Soderlund J, Schroder J, Nordin C, et al. Activation of brain interleukin-1beta in schizophrenia. Mol Psychiatry. England. 2009:1069–71. doi: 10.1038/mp.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nomenclature and research case definitions for neurologic manifestations of human immunodeficiency virus-type 1 (HIV-1) infection. Report of a Working Group of the American Academy of Neurology AIDS Task Force. Neurology. 1991 Jun;41(6):778–85. doi: 10.1212/wnl.41.6.778. [DOI] [PubMed] [Google Scholar]
- 12.Clinical confirmation of the American Academy of Neurology algorithm for HIV-1-associated cognitive/motor disorder. The Dana Consortium on Therapy for HIV Dementia and Related Cognitive Disorders. Neurology. 1996 Nov;47(5):1247–53. doi: 10.1212/wnl.47.5.1247. [DOI] [PubMed] [Google Scholar]
- 13.Fischer-Smith T, Croul S, Adeniyi A, et al. Macrophage/microglial accumulation and proliferating cell nuclear antigen expression in the central nervous system in human immunodeficiency virus encephalopathy. Am J Pathol. 2004 Jun;164(6):2089–99. doi: 10.1016/S0002-9440(10)63767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Che S, Ginsberg SD. Amplification of transcripts using terminal continuation. Lab Invest. 2004;84:131–7. doi: 10.1038/labinvest.3700005. [DOI] [PubMed] [Google Scholar]
- 15.Alldred MJ, Che S, Ginsberg SD. Terminal Continuation (TC) RNA amplification enables expression profiling using minute RNA input obtained from mouse brain. Int J Mol Sci. 2008 Nov;9(11):2091–104. doi: 10.3390/ijms9112091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alldred MJ, Che S, Ginsberg SD. Terminal continuation (TC) RNA amplification without second strand synthesis. J Neurosci Methods. 2009 Mar 15;177(2):381–5. doi: 10.1016/j.jneumeth.2008.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginsberg SD, Alldred MJ, Counts SE, et al. Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer’s disease progression. Biol Psychiatry. 2010 Nov 15;68(10):885–93. doi: 10.1016/j.biopsych.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alldred MJ, Duff KE, Ginsberg SD. Microarray analysis of CA1 pyramidal neurons in a mouse model of tauopathy reveals progressive synaptic dysfunction. Neurobiol Dis. 2012 Feb;45(2):751–62. doi: 10.1016/j.nbd.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alldred MJ, Lee SH, Petkova E, Ginsberg SD. Expression profile analysis of vulnerable CA1 pyramidal neurons in young-Middle-Aged Ts65Dn mice. J Comp Neurol. 2015 Jan 01;523(1):61–74. doi: 10.1002/cne.23663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginsberg SD. RNA amplification strategies for small sample populations. Methods. 2005 Nov;37(3):229–37. doi: 10.1016/j.ymeth.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Ginsberg SD. Transcriptional profiling of small samples in the central nervous system. Methods Mol Biol. 2008;439:147–58. doi: 10.1007/978-1-59745-188-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ginsberg SD, Che S, Wuu J, Counts SE, Mufson EJ. Down regulation of trk but not p75NTR gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J Neurochem. 2006 Apr;97(2):475–87. doi: 10.1111/j.1471-4159.2006.03764.x. [DOI] [PubMed] [Google Scholar]
- 23.Ginsberg SD, Che S, Counts SE, Mufson EJ. Single cell gene expression profiling in Alzheimer’s disease. NeuroRx. 2006;3(3):302–18. doi: 10.1016/j.nurx.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCulloch C. Joint modelling of mixed outcome types using latent variables. Stat Methods Med Res. 2008 Feb;17(1):53–73. doi: 10.1177/0962280207081240. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. (1995) [Google Scholar]
- 26.Efron B. Correlation and Large-Scale Simultaneous Significance Testing. Journal of the American Statistical Association. 2007 Mar;102(477):93–103. [Google Scholar]
- 27.Abassi M, Morawski BM, Nakigozi G, et al. Cerebrospinal fluid biomarkers and HIV-associated neurocognitive disorders in HIV-infected individuals in Rakai, Uganda. J Neurovirol. 2017 Jun;23(3):369–75. doi: 10.1007/s13365-016-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sisay S, Lopez-Lozano L, Mickunas M, et al. Untreated relapsing remitting multiple sclerosis patients show antibody production against latent Epstein Barr Virus (EBV) antigens mainly in the periphery and innate immune IL-8 responses preferentially in the CNS. J Neuroimmunol. 2017 May 15;306:40–5. doi: 10.1016/j.jneuroim.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Thelin EP, Tajsic T, Zeiler FA, et al. Monitoring the Neuroinflammatory Response Following Acute Brain Injury. Front Neurol. 2017;8:351. doi: 10.3389/fneur.2017.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coughlin JM, Wang Y, Ma S, et al. Regional brain distribution of translocator protein using [(11)C]DPA-713 PET in individuals infected with HIV. J Neurovirol. 2014 Jun;20(3):219–32. doi: 10.1007/s13365-014-0239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vera JH, Guo Q, Cole JH, et al. Neuroinflammation in treated HIV-positive individuals: A TSPO PET study. Neurology. 2016 Apr 12;86(15):1425–32. doi: 10.1212/WNL.0000000000002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thornberry NA, Bull HG, Calaycay JR, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992 Apr 30;356(6372):768–74. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 33.Gerngross L, Fischer T. Evidence for cFMS signaling in HIV production by brain macrophages and microglia. J Neurovirol. 2015 Jun;21(3):249–56. doi: 10.1007/s13365-014-0270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerngross L, Lehmicke G, Belkadi A, Fischer T. Role for cFMS in maintaining alternative macrophage polarization in SIV infection: implications for HIV neuropathogenesis. J Neuroinflammation. 2015;12:58. doi: 10.1186/s12974-015-0272-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Li Y, Yu M, Chen B, Shen B. Lineage-dependent NF-kappaB activation contributes to the resistance of human macrophages to apoptosis. Hematol J. 2003;4(4):277–84. doi: 10.1038/sj.thj.6200252. [DOI] [PubMed] [Google Scholar]
- 36.He C, Ryan AJ, Murthy S, Carter AB. Accelerated development of pulmonary fibrosis via Cu, Zn-superoxide dismutase-induced alternative activation of macrophages. J Biol Chem. 2013 Jul 12;288(28):20745–57. doi: 10.1074/jbc.M112.410720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993 Aug 13;261(5123):921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 38.Corder EH, Robertson K, Lannfelt L, et al. HIV-infected subjects with the E4 allele for APOE have excess dementia and peripheral neuropathy. Nat Med. 1998 Oct;4(10):1182–4. doi: 10.1038/2677. [DOI] [PubMed] [Google Scholar]
- 39.Frikke-Schmidt R, Nordestgaard BG, Thudium D, Moes Gronholdt ML, Tybjaerg-Hansen A. APOE genotype predicts AD and other dementia but not ischemic cerebrovascular disease. Neurology. 2001 Jan 23;56(2):194–200. doi: 10.1212/wnl.56.2.194. [DOI] [PubMed] [Google Scholar]
- 40.Ali K, Middleton M, Pure E, Rader DJ. Apolipoprotein E suppresses the type I inflammatory response in vivo. Circ Res. 2005 Oct 28;97(9):922–7. doi: 10.1161/01.RES.0000187467.67684.43. [DOI] [PubMed] [Google Scholar]
- 41.Janssen CI, Jansen D, Mutsaers MP, et al. The Effect of a High-Fat Diet on Brain Plasticity, Inflammation and Cognition in Female ApoE4-Knockin and ApoE-Knockout Mice. PLoS One. 2016;11(5):e0155307. doi: 10.1371/journal.pone.0155307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cammarota M, Paratcha G, Bevilaqua LR, et al. Cyclic AMP-responsive element binding protein in brain mitochondria. J Neurochem. 1999 Jun;72(6):2272–7. doi: 10.1046/j.1471-4159.1999.0722272.x. [DOI] [PubMed] [Google Scholar]
- 43.Ryu H, Lee J, Impey S, Ratan RR, Ferrante RJ. Antioxidants modulate mitochondrial PKA and increase CREB binding to D-loop DNA of the mitochondrial genome in neurons. Proc Natl Acad Sci U S A. 2005 Sep 27;102(39):13915–20. doi: 10.1073/pnas.0502878102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu S, Xie Z, Onishi A, et al. Profiling the Human Protein-DNA Interactome Reveals MAPK1 as a Transcriptional Repressor of Interferon Signalling. Cell. 2009 Oct 30;139(3):610–22. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Counts SE, He B, Che S, et al. Alpha7 nicotinic receptor up-regulation in cholinergic basal forebrain neurons in Alzheimer disease. Arch Neurol. 2007 Dec;64(12):1771–6. doi: 10.1001/archneur.64.12.1771. [DOI] [PubMed] [Google Scholar]
- 46.Counts SE, He B, Che S, Ginsberg SD, Mufson EJ. Galanin hyperinnervation upregulates choline acetyltransferase expression in cholinergic basal forebrain neurons in Alzheimer’s disease. Neurodegener Dis. 2008;5(3–4):228–31. doi: 10.1159/000113710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Counts SE, He B, Che S, Ginsberg SD, Mufson EJ. Galanin fiber hyperinnervation preserves neuroprotective gene expression in cholinergic basal forebrain neurons in Alzheimer’s disease. J Alzheimers Dis. 2009;18(4):885–96. doi: 10.3233/JAD-2009-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao ML, Kim MO, Morgello S, Lee SC. Expression of inducible nitric oxide synthase, interleukin-1 and caspase-1 in HIV-1 encephalitis. J Neuroimmunol. 2001 Apr 2;115(1–2):182–91. doi: 10.1016/s0165-5728(00)00463-x. [DOI] [PubMed] [Google Scholar]
- 49.Tyor WR, Glass JD, Griffin JW, et al. Cytokine expression in the brain during the acquired immunodeficiency syndrome. Ann Neurol. 1992 Apr;31(4):349–60. doi: 10.1002/ana.410310402. [DOI] [PubMed] [Google Scholar]