Abstract
Purpose
Transcriptional pathway activity and the molecular subtypes of breast cancer metastases have been shown to significantly influence patient postrelapse survival. Here, we further determine the relevance of clinically employed gene signatures in the advanced breast cancer (ABC) setting.
Experimental Design
Sufficient RNA for expression profiling was obtained from distant metastatic or inoperable locoregional relapse tissue by fine-needle aspiration from 109 patients of the Swedish TEX clinical trial. Gene signatures (GGI, 70 gene, recurrence score, cell-cycle score, risk of recurrence score, and PAM50) were applied to all metastases, and their relationship to long- (5-year) and short-term (1.5-year) postrelapse survival at all and locoregional lymph nodes (n = 40) versus other metastatic sites (n = 69) combined was assessed using Kaplan-Meier and/or multivariate Cox regression analyses.
Results
The majority of metastases were classified into intermediate or high-risk groups by all signatures, and a significant association was found between metastatic signature subgroups and primary tumor estrogen receptor status and histologic grade (P < 0.05). When considering all sites of metastasis, only PAM50 was statistically significant in Kaplan–Meier analysis (Log-rank P = 0.008 and 0.008 for long- and short-term postrelapse breast cancer–specific survival, respectively). This significance remained in both uni- and multivariate models when restricting analyses to lymph node metastases only, and a similar trend was observed in other metastatic sites combined, but did not reach formal significance.
Conclusions
Our findings are the first to demonstrate that the PAM50 signature can provide prognostic information from the lymph node metastases of ABC patients.
Introduction
The widespread availability and focused application of large-scale “Omic” technologies have greatly contributed to our understanding of the molecular heterogeneity of primary breast tumors (1). We now know that breast cancer should no longer be considered a single disease but one comprised of five (2) to 10 (3) individual subgroups each of which corresponds to a different underlying biology (4), survival rate, and response to treatment (5, 6).
Recent clinical trial data have served to highlight the capacity of gene-expression signatures to select subgroups of breast cancer patients who could safely be spared from chemotherapy (7, 8) and who demonstrate a better response to neoadjuvant treatment with trastuzaumab (9). While much data have been accrued on the prognostic and treatment predictive capacity of gene signatures in primary breast tumors, their applicability in the advanced breast cancer (ABC) setting remains unclear. Given that ABC3 clinical guidelines recommend reevaluation of metastatic lesions for estrogen (ER), progesterone (PR), and HER2 expression (10), and that data from primary tumors have shown the ability of gene signatures to compete well with the same IHC markers plus Ki67 (11), an assessment of whether gene expression signatures can provide prognostic information in breast cancer metastases is warranted. Related to this, a number of studies have demonstrated differences in protein expression (refs. 12, 13; ER, PR, HER2), DNA mutations, and gene copy numbers (14–16) between matched primary and metastatic tumors, serving to reaffirm the importance of biomarker assessment in metastatic lesions, in particular if a targeted breast cancer treatment is to be administered.
We have previously demonstrated that transcriptional pathway activity and the molecular subtypes (PAM50) of breast cancer metastases significantly influence patient postrelapse survival (17). Here, we aim to extend these findings through Kaplan–Meier and Cox regression analysis of five routinely employed gene expression signatures, along with a simple cell-cycle classifier, with specific focus on site of metastatic relapse.
Patients and Methods
Cohort description
A full description for this cohort, including patient characteristics, treatments received, and clinical endpoints, has been previously published (18). Briefly, the TEX trial (18) was a Swedish multicenter randomized clinical trial (ClinicalTrials.gov identifier NCT01433614) that compared the efficacy of epirubicin and paclitaxel alone or in combination with capecitabine as a firstline treatment in the locally advanced inoperable or metastatic breast cancer setting. A total of 304 patients were enrolled in the trial from December 2002 to June 2007 with morphologically confirmed advanced locoregional or distant breast cancer relapse. As part of the TEX translational study, patients were asked to give a sample of metastatic lesions if accessible by either a fine-needle aspiration (FNA) or a core biopsy, but sampling was optional. After exclusion of patients who did not provide a biopsy along and samples that did not pass quality controls regarding tumor cell purity and cellularity, 109 patients remained with whole-genome gene expression data from array profiling, detailed clinical information and complete follow-up. A CONSORT diagram is shown in Supplementary Fig. S1. The clinicopathologic characteristics of the patients included in the translational TEX trial were representative of the original TEX trial and the primary tumor characteristics for these 109 patients are shown in Supplementary Table S1. Sites of metastasis/relapse biopsy for these patients were as follows: lymph node (37%), liver (24%), skin (18%), breast (14%), skeleton (4%), lung/pleura (2%), and other (1%). The breakdown of the lymph node biopsy sites were: supraclavicular (23, 57.5%), axilla (10, 25%), neck (3, 7.5%), retrosternal (1, 2.5%), inguinal (1, 25%), infraclavicular (1, 2.5%), and lymph node unspecified (1, 2.5%). For the sake of brevity, we henceforth call all samples metastases regardless of whether they were taken from an inoperable locoregional or distant metastatic site. The clinical study was approved by the ethics committee at Karolinska Institutet, which had jurisdiction for all participating centers, and by the Swedish Medical Product Agency. All patients received oral and written information and consented to participate.
Expression array profiling and data normalization
All metastases were profiled on the Rosetta/Merck Human RSTA Custom Affymetrix 2.0 microarray (GEO: GPL10379) and background corrected/normalized using the aroma.affymetrix R package. Data can be retrieved from NCBI GEO under accession number GSE56493. Because this cohort contains more clinically aggressive and highly proliferative tumors relative to a population-based primary breast cancer cohort, we normalized our gene expression arrays from the TEX metastatic material with 623 primary breast tumors (NCBI GEO reference: GSE48091) run on the same array platform. A full description can be found in ref. 17.
Gene expression signatures
Research versions of the Genomic Grade Index (GGI), 70 gene (commercially Mammaprint), Recurrence Score (RS, commercially OncotypeDx), Risk of Recurrence - Subtype (ROR-S), and prediction analysis of microarray 50 (PAM50) signatures were applied as described in the original publications, and we have previously published our R code for these classification calls (19). Note that the ROR-S signature is derived from the PAM50 tumor calls. Tumors classified as normal-like by PAM50 were excluded from analyses. Signatures were chosen on the basis of their relevancy in an on-going Swedish clinical trial (20) and owing to their use in a routine clinical setting. The cell-cycle score (CCS) was derived by adding the expression of cell-cycle genes identified from three different databases (KEGG, HGNC, Cyclebase) and splitting the resulting continuous variable into tertiles of low, intermediate, and high cell-cycle activity, further details here (21).
Statistical analysis
All statistical analyses were performed using R statistical software version 3.3.1. Kaplan–Meier and multivariate proportional hazard (Cox) analyses were performed adjusting the latter for calendar year and age at diagnosis in addition to the TEX clinical study treatment arms. We did not adjust for additional tumor characteristics due to sample size and no significant deviation was noted for the proportional hazard assumption in the survival model. The likelihood ratio (LR) for all signatures was also calculated from this multivariate model and used as a measure of signature prognostic capacity. Postrelapse breast cancer–specific survival (BCSS) was defined as long- (up to 5 years or more) and short-term (up to 1.5 years postrelapse survival). Of note, this short-term cutoff was defined retrospectively to capture the visually apparent distribution in postrelapse BCSS (41% of patients died within 1.5 years) and was determined by applying a model of two normal distributions to the survival time variable. A comprehensive description of this cutoff and associated methods has been previously published (17). To assess differences between primary tumor clinicopathologic variables and metastatic tumor gene-expression subtypes statistical tests were chosen based on the class of variables being compared: ordinal versus nominal (e.g., RS vs. ER)—Wilcoxon/Mann–Whitney test; categorical versus nominal (e.g., PAM50 vs. ER)—χ2 or Fisher exact tests; categorical versus ordinal—Kruskal–Wallis test; ordinal versus ordinal—Spearman rank correlation test. Tests used are indicated in table legends.
Results
Gene signature clinicopathologic characteristics
We applied the GGI, 70 gene, CCS, RS, ROR-S and PAM50 signatures to expression array data from 109 metastatic breast cancer biopsies, taken by fine-needle aspiration. As expected, given the aggressive nature of metastases, few tumors were classified as low risk/good prognosis by gene signatures (Fig. 1, blue bars. GGI: 27%, 70 gene: 21%, CCS: 8%, RS: 3%, ROR-S: 4%, PAM50, Luminal A: 11%).
Figure 1.
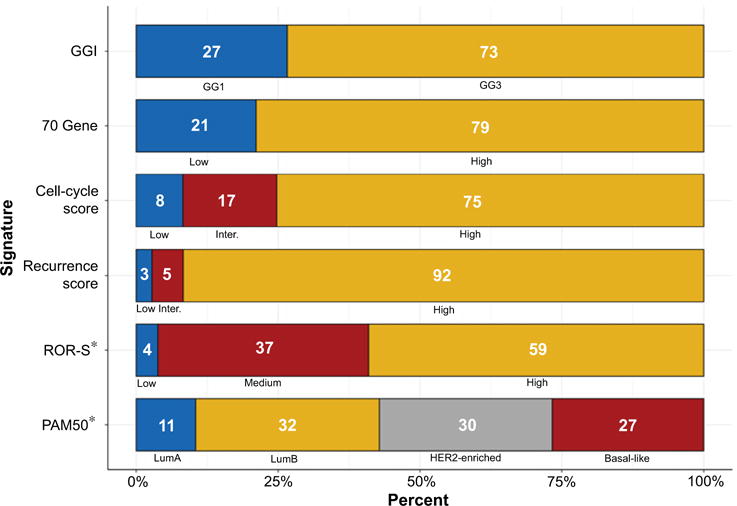
Distribution of gene expression signature subgroups in breast cancer metastases (N = 109). The distribution of gene expression signature subgroups across all 109 breast cancer metastases; numbers represent the percentage of each signature subgroup. For example, GGI: GG127%, GG3 73%. Blue shading represents expected good prognosis tumors. GGI, Genomic grade index; ROR-S, Risk of recurrence subtype score; *, Reduced numbers (N = 105).
The relationship between the signature subgroups of the metastases and patient/tumor characteristics from their matching primary breast tumor are shown in Table 1 and Supplementary Table S2. For all signatures, we found a statistically significant association between metastatic tumor gene signature subtype and the ER status (Table 1, left hand side) or histologic grade (Table 1, right hand side) of its matching primary tumor. Similar results were noted for PR (Supplementary Table S2, no adjustment was made for multiple-testing).
Table 1.
Metastatic tumor signature subtype split by primary tumor ER status and grade
| Signature | ERa N (%) |
P | Gradeb N (%) |
P | |||
|---|---|---|---|---|---|---|---|
| Positive 64 (62) | Negative 39 (38) | 1 4 (5) |
2 35 (43) |
3 42 (52) |
|||
| Genomic grade | |||||||
| GG1 | 26 (41) | 2 (5) | 1 (25) | 16 (46) | 2 (5) | ||
| GG3 | 38 (59) | 37 (95) | <0.001 | 3 (75) | 19 (54) | 40 (95) | <0.001 |
| 70 gene | |||||||
| Low | 20 (31) | 1 (3) | 0 (0) | 12 (34) | 4 (10) | ||
| High | 44 (69) | 38 (97) | <0.001 | 4 (100) | 23 (66) | 38 (90) | 0.043 |
| Recurrence score | |||||||
| Low | 3 (5) | 0 (0) | 0 (0) | 2 (6) | 0 (0) | ||
| Intermediate | 6 (9) | 0 (0) | 0 (0) | 5 (14) | 0 (0) | ||
| High | 55 (86) | 39 (100) | 0.015 | 4 (100) | 28 (80) | 42 (100) | 0.011 |
| CCS | |||||||
| Low | 7 (11) | 1 (3) | 0 (0) | 4 (11) | 0 (0) | ||
| Intermediate | 14 (22) | 4 (10) | 0 (0) | 8 (23) | 3 (7) | ||
| High | 43 (67) | 34 (87) | 0.022 | 4 (100) | 23 (66) | 39 (93) | 0.015 |
| ROR-S | |||||||
| Low | 3 (4) | 0 (0) | 0 (0) | 3 (10) | 0 (0) | ||
| Medium | 29 (48) | 8 (21) | 1 (25) | 15 (48) | 8 (19) | ||
| High | 29 (48) | 30 (79) | 0.002 | 3 (75) | 13 (42) | 34 (81) | 0.002 |
| PAM50c | |||||||
| Luminal A | 10 (17) | 0 (0) | 0 (0) | 6 (19) | 0 (0) | ||
| Luminal B | 30 (49) | 2 (5) | 2 (50) | 17 (55) | 9 (22) | ||
| HER2-enriched | 19 (31) | 12 (32) | 1 (25) | 6 (19) | 14 (33) | ||
| Basal-like | 2 (3) | 24 (63) | <0.001d | 1 (25) | 2 (7) | 19 (45) | <0.001e |
NOTE: Correlations were calculated using Wilcoxon/Mann-Whitney (ER) and Spearman rank (Grade) unless otherwise specified. Bold values indicate P < 0.05.
Primary tumor ER status unknown in 6 cases, ≥10% cutoff value for positivity.
Primary tumor grade unknown in 28 cases.
Reduced numbers (n = 105).
The Fisher exact test.
The Kruskal-Wallis test; n, number of patients.
PAM50 predicts long- and short-term postrelapse breast cancer–specific survival
We next assessed the capacity of gene-expression signatures to predict postrelapse BCSS using Kaplan–Meier and Cox regression analyses. Neither of the binary GGI nor 70 gene signatures demonstrated statistical significance in Kaplan–Meier analysis (Supplementary Figs. S2 and S3, A and B, log-rank P values: GGI = 0.655 and 0.368, 70 gene = 0.389 and 0.188 for long/short-term survival, respectively). The same was true for the multilevel RS, CCS, and ROR-S signatures (Supplementary Figs. S2 and S3C–S3E, Log-rank P values: RS = 0.403 and 0.461, CCS = 0.181 and 0.450 and ROR-S = 0.221 and 0.148, for long/short-term survival, respectively). Only PAM50 provided statistically significant prognostic information when considering all metastatic sites whereby tumors classified as Basal-like, Luminal B, and HER2-enriched have a worse prognosis relative to those classified as Luminal A (Supplementary Figs. S2 and S3, F, log-rank P: PAM50 = 0.008 and 0.008 for long/short-term BCSS, respectively), this significance remained in multivariate analysis (Table 2, left-hand column, “All Sites”). Long- and short-term univariate HRs for all signatures are shown in Supplementary Table S3.
Table 2.
Long and short-term multivariate analysis for PAM50 at specific metastatic sites, postrelapse BCSS as clinical endpoint
| Signature subgroup | N (%) | All sites (N = 105)a
|
N (%) | Lymph nodes (N = 40)a
|
N (%) | Other sites (N = 65)a
|
|||
|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||||
| Long-term BCSS | |||||||||
| PAM50 | |||||||||
| Luminal B (Ref.) | 34 (32) | 1.0 (−) | – | 11 (28) | 1.0 (−) | – | 23 (35) | 1.0 (−) | – |
| Luminal A | 11 (11) | 0.4 (0.1–1.3) | 0.121 | 2 (5) | − (0.0–Inf.) | 0.998 | 9 (14) | 0.3 (0.1–1.1) | 0.063 |
| HER2-enriched | 32 (30) | 1.9 (1.1–3.3) | 0.031 | 12 (30) | 3.7 (1.2–11.6) | 0.027 | 20 (31) | 2.0 (1.0–4.1) | 0.061 |
| Basal-like | 28 (27) | 1.6 (0.9–2.9) | 0.132 | 15 (37) | 7.9 (2.2–28.2) | 0.001 | 13 (20) | 0.9 (0.4–2.3) | 0.832 |
| Short-term BCSS | |||||||||
| PAM50 | |||||||||
| Luminal B (Ref.) | 34 (32) | 1.0 (−) | – | 11 (28) | 1.0 (−) | – | 23 (35) | 1.0 (−) | − |
| Luminal A | 11 (11) | 0.4 (0.1–3.5) | 0.421 | 2 (5) | − (0.0–Inf.) | 0.998 | 9 (14) | 0.3 (0.0–2.4) | 0.252 |
| HER2-enriched | 32 (30) | 3.2 (1.3–7.9) | 0.013 | 12 (30) | 7.2 (0.8–64.1) | 0.078 | 20 (31) | 3.6 (1.2–10.8) | 0.019 |
| Basal-like | 28 (27) | 3.0 (1.2–7.5) | 0.019 | 15 (37) | 2.6 (2.7–247.5) | 0.005 | 13 (20) | 1.6 (0.5–5.4) | 0.466 |
NOTE: Bold values indicate P < 0.05.
Adjusted for age at diagnosis, diagnosis date, and clinical trial treatment received.
The prognostic capacity of signatures is recurrence site dependent
On the basis of previous publications showing that breast cancer subtypes display preferential sites of metastasis (22–24), we examined whether gene signature subtype is influenced by metastatic site (Table 3). Interestingly, no liver metastases were classified as Basal-like by PAM50, in line with the work of Kennecke and colleagues (23) demonstrating a lower rate of liver metastasis for this tumor subtype (Table 3, see “Liver”).
Table 3.
Metastatic tumor signature subtype split by relapse site (N = 109)
| Signature | Lymph node 40 (37%) | Relapse site N (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Other sites (combined) 69 (63%) | Liver 26 (24%) | Skin 20 (18%) | Breast 15 (14%) | Skeleton 5 (4%) | Lung/Pleura 2 (2%) | Other 1 (1%) | Pa | ||
| Genomic grade | |||||||||
| GG1 | 6 (15) | 23 (33) | 13 (50) | 4 (20) | 4 (27) | 1 (20) | 0 (0) | 1 (100) | |
| GG3 | 34 (85) | 46 (67) | 13 (50) | 16 (80) | 11 (73) | 4 (80) | 2 (100) | 0 (0) | 0.062 |
| 70 gene | |||||||||
| Low | 5 (12) | 18 (26) | 10 (38) | 2 (10) | 3 (20) | 1 (20) | 1 (50) | 1 (100) | |
| High | 35 (88) | 31 (74) | 16 (62) | 18 (90) | 12 (80) | 4 (80) | 1 (50) | 0 (0) | 0.152 |
| Recurrence score | |||||||||
| Low | 1 (2) | 2 (3) | 1 (4) | 1 (5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Intermediate | 2 (5) | 4 (6) | 1 (4) | 2 (10) | 1 (7) | 0 (0) | 0 (0) | 0 (0) | |
| High | 37 (93) | 63 (91) | 24 (92) | 17 (85) | 14 (93) | 5 (100) | 2 (100) | 1 (100) | 1.000b |
| CCS | |||||||||
| Low | 3 (8) | 6 (9) | 3 (11) | 3 (15) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Intermediate | 3 (8) | 15 (22) | 7 (27) | 2 (10) | 5 (33) | 0 (0) | 0 (0) | 1 (100) | |
| High | 34 (84) | 48 (69) | 16 (62) | 15 (77) | 10 (67) | 5 (100) | 2 (100) | 0 (0) | 0.107b |
| ROR-Sc | |||||||||
| Low | 1 (2) | 3 (4) | 1 (4) | 1 (6) | 1 (7) | 0 (0) | 0 (0) | 0 (0) | |
| Medium | 8 (20) | 31 (48) | 18 (69) | 5 (29) | 5 (33) | 2 (40) | 1 (50) | 0 (0) | |
| High | 31 (78) | 31 (48) | 7 (27) | 11 (65) | 9 (60) | 3 (60) | 1 (50) | 0 (0) | 0.006b |
| PAM50c | |||||||||
| Luminal A | 2 (5) | 9 (14) | 6 (23) | 1 (6) | 2 (13) | 0 (0) | 0 (0) | 0 (0) | |
| Luminal B | 11 (28) | 23 (35) | 12 (46) | 3 (18) | 4 (27) | 3 (60) | 1 (50) | 0 (0) | |
| HER2-enriched | 12 (30) | 20 (31) | 8 (31) | 4 (23) | 6 (40) | 2 (40) | 0 (0) | 0 (0) | |
| Basal-like | 15 (37) | 13 (20) | 0 (0) | 9 (53) | 3 (20) | 0 (0) | 1 (50) | 0 (0) | 0.177b |
NOTE: Bold values indicate P < 0.05.
P values are based on the χ2 or Fisher exact test comparison of lymph node versus other sites combined columns.
The Fisher exact test.
Reduced numbers (n = 105).
As lymph node metastases are often the most accessible/practical biopsy site in metastatic cancer patients, we next wanted to determine whether a signature could also predict patient postrelapse survival from a lymph node biopsy. For this analysis we divided our metastases into two groups of lymph node versus other metastatic sites combined (liver, skin, breast, skeleton, lung/pleura and other) and focused on the PAM50 signature. We found no statistically significant difference in subtype distribution when comparing these two groups (Table 3, compare “Lymph node” with “Other sites” for PAM50, P = 0.177) and Kaplan–Meier curves were also comparatively similar, in particular the Luminal B and HER2-enriched subtypes, although statistical significance was not reached in long-term survival analysis (Supplementary Fig. S4, compare A with B, P = 0.055 and P = 0.111, respectively). The corresponding multivariate analysis showed an increased HR for HER2-enriched tumors in both groups, but only reached formal significance in lymph node metastases [Table 2, compare lymph nodes with other sites, HER2-enriched subgroup HR, 3.7; 95% confidence interval (CI), 1.2–11.6 and HR, 2.0; 95% CI, 1.0–4.1, respectively, long-term BCSS]. The Basal-like subgroup provided statistically significant information in lymph node metastasis only (Table 2, Lymph nodes, Basal-like subgroup HR, 7.9; 95% CI, 2.2–28.2; long-term BCSS). Finally, using the LR as a measure of the prognostic capacity, we found that PAM50 provides prognostic information in both groups after adjusting for calendar year, age at diagnosis and the TEX clinical study treatment arms (Supplementary Table S4. Long-term LR: PAM50 = 20.0 and 10.4; P < 0.001 and P = 0.015 for Lymph nodes and Other sites, respectively).
Taken together these findings indicate that assessment of PAM50 on a lymph node biopsy can provide significant prognostic information on patient postrelapse survival.
Discussion
In this study, we applied six gene signatures to expression array data from 109 patients with ABC and determined the prognostic capacity of each signature across all sites, before comparing lymph node versus all other sites using postrelapse survival as the clinical endpoint. Our analyses yielded three main findings; first, that the majority of breast cancer metastases are classified as poor prognosis by gene expression signatures, second, that there is a significant relationship between metastatic tumor signature subtype and the ER status or grade of its matching primary tumor, and third, that PAM50 can predict postrelapse survival in lymph node metastases. To our knowledge this is the first time these signatures have been applied and compared in the ABC setting and importantly, the first clear demonstration of the prognostic utility of PAM50 at a specific metastatic site. It is also worth noting that we found a trend toward significance for PAM50 in multivariate analysis of our “other metastatic sites combined” grouping, indicating that this signature may also be informative at other metastatic sites. However, we lack the statistical power to draw any significant conclusions regarding the individual sites within this grouping.
Regarding our finding of an association between metastatic tumor signature subtype and primary tumor characteristics (ER, PR, and histologic grade), a recent comparison of the PAM50 subtypes between 123 paired primary and metastatic samples showed that tumor molecular subtype is generally maintained at recurrence except in Luminal A tumors which changed to a different subtype in up to 55% of cases (25). As PAM50 defined Luminal A/B tumors are predominantly ER-positive by IHC analysis (26) our finding of less metastatic tumors with these tumor subtypes among patients who had an ER-negative primary tumor (Table 1) was anticipated. Molecular subtype concordance between different tumors in the same patient has also been demonstrated by Hoadley and colleagues (27) in a study of two triple-negative breast cancer patients where the PAM50 Basal-like subtype was maintained across multiple metastatic sites. These results further support the potential of PAM50 as determined from a lymph node metastasis to provide an accurate representation of postrelapse survival in ABC patients.
Although current ABC and ASCO guidelines (10, 28) recommend the reassessment of ER, PR, and HER2 at relapse, the utility of Ki67 is less clear. In the primary tumor setting, Ki67 is used to differentiate better prognosis, low proliferation luminal A tumors from highly proliferative and aggressive luminal B tumors. This distinction has direct treatment implications as luminal A tumors do not derive benefit from adjuvant chemotherapy (6) and show the least pathologic complete response (pCR) following treatment with chemotherapy in the neoadjuvant setting (5). Assessing Ki67 expression in ABC patients is, however, hampered as scoring the protein at the invasive edge of the tumor (which is recommended for accuracy; ref. 29) is not possible if the sample is taken by fine-needle aspiration owing to loss of tissue architecture. Moreover, even if the samples were to be taken by core needle biopsy, the prognostic relevance of Ki67 in breast cancer metastases and indeed the most appropriate cutoffs for good versus poor prognosis have not been sufficiently evaluated. Taken together, these issues point to the potential utility of a gene signature such as PAM50 over routine IHC biomarker analysis to aid in treatment decisions in patients with ABCs.
The main limitations of our study are as follows; first, this study is retrospective, unplanned, and as such is exploratory in nature. Second, sample size is limited with very few samples classified as good prognosis. The RS signature offers the best illustration of this where only three metastases were designated as “Low risk.” Although there is most certainly a biological rationale for this dearth of low-risk samples (metastatic tumors are inherently aggressive, poor prognosis tumors), it may be the case that the study is not powered to detect prognostic differences between some signature subgroups. Given the exploratory nature of our work, power calculations have not been performed and as such further larger studies with long-term postrelapse follow-up data will be required to confirm our findings. Third, we are using the research versions of gene-expression signatures rather than their commercial counterparts and fourth, our results have not been validated in a second independent dataset; however, we are not aware of any another dataset where gene expression profiling has been performed on such a large number of metastatic tumors coupled with long-term complete clinical follow-up data.
In summary, we are the first to apply and directly assess the ability of several clinically relevant gene expression signatures to predict postrelapse survival in metastatic biopsies from breast cancer patients and to demonstrate the prognostic strength of the PAM50 signature in lymph node metastases.
Supplementary Material
Translational Relevance.
Gene-expression signatures have been shown to provide prognostic and treatment predictive capacity in primary breast tumors; however, their applicability in the advanced breast cancer (ABC) setting is unknown. Here, we apply and directly assess the ability of several clinically relevant gene-expression signatures to predict postrelapse survival in metastatic biopsies from ABC patients. Our findings demonstrate not only that the majority of metastases are classified as high-risk/poor prognosis by all signatures, but also that PAM50 can provide prognostic information when applied to lymph node metastases. Given the difficulty in assessing proliferative biomarkers such as Ki67 at metastatic sites, this research highlights the potential utility of a gene signature to aid in treatment decisions in patients with ABC.
Acknowledgments
The TEX Trialists Group: Coordinating Investigator: Thomas Hatschek; Translational research: Mårten Fernö, Linda Lindström, Ingrid Hedenfalk; HRQoL: Yvonne Brandberg; Statistics: John Carstensen; Laboratory: Suzanne Egyhazy, Marianne Frostvik Stolt, Lambert Skoog; Clinical Trial Office: Mats Hellström, Maarit Maliniemi, Helene Svensson; Radiology: Gunnar Åström; Karolinska University Hospital, Stockholm: Jonas Bergh, Judith Bjöhle, Elisabet Lidbrink, Sam Rotstein, Birgitta Wallberg; Sahlgrenska University Hospital, Gothenburg: Zakaria Einbeigi, Per Carlsson, Barbro Linderholm; Linköping University Hospital: Thomas Walz; Skåne University Hospital Lund/Malmö: Niklas Loman, Per Malmström, Martin Söderberg; Helsingborg General Hospital: Martin Malmberg; Sundsvall General Hospital: Lena Carlsson; Umeå University Hospital: Birgitta Lindh; Kalmar General Hospital: Marie Sundqvist; Karlstad General Hospital: Lena Malmberg.
Grant Support
This work was supported by BRECT, the Swedish Cancer Society, the Cancer Society in Stockholm, the King Gustaf V Jubilee Foundation, the Swedish Breast Cancer Association (BRO), and the Swedish Research Council (J. Bergh); unrestricted grants from Bristol-Myers Squibb Sweden AB, Pfizer Sweden AB, and Roche Sweden AB for the TEX trial (to T. Hatschek); and the Swedish Research Council (grant 521-2014-2057 to L.S. Lindström). CM. Perou and J.C. Harrell were supported by funds from the NCI Breast SPORE program (P50-CA58223-09A1), by R01-CA195754-01, and the Breast Cancer Research Foundation (to CM. Perou).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
M. Fernö reports receiving speakers bureau honoraria from AstraZeneca, and is a consultant/advisory board member for Pfizer. C.M. Perou holds ownership interest (including patents) in and is a consultant/advisory board member for Bioclassifier LLC. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: N.P. Tobin, L.S. Lindström, L. Carlsson, Z. Einbeigi, M. Fernö, T. Hatschek
Development of methodology: N.P. Tobin, M. Malmberg, C.M. Perou
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): T. Foukakis, L. Carlsson, Z. Einbeigi, N. Loman, M. Malmberg, M. Fernö, T. Hatschek
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): N.P. Tobin, A. Lundberg, L.S. Lindström, T. Foukakis, M. Malmberg, K. Czene, C.M. Perou
Writing, review, and/or revision of the manuscript: N.P. Tobin, A. Lundberg, L.S. Lindström, J.C. Harrell, T. Foukakis, L. Carlsson, B.K. Linderholm, M. Malmberg, M. Fernö, K. Czene, C.M. Perou, J. Bergh, T. Hatschek
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): N.P. Tobin, M. Malmberg, T. Hatschek
Study supervision: N.P. Tobin, M. Malmberg, T. Hatschek
References
- 1.The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson S-J, Rueda OM, Aparicio S, Caldas C. A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 2013;32:617–28. doi: 10.1038/emboj.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prat A, Fan C, Fernández A, Hoadley KA, Martinello R, Vidal M, et al. Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy. BMC Med. 2015;13:303. doi: 10.1186/s12916-015-0540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen TO, Jensen MB, Burugu S, Gao D, Jorgensen CLT, Balslev E, et al. High risk premenopausal Luminal A breast cancer patients derive no benefit from adjuvant cyclophosphamide-based chemotherapy: results from the DBCG77B clinical trial. Clin Cancer Res. 2017;23:946–53. doi: 10.1158/1078-0432.CCR-16-1278. [DOI] [PubMed] [Google Scholar]
- 7.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med. 2015;373:2005–14. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. 2016;375:717–29. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 9.Carey LA, Berry DA, Cirrincione CT, Barry WT, Pitcher BN, Harris LN, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol. 2016;34:542–9. doi: 10.1200/JCO.2015.62.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardoso F, Costa A, Senkus E, Aapro M, André F, Barrios CH, et al. 3rdESO–ESMO international consensus guidelines for advanced breast cancer (ABC 3) Ann Oncol. 2017;28:16–33. doi: 10.1093/annonc/mdw544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31:2783–90. doi: 10.1200/JCO.2012.46.1558. [DOI] [PubMed] [Google Scholar]
- 12.Lindstrom LS, Karlsson E, Wilking UM, Johansson U, Hartman J, Lidbrink EK, et al. Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol. 2012;30:2601–8. doi: 10.1200/JCO.2011.37.2482. [DOI] [PubMed] [Google Scholar]
- 13.Niikura N, Liu J, Hayashi N, Mittendorf EA, Gong Y, Palla SL, et al. Loss of human epidermal growth factor receptor 2 (HER2) expression in metastatic sites of HER2-overexpressing primary breast tumors. J Clin Oncol. 2011;30:593–9. doi: 10.1200/JCO.2010.33.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown D, Smeets D, Székely B, Larsimont D, Szász AM, Adnet P-Y, et al. Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nat Commun. 2017;8:14944. doi: 10.1038/ncomms14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobin NP, Harrell JC, Lövrot J, Brage SE, Stolt MF, Carlsson L, et al. Molecular subtype and tumor characteristics of breast cancer metastases as assessed by gene expression significantly influence patient post-relapse survival. Ann Oncol. 2015;26:81–8. doi: 10.1093/annonc/mdu498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatschek T, Carlsson L, Einbeigi Z, Lidbrink E, Linderholm B, Lindh B, et al. Individually tailored treatment with epirubicin and paclitaxel with or without capecitabine as first-line chemotherapy in metastatic breast cancer: a randomized multicenter trial. Breast Cancer Res Treat. 2012;131:939–47. doi: 10.1007/s10549-011-1880-9. [DOI] [PubMed] [Google Scholar]
- 19.Tobin NP, Lindström LS, Carlson JW, Bjöhle J, Bergh J, Wennmalm K. Multi-level gene expression signatures, but not binary, outperform Ki67 for the long term prognostication of breast cancer patients. Mol Oncol. 2014;8:741–52. doi: 10.1016/j.molonc.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saal LH, Vallon-Christersson J, Häkkinen J, Hegardt C, Grabau D, Winter C, et al. The Sweden Cancerome Analysis Network - Breast (SCAN-B) Initiative: a large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015;7:20. doi: 10.1186/s13073-015-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundberg A, Lindström LS, Harrell JC, Falato C, Carlson JW, Wright PK, et al. Gene expression signatures and immunohistochemical subtypes add prognostic value to each other. Clin Cancer Res. 2017 Sep 29; doi: 10.1158/1078-0432.CCR-17-1535. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JGM, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–14. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 23.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–7. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 24.Harrell JC, Prat A, Parker JS, Fan C, He X, Carey L, et al. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat. 2012;132:523–35. doi: 10.1007/s10549-011-1619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cejalvo JM, de Dueñas EM, Galvan P, García-Recio S, Gasión OB, Paré L, et al. Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer. Cancer Res. 2017;77:2213–21. doi: 10.1158/0008-5472.CAN-16-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoadley KA, Siegel MB, Kanchi KL, Miller CA, Ding L, Zhao W, et al. Tumor evolution in two patients with basal-like breast cancer: a retrospective genomics study of multiple metastases. PLOS Med. 2016;13:e1002174. doi: 10.1371/journal.pmed.1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Poznak C, Somerfield MR, Bast RC, Cristofanilli M, Goetz MP, Gonzalez-Angulo AM, et al. Use of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2015;33:2695–704. doi: 10.1200/JCO.2015.61.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–64. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


