Abstract
Drug resistance is a serious impediment in treating cancer. However, the mechanisms involved remain poorly understood. While it is widely held that the phenomenon is genetic in nature, emerging evidence suggests that non-genetic mechanisms may also be important. Furthermore, at least in some cases, refractoriness to treatment can be reversed by epigenetic reprogramming, and combination and intermittent therapy, as opposed to sustained monotherapy, appear more effective in attenuating it. Here, we iterate the confusion in understanding the phenomenon by which cancer cells evade drug response, and underscore the need to recognize the genetic/non-genetic duality of drug resistance in cancer. We discuss how ecological and evolutionary principles may help reconcile the duality, and may even offer new treatment strategies.
Keywords: Drug resistance, duality, cancer, epigenetic mechanism, intermittent therapy
The clinician’s dilemma - one size does not fit all
A growing trend in medicine, especially with regard to medical oncology, is targeted therapy which is very often combined with other cancer treatments in patients with advanced disease (1–3). While patients respond fairly well initially, in most cancers, sustained treatment typically results in the failure of response to treatment (typically referred to as drug resistance) with poorer prognosis. Furthermore, challenges and complexities regarding toxicity when combining these therapies and how to personalize medicine - select the right patient population who can benefit from them and not develop drug resistance - are still a concern.
Understanding how cancer cells become refractory to drug treatment
Although, today, we have a much greater understanding of cancer biology and genetics, one of the main reasons for our failure to overcome the so-called drug resistance in cancer may have to do with the difficulty in how we perceive the phenomenon (see Text Box 1 for the various mechanisms by which organisms can evade drug treatment. For sake of simplicity, here, we shall refer to it as drug resistance). Is the phenomenon driven solely by genetic mechanisms i.e., is it irreversible and deterministic? Or, are other non-genetic mechanisms such as stochastic phenotypic switching or epigenetic factors that promote intrinsic diversity and phenotypic plasticity involved? If so, can this information help in guiding treatment decisions which are currently based solely on genetic biomarkers? Unfortunately, it appears that the phenomenon is much more nuanced (4). The water is muddied further by seemingly conflicting reports in the literature (5); also see examples discussed below) and the erroneous assumption that drug resistance, tolerance and persistence are synonymous or equivalent albeit inadvertently (see Text Box 1). Here, we consider two recent studies as a case in point; while one emphasizes the genetic underpinning of drug resistance, the other points to a non-genetic mechanism in the same cancer type. Using these two cases as recent examples, we stress the need to recognize the genetic/non-genetic duality of drug resistance in cancer, and discuss how ecological and evolutionary principles may help reconcile the duality and may even offer new strategies for treatment and prevention of drug resistance. However, the reader is referred to several excellent reviews for an in-depth discussion (6–8), and recent reports underscoring non-genetic mechanisms underlying drug resistance in cancer (9).
Text Box 1. Drug resistance, persistence and tolerance.
The term resistance is used (almost colloquially) to mean the patient’s tumor no longer responds to a given drug/hormone therapy. However, as has been elegantly demonstrated in microbiology, resistance is typically due to mutations and is defined the inherited ability of an organism to grow at high concentrations of a drug. It has a strong genetic underpinning. The terms ‘tolerance’ and ‘persistence’ are used to distinguish these modes of survival from resistance. Tolerance is more generally used to describe the ability, whether inherited or not, to survive transient exposure to high concentrations of a drug. Persistence, on the other hand, is the ability of a subpopulation of a clonal population to survive exposure to high concentrations of a drug. Persistence is typically observed when the majority of the population is rapidly killed when exposed to the drug while a subpopulation persists for a much longer period of time, despite the population being clonal. The three phenomena have been well documented and are robust in characterizing the response of micro-organisms to antibiotics [for an excellent review, see (37)]. However, the definitions of these different terms, and their distinction from one another, have remained somewhat ambiguous insofar as cancer cells are concerned and are often used synonymously albeit inadvertently.
It is widely, perhaps invariably, held that cancer is a genetic disease (10, 11). Further, it is also believed by many that, cancers evolve by a reiterative process of clonal expansion, genetic diversification and selection within the adaptive landscapes of the tissue ecosystems they inhabit (12). Therefore, while it may seem obvious that therapeutic intervention can destroy cancer clones and erode their habitats, it may not seem as obvious to many that the same intervention, especially when administered continuously, may also provide a potent selective pressure for the expansion of drug-resistant individuals albeit inadvertently. The first study by Xue et al (13) serves as a good example of this phenomenon.
A genetic basis for drug resistance
Xue et al (13) modeled the selection and propagation of an amplification of the mutant B-Raf proto-oncogene (BRAFamp) in patient-derived tumor xenograft mouse models of both lung cancer and melanoma that harboured the BRAFV600E mutation and were treated with a direct inhibitor of the ERK kinase, either alone or in combination with other ERK signaling inhibitors. Using single-cell sequencing and multiplex fluorescence in situ hybridization (FISH) analyses, they mapped the emergence of extra-chromosomal amplification in parallel evolutionary trajectories that arose in the same tumor shortly after treatment. Consistent with the idea of therapy acting as a selection pressure, the authors observed that, when treated continuously with a single drug, tumor cells appeared to acquire extensive genetic alterations that can be expanded through parallel evolution, enabling tumor cells to adapt while maintaining their intratumoral heterogeneity. Thus, the evolutionary selection of BRAFamp in this case was determined by the fitness threshold (see Glossary), the barrier that subclonal populations need to overcome to regain fitness in the presence of therapy. This differed for inhibitors of ERK signaling, suggesting that sequential monotherapy is ineffective and selects for a progressively higher BRAF copy number underscoring the genetic underpinning of drug resistance in cancer.
On the other hand, concurrent targeting of multiple kinases that are active in lung cancer and melanoma, and when administered on an intermittent rather than a continuous schedule, inhibited tumor growth with 100% efficiency presumably due to the inability of tumor cells to adapt well to the changing fitness threshold imposed by selection (13). Thus, as concluded by the authors, gene amplification can be acquired and expanded through parallel evolution, enabling tumors to adapt while maintaining their intratumoral heterogeneity. However, when the authors turned the table on cancer, they found that the cancer cell’s adaptive strategy could be exploited and treatments which imposed the highest fitness threshold also prevented the evolution of resistance-causing alterations highlighting the ecological and evolutionary principles underlying drug resistance.
A non-genetic mechanism enables cancer cells to evade drug treatment
In contrast to the study by Xue et al illuminating the deterministic (genetic) perception, the study by Shaffer et al (14) raises the possibility of alternative, non-genetic mechanisms contributing to the emergence of cells that can evade drug treatment and persist. Shaffer et al (14) also addressed drug resistance in human melanoma and harbouring the same BRAF mutation, BRAFV600E, that Xue et al investigated (13). However, here the authors treated with the BRAF inhibitor vemurafenib; but unlike Xue et al who followed BRAF amplification (BRAFamp), Shaffer et al discerned mutations in the BRAF gene. Vemurafenib which only inhibits the mutated BRAF V600E protein nearly eradicated tumor cells in a population; however, a small subset of cancer cells developed drug resistance and persisted.
To understand resistance at the single-cell level, the authors considered two models namely, a genetic ‘mutation’ model and a transient, non-heritable model. They hypothesized that in the mutation model that is heritable, a cell in the resistant state cannot revert to being non-resistant. In contrast, in the transient model, they conjectured that cells transition between pre-resistant and non-resistant states, with pre-resistant cells defined as those that give rise to resistant colonies upon addition of drug. To test these hypotheses the authors applied the Luria-Delbrück ‘fluctuation analysis’ (15). Surprisingly, Shaffer et al found no evidence for the heritable, pre-resistant phenotype meaning there were no mutations in response drug treatment. Instead, what they observed was that these cells can display profound transcriptional variability at the single-cell level. This variability even allowed them to predict which cells will ultimately resist drug treatment. This variability involves infrequent, semi-coordinated transcription of a number of resistance markers at high levels in a very small percentage of cells. The addition of drug then induces epigenetic reprogramming in such cells converting the transient transcriptional state to a stably resistant state that is heritable (14). However, from the data shown in the paper, it may also be argued that, what the authors observed was indeed therapy-induced selection that acts on epigenetic heterogeneity which can get hardwired over time. Nonetheless, these data underscore the non-genetic aspects of drug resistance in cancer.
Epigenetic reprogramming and drug resistance
Perhaps, it is worth pointing out that, conceptually similar effects of epigenetic reprogramming were observed by Sharma et al (16) and more recently by Vaz et al (17) in human lung cancer cells. Sharma et al (16) modeled the response to epidermal growth factor receptor (EGFR) inhibitors in an EGFR mutant lung cancer-derived cell line (PC9) that demonstrates exquisite EGFR tyrosine kinase inhibitor sensitivity. The authors found that, whereas the vast majority of cells are killed within a few days of exposure to a drug concentration 100-fold greater than the IC50 value, a small fraction of viable, largely quiescent cells could still be detected 9 days later. However, treating these persisters with a histone deacetylase inhibitor reversed the situation making them drug sensitive again suggesting that cancer cell populations employ a dynamic survival strategy in which individual cells transiently assume a reversibly drug-tolerant state to protect the population from eradication by potentially lethal exposures (16).
Although Vaz et al (17) addressed transformation rather than drug resistance per se their data showed that epigenetic changes may predispose cells to single step transformation mediated by a single oncogene in the absence of other driver genetic changes. More specifically, the authors showed that long-term exposure of untransformed human bronchial epithelial cells to cigarette smoke condensate (CSC) induces epigenetic changes, consistent with those commonly seen in smoking-related non-small cell lung cancer, that sensitize the cells to transformation with a single KRAS mutation. Strikingly, whole-exome and targeted sequencing revealed that these changes occurred in the absence of any detectable driver mutations, including in a suite of genes commonly mutated in lung cancer. In contrast, CSC exposure had led to progressive and extensive changes in DNA methylation, implicating epigenetic mechanisms as key drivers of the pro-oncogenic changes induced by CSC. Therefore, it may be surmised that environmentally induced epigenetic change (Lamarckism induction rather than Darwinian selection) can substitute for genetically driven alteration in cancer and provide a fertile ground for oncogenic transformation.
The duality of drug resistance
To reconcile these seemingly opposing observations in the same cancer types, it is important to understand that the genetic and non-genetic mechanisms underlying drug resistance need not be mutually exclusive and that, both Darwinian selection as well as Lamarckian induction may be operational. Thus, transient effects may provide initial resistance, allowing a small subpopulation of tumor cells to escape the fitness threshold and survive until some acquire epigenetic changes and/or secondary mutations that drive the progression to relapse and get ‘hard-wired’ for transgenerational inheritance. In that case, perhaps targeting multiple signaling pathways concomitant with an intermittent dosing strategy may preclude individuals in this subpopulation to adapt well to the changing fitness threshold imposed by selection. This may prevent the completion of the hardwiring process, allowing them to revert to a drug-sensitive state (14). Furthermore, phenotypic plasticity in cancer may also arise due to the stochastic ‘rewiring’ of the regulatory networks uncovering/activating latent/alternate pathways in response to perturbations (e.g., drug treatment) that involves epigenetic chromatin modifications (18). While some stochastic rewiring events will be inconsequential (the so-called ‘passengers’ in cancer genetic parlance), others will confer fitness and be selected (the so-called ‘drivers’), and can give rise to persisters (17, 19). Similar observations in other systems based on mathematical modeling lend further credence to this argument. For example, while Goldman et al (20) found that administering anticancer drugs in the right temporal sequence can overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition, Zhou et al (21), found that changing either the growth rates of the subpopulations or by environment-instructed transitions, the cell fraction ratio in a population can be altered thus paving the way for new strategies to overcome cancer drug resistance.
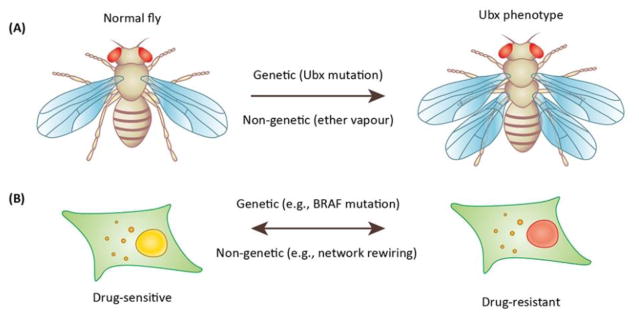
Classical work in the fruit fly may further help to reconcile the apparent dichotomy (Fig. 1). In most insect orders, the second and third thoracic segments (T2 and T3, respectively) each carry a pair of wings. However, the fruit fly Drosophila melanogaster, which belongs to the insect order diptera, only has a single pair of wings on T2. Edward Lewis (22) discovered the famous four-winged fly that resulted due to mutations in the regulatory region of the Ultrabithorax (Ubx) gene whereby the halters, reduced hind wings that evolved into gyroscopic sensors in T3, are transformed into an additional pair of wings.
Fig. 1. Both genetic and non-genetic mechanisms may underlie phenotypic switching.
A) The normal fruit fly (left) has one pair of wings on thoracic segment 2 and a pair of halteres on thoracic segment 3. Edward Lewis discovered the ubx phenotype (right) in which the halteres are transformed into an extra pair of wings (reproduced with permission from the Achieves, California Institute of Technology). The mechanism involved mutations in the regulatory region of the Ubx gene. Conrad Waddington on the other hand also discovered the same phenotype but by exposing fly larvae to ether vapour that is not known to cause mutations. B) A cancer cell that is sensitive to a drug (left) can develop resistance via (right) a genetic mechanism that involves mutations (e.g., BRAF) and is transgenerationally heritable. In contrast it can also develop resistance via non-genetic mechanisms by rewiring the regulatory network (35) and such changes can be transferred to the DNA via epigenetic mechanisms. Thus, the process can be reversed and cancer cells can switch from one phenotype to the other.
Amazingly enough, Conrad Waddington (23) also discovered that the halters can be transformed into wings to generate the four-winged fly; however, he did so by exposing developing flies to ether vapour that does not cause mutations. Three important outcomes were apparent in this experiment by Waddington. First, the proportion of adult flies with the desired phenotype (halteres to wings) kept rising from one generation to the next. Second, they began more and more to resemble four-winged flies. Third, the intensity of the environmental shock required to get the desired effect kept falling from one generation to the next. In fact, after about 15 generations or so, Waddington discovered that there was no need to provide the environmental shock at all, because from then on the four-winged ‘phenocopies‘ began to breed true. To explain this remarkable phenomenon, Waddington invoked genetic assimilation, a process whereby the trait is first canalized and subsequently assimilated (23–25). In Waddington’s terminology, canalization is a measure of the ability of a population to produce the same phenotype regardless of variability of its environment or genotype (heterogeneity) and genetic assimilation is a process by which a phenotypic character which initially is produced only in response to an environmental influence, through a process of selection, is taken over by the genotype, so that it is formed even in the absence of the environmental influence which had at first been necessary (25).
A more recent study on drug resistance due to continuous BRAFV600 inhibition also in melanoma by Su et al (2017) (26), lends further credence to the duality argument. Using genome-wide transcriptomics and single-cell phenotyping, the authors found a subset of plastic cell lines, which followed a trajectory covering multiple known cell state transitions. Indeed, Markov modeling revealed that the cell state transitions were reversible and mediated by both Lamarckian induction and Darwinian albeit non-genetic, selection of drug-tolerant states. Taken together, the data presented suggest that the adaptation in this case is influenced by cell phenotype-specific drug selection and cell state interconversion, but not selection of genetically resistant clones.
As this field is burgeoning, another paper was recently published by Chen et al (2017) (27) that also signifies the mechanisms of resistance and survival as non-genetic. In this study, the authors showed that pancreatic cancer is KRAS driven; however, there can be compensatory mechanisms with KRAS inhibition as related to the focal adhesion. The focal adhesion kinase does not appear to be the solitary cause of this since inhibition of FAK did not lead to cell death. It is possible that other proteins such as paxillin may be central players for these effects (28). As was demonstrated, PXN can potentially be a mechanism of resistance to cisplatin (29). As we learn more about the genotype/phenotype relation, focal adhesion machinery may also emerge as important in carcinogenesis and mechanisms of resistance to therapeutics.
Ecology, evolution and drug resistance in cancer
Traditionally, drug resistance has been perceived as a binary decision in cell fate specification. Tumor cells are viewed as drug-sensitive or resistant and the two states are thought to be mutually exclusive since resistance, whether intrinsic or acquired, is believed to be irreversible arising from accumulating alterations within or outside the target to promote cancer cell survival. Thus, current treatment protocols typically apply the same drugs and doses through multiple cycles. This strategy is based on the principle that a tumor must be eradicated as quickly as possible to prevent evolution of resistance and dissemination to other organs. However, such a view may very likely be an oversimplification; as it turns out, maximum dose treatment may in fact be evolutionarily unwise.
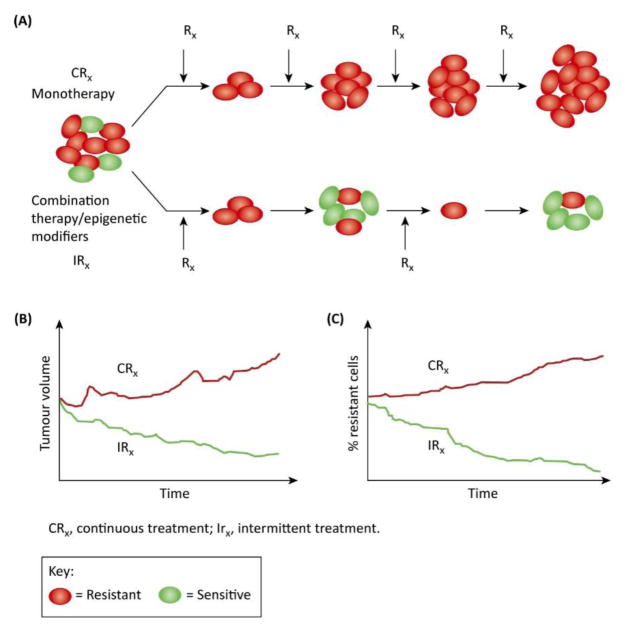
In cancer cells, phenotypic heterogeneity ensures that resistant clones are present prior to therapy. This has been well documented and should be fairly obvious given the fact that maximum dose density therapy has been practice for > 50 years but does not cure metastatic epithelial cancers. In fact, albeit ironically, maximum dose density therapy actually promotes the growth of resistant population because it both strongly selects for adapted phenotypes and eliminates all potentially competing populations (30). On the other hand, since evolving populations can only adapt to current and local conditions but cannot anticipate future or distant environmental factors, the evolutionary tenacity of a cancer cell could potentially be exploited to treat cancer. For instance, treatment protocols can be designed that strategically use initial therapies to induce ‘adaptive strategies’ to change the tumor environment in such a way that proliferation of resistant clones can be suppressed for prolonged periods of time. In this paradigm, therapy is applied in small doses to reduce the tumor population only sufficiently to improve symptoms. In other words, treatment should be administered at a dose that is not the maximum possible but the minimum necessary. Furthermore, treatment is then withdrawn for a period of time (intermittent treatment) so that drug-sensitive cells will proliferate at the expense of the resistant ones. Although the tumor will increase in size between treatments, the extant tumor cells will continue to be sensitive to therapy (30, 31) (Fig. 3).
Fig. 3. Continuous monotherapy versus intermittent combination therapy.
A) In continuous monotherapy, the idea is to eradicate the tumor as quickly as possible. However this strategy can give rise resistance and resistant cells are expected to propagate over time (top). In contrast, combination therapy applied intermittently (bottom) could to induce ‘adaptive strategies’ to change the tumor environment in such a way that proliferation of resistant clones can be suppressed for prolonged periods of time. Therapy is applied in small doses to reduce the tumor population only sufficiently to improve symptoms. Furthermore, treatment is intermittent so that drug-sensitive cells will proliferate at the expense of the resistant ones. B & C). Although the tumor will increase in size between treatments, the extant tumor cells will continue to be sensitive to therapy. Modified from (30).
Nonetheless, it should be pointed out that even though the adaptive therapy that is based on evolutionary principles may appear promising, it may be advisable to exercise caution. For example, when a tumor is sensitive to two or more drugs, evolutionary principles have demonstrated that the application of these drugs at the same time will result in the emergence of cells resistant to both therapies. However, if these drugs are applied one at a time, a subpopulation of cells will be sensitive to one or the other drug and, delaying the emergence of the double-resistant cell clone (30, 31). In stark contrast, as noted above in the work by Xue et al (13), concurrent targeting of multiple kinases that are active in lung cancer rather than with the ERK kinase inhibitor alone, inhibited tumor growth with 100% efficiency presumably due to the inability of tumor cells to adapt well to the changing fitness threshold imposed by selection. Notwithstanding these contradictions however, it is important to note that the latter strategy was successful only in the case of intermittent but not continuous treatment. Consistent with this fractionated treatment approach, when 50 years later Gibson & Hogness repeated the Waddington experiment and applied the ether treatment for brief periods of time and adult flies with T3 abnormalities were selected and bred, there was a steady increase in the frequency of thoracic abnormalities in each generation. Conversely, selectively breeding non-transformed flies resistant to ether treatment exhibited a steady decline in the frequency of thoracic abnormalities (32). In light of the fact that humans and the fruit fly diverged from a common ancestor >700 million years ago (33), the parallelism suggests that multiple mechanisms that are both genetic and non-genetic in nature drive phenotypic plasticity and adaptive evolution.
Concluding Remarks
Tumor cells are complex adaptive systems governed by nonlinear dynamics. Recent studies integrating mathematics, physics and the biology of such systems have underscored the multifaceted, heterogeneous nature of drug resistance which evolves dynamically with changes in therapy (34, 35). The results from these thought-provoking theoretical and empirical studies collectively demonstrate that multiple mechanisms regulating phenotypic switching exist even within a given cancer type that can be genetic or non-genetic in nature. Thus, it may be prudent to understand the mechanism involved before considering a therapeutic approach (see Outstanding Questions). For example, including epigenetic modifiers in combination with targeted therapies may help alter the ability of the cancer cell to switch phenotypes to acquire a drug-resistant state while rendering it more susceptible to adaptive therapy. Although several questions remain (see Outstanding Questions box) and a deeper understanding is required, incorporating this new thinking in treatment protocols may help enhance the precision with which we deliver personalized medicine.
Outstanding Questions.
How to objectively define what we mean by drug resistance: resistant, tolerant or persistent?
How do cancer cells ‘decide’ which mechanism (genetic or non-genetic) to adapt?
What are the (local) environmental cues that guide a given mechanism?
Can the success with reprogramming seen in the laboratory be reproduced in the clinic?
It is now increasingly evident that there is intercellular communication, and thus information transfer, between cancer cells (or example via exosomes or tunneling nanotubes and tunneling microtubes). If so, can CTCs isolated from a drug resistant patient be ‘cajoled’ to rewire their regulatory networks (reprogrammed), and re-injected into the same patient so that they can ‘instruct’ drug resistant cells to revert to sensitivity?
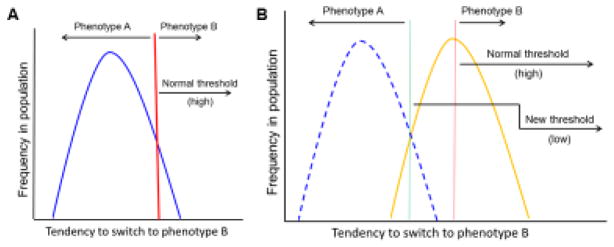
Fig. 2. Phenotypic switching and drug resistance in cancer.
A) As envisaged by Waddington in his famous epigenetic landscape analogy (40), a phenotype is canalized and buffered against minor fluctuations such that the majority of the individuals in the population exhibit a similar phenotype (phenotype A). Selection pressure acts as a threshold (red vertical bar). B) In response to changes in the environment (e.g., drug treatment), if the threshold is lowered (green vertical bar), then the individuals in the population breach the new threshold and a majority will exhibit the new phenotype (phenotype B).
Trends Box.
Evading drug response (so-called drug resistance) is a serious impediment in treating cancer but the mechanisms involved remain poorly understood.
While prevailing wisdom strongly suggests that the phenomenon (resistance) is driven by mutations, emerging evidence suggests that non-genetic/epigenetic mechanisms (tolerance and persistence) may also be important in acquiring drug resistance.
Refractoriness to drug treatment, at least in some cases, can be reversed by epigenetic reprogramming.
New data also indicate that combination and intermittent therapy, as opposed to sustained monotherapy, may be more effective in attenuating recalcitrance.
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572 (RS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. PK thanks Dr. Mohit Jolly (Rice University) and Prof. Abhyudai Singh (University of Delaware) for their thoughtful comments on an earlier draft of the manuscript as well the three anonymous reviewers for their constructive comments. PK would like to dedicate this paper to Prof. M.R.S. Rao, Jawaharlal Nehru Center for Advanced Scientific Research, Bangalore, on the occasion of his 70th birthday.
Glossary Box
- Adaptive therapy:
The emergence of drug resistance in cancer reflects the temporal and spatial heterogeneity of the tumor microenvironment as well as the evolutionary capacity of cancer phenotypes to adapt to therapeutic perturbations. However, cancer therapy is typically administered according to a fixed, linear protocol. When resistant phenotypes arise in the untreated tumor, they are typically present in small numbers because they are less fit than the sensitive population. Thus, the fitter chemosensitive cells will ordinarily proliferate at the expense of the less fit chemoresistant cells. However, if resistant populations are present before administration of therapy, treatments designed to kill maximum numbers of cancer cells remove this inhibitory effect and actually promote more rapid growth of the resistant populations. The goal of adaptive therapy is to enforce a stable tumor burden by permitting a significant population of chemosensitive cells to survive so that they, in turn, suppress proliferation of the less fit but chemoresistant subpopulations. For detailed discussion see (36)
- Epigenetic reprogramming
Epigenetic modifications commonly include several covalent modifications to chromatin at the DNA and/or protein level, and are important for ‘programming’ lineage determination and cellular identity during development and differentiation thereby progressively restricting the pluripotency of the cell. Such modifications that constitute the ‘epigenetic landscape’ manifest in the ‘wiring’ of the cell’s regulatory network which in turn defines the phenotype of a given cell type. Epigenetic reprogramming refers to the conversion of differentiated cells to pluripotent or even totipotent states by erasing/changing the covalent marks and ‘rewiring the regulatory networks.
- Fitness
Is the ability of an organism to survive and reproduce in the environment in which it finds itself. Fitness can be defined either with respect to a genotype or to a phenotype in a given environment. In either case, it describes the individual reproductive success in a population, and is equal to the average contribution to the gene pool of the next generation that is made by individuals of the specified genotype or phenotype (38).
- Fitness threshold
It is the barrier that subclonal populations need to overcome to regain fitness in the presence of therapy.
- Luria-Delbrück fluctuation analysis
A mathematical model developed primarily by Salvador Luria and Max Delbrück (1943) as a means to elucidate the timing of mutation in relation to the imposition of selective conditions by addressing whether mutations arise randomly over time or are they induced by unfavorable environments? In other words, do the mutations preexist or do they arise de novo in response to environmental insult?
- Phenocopy
Typically refers to an individual (such as the 4-winged fly) showing features characteristic of a genotype (for example, Ubx mutations) other than its own, but produced environmentally rather than genetically. It may also be defined as a phenotypic trait or disease that resembles the trait expressed by a particular genotype, but in an individual who is not a carrier of that genotype.
- Genetic assimilation
Genetic assimilation is a process whereby environmentally induced phenotypic variation becomes constitutively produced i.e. no longer requires the environmental signal for expression. The main proponents of genetic assimilation, Conrad Waddington and Ivan Schmalhausen envisioned the phenomenon is a means of facilitating phenotypic evolution [for an excellent review, see (39)].
Footnotes
Conflict of interests: The authors declare they have no conflict of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Santa-Maria CA, Gradishar WJ. Changing Treatment Paradigms in Metastatic Breast Cancer: Lessons Learned. JAMA Oncol. 2015;1:528–34. doi: 10.1001/jamaoncol.2015.1198. [DOI] [PubMed] [Google Scholar]
- 2.Salgia R. Mutation testing for directing upfront targeted therapy and post-progression combination therapy strategies in lung adenocarcinoma. Expert Rev Mol Diagn. 2016;16:737–749. doi: 10.1080/14737159.2016.1181545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez JS, Banerji U. Combine and conquer: challenges for targeted therapy combinations in early phase trials. Nat Rev Clin Oncol. 2017;14:57–66. doi: 10.1038/nrclinonc.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee MC, et al. Single-cell analysis of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc Natl Acad Sci U S A. 2014;111:E4726–35. doi: 10.1073/pnas.1404656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillman AR, Schneider DS. defining resistance and tolerance to cancer. Cell Rep. 2015;13:884–7. doi: 10.1016/j.celrep.2015.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabassum DP, Polyak K. Tumourigenesis: It takes a village. Nat Rev Cancer. 2015;15:473–83. doi: 10.1038/nrc3971. [DOI] [PubMed] [Google Scholar]
- 7.Scott J, Marusyk A. Somatic clonal evolution: A selection-centric perspective. Biochim Biophys Acta. 2017;1867:139–150. doi: 10.1016/j.bbcan.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–91. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Hangauer MJ, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stratton MR, et al. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelstein B, et al. Cancer Genome Landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue Y, et al. An approach to suppress the evolution of resistance in BREAFV600E-mutant cancer. Nat Med. 2017;23:929–937. doi: 10.1038/nm.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaffer SM, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431–435. doi: 10.1038/nature22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luria SE, Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaz M, et al. Chronic Cigarette Smoke-Induced Epigenomic Changes Precede Sensitization of Bronchial Epithelial Cells to Single-Step Transformation by KRAS Mutations. Cancer Cell. 2017;32:360–376. e6. doi: 10.1016/j.ccell.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahmoudabadi G, et al. Intrinsically disordered proteins and conformational noise: implications in cancer. Cell Cycle. 2013;12:26–31. doi: 10.4161/cc.23178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flavahan WA, et al. Epigenetic plasticity and the hallmarks of cancer. Science. 2017:357. doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman A. Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat Commun. 2015;6:6139. doi: 10.1038/ncomms7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou JX. Non-equilibrium population dynamics of phenoptypic conversion of cancer cells. PLoS One. 2014;9(12):e110714. doi: 10.1371/journal.pone.0110714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 23.Waddington CH. Genetic assimilation of the bithorax phenotype. Evolution. 1956;10:1–13. [Google Scholar]
- 24.Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183:1654–1655. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 25.Waddington CH. Genetic assimilation. Adv Genet. 1961;10:257–93. doi: 10.1016/s0065-2660(08)60119-4. [DOI] [PubMed] [Google Scholar]
- 26.Su Y, et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc Natl Acad Sci U S A. 2017;114:13679–13684. doi: 10.1073/pnas.1712064115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen PY. Adaptive and reversible resistance to Kras inhibition in pancreatic cancer cells. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-17-2129. pii: canres.2129.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salgia R. Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by P210/BCR/ABL. J Biol Chem. 1995;270:5039–47. doi: 10.1074/jbc.270.10.5039. [DOI] [PubMed] [Google Scholar]
- 29.Kawada I, et al. Paxillin mutations affect focal adhesions and lead to altered mitochondrial dynamics: relevance to lung cancer. Cancer Biol Ther. 2013;14:679–91. doi: 10.4161/cbt.25091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enriquez-Navas PM, Gatenby RA. Applying Tools From Evolutionary Biology to Cancer Research. In: Ujvari B, Roche B, Thomas F, editors. Ecology, Evolution and Cancer. Academic Press; 2017. pp. 193–201. [Google Scholar]
- 31.Enriquez-Navas PM, et al. Application of Evolutionary Principles to Cancer Therapy. Cancer Res. 2015;75:4675–4680. doi: 10.1158/0008-5472.CAN-15-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson G, Hogness DS. Effect of polymorphism in the Drosophila regulatory gene Ultrabithorax on homeotic stability. Science. 1996;271:200–3. doi: 10.1126/science.271.5246.200. [DOI] [PubMed] [Google Scholar]
- 33.Shih J, et al. Comparison of inter- and intraspecies variation in humans and fruit flies. Genome Data. 2015;3:49–54. doi: 10.1016/j.gdata.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delsanto PP, et al. Analysis of a “phase transition” from tumor growth to latency. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 2000;62:2547–54. doi: 10.1103/physreve.62.2547. [DOI] [PubMed] [Google Scholar]
- 35.Kulkarni P, et al. Phosphorylation-induced conformational dynamics in an intrinsically disordered protein and potential role in phenotypic heterogeneity. Proc Natl Acad Sci U S A. 2017;114:E2644–E2653. doi: 10.1073/pnas.1700082114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gatenby RA, et al. Adaptive therapy. Cancer Res. 2009;69:4894–4903. doi: 10.1158/0008-5472.CAN-08-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brauner A, et al. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol. 2016;14:320–30. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 38.Orr HA. Fitness and its role in evolutionary genetics. Nat Rev Genet. 2009;10:531–9. doi: 10.1038/nrg2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pigliucci M, et al. Phenotypic plasticity and evolution by genetic assimilation. J Exp Biol. 2006;209:2362–7. doi: 10.1242/jeb.02070. [DOI] [PubMed] [Google Scholar]
- 40.Waddington CH. The strategy of the genes. George Allen & Unwin Ltd; London: 1957. [Google Scholar]