Abstract
The pace of genomic and immunologic breakthroughs in oncology is accelerating, making it likely that large randomized trials will increasingly become outdated before their completion. Traditional clinical research/practice paradigms must adapt to the reality unveiled by genomics, especially the need for customized drug combinations, rather than one-size-fits-all monotherapy. Precision oncology’s raison-d'être is to offer “the right drug for the right patient at the right time, ” a process enabled by transformative tissue and blood-based genomic technologies. Genomically-targeted therapies are most suitable in early disease, when molecular heterogeneity is less pronounced, while immunotherapy is most effective against tumors with unstable genomes. Next-generation cancer research/practice models will need to overcome the tyranny of tradition and emphasize an innovative, precise and personalized patient-centric approach.
Keywords: Precision oncology, Immunotherapy, targeted therapy, genomics, personalized medicine
Clinical trial paradigms in the era of targeted therapies and immunotherapies
“Victorious warriors win first and then go to war, while defeated warriors go to war first and then seek to win”
Between 2003 and 2013, new cancer drugs approved by the European Medicines Agency (EMA) or the Food and Drug Administration (FDA) produced a total mean improvement in overall survival of only 3.4 months relative to the treatments that were available in 2003[1]. Routinely, new medicines that confer an additional survival of mere weeks with statistical p value victories are hailed as major breakthroughs in oncology. The randomized controlled clinical trial (RCT), considered the gold standard for cancer clinical trials, has failed to render cures or long-term survival for the majority of patient suffering from advanced malignancies. In diseases such as metastatic pancreatic cancer, over 90% of patients are dead at two years, despite a multitude of traditional trials[2]. The very high costs of conventional trials, the large number of patients receiving futile therapy on control arms, and the lack of biomarker selection hampers progress. In this opinion piece, we critically appraise the state of “standard-of-care” therapies, and present an overview of current clinical trial design paradigms in the era of genomically targeted therapies and immunotherapy.
Targeted Therapies
Over a hundred years ago, Paul Ehrlich introduced the concept of “magic bullet cures” in oncology [3]. Realization of this idea remained elusive until the last decade, with the advent of drugs such as imatinib targeting the altered Bcr-Abl tyrosine kinase, which is pathognomonic of chronic myelogenous leukemia (CML). CML became a poster-child for precision oncology. Before the imatinib era, median survival was about four years; today, life expectancy for patients with CML approaches normal, provided that treatment is started at the time of diagnosis [4]. Delaying treatment until late-stage disease (as is standard in solid tumors) renders even the breakthrough targeted therapies for CML ineffective. Other early examples of precision oncology efforts included the success of trastuzumab in Her2-positive breast cancer, and EGFR and ALK inhibitors in EGFR- and ALK-aberrant lung cancers [5–7], all of which have significantly impacted outcome, albeit not to the extent seen in CML.
In parallel, massive sequencing efforts have mapped the genome. The sequencing costs of a single human genome have dropped in a breathtaking manner, from 3 billion dollars over a decade ago to about 1000 dollars today. Hundreds of actionable genes have been discovered and thousands of new drugs with novel mechanisms of action, including gene-targeted agents and immunotherapy, are being identified. Yet, although we have witnessed a few remarkable triumphs by utilizing genomics, other high throughput “omics” technologies such as proteomics, transciptomics, and metabolomics are in nascent stages.
Immunotherapies
Immunotherapy may be the ultimate example of a precision treatment. Checkpoint inhibitors, for instance, activate the immune machinery, enabling its innate ability to recognize and destroy tumors[8, 9]. The immune system is both personalized and precise. Further, we now realize that the immune apparatus distinguishes malignant cells from their normal counterparts because the cancer cells present neo-antigens, which are produced as a result of the mutanome[10]. Additionally, specific genomic alterations, such as PD-L1 amplification (associated with almost a 90% response rate in refractory Hodgkin disease treated with anti-PD-1 checkpoint inhibitors) and high tumor mutational burden are greatly predictive or response [9, 11–13]. Most striking is the ability of immunotherapy to induce durable complete remissions, even in patients with advanced metastatic cancer. The recent US FDA approval of pembrolizumab, an immune checkpoint inhibitor for microsatellite instability high (MSI-H) cancers across all solid tumor types (histology-agnostic approval) in pediatric and adult patients is an attestation to the power of precision medicinei [14–16]. This approval also demonstrates that genomics and immunotherapy are wedded to each other, and their successes epitomize the power and potential of this marriage.
Conventional Clinical Trial Paradigms
Unfortunately, conventional clinical trial strategies may not be the best way to evaluate the new generation of genomically or immune-targeted agents. Indeed, genomics has unveiled a reality that is incompatible with canonical trial design-- every metastatic tumor is both unique and complex at the molecular level [17–20] (KEY FIGURE 1, Table 1). Further, drugs that are highly effective in small sub-populations of patients are not amenable to randomized trials in unselected patient populations. Under such circumstances, trials must first identify response biomarkers and then individualized combination therapy needs to be given.
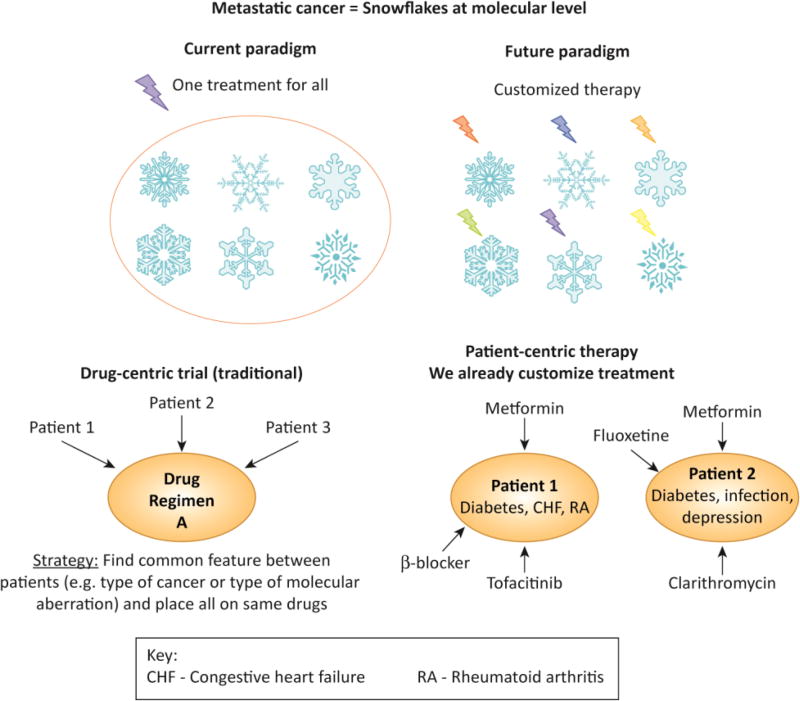
Key Figure 1. The snowflake theory and changing drug development paradigms.
Top panel: Cancers are akin to malignant snowflakes: No two snowflakes are identical, and it seems that it is also extremely unusual for two metastatic tumors to have the same genomic fingerprint. As it turns out, if metastatic tumors are akin to malignant snowflakes in their distinctiveness, individual tumors become the ultimate extrapolation of rare and ultra-rare tumors--N-of-one malignancies.
Bottom panel: Moving from drug-centric (top panel) to patient-centric trials and care (middle and bottom panels). If each cancer is unique and complex, precisely targeting it requires personalized combination therapy regimens. Bottom panel shows that personalized therapy is already routine in patient care outside the oncology setting.
Abbreviations: CHF = congestive heart failure; RA = rheumatoid arthritis
Table 1.
Redefining clinical trial paradigms and standard of care
| Subject matter | Solution | Challenge |
|---|---|---|
| The definition of “personalized” treatment is inconsistent with canonical trial/practice paradigms, where patients are grouped together based on a biologic commonality. | A patient-centered, N-of-one approach is needed to optimize therapy. | Current treatment paradigms, including precision oncology trials, are drug centered rather than patient centered. |
| Mono-therapy is unlikely to cure patients with advanced/complex malignancies. | Combination therapies needed | Matched customized combinations for N-of-one tumors require evaluation of the strategy of personalization or an algorithm for matching, rather than the drug regimens themselves |
| The inimitability of tumors means that each cancer is akin to a malignant snowflake—both unique and complex in its genomic portrait. | Unique/complex tumors require individualized combination regimens | With 300 drugs, there are about 4.5 million three-drug regimens. |
| Dosing of combinations of anti-cancer drugs has traditionally required a phase I study. | Outside of oncology, patients regularly receive de novo combinations of drugs based on understanding impact on metabolic enzymes etc. The average oncology patient is already on eight medications, which have not been assessed together in a phase I study, but are given safely together. Dosing algorithms for anti-cancer drug combinations can be similarly derived from a variety of sources including the literature[57–60] | The pathway to approval and payor acceptance of drug combinations is unclear |
| If tumors are defined by their molecular makeup, advanced molecular tests should be considered a standard diagnostic tool for patients with cancer. | Universal genomic testing of cancers | Points and counterpoints in Table 2 |
The central premise of precision oncology is to offer “the right drug(s) for the right patient at the right time.” Ironically, traditional models for clinical research are almost diametrically opposed to those needed based on the science of precision medicine: (i) in conventional models, commonalities are found between patients in order for them to receive the same drug regimen, instead of individualizing therapy; and (ii) targeted monotherapies are matched to one specific molecular alteration in a patient’s tumor, rather than giving combination treatment optimally tailored to the entirety of the tumor genomic portrait. Regarding timing of therapy, genomically-targeted agents are often applied to heavily-pretreated patients, rather than early in the course of the disease, when tumors are less heterogeneous, and the targeted drugs are more likely to be effective[21, 22]. Tumor mutational burden and complexity, on the other hand, may be an advantage for immunotherapy. Importantly, “standard-of-care” therapies deny and/or delay evaluation of new drugs in patients with lethal cancers by making the tumors more drug resistant, impairing the immune system, and/or rendering the patients “too sick” to be eligible for innovative treatment.
In order to unlock the potential of precision oncology, profound changes in our traditional approaches need to occur. These changes start with universal genomic testing at the time of diagnosis of cancer[23] (Table 2) and include customizing drug combinations, with genomically targeted treatments given early in a patient’s disease course, and immunotherapy using checkpoint inhibitors administered to patients with evolved cancers harboring high mutational burdens or microsatellite instability.
Table 2.
The case for universal genomic testing of tumors: points and counterpoints
| Points | Counterpoints | References |
|---|---|---|
| Obtaining knowledge of genetic aberrations is not worthwhile if no action can be taken in terms of treatment. | Genomics IS the diagnosis. Every patient with cancer deserves a diagnosis. Genetic abnormalities also predict prognosis. Genomics can also predict contraindicated drugs, e.g., EGFR therapy in KRAS-mutant colorectal cancer | [23] |
| The prohibitive cost precludes universal genomic testing. | Cost of testing has decreased precipitously. Financial burden of cancer therapy is massive. The cost of testing for a complete diagnosis and to select appropriate therapy is tiny compared with the money squandered on ill-chosen treatments. | |
| Genomic testing has not been validated in prospective trials. | In comprehensive meta-analyses of ~85,000 patients treated on clinical trials, genomic biomarkers were an independent factor associated with improvement of all outcome variables | [33–35] |
| Genomic testing may benefit only a subgroup of patients or may be germane to only rare diseases. | Virtually impossible to know in advance of testing who will benefit. Options that may not exist at the time of a patient’s initial diagnosis may become available before the patient’s disease progresses. Universal genomic testing of malignancies will enable curating clinically relevant data in large databases |
Standard of Care, Standard of Proof and Proof of Standards
Evidenced-based, standard-of-care guidelines/pathways are promulgated by a variety of organizations and emphasize consistencyii [24, 25]. Departure from these guidelines may leave the physician legally liable and justify insurers’ refusal to pay. Yet, the “standard-of-care” oncology treatments are associated with over 90 percent mortality at two years for some metastatic cancers.
Importantly, in their present rendition, standard-of-care pathways, by virtue of their emphasis on uniformity of management, are antithetical to precision oncology, which requires personalization of therapy. Indeed, if each patient’s tumor is complex and unique, then, in order to “precisely” target that tumor, one must apply medicines that impact the tumor’s distinct alterations, and this requires customized treatment.
Moving Precision Oncology Forward
Precision oncology trials test feasibility of matching drugs to targeted therapy [26–29]. The evidence for this matching strategy is rapidly accumulating, both from these trials and from literature data mining[30, 31]. Indeed, large-scale meta-analysis of approximately 85,000 participants in Phase 1, 2, and 3 studies demonstrated that biomarker selection was the single most significant independent factor predicting improvement in all outcome parameters. Of equal importance, the use of genomically-targeted therapy without a biomarker produced negligible response rates, which were also worse than the results with cytotoxic agents[32–35].
The right drug(s) at the right time for the right patient
The right drug(s)
The discovery of BRAFV600E mutations as a bona-fide oncogenic driver in 50% of melanomas led to a “drug development race” in order to target this gene’s product. Treatment with the potent BRAF inhibitor vemurafenib showed high response rates leading to FDA approval in 2011. [36] [37] Since then, the BRAF inhibitor dabrafenib and two MEK inhibitors (trametinib and cobimetinib) have also been approved [38–40]. Yet, most patients fail to achieve complete remissions or long-term partial remissions. This is likely due to the fact that the majority of metastatic melanomas harbor several genomic alterations[41]. Hence, patients will require combination therapy tailored to their tumor’s biomarker portfolio. Indeed, a recent study demonstrates that higher matching scores (number of matches divided by number of alterations) independently correlates with better outcomes [26].
The right time
Timing is vital in cancer therapy. Tumor complexity increases with time and under the pressure of therapy. CML epitomizes this evolution with three well-defined stages: chronic phase, accelerated phase, and blast crisis. Other cancers almost certainly undergo a similar evolution, but it is not as well delineated clinically[42]. In recent years, the clinical outcome of CML has been transformed. Three major steps enabled this transformation: (i) discovery of the underlying genetic defect (BCR-ABL); (ii) identification of a targeted agent (imatinib) that obviated the aberrant enzymatic activity of Bcr-Abl; and (iii) administration of imatinib to patients with newly diagnosed disease. The third step, that is treating early disease, is the one that is most frequently not addressed in solid tumors.
As an example, BRAF inhibitors in patients with BRAF-mutant melanoma can result in responses so remarkable that they have been designated as the oncologic equivalent of the Lazarus syndrome[43]. This syndrome refers to the spontaneous return of circulation after failed attempts at resuscitation. Patients near death from melanoma can experience dramatic tumor reduction. Unfortunately, these patients are not usually cured, and the disease almost inevitably returns after a few months and results in the patient’s demise. If the experience with CML holds true, durable responses in solid tumors will require either administration of targeted agents such as BRAF inhibitors to newly-diagnosed disease and/or giving customized combinations of drugs to patients with advanced disease in order to block resistance pathways.
The right patient (and the right cancer)
Most novel drugs are tested in patients who have exhausted standard-of-care therapies. At this time, not only is the cancer refractory, but the patient’s performance status and biological/immune reserve may also be too poor to realistically expect the best outcomes. For these reasons, patients should be treated with novel therapies earlier in their disease course.
Advanced cancers are akin to malignant snowflakes--complex and unique
No two snowflakes are identical, and it seems that it is also extremely unusual for two metastatic tumors to have the same genomic fingerprint.[17–20, 44] (KEY figure 1) For example, in 57 patients with advanced breast cancer, 216 somatic aberrations were observed (131 being distinct) in 70 different genes; no two patients had the same molecular signature [17]. A study in advanced osteosarcoma with multiple molecular profiling technologies showed similar results [20]. Further, we may be viewing only the tip of the iceberg. As new technologies emerge beyond limited panel genomic sequencing, both the complexity and the individuality of tumors are likely to be amplified (Figure 2).
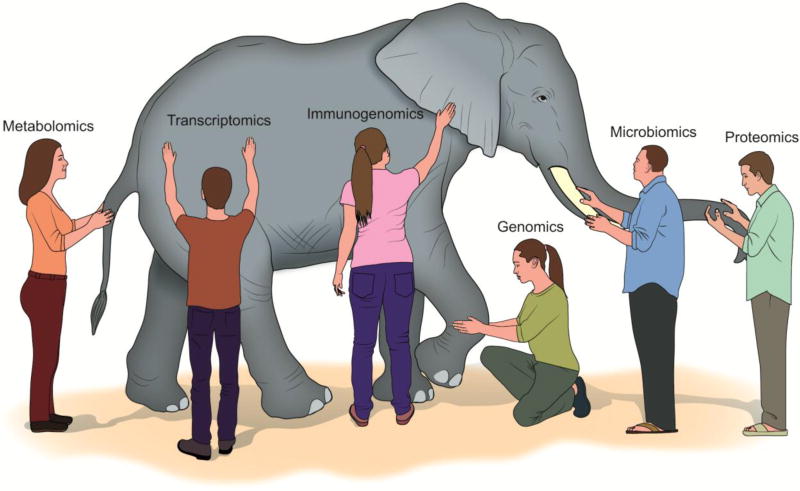
Figure 2. Six blind men and elephants.
Beyond genomics—transcriptomics, proteomics and more: The comprehensive molecular profile of the not-too-distant future may include genomics, transcriptomics, proteomics, metabolomics, microbiomics epigenomics, mutanomics, lipidomics, and immunogenotyping, and may hence predict response to multiple modalities including immunotherapy and chemotherapy [47–56]. Each of these modalities gives us a piece of the puzzle, akin to the parable of the six blind men who each touch a different part of the elephant, such as the tusk versus the trunk, and therefore have vastly different views of the elephant. “Panomics” testing is a requisite of comprehensive analysis and may require complex computer algorithms for data integration and computation.
Customized combination therapy- drug-centric to patient-centric research and care
One of the major stumbling blocks in precision oncology is that there are intrinsic and acquired resistance mechanisms to targeted therapy. One drug matched to a driver aberration may not realistically be expected to cure patients or achieve remissions if each tumor has distinct and complex alterations [31, 41]. Other drugs must be added to overcome resistance [31, 45, 46]
A paradigm of individualized therapy means that the traditional way that drugs/drug regimens become standard of care no longer works. Canonical drug development paradigms are “drugcentered.” (KEY Figure 1) The drug(s) are the focus of the trial and each patient enrolled receives the same regimen, regardless of their genomic and phenotypic heterogeneity. But, if each tumor is different, we may need to test thousands of regimens in increasingly small subsets of patients. Indeed, if there are about 300 drugs in oncology, there are approximately 45,000 two-drug regimens and about 4.5 million three-drug regimens. The traditional clinical trial design model breaks down. However, the conundrum is solvable. Precision medicine implies “patient-centered” trials and care. The patient is the focus and the drugs can therefore vary from patient to patient. In this model, it is not the drug regimen that is evaluated, but rather the strategy of individualization. The question then becomes what is the standard of proof for this strategy? In the era of precision oncology, new clinical trial designs need to evaluate personalized care performance so that “standard-of-care” guidelines can include, emphasize, or even mandate individualized treatment.
The one-size-fits all treatment model in oncology is an anomaly
In daily medical practice, physicians already use customized combinations to treat non-malignant conditions. A patient with diabetes, congestive heart failure, and rheumatoid arthritis will receive a different set of drugs than a patient with diabetes, infection, and depression (KEY Figure 1). The drug doses are adjusted to prevent drug-drug interactions based on known factors such as impact on metabolic enzymes. The average patient enters the oncology clinic on approximately eight drugs tailored to their specific health problems. These individualized drug combinations have never been formally tested in phase I studies; yet physicians safely and effectively administer them on a regular basis to the benefit of their patients.
In oncology, however, there is a cultural precept that, if a new drug combination has not been tested in phase I studies, it should not be used because its safety is unknown. This precept may be a legacy of the cytotoxic era, since combining cytotoxics could have serious safety concerns. However, modern anti-cancer agents have less prohibitive side effects and our understanding of drug combinations has grown. One-size-fits all is not the norm in medicine, and, since advanced cancers are heterogeneous, it should cease to be the norm in oncology care.
Immunotherapy: Yet another paradigm shift
One of the most important mechanisms by which cancer cells evade the immune system is the tumor’s exploitation of checkpoints to disable T-cells. The PD-1/PD-L1 axis is of particular interest because of rapidly emerging data suggesting that inhibition of this checkpoint can restore anti-cancer immunity. Impressively, clinical responses with checkpoint inhibitors have been observed in multiple different malignancies. Remarkably, some patients with advanced tumors can achieve durable complete remission.
The marriage of genomics and immunotherapy
The major predictive markers for checkpoint inhibitor response include high tumor mutational burden, either associated with microsatellite instability or not, CD8 infiltrates, and PD-L1 overexpression or amplification [9, 11, 12]. These markers reflect the coupling of the immune system and genomics. Once the immune system is reactivated with the use of checkpoint inhibitors, T cells must still be able to differentiate tumor cells from normal elements. T cells distinguish tumor cells from normal self in large part through presentation of neo-antigens created by the mutanome. The more neo-antigens, the better the chance of immune recognition. Hence, high tumor mutational burden correlates with favorable outcome after checkpoint inhibitor treatment [13]. In contrast, patients with lower number of genomic alterations appear to respond better to gene-targeted therapy[26], presumably because, in malignancies with more genomic alterations, the presence of resistance mutations abrogate the effects of treatment.
CONCLUDING REMARKS
Breathtaking advances in our understanding of genomics and the immune system have brought us to the threshold of a tipping point in cancer treatment. It appears, however, that our established models for clinical research and practice are a suboptimal fit for the reality of tumor heterogeneity (see outstanding questions). In order to overcome the cancer problem, it is important to break free from the tyranny of tradition, and construct novel paradigms for the management of neoplastic disease.
Outstanding questions BOX.
Genomic sequencing is a basic diagnostic tool that delineates the underpinnings of malignancy and is therefore crucial for classifying disease, predicting prognosis, and directing therapy. If a basic precept of medicine is that each patient deserves an accurate diagnosis, shouldn’t universal genomic testing of tumors be necessary?
What adjustments to clinical trial design and regulatory and care structures are needed to move from a “drug-centric” approach, to a “patient-centric” approach, wherein each tumor is prosecuted with a customized combination of drugs?
Would finding patients with identical or near-identical tumors treated in the same manner still be feasible with a new form of interrogation based on mining of large, well-annotated databases using computerized and artificial intelligence algorithms?
What is the optimal approach to identifying immunogenic, mutanome-derived neo-antigens that induce a T-cell response?
TRENDS BOX.
The central tenet of the precision oncology paradigm requires the delivery of the right drug(s) at the right time to the right patient.
The current model for precision oncology usually matches single agents to patients with late-stage, refractory, molecularly complex disease. This is sub-optimum.
Optimizing targeted therapy requires a departure from traditional paradigms: (i) deploying gene-targeted agents early in the disease course when the tumor is less complicated at the genomic level; (ii) administration of immune-targeted therapies to patients with complex cancers harboring high tumor mutational burdens; and (ii) moving from monotherapy to customized combinations.
Genomics represents the tip of the iceberg. In the future, “panomic” testing that includes transcriptomics, proteomics, metabolomics, and immunogenomics will paint a more complete portrait of each tumor.
Acknowledgments
Competing interests’ statement:
Vivek Subbiah receives research funding for clinical trials from Novartis, Bayer, GSK, Nanocarrier, Vegenics, Northwest Biotherapeutics, Berghealth, Incyte, Fujifilm, Pharmamar, D3, Pfizer, Multivir, Amgen, Abbvie, Bluprint medicines, LOXO and Roche/ Genentech. Razelle Kurzrock receives consultant fees from X-biotech, Actuate Therapeutics, and Roche as well as research funds from Genentech, Pfizer, Sequenom, Guardant, Foundation Medicine and Merck Serono, and has an ownership interest in CureMatch Inc.
Glossary Box
- Precision medicine
An emerging approach for disease treatment and prevention that takes into account individual variability in genes environment, and lifestyle for each person’ (definition of the National Institutes of Health, NIH); ‘a form of medicine that uses information about a person' s genes, proteins, and environment to prevent, diagnose, and treat disease’ (definition of the National Cancer Institute, NCI).
- Precision Oncology
Field in oncology defined by customizing treatment to an individual’s molecular profile.
- Biomarker
Characteristic that is objectively measured or evaluated as an indicator of abnormal biological processes or pharmacologic/biologic responses to a therapeutic intervention.
- Randomized controlled clinical trial
Randomised clinical trials (RCTs) are clinical trials wherein two treatment groups (an experimental group versus control group (sometimes given a placebo or a traditional therapy regimen) are compared. The only expected difference between the control and experimental groups in RCTs is the treatment effect of the experimental therapy being studied.
- Genomics
Study of genes
- Targeted Therapy
drugs that either target molecular alterations specific to cancer cells (e.g., mutated, amplified or epigenetically up- and/or downregulated signaling proteins), or target immune cells to increase anticancer immunity.
- Immunotherapy
The prevention or treatment of disease with agents that stimulate the immune response of the host.
- Tumor mutational burden
Number of mutations in a tumor
- Vemurafenib, Dabrafenib
Tyrosine kinase inhibitor of aberrant BRAF
- Trametinib, Cobimetinib
MEK inhibitor.
- Panomics
informal name for technological fields in biology that end in ‘omics’, such as genomics, proteomics, and metabolomics.
- Proteomics
Study of proteins
- Transcriptomics
Study of transcripts
- Metabolomics
Study of metabolism
- Drug-centric approach
An approach to treatment centered on a drug or drug regimen
- Patient centric approach
An approach to treatment centered on the patient
- Checkpoint inhibitor
Agent that inhibits an immune checkpoint and hence can reactivate the immune system
- Pembrolizumab
An antibody that works as a checkpoint inhibitor.
- Microsatellite instability
Microsatellites represent repeated sequences of DNA that are one to six base pairs in length. Microsatellite instability is a condition of genetic predisposition to mutation in microsatellites that results from an impaired DNA mismatch repair gene.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
VS and RK researched data for the article, wrote the manuscript, made a substantial contribution to discussion of content and reviewed/edited the manuscript before submission.
RESOURCES
https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm. FDA (2017) FDA approves first cancer treatment for any solid tumor with a specific genetic feature
https://www.nccn.org/professionals/physician_gls/f_guidelines.asp NCCN (2016) NCCN Guidelines
References
- 1.Salas-Vega S, et al. Assessment of overall survival, quality of life, and safety benefits associated with new cancer medicines. JAMA Oncology. 2016 doi: 10.1001/jamaoncol.2016.4166. [DOI] [PubMed] [Google Scholar]
- 2.Stewart DJ, Kurzrock R. Fool's gold, lost treasures, and the randomized clinical trial. BMC Cancer. 2013;13:193. doi: 10.1186/1471-2407-13-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strebhardt K, Ullrich A. Paul Ehrlich's magic bullet concept: 100 years of progress. Nat Rev Cancer. 2008;8(6):473–480. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 4.Gambacorti-Passerini C, et al. Multicenter independent assessment of outcomes in chronic myeloid leukemia patients treated with imatinib. J Natl Cancer Inst. 2011;103(7):553–61. doi: 10.1093/jnci/djr060. [DOI] [PubMed] [Google Scholar]
- 5.Piccart-Gebhart MJ, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–72. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 6.Ciardiello F, Tortora G. EGFR Antagonists in Cancer Treatment. New England Journal of Medicine. 2008;358(11):1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 7.Shaw AT, et al. Crizotinib versus Chemotherapy in Advanced ALK-Positive Lung Cancer. New England Journal of Medicine. 2013;368(25):2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 8.Farkona S, et al. Cancer immunotherapy: the beginning of the end of cancer? BMC Medicine. 2016;14(1):73. doi: 10.1186/s12916-016-0623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman A, et al. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev Clin Oncol. 2016 doi: 10.1038/nrclinonc.2016.168. [DOI] [PubMed] [Google Scholar]
- 10.Champiat S, et al. Exomics and immunogenics: Bridging mutational load and immune checkpoints efficacy. Oncoimmunology. 2014;3(1):e27817. doi: 10.4161/onci.27817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khagi Y, et al. Next generation predictive biomarkers for immune checkpoint inhibition. Cancer Metastasis Rev. 2016 doi: 10.1007/s10555-016-9652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14(4):847–56. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 13.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le DT, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017 doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemery S, et al. First FDA Approval Agnostic of Cancer Site - When a Biomarker Defines the Indication. N Engl J Med. 2017;377(15):1409–1412. doi: 10.1056/NEJMp1709968. [DOI] [PubMed] [Google Scholar]
- 16.First Tissue-Agnostic Drug Approval Issued. Cancer Discov. 2017;7(7):656. doi: 10.1158/2159-8290.CD-NB2017-078. [DOI] [PubMed] [Google Scholar]
- 17.Wheler JJ, et al. Unique molecular signatures as a hallmark of patients with metastatic breast cancer: implications for current treatment paradigms. Oncotarget. 2014;5(9):2349–54. doi: 10.18632/oncotarget.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurzrock R, Giles FJ. Precision oncology for patients with advanced cancer: the challenges of malignant snowflakes. Cell Cycle. 2015;14(14):2219–21. doi: 10.1080/15384101.2015.1041695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheler J, et al. Unique molecular landscapes in cancer: implications for individualized, curated drug combinations. Cancer Res. 2014;74(24):7181–4. doi: 10.1158/0008-5472.CAN-14-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egas-Bejar D, et al. Theranostic Profiling for Actionable Aberrations in Advanced High Risk Osteosarcoma with Aggressive Biology Reveals High Molecular Diversity: The Human Fingerprint Hypothesis. Oncoscience. 2014;1(2):167–179. doi: 10.18632/oncoscience.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westin JR, Kurzrock R. It's about time: lessons for solid tumors from chronic myelogenous leukemia therapy. Mol Cancer Ther. 2012;11(12):2549–55. doi: 10.1158/1535-7163.MCT-12-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westin JR, et al. Treatment of chronic myelogenous leukemia as a paradigm for solid tumors: how targeted agents in newly diagnosed disease transformed outcomes. Am Soc Clin Oncol Educ Book. 2012:179–85. doi: 10.14694/EdBook_AM.2012.32.60. [DOI] [PubMed] [Google Scholar]
- 23.Subbiah V, Kurzrock R. Universal Genomic Testing Needed to Win the War Against Cancer: Genomics IS the Diagnosis. JAMA Oncol. 2016;2(6):719–20. doi: 10.1001/jamaoncol.2016.0078. [DOI] [PubMed] [Google Scholar]
- 24.Moffett P, Moore G. The standard of care: legal history and definitions: the bad and good news. West J Emerg Med. 2011;12(1):109–12. [PMC free article] [PubMed] [Google Scholar]
- 25.Markman M. Standard of Care Versus Standards of Care in Oncology: A Not So Subtle Distinction. Journal of Oncology Practice. 2007;3(6):291–291. doi: 10.1200/JOP.0761502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheler JJ, et al. Cancer Therapy Directed by Comprehensive Genomic Profiling: A Single Center Study. Cancer Res. 2016;76(13):3690–701. doi: 10.1158/0008-5472.CAN-15-3043. [DOI] [PubMed] [Google Scholar]
- 27.Tsimberidou AM, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18(22):6373–83. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Von Hoff DD, et al. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28(33):4877–83. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 29.Roychowdhury S, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3(111):111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diamond EL, et al. Vemurafenib for BRAF V600-Mutant Erdheim-Chester Disease and Langerhans Cell Histiocytosis: Analysis of Data From the Histology-Independent, Phase 2, Open-label VE-BASKET Study. JAMA Oncol. 2017 doi: 10.1001/jamaoncol.2017.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyman DM, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med. 2015;373(8):726–36. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwaederle M, et al. Precision Oncology: The UC San Diego Moores Cancer Center PREDICT Experience. Mol Cancer Ther. 2016;15(4):743–52. doi: 10.1158/1535-7163.MCT-15-0795. [DOI] [PubMed] [Google Scholar]
- 33.Schwaederle M, et al. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J Clin Oncol. 2015;33(32):3817–25. doi: 10.1200/JCO.2015.61.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwaederle M, et al. Association of Biomarker-Based Treatment Strategies With Response Rates and Progression-Free Survival in Refractory Malignant Neoplasms: A Meta-analysis. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.2129. [DOI] [PubMed] [Google Scholar]
- 35.Jardim DL, et al. Impact of a Biomarker-Based Strategy on Oncology Drug Development: A Meta-analysis of Clinical Trials Leading to FDA Approval. J Natl Cancer Inst. 2015;107(11) doi: 10.1093/jnci/djv253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falchook GS, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379(9829):1893–901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falchook GS, et al. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol. 2012;13(8):782–9. doi: 10.1016/S1470-2045(12)70269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ascierto PA, et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol. 2016;17(9):1248–60. doi: 10.1016/S1470-2045(16)30122-X. [DOI] [PubMed] [Google Scholar]
- 41.Sen S, et al. Co-occurring Genomic Alterations and Association With Progression-Free Survival in BRAFV600-Mutated Nonmelanoma Tumors. J Natl Cancer Inst. 2017;109(10) doi: 10.1093/jnci/djx094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 43.Furmark L, Pavlick AC. BRAF Inhibitors and the "Lazarus Syndrome" -- An Update and Perspective AJHO.COM 2015 [Google Scholar]
- 44.Groisberg R, Subbiah V. The big, the bad, and the exon 11: adjuvant imatinib for all gastro-intestinal stromal tumors or just the ugly? Transl Gastroenterol Hepatol. 2017;2:81. doi: 10.21037/tgh.2017.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong DS, et al. Phase IB Study of Vemurafenib in Combination with Irinotecan and Cetuximab in Patients with Metastatic Colorectal Cancer with BRAFV600E Mutation. Cancer Discov. 2016;6(12):1352–1365. doi: 10.1158/2159-8290.CD-16-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subbiah V, et al. Dabrafenib and Trametinib Treatment in Patients With Locally Advanced or Metastatic BRAF V600-Mutant Anaplastic Thyroid Cancer. J Clin Oncol. 2017 doi: 10.1200/JCO.2017.73.6785. JCO2017736785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desai N, et al. SPARC Expression Correlates with Tumor Response to Albumin-Bound Paclitaxel in Head and Neck Cancer Patients. Transl Oncol. 2009;2(2):59–64. doi: 10.1593/tlo.09109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Ahmadie H, et al. Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy. Cancer Discov. 2014;4(9):1014–21. doi: 10.1158/2159-8290.CD-14-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komiya K, et al. SPARC is a possible predictive marker for albumin-bound paclitaxel in non-small-cell lung cancer. Onco Targets Ther. 2016;9:6663–6668. doi: 10.2147/OTT.S114492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohni KN, et al. A Synthetic Lethal Screen Identifies DNA Repair Pathways that Sensitize Cancer Cells to Combined ATR Inhibition and Cisplatin Treatments. PLoS One. 2015;10(5):e0125482. doi: 10.1371/journal.pone.0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subbiah V, et al. Targeting the apoptotic pathway in chondrosarcoma using recombinant human Apo2L/TRAIL (dulanermin), a dual proapoptotic receptor (DR4/DR5) agonist. Mol Cancer Ther. 2012;11(11):2541–6. doi: 10.1158/1535-7163.MCT-12-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subbiah V, et al. Targeted morphoproteomic profiling of Ewing's sarcoma treated with insulin-like growth factor 1 receptor (IGF1R) inhibitors: response/resistance signatures. PLoS One. 2011;6(4):e18424. doi: 10.1371/journal.pone.0018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subbiah V, et al. STUMP un"stumped": anti-tumor response to anaplastic lymphoma kinase (ALK) inhibitor based targeted therapy in uterine inflammatory myofibroblastic tumor with myxoid features harboring DCTN1-ALK fusion. J Hematol Oncol. 2015;8:66. doi: 10.1186/s13045-015-0160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jardim DL, et al. Comprehensive characterization of malignant phyllodes tumor by whole genomic and proteomic analysis: biological implications for targeted therapy opportunities. Orphanet J Rare Dis. 2013;8:112. doi: 10.1186/1750-1172-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding K, et al. Leveraging a Multi-Omics Strategy for Prioritizing Personalized Candidate Mutation-Driver Genes: A Proof-of-Concept Study. Sci Rep. 2015;5:17564. doi: 10.1038/srep17564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roychowdhury S, Chinnaiyan AM. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin. 2016;66(1):75–88. doi: 10.3322/caac.21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nikanjam M, et al. Dosing targeted and cytotoxic two-drug combinations: Lessons learned from analysis of 24,326 patients reported 2010 through 2013. Int J Cancer. 2016;139(9):2135–41. doi: 10.1002/ijc.30262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu S, et al. Dosing de novo combinations of two targeted drugs: Towards a customized precision medicine approach to advanced cancers. Oncotarget. 2016;7(10):11310–20. doi: 10.18632/oncotarget.7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nikanjam M, et al. Dosing immunotherapy combinations: Analysis of 3,526 patients for toxicity and response patterns. Oncoimmunology. 2017;6(8):e1338997. doi: 10.1080/2162402X.2017.1338997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nikanjam M, et al. Dosing Three-Drug Combinations That Include Targeted Anti-Cancer Agents: Analysis of 37,763 Patients. Oncologist. 2017;22(5):576–584. doi: 10.1634/theoncologist.2016-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]