Abstract
The ability to study rapidly evolving viral populations has been constrained by the read length of next-generation sequencing approaches and the sampling depth of single-genome amplification methods. Here, we develop and characterize a method using Pacific Biosciences’ Single Molecule, Real-Time (SMRT®) sequencing technology to sequence multiple, intact full-length human immunodeficiency virus-1 env genes amplified from viral RNA populations circulating in blood, and provide computational tools for analyzing and visualizing these data.
Keywords: HIV envelope, deep sequencing
1. Introduction
Next-generation sequencing (NGS) has greatly improved our understanding of the evolutionary dynamics of rapidly evolving viral pathogens and the within-host genetic diversity of chronic viral infections such as HIV (Archer et al. 2012; Giallonardo et al. 2014). A major limitation of most available platforms is their inability to reliably and accurately sequence long (≥1 kb) stretches of viral genes as single molecules. In these cases, relying upon computational inference of full-length viral haplotypes from shorter overlapping sequences fragments has proven insufficient (Giallonardo et al. 2014). More traditional approaches, like single-genome amplification (SGA) followed by Sanger sequencing, can generate single sequences that span the entire gene, but sampling depth is low, and SGA requires substantial effort at low throughput. Neither approach is ideal for studying diverse populations of long genes.
The HIV-1 envelope gene (env) is a ±2.6-kb region (Fig. 1) that encodes a polyprotein that enables the virus to target and infect cells. Small molecule inhibitors of env have been approved for clinical use (Kuritzkes 2009), and it is the only possible target of an antibody-mediated HIV-1 vaccine (Burton and Mascola 2015), as well as antibody passive immunotherapy (Barouch et al. 2013; Moldt et al. 2012) and gene therapy (Gardner et al. 2015). Even within a single host, HIV env is highly genetically diverse, and this diversity plays a role in disease progression (Richman and Bozzette 1994). Due to the technical insufficiencies outlined above, both traditional (bulk or SGA) and existing NGS approaches are limited for the high-throughput sampling of a gene with the intrahost diversity and length of HIV-1 envelope (env). One recent way around this is to amplify and sequence many different short regions that together span the length of the region of interest, and this has recently been used to study the longitudinal evolution in near full-length HIV (Zanini et al 2015). Here, we focus on sequencing entire HIV env molecules, preserving the linkage across the length of the gene.
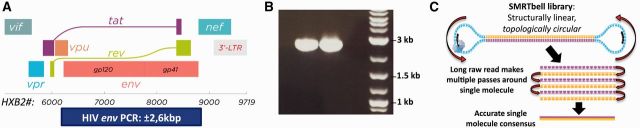
Figure 1.
HIV NL4-3 env amplification, SMRT sequencing and quality assessment. HIV-1 env protocols were first tested using the HIV NL4-3 infectious clone. (A) Location of the ±2.6 kb env amplicon at the 3′ end of the HIV genome, spanning the entire envelope region (figure adapted from Thomas Splettstoesser, www.scistyle.com, CC BY-SA 3.0). (B) Efficient and specific amplification of the full-length NL4-3 env gene. (C) Multiple passes around the same DNA template can be collapsed into a single, accurate CCS read.
Pacific Biosciences’ Single Molecule, Real-Time (SMRT) DNA sequencing technology can interrogate the long coding regions of individual viral genes. This platform currently has an average raw read length of >10 kb and, for the case of mixed DNA populations, the ability to generate accurate sequences from individual molecules through repeated sequencing of circularized templates, termed circular consensus sequencing (CCS) (Travers et al. 2010). Here, we demonstrate how this approach can provide an unprecedented view of high-quality, full-length HIV-1 env sequence populations derived from clinical samples. We used three different sets of samples: 1) the HIV infectious clone, NL4-3, to validate sequence quality, 2) a cross-sectional cohort to investigate amplification and sequencing performance, and 3) longitudinal samples to demonstrate the utility of our molecular methods and bioinformatics analyses.
2. Materials and methods
2.1 Study participants and sample collection
Thirty-six previously collected HIV-1-positive blood plasma samples were selected, representing twelve individuals from two geographically and HIV subtype-distinct cohorts (Table 1). Multiple time points were available from four of twelve subjects examined. HIV-1 subtype B plasma samples were collected from participants enrolled in the San Diego HIV-1 Primary Infection Research Consortium (SD-PIRC) between January 1998 and January 2007 (Morris et al. 2010). HIV-1 subtype A plasma samples were collected from donor PC64, enrolled in the International AIDS Vaccine Initiative (IAVI) Protocol C program. The IAVI-sponsored Protocol C cohort participants were selected through rapid screening of individuals with a recent history of HIV exposure for HIV antibodies in Uganda, Rwanda, Zambia, Kenya, and South Africa (Amornkul et al. 2013; Landais et al. 2016). The study was reviewed and approved by the Ethics Committees in each participating country.
Table 1.
Sample information and HIV-1 env SMRT sequencing metrics.
| Subject information |
Raw read metrics |
≥3-pass CCS filter | >99 per cent accuracy | HQCS | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID | MPIa | HIV-1 subtype | Cohort enrollmentc | Viral load (RNA copies/ml) | SMRT cell loading (nM)b | Total reads | Mean read length (bp) | CCS reads | CCS reads | Counts |
| NL4-3 | N/A | B | N/A | 2000 | 0.05 | 64,581 | 10,298 | 19,902 | 10694 | N/A |
| PC64 | 0 | A | IAVI Protocol C | 25,200 | 0.2 | 8,521 | 9,306 | 2,616 | 1841 | 66 |
| 2 | 25,500 | 0.075 | 23,249 | 8,761 | 6,256 | 4698 | 194 | |||
| 3 | 16,300 | 0.2 | 37,669 | 9,828 | 12,084 | 8244 | 323 | |||
| 6 | 150,000 | 0.125 | 46,325 | 9,800 | 15,065 | 8931 | 518 | |||
| 9 | 169,000 | 0.125 | 15,166 | 10,276 | 4,795 | 3273 | 260 | |||
| 12 | 64,000 | 0.2 | 26,068 | 10,747 | 9,046 | 5246 | 364 | |||
| 18 | 59,300 | 0.125 | 61,688 | 9,694 | 20,751 | 12173 | 789 | |||
| 24 | 46,300 | 0.125 | 34,718 | 9,749 | 11,476 | 7073 | 459 | |||
| 30 | 79,600 | 0.05 | 44,387 | 9,891 | 14,938 | 10081 | 515 | |||
| 36 | 81,543 | 0.05 | 67,307 | 10,055 | 23,301 | 14907 | 780 | |||
| 42 | 109,506 | 0.05 | 41,175 | 9,852 | 13,814 | 9405 | 554 | |||
| 48 | 303,680 | 0.125 | 54,157 | 10,131 | 20,323 | 10684 | 651 | |||
| P018 | 2 | B | SD-PIRC | 446,684 | 0.05 | 41,469 | 8,920 | 11,104 | 5020 | 178 |
| 6 | 50,118 | 0.05 | 47,887 | 10,056 | 17,919 | 11715 | 585 | |||
| 12 | 35,481 | 0.05 | 43,090 | 10,471 | 14,451 | 8548 | 392 | |||
| 22 | 35,481 | 0.1 | 42,294 | 10,812 | 15,463 | 10114 | 315 | |||
| 33 | 34,684 | 0.075 | 34,514 | 10,368 | 11,546 | 7344 | 328 | |||
| 37 | 40,738 | 0.075 | 43,151 | 9,345 | 12,757 | 7453 | 394 | |||
| K453 | 2 | B | SD-PIRC | 5,370 | 0.2 | 16,646 | 7,200 | 4,000 | 2062 | 136 |
| 6 | 5,623 | 0.2 | 4,919 | 8,680 | 1,360 | 1018 | 40 | |||
| 12 | 37,153 | 0.2 | 28,102 | 10,017 | 9,191 | 4963 | 226 | |||
| 14 | 26,915 | 0.05 | 26,280 | 9,388 | 7,823 | 4252 | 196 | |||
| H497 | 5 | B | SD-PIRC | 9,550 | 0.05 | 4,572 | 9,926 | 1,361 | 985 | 44 |
| 6 | 16,596 | 0.2 | 14,498 | 9,936 | 4,566 | 3328 | 257 | |||
| 10 | 10,471 | 0.2 | 14,959 | 11,226 | 5,537 | 4109 | 213 | |||
| 14 | 13,183 | 0.2 | 25,114 | 10,642 | 9,090 | 5942 | 320 | |||
| 26 | 117,490 | 0.1 | 36,404 | 11,350 | 13,968 | 9409 | 597 | |||
| 28 | N/A | 0.05 | 38,278 | 10,087 | 13,626 | 9450 | 414 | |||
| S068 | 3 | B | SD-PIRC | 204,174 | 0.1 | 41,379 | 9,710 | 13,067 | 8274 | 412 |
| G041 | 9 | B | SD-PIRC | 117,490 | 0.05 | 39,717 | 9,878 | 13,555 | 8988 | 412 |
| M964 | 2 | B | SD-PIRC | 346,737 | 0.05 | 44,665 | 9,686 | 14,389 | 8798 | 521 |
| M856 | 2 | B | SD-PIRC | 1,096,478 | 0.05 | 66,065 | 8,791 | 19,397 | 10645 | 688 |
| L342 | 4 | B | SD-PIRC | 190,546 | 0.05 | 59,155 | 9,496 | 18,620 | 11149 | 585 |
| N490 | 2 | B | SD-PIRC | 39,804 | 0.05 | 41,684 | 9,504 | 13,091 | 8505 | 335 |
| N670 | 2 | B | SD-PIRC | 23,442 | 0.05 | 40,070 | 9,376 | 12,145 | 7479 | 371 |
| P767 | 3 | B | SD-PIRC | 75,857 | 0.05 | 39,049 | 9,632 | 12,686 | 8041 | 466 |
MPI: months post infection.
SMRT cell loading concentration reported as concentration of MagBead-bound SMRTbell complex on sequencing plate.
Samples were collected from donors enrolled in one of two HIV longitudinal cohorts: 1) SD-PIRC = San Diego Primary Infection Research Consortium or 2) IAVI Protocol C = International AIDS Vaccine Initiative Protocol C consortium.
2.2 Sample processing, HIV-1 RNA extraction and cDNA generation
Aliquots of previously stored plasma samples (500 μl) were thawed on ice. Once defrosted, plasma was layered onto 200 μl of 20 per cent sterile-filtered sucrose in 2-ml screw-cap microcentrifuge tubes. Virions were pelleted through the sucrose cushion at 23,500 g for 1 hour at 4 °C. Following the spin, supernatants were removed from the viral pellet and discarded. While on ice, 140 μl sterile phosphate-buffered saline (pH = 7.4) was applied to the remaining viral pellet and allowed to sit for 1 hour. After this, the loosened viral pellets were resuspended in this volume and used as input into the QIAamp Viral RNA Mini Kit (part no. 52906; Qiagen, Valencia, CA, USA), which was used to extract total HIV-1 RNA according to the manufacturer’s instructions. Viral RNA was eluted from the QIAmp columns in 55 µl AVE buffer. An 8 μl sub-aliquot of the eluted viral RNA was added directly to cDNA synthesis reactions to avoid any degradation from freeze/thaw cycles. Remaining viral RNA was aliquoted and stored at −80 °C for later use.
Blood plasma HIV-1 RNA levels (Roche Molecular Systems, Inc., Pleasanton, CA, USA) were quantified for each sample. All samples were collected prior to the initiation of any antiretroviral therapy. Estimated dates of infection were calculated at baseline for each participant based on serologic and virologic criteria per established protocols (Morris et al. 2010).
Total HIV-1 cDNA was generated using the SuperScript III First Strand Synthesis System for RT-PCR (part no. 18080-051; Thermo Fisher, Fremont, CA, USA) using the provided oligo (dT) to prime first-strand synthesis and according to the manufacturer’s protocol. Aliquots of cDNA were stored at − 20 °C until needed for targeted HIV-1 env amplification.
2.3 HIV-1 env amplification
Targeted HIV-1 env amplification was performed using polymerase chain reaction (PCR) with High Performance Liquid Chromato- graphy (HPLC)-purified primers: Env-F: GAGCAGAAGACAGT- GGCAATGA (corresponding to positions 6,207–6,228 in HXB2); and Env-R: CCACTTGC CACC C ATB TT AT AG CA (corresponding to positions 8,788–8,811 in HXB2). The primers were purchased from Integrated DNA Technologies (San Diego, CA) and diluted to 20 pmol in 0.1× TE buffer before use. Each reaction consisted of 2 μl total HIV-1 cDNA and 48 μl of Advantage 2 PCR reaction mixture (Advantage 2 PCR Kit, catalog no. 639206; Clontech, Mountain View, CA, USA). The reaction mixture comprised 5 μl 10× SA PCR Buffer, containing 2 mM magnesium acetate, 1 μl of 10 mM dNTP mix, 1 μl each of Env-F and Env-R (at 20 pmol), 39 μl of nuclease free water, and 1 μl of Advantage 2 Polymerase Mix. Reactions were heated to 95 °C for 1 minute and then subjected to 35 cycles of PCR using the following parameters: 15-sec denaturation at 95 °C, followed by 30-sec annealing at 64°C, followed by 3-min extension at 68 °C. After the 35th cycle, the reactions were incubated for 10 min at 68 °C and then held at 4 °C.
HIV-1 env amplicons were purified from PCR reactions using the QIAquick PCR Purification Kit (part no. 28106; Qiagen, Valencia, CA, USA) as described by the manufacturer, and eluted in 30 μl EB buffer (10 mM Tris, pH 8). HIV-1 env amplicons were visualized by gel electrophoresis and quantified using the 2100 Bioanalyzer System with the DNA 12000 kit (Agilent Biosciences, Mountain View, CA, USA). Replicate PCR reactions for each sample were visualized, quantitated, and pooled by sample, until a final mass of >250-ng HIV-1 env amplicon was achieved. To remove any residual PCR reagents and primer dimers, the 250 ng of sample was then purified with a 1× volume of AMPure PB beads (part no. 100-265-900), as described by the manufacturer (Pacific Biosciences, Menlo Park, CA, USA).
2.4 PacBio library preparation and sequencing
SMRTbell template libraries of ∼2.6-kb insert size were prepared according to the manufacturer’s instructions using the SMRTbell Template Prep Kit 1.0 (part no. 100-259-100; Pacific Biosciences). A total of 250 ng of AMPure PB bead-purified HIV-1 env amplicon was added directly into the DNA damage repair step of the 10-kb Template Preparation and Sequencing (with low-input DNA) protocol. Library quality and quantity were assessed using both the Agilent 12000 DNA Kit and the 2100 Bioanalyzer System (Santa Clara, CA, USA), as well as the Qubit dsDNA BR Assay kit and Qubit Fluorometer (Thermo Fisher). Sequencing primer annealing was performed using the recommended 20:1 primer:template ratio, whereas P5 polymerase binding was performed at a modified polymerase:template ratio of 3:1. HIV-1 env SMRTbell libraries were immobilized onto SMRT cells at a starting concentration of 10 pM on chip, loading titrations were performed to achieve optimal sequencing conditions for particular samples as necessary. Final loading conditions for all samples tested are found in Table 1. SMRT sequencing was performed on the PacBio RS II using the C3 sequencing kit with magnetic bead loading and 180-minute movies.
2.5 Sanger sequencing of HIV-1 Env clones
Full-length envelope genes were isolated by reverse transcription-PCR (RT-PCR) from cryopreserved longitudinal plasma samples from IAVI donor PC64. The swarm analysis protocol was described previously and is an application of the clonal amplicon analysis procedure developed by Monogram Biosciences (South San Francisco, CA, USA) (Giallonardo et al. 2014). Briefly, for each sample, the population of viral envelope genes present in the patient plasma was bulk amplified by RT-PCR and then ligated in bulk into pCXAS-env expression vectors. Individual clones comprising the pool were selected from (plated) transformed bacteria. Human Embryonic Kidney (HEK) 293 cells were co-transfected with purified clonal HIV-1 env expression vectors and an HIV genomic vector that contained a firefly luciferase indicator gene replication-incompetent HIV-1 genomic vector. Pseudovirion particles (i.e. env outside, replication incompetent-genome inside) containing cloned envelope genes were harvested from transfected cells and used to infect U87 cells expressing either CXCR4 or CCR5 co-receptors. Individual clones screening positive for infectivity and chemokine receptor tropism were Sanger sequenced on an 3730xl DNA Analyzer (Life Technologies, Foster City, CA, USA) using sixteen overlapping sequencing primers and assembled using Monogram’s proprietary pipeline. Sequences showing mixed nucleotides were excluded from downstream analysis.
2.6 CCS generation
CCS reads were generated using Quiver (Chin et al. 2013) and the Reads of Insert (Larsen, Heilman, and Yoder 2014) protocol as a part of SMRT analysis version 2.3, and .fastq files were used for downstream analyses.
2.7 Full-length envelope analysis
Full-length envelope analysis (FLEA) is structured into two sections: a back-end that performs the computationally intensive processing components, intended to run on a cluster environment, and a web-interface front-end that allows the user to explore the data interactively.
2.7.1 FLEA back-end
A) Using the QV scores provided in the .FASTQ files, the expected number of errors per sequence is calculated using USEARCH (Edgar 2010), and CCS reads are filtered on a user-selectable threshold (we recommend ≥99 per cent expected accuracy). CCS reads are then aligned to a potential contaminant reference database containing NL4-3 and HXB2 env sequences, and any reads with ≥98 per cent identity to these are removed. CCS reads are then mapped to the nearest sequence in the Los Alamos National Laboratory subtype reference alignment, retaining reads with 80 per cent identity to an env reference sequence. This provides a filtered but unaligned set of env CCS reads.
A) The workflow outlining the FLEA back-end is shown in Supplementary Figure S1. It can be divided into five sections:
B) Quality and contaminant filtering.
C) Generating High Quality Consensus Sequences (HQCSs).
D) Aligning HQCSs.
E) Preparing HQCSs data for visualization.
F) Mapping CCS reads into HQCS alignment.
B) Filtered CCS reads are clustered by USEARCH, using a default identity threshold of 99 per cent and with USEARCH parameters max_accepts = 300 and max_rejects = 600 to prevent premature cluster assignment. Highly similar sequences from each cluster are aligned with MAFFT, and the consensus of each cluster is defined as the HQCS. At this point, a frameshift correction is applied, and the number of CCS sequences that belonged to the cluster is recorded.
C) The HQCSs are translated into amino acid sequences, and these are aligned using MAFFT. Once aligned, the procedure can be halted for manual alignment curation, which is sometimes required for variable regions where duplications produce ambiguous alignments. Each nucleotide HQCS is back-translated into the alignment, producing a codon-preserving multiple alignment of all HQCSs.
D) HXB2 is aligned to the consensus of the earliest time point to provide HXB2 numbering for the entire alignment, and a number of site-specific evolutionary metrics are computed for each site in the alignment. These include
The amino acid distribution entropy, both at single time points and across all time points, highlighting sites with increased diversity.
The Jensen–Shannon divergence between the amino acid distributions for each site at successive time points. This gives a value of 0 when no change has occurred, and 1 when the t + 1th distribution is entirely different to the tth distribution, such as after a total selective sweep.
Site-wise dN/dS ratios, quantifying the extent of positive selection, computed using FUBAR, an ultra-fast Bayesian approach to selection inference (Murrell et al. 2013). These are computed both from individual time points, quantifying selection in the recent evolutionary history leading up to that time point, and from all time points collectively, providing an overall index of selection at each site.
Besides the site-specific metrics, we also infer a phylogeny by maximum-likelihood, using FastTree (Price, Dehal, and Arkin 2010).
E) Finally, each original CCS read is aligned to the closest HQCS and projected into the HQCS alignment. For longitudinal sampling, this typically represents an extremely large number of sequences (approximately 8,000 per time point), which is too large for many kinds of analyses, but which can be used for very accurate amino acid frequency quantification, thereby avoiding the possibility of introducing sampling variability via clustering and HQCS construction.
2.7.2 FLEA front-end
The FLEA web-interface front-end provides four panels of visualization, all tailored to HIV env sequences, which, together, provide a comprehensive picture of viral evolution over time. A live example can be found at http://test.datamonkey.org/veg/FLEA/demo.html
The first panel, ‘Evolutionary Trajectory’, displays a number of metrics of diversity, divergence, and other macroscopic sequence properties that show general features of the population structure and how these change over time. These metrics can be computed from full-length gp160 sequences or from pre-selected genetic regions (C1…C5, V1…V5, MPER, etc.).
Second, the ‘Gene-wide information’ panel shows the residue-level properties described in step D above from a big-picture, whole-genome view, both mapped to the Env trimer structure and displayed as a linear sequence. The 3D Env trimer view (shown in Supplementary Fig. S2) is fully interactive, and the properties can be animated to visualize how they change over the sample time points. Further, we have augmented the Env trimer structure to contain some of the loops that are not resolved in the crystal structure, and regions that are missing from the SOSIP.664 structure entirely (MPER). These are not intended to be structurally realistic, but having them present and incorrect is preferable, in this context, to having them absent.
Third, the ‘Amino acid sequences’ panel (shown in Supplementary Fig. S3) is for viewing the protein sequences themselves. Two views are provided: 1) a set of HXB2 sites can be selected and frequency curves over time are plotted for each unique combination of residues. 2) A region (or regions) of the genome are selected, and all identical amino sequences within each time point are collapsed to provide a summarized view of the AA sequences with each region and time point.
Finally, the ‘Trees’ panel (Supplementary Fig. S4) displays the inferred phylogeny. HXB2 positions selected in the ‘Amino acid sequences’ viewer may optionally be displayed on the leaf nodes of the phylogeny, allowing the user to examine the phylogenetic placement of sequence features.
2.8 Accessing data
Data for several samples presented here have been deposited into the NCBI Sequence Read Archive under BioProject PRJNA320111, including the NL4-3 clone, the NL4-3/CH040 mixture to evaluate PCR recombination, and six time points from donor P018. FLEA pages for donors P018, K453, and H497 can be accessed at http://test.datamonkey.org/flea-demo/P018_kinetics/, http://test.datamonkey.org/flea-demo/K453_kinetics/, and http://test.datamonkey.org/flea-demo/H497_kinetics/.
3. Results
3.1 Amplification
The protocol developed consists of five steps: 1) extraction of viral RNA from primary samples; 2) generation of cDNA libraries from total viral RNA; 3) replicate PCR amplification of the ∼2.6-kb env gene from cDNA libraries to generate the needed input quantity for library preparation; 4) library preparation and sequencing on a single SMRT cell; 5) analysis of HIV env data using novel bioinformatics tools. Technical details and reagents used for each step are described in the Section 2. Our amplification protocol works across HIV-1 subtypes A, B, and C with a single primer set. These primers (forward: HXB2 positions 6,207–6,228; reverse: HXB2 positions 8,788–8,811) are well conserved across subtypes A, B, and C with more variation seen at primer binding sites in subtype D (Supplementary Fig. S5). Amplification of all samples from nineteen of twenty-one donors tested was specific, efficient, and robust.
At the average read length of 10 kb, a 2.6-kb env molecule will have ±4× CCS coverage. However, the read length distribution of SMRT sequencing data is highly asymmetric, with a median length of ±7 kb, but with the top 15 per cent of reads having lengths >22 kb. This equates to over 8× CCS coverage in the top 15 per cent subset. This means that the CCS accuracy can be meaningfully traded against sequencing depth, and that additional SMRT cells can provide increased depth for experiments that require more stringent accuracy thresholds.
3.2 Sequencing depth, accuracy, and error profiles
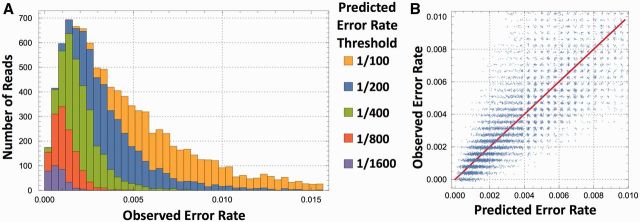
First, we generated SMRT sequencing data from the well-characterized, lab-adapted infectious clone NL4-3 to determine sequence yields at various accuracy cutoffs. We binned sequences based on the quality scores provided by the Quiver algorithm (Chin et al. 2013) and then plotted the empirical error rates (as measured against the known NL4-3 sequence) and read counts at the various filtering thresholds (Fig. 2). From a single SMRT cell, we obtained either 1) 10,000 sequences with a per-base error rate of one in two hundred, 2) 6,000 sequences with an error rate of one in four hundred, or 3) 1,800 sequences with an error rate of 1 in 1,000. See Supplementary Figure S6 for further details about the relationship among filtering threshold, CCS yield, and empirical error rate.
Figure 2.
Accuracy of full-length env sequences. At the per-CCS-read level, we show (A) distribution of observed error rates for different thresholds of the predicted error rates. (B) Predicted and true error rates plotted pairwise, showing that QV scores are highly informative and useful for quality filtering of CCS reads.
Considering an accuracy threshold of 1 per cent, which yields an empirical 0.47 per cent error rate (the ±1 in two hundred in the above paragraph), 84 per cent of these errors were indel errors, and the rest were substitution errors. Further, indel rates increase with longer homopolymer runs. Supplementary Figure S7X shows the error rates for homopolymers of different base lengths, with the per-base error rate, e.g. being approximately five times greater for homopolymers of length 5 than for those of length 2. Note that these error rates are counted from multiple noisy observations of the same amplicon, and thus will not necessarily generalize to different amplicons, which might have homopolymers that occur in different contexts. Supplementary Figure S7 contains further analysis of homopolymer error statistics.
Next, we examined the ability of PacBio’s per-base QV scores to predict empirical error rates. In summary, the QV scores do an admirable job: the average QV scores (transformed into the probability domain) at a base position strongly predict the average empirical errors for that base (ρ = 0.85, see Supplementary Fig. S8A for the scatter plot), and even for individual bases on individual CCS reads, the QV scores correlate with the presence of errors (see Supplementary Fig. S8B for the histogram of these correlations). In both cases, we used a sliding window of size 3 to average the QV scores, accounting for the fact that errors can distribute arbitrarily depending on the alignment to reference (Supplementary Fig. S8 contains a more detailed explanation of how errors were counted). Supplementary Figure S8C shows that strand orientation relative to the reference affects the site-wise mean QV value, and Supplementary Figure S8D shows that the site-wise mean QV value is almost, but not entirely, explained by the homopolymer content, indicating that PacBio’s QV scores are sensitive to the greater homopolymer error rates.
3.3 PCR recombination
Because high rates of PCR recombination could potentially diminish the importance of long reads, we performed a preliminary investigation into the PCR recombination rate of our amplification strategy. Input template material was derived from plasmid-encoded infectious molecular clones NL4-3 and CH040, and 2,000 DNA copies per isolate were mixed prior to amplification. All env-specific amplification was performed according to the protocol presented here. Because this experiment was performed later than the rest, sequencing was performed using the updated PacBio P6/C4 chemistry and 6-hour movie lengths. Although this change will yield better sequence quality (which we did not further investigate), it will not affect the PCR recombination rates, and so is not relevant to the data presented here.
We adapted a hidden Markov model (HMM) approach to identify recombinant sequences (Martin et al. 2015), using the Akaike Information Criterion (AIC) to select between a model that allows recombination (i.e. template switching between the two known parents) and a model that does not. Like any statistical approach, in the presence of noise, there will be some false-positive identifications, and to get around this, we titrated the sequence quality at progressively more stringent thresholds: 99 per cent accuracy (yielding 14,705 CCS reads), 99.9 per cent accuracy (6,531 CCS reads), and 99.99 per cent accuracy (928 CCS reads) (these increased read counts relative to the pure NL4-3 sequencing we performed are due to the newer PacBio chemistry and longer movie lengths, which produce a longer raw read distribution). We additionally filtered out a small proportion of reads that were only part env (including tandem repeats, which are often present at low frequencies in these datasets), but could confuse the recombination analysis. To count recombination events, each CCS read was aligned to a combination of both NL4-3 and CH040, and parameters for the HMM were estimated, likelihoods computed, and AIC-based model selection performed. In the 99 per cent accuracy condition, we identify 0.87 per cent of the CCS reads as recombinants (Wilson’s 95 per cent CI: 0.72–1.05), 0.63 per cent as recombinants in the 99.9 per cent accuracy condition (Wilson’s 95 per cent CI: 0.46–0.85), and 0.22 per cent as recombinants in the 99.99 per cent condition (Wilson’s 95 per cent CI: 0.06–0.79). The fact that the inferred recombination rate decreases as the accuracy cutoff becomes more stringent suggests that the HMM, which is extremely sensitive, has an appreciable false-positive rate at the higher noise levels. The recombination rate, however, is too low for reliable inference from the 99.99 per cent condition, as only two sequences are recombinants. The 99.9 per cent condition thus likely provides the best estimate, and by inspecting the HMM results by eye, we suspect the true rate is somewhere between the 0.46 per cent and 0.85 per cent confidence intervals we infer from this condition, but further investigation will be required to precisely narrow it down. These low PCR recombination rates are consistent with low rates observed in non-nested PCR in (Zanini et al. 2015). In any case, it is low enough to conclude that the overwhelming majority of sequenced species are derived from non-recombinants.
3.4 Amplification from primary isolates
Second, we assessed our amplification protocol using both HIV isolates from the NIH AIDS Reagent Program (subtypes A, C, and D; Supplementary Fig. S9) and from blood plasma samples collected from twelve individuals during primary infection, from the IAVI Protocol C Cohort (Amornkul et al. 2013) (IAVI, subtype A) and the San Diego Primary Infection Research Cohort (Le et al. 2013) (SD-PIRC, subtype B). Robust amplification of env genes was observed for HIV subtypes A (isolate 92RW021) (Gao et al. 1994) and primary IAVI subject PC64; 2/2, 100 per cent), B (primary SD-PIRC subjects; 14/15, 93.3 per cent), and C (isolates 96ZM651 and 97ZA003; 2/2, 100 per cent). Limited env amplification was observed when testing two subtype D env isolates (93UG086 and 92UG035; 1/2, 50 per cent).
3.5 Sequencing primary isolates
Third, we successfully sequenced the amplified HIV env product from thirty-six samples from twelve individuals with primary infection (IAVI and SD-PIRC) on the PacBio RSII. We obtained a median of 8,244 CCS reads after quality filtering (using a 99 per cent expected accuracy threshold) (Table 1).
Designing the analysis strategy for thousands of closely related full-length HIV env sequences presents multiple computational challenges. An often-used solution for NGS data is to pairwise align each read to a reference sequence to derive numerical metrics that can be compared; however, the high frequency and biological importance of insertions and deletions during the course of env evolution precludes this strategy, mainly because the homology of insertions relative to the reference would be lost. To address these and other issues, we have developed the FLEA, which is a freely available web application (hosted at www.datamonkey.org/flea). This analyzer processes PacBio .FASTQ files and provides interactive web visualizations (Supplementary Figs S2 and S3, or visit any of the URLs we provide in Section 2.8), including condensed views of the sequence alignment and the frequencies of residues (alone or in combination) over time in longitudinally examined samples. The application also provides phylogenies, and visualizations of viral evolution mapped onto the HIV Env trimer structure (Pancera et al. 2014).
Overall, FLEA (A) condenses sequences into clusters of high identity (99 per cent similar), (B) generates high-quality consensus sequences (HQCSs) from each cluster, (C) multiply aligns these HQCSs, and (D) pair-wise aligns each original CCS read to the closest HQCS, placing them all in the same large multiple alignment (Supplementary Fig. S1). Amino acid frequencies, which are often of primary interest to assess viral protein evolution, can be counted from the full set of aligned CCS reads. Thus, phylogenetic trees of HQCSs can provide useful insight into the population structure, without having to view tens of thousands of raw reads, which is currently infeasible.
For three longitudinal donors from the First Choice cohort, maximum likelihood phylogenies (which are obtained from screenshots of the FLEA phylogeny viewing tool) are presented in Figure 3. The expected pattern of low initial diversity followed by rapid diversification is apparent in all three. We note, as a caveat, that in vivo recombination prevents the true ancestral history from being represented on a single phylogeny, so these trees need to be carefully interpreted, but they still provide a useful overall picture of evolution and diversification. Detailed study of evolution within and comparison between these donors is beyond the scope of the present work.
Figure 3.
Phylogenies from three donors sequenced longitudinally. Maximum likelihood phylogenetic trees of HQCS sequences obtained from three different donors. Each bubble represents the number of CCS reads that are assigned to an HQCS, and the bubbles are colored by time point. As can be seen, in all cases the diversity starts off low in early infection, and expands as time progresses. These figures are generated from the interactive online FLEA pages: http://test.datamonkey.org/flea-demo/K453_kinetics/, http://test.datamonkey.org/flea-demo/H497_kinetics/, and http://test.datamonkey.org/flea-demo/P018_kinetics/.
3.6 Agreement with Sanger sequencing
Twelve plasma samples we sequenced were collected longitudinally from a single HIV-1 subtype A infected donor. Clonal data derived from ten of the twelve time points were also available, comprising ten individual Sanger sequences for each time point (n = 100). These clonal data were generated using an orthogonal method of bulk RT-PCR, amplicon cloning, and Sanger sequencing as previously described (Richman et al. 2003). The 100 clonally derived Sanger sequences were intercalated into the HQCS read alignment provided by FLEA and a phylogeny was inferred using FastTree (Price, Dehal, and Arkin 2010). The phylogeny, which had a very similar structure to the other three longitudinal donors presented in Figure 3, also showed that each clone sequence had a close HQCS neighbor (Supplementary Fig. S10), and pairwise distance calculations comparing each clonal sequence to the closest HQCS demonstrated a median similarity of 99.5 per cent (Quartiles: 99.1–99.8).
4. Discussion
One limitation of the present study is that we do not address possible recombination that may occur during cDNA synthesis. To our knowledge, no widely used sequencing method prevents recombination here, and the exact rates of RT-mediated recombination will be investigated in future studies. We also note that, for all samples sequenced here, the input viral loads were very high, and, as a result, we made no attempt to quantify the number of input templates and template recovery. For sequencing applications from more restricted input material, template quantification could be critical, as low observed diversity could be confounded with template resampling. We are also presently investigating primer ID molecular barcoding strategies (Jabara et al. 2011; Sheward, Murrell, and Williamson, 2012), which would solve the template recovery problem during sequencing, thus making independent template quantification unnecessary, as well as reducing the PCR recombination rate (at the barcode consensus level). Finally, as with any PCR-based sequencing assay, it is possible that variation in the primer region causes some variants to be undersampled or missed entirely. Because our forward primer is extremely conserved, one option to mitigate mismatches in the reverse primer region is to perform non-specific amplification off the natural poly(A) tail, producing a longer amplicon, but sequencing over the reverse primer region and discovering potential mismatches. Preliminary investigation (data not shown here) suggests that this approach works, but comes at the expense of some non-HIV contaminant sequences, as the amplification is less specific.
When entire genes need to be sampled, our proposed approach to sequencing full-length Env provides a valuable complement to, and in some cases can replace, SGA and Sanger sequencing, especially in applications where a little additional sequencing noise and PCR recombination can be tolerated in exchange for 1) orders of magnitude greater depth, 2) reduced cost, and 3) reduced turn-around time. Moreover, as the cost of gene synthesis decreases, our full-length env protocol allows viral populations to be comprehensively screened and appropriate candidate sequences rationally selected for gene synthesis and subsequent functional interrogation for therapeutic use and other research applications. Together with FLEA, the targeted, full-length HIV env amplification and single-molecule sequencing protocols shown here provide considerable improvements to the existing strategies for studying HIV env in infected individuals. Moreover, this study provides the methodological blueprint for the development of sequencing and analysis protocols for the interrogation of full-length genes of other rapidly evolving pathogens.
Acknowledgements
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1s 92RW021, 92UG035, 93UG086 from The UNAIDS Network for HIV Isolation and Characterization; HIV-1 96ZM651 from Dr. Feng Gao and Dr. Beatrice Hahn. We are grateful to Kay Limoli from Monogram Biosciences for assistance generating clonal env sequences. The IAVI Protocol C Investigators credited in this study are Eric Hunter, Susan Allen, Etienne Karita, Julien Nyombayire, Rosine Ingabire, Jeannine Mukamuyango, Jean Bizimana, Gisele Umviligihozo and Jean-Nepo Nduwamungu.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases [grant numbers U19AI090970, K99AI120851, AI120009, AI100665, and AI036214], the National Library of Medicine [grant number T15LM007092 (K.E.)], National Institute of General Medical Sciences [grant number U01GM110749], and the International AIDS Vaccine Initiative with generous support of USAID and other donors (a full list of IAVI donors is available at www.iavi.org). The content presented here is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy And Infectious Diseases, USAID or the US Government.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
References
- Amornkul P. N. et al. (2013) ‘Disease Progression by Infecting HIV-1 Subtype in a Seroconverter Cohort in sub-Saharan Africa’, AIDS, 27: 2775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer J. et al. (2012) ‘Use of Four next-Generation Sequencing Platforms to Determine HIV-1 Coreceptor Tropism’, PLoS One, 7: e49602.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D. H. et al. (2013) ‘Therapeutic Efficacy of Potent Neutralizing HIV-1-Specific Monoclonal Antibodies in SHIV-Infected Rhesus Monkeys’, Nature, 503: 224–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D. R., Mascola J. R. (2015) ‘Antibody Responses to Envelope Glycoproteins in HIV-1 Infection’, Nature Immunology, 16: 571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S. et al. (2013) ‘Nonhybrid, Finished Microbial Genome Assemblies from long-Read SMRT Sequencing Data’, Nature Methods, 10: 563–9. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2010) ‘Search and Clustering Orders of Magnitude Faster than BLAST’, Bioinformatics, 26: 2460–1. [DOI] [PubMed] [Google Scholar]
- Gao F. et al. (1994) ‘Genetic Variation of HIV Type 1 in Four World Health Organization-Sponsored Vaccine Evaluation Sites: Generation of Functional Envelope (Glycoprotein 160) Clones Representative of Sequence Subtypes a, B, C, and E. WHO Network for HIV Isolation and Characterization’, AIDS Research and Humun Retroviruses, 10: 1359–68. [DOI] [PubMed] [Google Scholar]
- Gardner M. R. et al. (2015) ‘AAV-Expressed eCD4-Ig Provides Durable Protection from Multiple SHIV Challenges’, Nature, 519: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giallonardo F. D. et al. (2014) ‘Full-Length Haplotype Reconstruction to Infer the Structure of Heterogeneous Virus Populations’, Nucleic Acids Research, 42: e115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabara C. B. et al. (2011) ‘Accurate Sampling and Deep Sequencing of the HIV-1 Protease Gene Using a Primer ID’, Proceedings of the National Academy of Sciences of the United States of America, 108: 20166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritzkes D. R. (2009) ‘HIV-1 Entry Inhibitors: An Overview’, Current Opinion in HIV and AIDS, 4: 82–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais E. et al. (2016) ‘Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort’, PLoS Pathogens, 12: e1005369.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P. A., Heilman A. M., Yoder A. D. (2014) ‘The Utility of PacBio Circular Consensus Sequencing for Characterizing Complex Gene Families in non-Model Organisms’, BMC Genomics, 15: 720.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. et al. (2013) ‘Enhanced CD4+ T-Cell Recovery with Earlier HIV-1 Antiretroviral Therapy’, The New England Journal of Medicine, 368: 218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P. et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, Virus Evolution, 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt B. et al. (2012) ‘Highly Potent HIV-Specific Antibody Neutralization in Vitro Translates into Effective Protection against Mucosal SHIV Challenge in Vivo’, Proceedings of the National Academy of Sciences of the United States of America, 109: 18921–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S. R. et al. (2010) ‘Evaluation of an HIV Nucleic Acid Testing Program with Automated Internet and Voicemail Systems to Deliver Results’, Annals of Internal Medicine, 152: 778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B. et al. (2013) ‘FUBAR: A Fast, Unconstrained Bayesian Approximation for Inferring Selection’, Molecular Biology and Evolution, 30: 1196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M. et al. (2014) ‘Structure and Immune Recognition of Trimeric pre-Fusion HIV-1 Env’, Nature, 514: 455–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010) ‘FastTree 2–Approximately maximum-Likelihood Trees for Large Alignments’, PLoS One, 5: e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D. D., Bozzette S. A. (1994) ‘The Impact of the syncytium-Inducing Phenotype of Human Immunodeficiency Virus on Disease Progression’, The Journal of Infectious Diseases, 169: 968–74. [DOI] [PubMed] [Google Scholar]
- Richman D. D. et al. (2003) ‘Rapid Evolution of the Neutralizing Antibody Response to HIV Type 1 Infection’, Proceedings of the National Academy of Sciences of the United States of America, 100: 4144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward D. J., Murrell B., Williamson C. (2012) ‘Degenerate Primer IDs and the Birthday Problem’, Proceedings of the National Academy of Sciences of the United States of America, 109: E1330; author reply E1331.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K. J. et al. (2010) ‘A Flexible and Efficient Template Format for Circular Consensus Sequencing and SNP Detection’, Nucleic Acids Research, 38: e159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanini F. et al. (2015) ‘Population Genomics of Intrapatient HIV-1 Evolution’, eLIFE, 4: e11282. [DOI] [PMC free article] [PubMed] [Google Scholar]