SUMMARY
YAP/TAZ are the major mediators of mammalian Hippo signaling; however, their precise function in the gastrointestinal tract remains poorly understood. Here we dissect the distinct roles of YAP/TAZ in endodermal epithelium and mesenchyme, and find that, although dispensable for gastrointestinal epithelial development and homeostasis, YAP/TAZ function as the critical molecular switch to coordinate growth and patterning in gut mesenchyme. Our genetic analyses reveal that Lats1/2 kinases suppress expansion of the primitive mesenchymal progenitors, where YAP activation also prevents induction of the smooth muscle lineage through transcriptional repression of Myocardin. During later development, zone-restricted down-regulation of YAP/TAZ provides the positional cue and allows smooth muscle cell differentiation induced by Hedgehog signaling. Taken together, our studies identify the mesenchymal requirement of YAP/TAZ in the gastrointestinal tract, and highlight the functional interplays between Hippo and Hedgehog signaling underlying temporal and spatial control of tissue growth and specification in developing gut.
Keywords: Hippo, Lats1/2, YAP/TAZ, Hedgehog, Gastrointestinal tract, Mesenchyme, Growth, Smooth muscle, Differentiation
In Brief
The precise function of the Hippo effectors, YAP/TAZ, in the gastrointestinal tract remains elusive. Cotton et al. identify a key role of YAP/TAZ in coordination of growth and patterning in gut mesenchyme, and highlight the functional interplay between Hippo and Hedgehog signaling underlying the spatial-temporal control of smooth muscle specification.
INTRODUCTION
The vertebrate gastrointestinal tract consists of two distinguished tissue layers: the endoderm-derived epithelia and mesoderm-derived mesenchyme (Kedinger et al., 1998). During embryonic development, the visceral endoderm recruits adjacent splanchnic mesoderm to form a primitive gut tube that is subsequently patterned into regional functional domains, including lung, stomach, and small and large intestine. Epithelial differentiation generates distinct self-renewing epithelia such as the characteristic crypt/villi unit of the intestine, while the mesenchyme underlying the epithelia is organized along a radial axis into distinct cell layers including the myofibroblasts surrounding the crypt and the orientated smooth muscles that control movement of the gastrointestinal tract (Kedinger et al., 1998).
Organogenesis and tissue homeostasis within the digestive tract require functional interactions between the epithelial and mesenchymal layers and are governed by several critical signaling pathways, including the Hedgehog (Hh) pathway. Hedgehog ligands such as Sonic hedgehog (Shh) and Indian hedgehog (Ihh), are secreted by the endodermal epithelia and signal to adjacent mesenchymal cells, where the Hh ligands bind to the receptor Patched (Ptch) and relieve the inhibition of Smoothened (Smo) resulting in activation of the Gli transcription factors and subsequent downstream gene transcription (Mao et al., 2010; Kolterud et al., 2009). This paracrine Hedgehog signaling is believed to play a critical role in regulating mesenchymal growth and promoting smooth muscle differentiation (Kolterud et al., 2009; Mao et al., 2010; Zacharias et al., 2011; Huang et al., 2013).
Hippo signaling, originally identified in Drosophila as an organ size control pathway, has emerged as a key pathway regulating development and homeostasis in a variety of mammalian tissues (Zhao et al., 2010; Pan, 2010; Halder and Johnson, 2011; Halder and Camargo, 2013; Varelas, 2014). In mammalian Hippo pathway, the core kinase cascade is comprised of Mst1/2 and Lats1/2, which leads to phosphorylation, cytosolic retention, and degradation of the transcriptional coactivators, YAP and TAZ. Upon Hippo pathway inactivation, YAP/TAZ translocate into the nucleus and interact with the Tead family of transcription factors, thereby regulating downstream gene transcription. Despite growing evidence points to a critical involvement of YAP/TAZ in injury or irradiation -induced intestinal cryptic regeneration as well as tumorigenesis (Cai et al., 2010; Barry et al., 2013; Taniguchi et al., 2015; Cai et al., 2015; Gregorieff et al., 2015), the function of YAP/TAZ in normal development and homeostasis is less clear. A prior study using in vivo delivery of siRNAs against YAP/TAZ suggests that they may regulate intestinal epithelial proliferation and goblet cell differentiation (Imajo et al., 2015); however several recent reports show that genetic removal of YAP and TAZ has no apparent phenotype in intestinal epithelium during homeostasis (Cai et al., 2015; Gregorieff et al., 2015).
In this study, we systemically dissected the role of YAP/TAZ in endodermal epithelium (lung, stomach and intestine) as well as mesoderm-derived gastrointestinal mesenchyme. We demonstrated that YAP/TAZ are dispensable for epithelial differentiation and Wnt signaling in both gastric and intestinal epithelium during development and homeostasis; however, they act as the key switchboard to coordinate growth and differentiation in gut mesenchyme. Our analysis also unraveled the functional interplay between YAP/TAZ and Hedgehog signaling that governs the specification of the smooth muscle lineage in a temporal and spatial manner.
RESULTS
Differential requirement of YAP/TAZ in endoderm-derived epithelia
To examine the role of YAP/TAZ in the developing endoderm and gastrointestinal tract, we crossed the Yap and Taz conditional alleles with two different Cre driver lines: ShhCre (Harfe et al., 2004) that drives Cre recombination as early as embryonic day 8.5 (E8.5) in the embryonic endodermal epithelia, including lung, esophagus, stomach, and intestine, and VillinCre (Madison et al., 2002) that directs Cre expression from E12.5 in the intestinal epithelia. Removal of both YAP and TAZ from early developing endoderm by ShhCre resulted in embryonic lethality at E18.5 and ShhCreYapflox/floxTazflox/flox mutant embryos exhibited severe lung epithelial differentiation defects (Figure 1A), resembling the YAP knockout phenotype previously reported in the developing lung (Mahoney et al., 2014). However, in contrast to the lung defect, loss of YAP/TAZ had no effect in the gastrointestinal epithelia such that both the stomach and intestine appeared normal at E18.5 (Figure 1A–1B and S1). Consistent with the results from ShhCreYapflox/floxTazflox/flox mutants and the previous reports (Cai et al., 2015; Gregorieff et al., 2015), removal of both Yap and Taz in the intestinal epithelia by VillinCre yielded no defects in either proliferation or differentiation, including the goblet cell lineage, at both embryonic and adult stages (Figure 1B–1C and 1E, and data not shown). Furthermore, we found that Wnt signal transduction in both gastric and intestinal epithelia was not affected in both ShhCre- and VillinCre-driven mutants. Nuclear localization of β-Catenin (Figure S1), as well as Wnt target expression, including CD44, Sox9, Axin2, and Lgr5, were unaffected by YAP/TAZ removal (Figure S1). Together, our analyses revealed that YAP/TAZ are required for lung development in embryonic endoderm, but likely dispensable for epithelial proliferation and differentiation in the gastrointestinal tract during development and homeostasis.
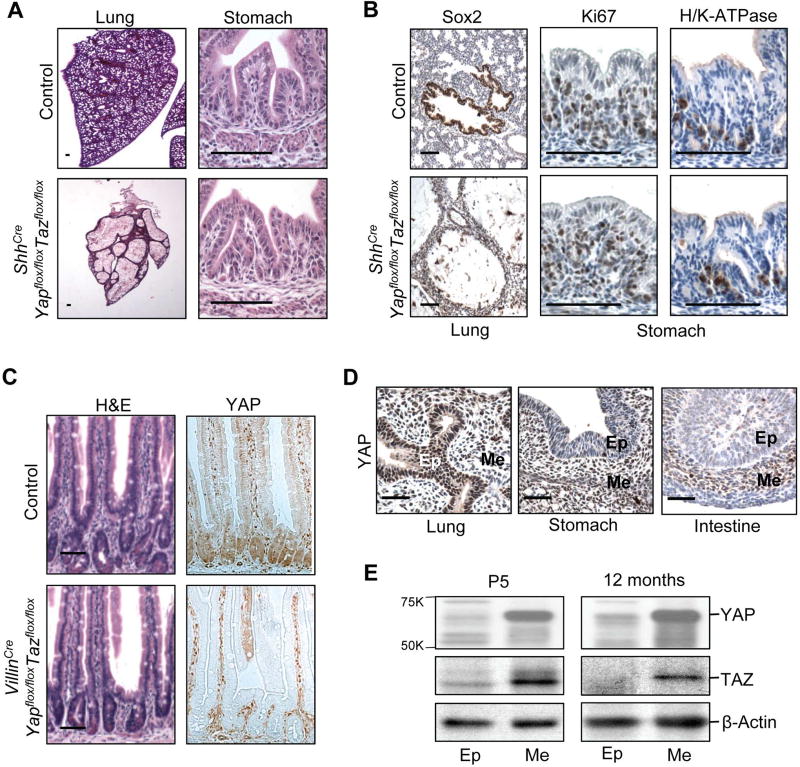
Figure 1. Expression and requirement of YAP/TAZ in endodermal epithelia and mesenchyme.
(A) Histology of lung and stomach in control and YAP/TAZ deficient endoderm (ShhCreYapflox/floxTazflox/flox) animals at E18.5. (B) Sox2 (lung epithelial marker), Ki67 (proliferation marker), and H/K-ATPase (gastric parietal cell marker) immunohistochemical staining in lung and stomach in control and ShhCreYapflox/floxTazflox/flox animals at E18.5. (C) Histology of intestine and immunohistochemical YAP staining in control and YAP/TAZ deficient intestinal epithelia (VillinCreYapflox/floxTazflox/flox) animals at 12 months old. Scale bar = 20 µM. (D) Immunohistochemical YAP staining in wild-type lung, stomach, and small intestine tissue at E13.5. Ep: Epithelium; Me: Mesenchyme. Scale Bar = 10µM. (E) Western blot analysis. Protein lysates from intestinal epithelia (Ep) and intestinal mesenchyme (Me) of wild-type postnatal day 5 (P5) mice and 12 month old mice probed with YAP and TAZ antibodies. See also Figure S1.
YAP and TAZ are highly expressed in the gastrointestinal mesenchyme
The specific requirement of YAP/TAZ in the different compartments of the endodermal epithelia prompted us to examine more closely YAP/TAZ expression. Immunohistochemical (IHC) staining showed that YAP is highly expressed in the nuclei of the embryonic lung epithelium, and the staining was significantly stronger than that in the gastric and intestinal epithelium (Figure 1D), which may provide a reason for the disparate requirement of YAP/TAZ in the developing endoderm. More interestingly, we found that YAP expression was much higher in the mesenchymal cells than that in the epithelial cells in both stomach and intestine during embryonic development (Figure 1D). This expression pattern of YAP/TAZ continued at perinatal and adult stages, with significantly higher mesenchymal expression (Figure 1E). The differential expression levels of YAP/TAZ in gastrointestinal epithelia and mesenchyme raises an intriguing possibility that YAP/TAZ may play a role in the gastrointestinal mesenchyme.
YAP and TAZ are essential for gastrointestinal mesenchymal development
To investigate the role of YAP/TAZ in the gastrointestinal mesenchyme, we used Nkx3.2Cre (Verzi et al., 2009) mice to specifically target the gastrointestinal mesoderm during development. Nkx3.2Cre is expressed from E8.5 in the lateral plate mesoderm which gives rise to the gastrointestinal mesenchyme(Verzi et al., 2009). Mesenchymal knockout of either YAP or TAZ had no apparent phenotype (data not shown); however homozygous knockout of both YAP and TAZ resulted in embryonic lethality at E18.5 and a dramatic mesenchymal growth defect (Figure 2). Consistent with an anterior-posterior time-dependent expression gradient for Nkx3.2, gastrointestinal mesenchymal reduction in the Nkx3.2CreYapflox/floxTazflox/flox animals was most pronounced in the stomach and less so in the intestine (Figure 2A–2B). As such, we focused our analysis on the stomach of mutant animals. Although YAP/TAZ double mutant animals exhibited a significant loss of overall gastrointestinal mesenchyme (Figure 2A–2B and 2D–2E), the different mesenchymal cell populations such as smooth muscle cells, myofibroblasts, and enteric neurons were still present, albeit greatly reduced (Figure 2D). Gastric epithelia adjacent to YAP/TAZ deficient mesenchyme exhibited much more shallow projections into the lumen and likely have differentiation defects, although the parietal cell differentiation marker, H/K-ATPase, was still present (Figure 2C), and the epithelial Wnt activity was also retained, measured by βCatenin and CD44 immunohistochemical staining (Figure 2C).
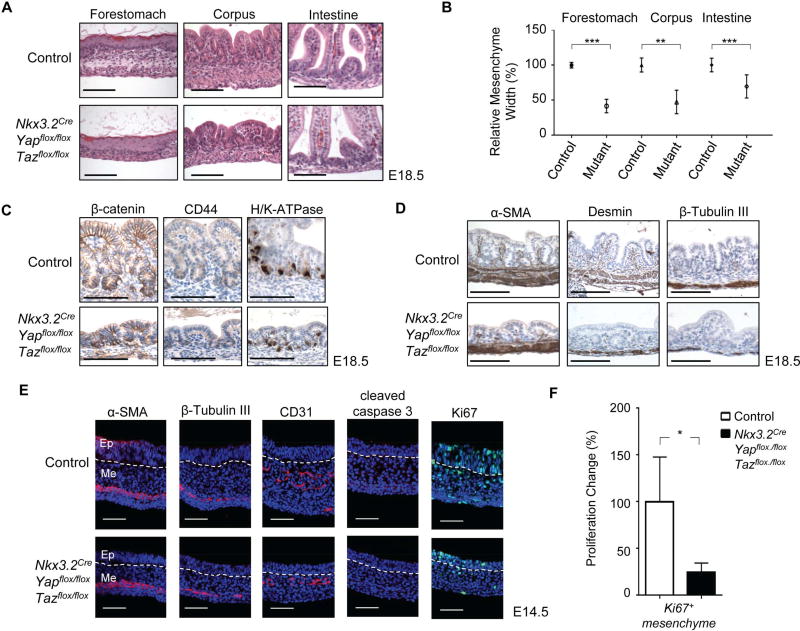
Figure 2. YAP and TAZ are required for gastrointestinal mesenchymal development.
(A) Histology of forestomach, corpus, and intestine of control and YAP/TAZ deficient mesenchyme (Nkx3.2CreYapflox/floxTazflox/flox) animals at E18.5. (B) Relative mesenchyme thickness quantification. Data are mean ± S.D., ** = p value ≤0.01; *** = p value ≤0.001. (C) Immunohistochemical βcatenin, CD44, and H/K-ATPase staining in stomach mesenchyme of control and Nkx3.2CreYapflox/floxTazflox/flox animals at E18.5. (D) Immunohistochemical α-smooth muscle actin (α-SMA), desmin, and β-Tubulin III staining in stomach mesenchyme of control and Nkx3.2CreYapflox/floxTazflox/flox animals at E18.5. (E) αSMA, β-Tubulin III, CD31, Ki67, and cleaved caspase 3 immunofluorescence staining in stomach mesenchyme of control and Nkx3.2CreYapflox/floxTazflox/flox animals at E14.5. Ep: Epithelium; Me: Mesenchyme. Scale bar = 20µM. (F) Quantification of fold change of Ki67+ cells in stomach mesenchyme of control and Nkx3.2CreYapflox/floxTazflox/flox animals at E14.5. Data are mean ± S.D., * = p value ≤0.05. See also Figure S2.
Further analysis of YAP/TAZ mutant animals at an earlier embryonic time point, E14.5, revealed similar phenotypic characteristics (Figure 2E). YAP/TAZ mutant embryos exhibited a smaller mesenchymal compartment as compared to controls (Figure 2E) yet the induction of the mesenchymal lineages, including smooth muscle cells (αSMA+), enteric neurons (βTubulin+), and endothelial cells (CD31+), still occurred (Figure 2E). Moreover, the mutant mesenchyme did not exhibit increased apoptosis, but did display a proliferation defect, as measured by Ki67 and phosphor-Histone 3 (pH3) staining (Figure 2E and S2). Together, our genetic analyses identified an essential requirement of YAP/TAZ in gastrointestinal mesenchymal development and showed that loss of YAP/TAZ results in overall mesenchymal reduction in the gut, suggesting that YAP/TAZ are critical for maintenance or expansion of early mesenchymal progenitor populations.
YAP activation drives mesenchymal growth
To further test this hypothesis, we generated a conditional Rosa26 allele, R26YAP5SA, which enables the in vivo expression of YAP5SA, a constitutively active form of YAP. YAP5SA has five canonical LATS phosphorylation sites mutated from serine to alanine to prevent Hippo/Lats mediated inhibition and degradation (Zhao et al., 2007). The R26YAP5SA allele also carries an N-terminal nuclear localization signal (NLS) and a C-terminal IRES-nuclear LacZ tag, under the control of a CMV enhancer/β–actin hybrid CAGGS promoter targeted into the Rosa26 locus (Figure 3A).
Figure 3. YAP activation promotes mesenchymal growth.
(A) Diagram of R26YAP5SA conditional mouse allele. (B) Gastrointestinal tract from control and mesenchymal YAP gain-of-function (Nkx3.2CreR26YAP5SA) mice at E13.5. St: Stomach; Int: Intestine. (C) Real-time PCR analysis of Ctgf and Ankrd1 mRNA levels in control and Nkx3.2CreR26YAP5SA mutant stomach at E13.5. (D) LacZ staining in control and Nkx3.2CreR26YAP5SA mutant stomach at E13.5. (E) YAP immunofluorescence staining in Nkx3.2CreR26YAP5SA mutant stomach at E13.5 (F) Histology of stomach and pancreas of control and mesenchymal Nkx3.2CreR26YAP5SA mutant animals at E13.5. Inserts show Sox9 immunohistochemical staining in the branching epithelia of pancreatic buds. (G) Immunohistochemical Ki67 and β-Tubulin III, and immunofluorescence CD31 and PDGFRα staining at E13.5. (H) Histology, immunohistochemical α-SMA, and immunofluorescence Ki67 staining in control and Nkx3.2CreR26Yap5SA stomach at E11.5. White dash lines delineate the boundary of gut epithelium and mesenchyme. (I) Quantification of fold change of Ki67+ mesenchymal cells at E11.5. Data are mean ± S.D., * = p value ≤0.05, *** = p value ≤0.001.
To investigate the function of this active YAP in the gastrointestinal mesenchyme, the R26YAP5SA allele was crossed to the Nkx3.2Cre mice. Nkx3.2Cre R26YAP5SA animals were embryonic lethal by E14.5. Upon dissection, we observed a striking enlargement of the gastrointestinal tract in mutant animals (Figure 3B and 3F). Expression of the R26YAP5SA transgene in the mesenchyme was detected by nuclear lacZ expression and nuclear YAP staining (Figure 3D–3E). As expected, transcription of the YAP target genes, such as Ctgf and Ankrd1, were significantly up-regulated (Figure 3C). We found that the massively expanded mesenchyme had even begun to engulf the developing pancreatic bud at E13.5 (Figure 3F, insert). Enteric neurons and endothelial cells were present, although overall cell organization was disrupted (Figure 3G). Not surprisingly, we find that mesenchymal cells with activated YAP were highly proliferative (Figure 3G).
We next examined embryos at E11.5, a developmental time point prior to induction of most mesenchymal lineages, including the smooth muscle progenitor cells that are first marked by the expression of α-smooth muscle actin (αSMA) around E12. In the E11.5 mesenchyme that lacked α-SMA expression (Figure 3H), YAP5SA-induced mesenchymal expansion was clearly evident and was likely due to elevated proliferation, as measured by Ki67 staining (Figure 3H–3I). Thus, consistent with the YAP/TAZ knockout studies, our R26YAP5SA analyses further supported the idea that YAP/TAZ play a critical role in the expansion of early gastrointestinal mesenchyme.
Lats1/2 kinases suppress gut mesenchymal growth
Next, we examined whether the endogenous Lats1/2 kinases, the key components of the upstream Hippo signaling, are responsible for controlling YAP/TAZ function in gut mesenchyme. We generated mesenchyme-specific Lats1/2 mutant embryos: Nkx3.2Cre;Lats1flox/flox;Lats2flox/flox. The individual Lats1 or Lats2 mutant animals had no apparent developmental phenotypes (data not shown). However, the Lats1/2 double mutant embryos died around E14–E14.5, and exhibited drastic mesenchymal overgrowth (Figure 4B–4C and S2), similar to the phenotype observed in the Nkx3.2Cre;R26YAP5SA mutant animals. Lats1/2 removal resulted in up-regulation of the YAP/TAZ downstream target genes, Ctgf, Cyr61 and Ankrd1 (Figure S2), and the mutant mesenchyme had the markedly increased level of cell proliferation, measured by pH3 staining (Figure S2). The phenotypic similarity between YAP5SA ectopic expression and Lats1/2 deletion pointed to an essential role of Lats1/2 kinases in gut mesenchyme and suggested that proper control of Lats1/2-dependent YAP/TAZ activity is critical for gut mesenchymal development.
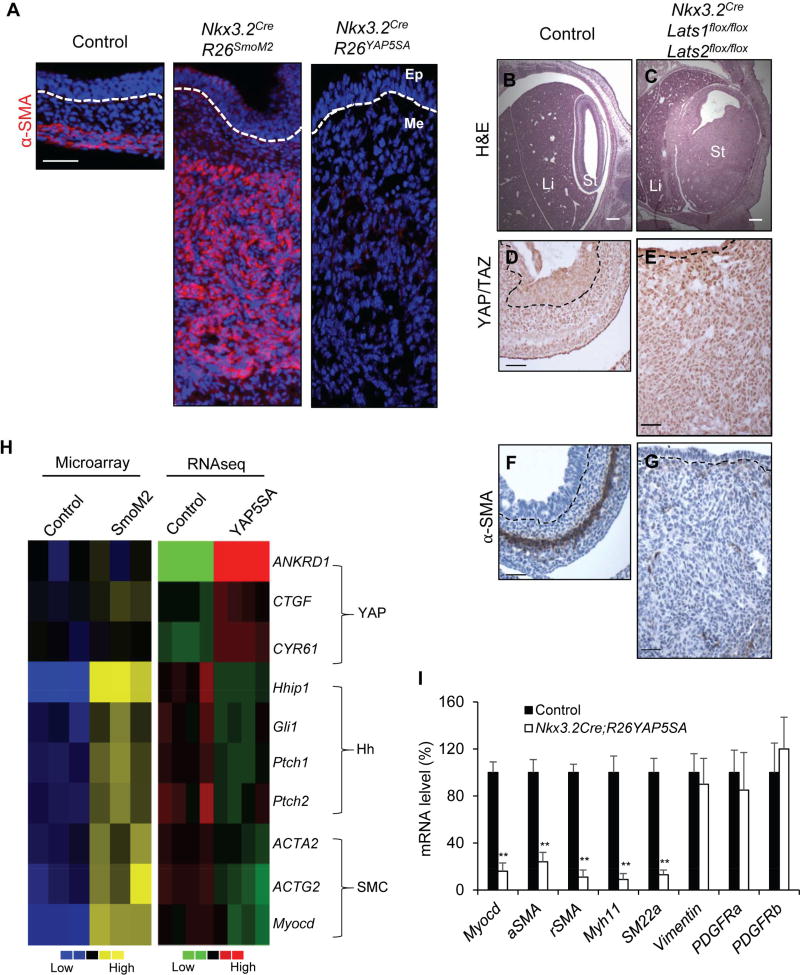
Figure 4. Lats1/2 removal and YAP activation block specification of the smooth muscle lineage.
(A) Immunofluorescence α-SMA staining in control, Hedgehog gain-of-function (Nkx3.2CreR26SmoM2) and YAP gain-of-function (Nkx3.2CreR26YAP5SA) stomach at E13.5. (B, C) Histology of stomach (St) and liver (Li) of control and Nkx3.2CreLats1flox/floxLats2flox/flox mutant animals at E13.5. (D–G) YAP/TAZ (D, E) and α-SMA (F, G) immunohistochemical staining in stomach of control and Nkx3.2CreLats1flox/floxLats2flox/flox mutant animals at E13.5. (H) Heat map analysis comparing mRNA expression of YAP/TAZ targets (Ankrd1, Ctgf, and Cyr61), Hedgehog pathway targets (Hhip1, Gli1, Ptch1 and Ptch2), and smooth muscle differentiation markers (Acta2, Actg2, and Myocd) in control, Nkx3.2CreR26SmoM2, and Nkx3.2CreR26YAP5SA stomach at E13.5, using the data from RNAseq and Affymetrix array. (I) Real-time PCR analysis of Myocd, αSMA, γSMA, Myh11, SM22α, Vimentin, PDGFRα, PDGFRβ mRNA levels in control and Nkx3.2CreR26YAP5SA stomach at E13.5. Data are mean ± S.D., ** = p value ≤0.01. See also Figure S2 and Table S1.
Lats1/2 removal and YAP activation block induction of smooth muscle lineage
The Lats1/2 and Yap/TAZ loss- and gain-of-function phenotypes in the gastrointestinal mesenchyme reminded us the phenotypes of Hedgehog pathway mutants. It has been previously demonstrated by us and others that paracrine Hedgehog signaling from gastrointestinal epithelia plays an important role in regulating mesenchymal growth and differentiation (Kolterud et al., 2009; Mao et al., 2010; Zacharias et al., 2011; Huang et al., 2013). Our prior studies showed that loss of Shh and Ihh ligands from the epithelium resulted in significant reduction of mesenchymal populations (Mao et al., 2010), while Hedgehog pathway over-activation by R26SmoM2 (Mao et al., 2006) in the mesenchyme generated dramatic overgrowth (Mao et al., 2010). R26SmoM2 is a Rosa26 conditional knock-in allele of SmoM2 that expresses a Hh ligand-independent constitutively active form of Smo fused with a C-terminal YFP tag upon Cre recombination (Mao et al., 2006).
One of the key features in SmoM2-induced mesenchymal overgrowth is the expansion of the α-SMA-expressing progenitor cells (Figure 4A), a mesenchymal lineage later giving rise to smooth muscle cells (SMC) and myofibroblasts. In wild-type gastrointestinal tract, the αSMA-expressing smooth muscle progenitor population is first detected as a tight band of cells located in the outer half of the mesenchyme (Figure 4A). Surprisingly, we found that αSMA expression was dramatically reduced in both Nkx3.2Cre;R26YAP5SA and Nkx3.2Cre;Lats1flox/flox;Lats2flox/flox mutants (Figure 4A, 4F, 4G, 4I, and S2). In the Lats1/2 and YAP5SA mutant animals, expression of other mesenchymal markers, such as PDGFRα and Vimentin, as well as the induction of enteric neuron and vasculature system were still present (Figure 3G, 4I, and S2); this indicated that the effect is specific for the smooth muscle lineage, and suggested that, in contrast to Hh/Smo’s role on promotion of differentiation, persistent YAP activation actually inhibits smooth muscle differentiation.
To understand the mechanism underlying YAP function in gut mesenchyme, we performed RNAseq analysis of wild-type and R26YAP5SA mutant stomach at E13.5 (Table S2), and intersected the data set with the Affymetrix array data we generated from E13.5 R26SmoM2 mutant stomach (Huang et al., 2013). Consistent with our phenotypic analysis, we found that transcription of the genes associated with smooth muscle differentiation, including Acta2 (encoding αSMA), Actg2 (encodingγSMA), Myocd (encoding Myocardin), Myh11 and SM22α, were up-regulated in R26SmoM2 mutants, but down-regulated in R26YAP5SA mutants (Figure 4H–4I and Table S2). It is not surprising that the Hippo pathway targets, such as Ankrd1, Ctgf, and Cyr61 were up-regulated in the R26YAP5SA mutants, while they were largely unaffected in the R26SmoM2 mutants (Figure 4H). Interestingly, in the R26YAP5SA mutants, expression of the genes related to the Hh pathway, including Hh ligands (Shh, Ihh), and the pathway components (Hhip1, Gli1, and Ptch1/2) that are also the Hh pathway targets, were down-regulated (Figure 4H, 5A, and Table S2). Decreased expression of Shh and Ihh is due to the reciprocal mesenchymal-epithelial crosstalk as these ligands are exclusively produced in gut epithelium while YAP5SA was only expressed in the Nkx3.2Cre;R26YAP5SA mutant mesenchyme. However, these data raised an intriguing possibility that Hippo/YAP and Hh signaling may functionally interact in gut mesenchyme.
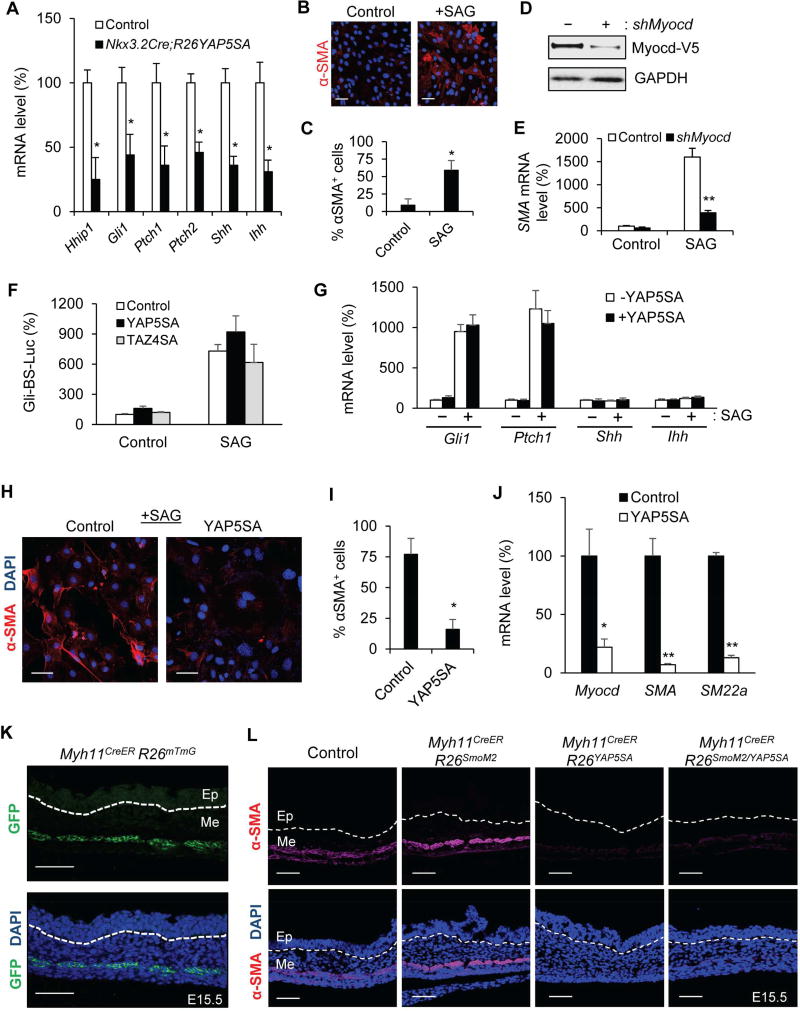
Figure 5. YAP inhibits Hh/Smo-induced cell differentiation, but not signal transduction.
(A) Real-time PCR analysis of Hhip1, Gli1, Ptch1, Ptch2, Shh and Ihh mRNA levels in control and Nkx3.2CreR26YAP5SA stomach at E13.5. (B) αSMA immunofluorescence staining in C3H10T1/2 cells with or without treatment of the Smoothened agonist SAG. (C) Quantification of change of α-SMA+ cells with or without SAG treatment. (D) Immunoblot analysis of V5-tagged Myocd ectopically expressed in C3H10T1/2 cells with or without lentiviral mediated Myocd knockdown. (E) Real-time PCR analysis of SMA mRNA level in control or SAG-treated C3H10T1/2 cells with or without Myocd knockdown. (F) Relative activity of the Gli-BS-Luciferase reporter in control or SAG-treated C3H10T1/2 cells transiently transfected with YAP5SA or TAZ4SA. (G) Real-time PCR analysis of Gli1, Ptch1, Shh and Ihh mRNA levels in control or SAG-treated C3H10T1/2 cells with or without stable expression of YAP5SA. (H) αSMA immunofluorescence staining in SAG-treated C3H10T1/2 cells with or without YAP5SA expression. (I) Quantification of change of αSMA+ cells with or without YAP5SA expression. (J) Real-time PCR analysis of Myocd, αSMA, and SM22α mRNA levels in SAG-treated C3H10T1/2 cells with or without stable expression of YAP5SA. (K) GFP images of the Myh11CreERR26mT/mG stomach at E15.5, following intraperitoneal tamoxifen injection at E12.5. (L) αSMA immunofluorescence staining in control and Myh11CreERR26YAP5SA stomach at E15.5, following intraperitoneal tamoxifen injection at E12.5. White dash lines delineate the boundary of gut epithelium (Ep) and mesenchyme (Me). Data are mean ± S.D., * = p value ≤0.05; ** = p value ≤0.01. See also Table S1.
YAP inhibits Hedgehog-mediated smooth muscle cell differentiation, but not signal transduction
To further explore this potential functional interaction between two pathways, we utilized the in vitro C3H10T1/2 cell differentiation model. The Hedgehog-responsive C3H10T1/2 cells are the mouse embryonic pluripotent mesenchymal cells that are capable of further differentiation into αSMA-positive smooth muscle progenitors upon Hh pathway activation, and these cells have been used as a surrogate system to study the Hhinduced gut mesenchymal differentiation (Zacharias et al., 2011; Huang et al., 2013).
In agreement with the prior report (Zacharias et al., 2011), we showed that treatment of SAG, a small molecule Smoothened agonist, was able to induce Hh pathway activation and αSMA expression in C3H10T1/2 cells, and the induction of SMA transcription was dependent on Myocd (Figure 5B–5G). SAG-mediated Hh pathway activation was measured by a Gli-dependent luciferase reporter, Gli-BS-Luc (Figure 5F), as well as transcription of the Hh direct downstream targets, Gli1 and Ptch1 (Figure 5G). We found that ectopic expression of YAP5SA in C3H10T1/2 cells effectively blocked SAG-induced smooth muscle cell differentiation, measured by transcription and expression of the smooth muscle differentiation markers, including Myocd, αSMA and SM22α (Figure 5H–5J). However, both YAP5SA and TAZ4SA, a constitutively active form of TAZ, were not able to inhibit Gli-BS-Luc reporter activity (Figure 5F) and transcriptional up-regulation of Gli1 and Ptch1 (Figure 5G) induced by SAG treatment. These results suggested that YAP/TAZ inhibition of smooth muscle cell differentiation is unlikely through directly affecting Hh/Smo-mediated signal transduction.
Further, our data suggested that YAP’s inhibitory effect on differentiation is independent of epithelial production of Hh ligands. In the mesenchymal C3H10T1/2 cells, YAP5SA expression or SAG treatment did not induce or affect Shh and Ihh transcription (Figure 5G). More importantly, our genetic analysis demonstrated that YAP5SA is capable of inhibiting smooth muscle differentiation in vivo when ligand-independent SmoM2 is activated. Due to the embryonic lethality of the Nkx3.2Cre;R26SmoM2 and Nkx3.2Cre;R26YAP5SA mutant animals, we utilized an inducible Myh11CreER allele, which drives efficient Cre recombination in the αSMA-expressing cells in the gut mesenchyme by Tamoxifen injection at E12.5 (Figure 5K). We crossed the Myh11CreER allele to the R26SmoM2 and R26YAP5SA mice to generate the Myh11CreER;R26SmoM2/YAP5SA embryos and showed that expression of YAP5SA in these committed smooth muscle progenitor cells was still able to effectively inhibit αSMA expression even with concomitant SmoM2 activation (Figure 5L). Together, these data showed that Hh ligand-independent pathway activation by SmoM2 cannot override YAP inhibitory effect and suggested that YAP acts in a cell autonomous fashion to block smooth muscle differentiation.
YAP regulates smooth muscle cell differentiation via Myocardin repression
To further elucidate the molecular mechanism mediating YAP’s inhibitory effect, we focused on Myocd, the master regulator of smooth muscle differentiation. Myocd is a potent co-activator of the transcription factor serum response factor (SRF) (Pipes et al., 2006). The Myocd/SRF complex is responsible for transactivating the expression of many key downstream smooth muscle differentiation genes, including SMA, SM22 and smooth muscle myosin heavy chain (smMHC), during smooth muscle differentiation (Pipes et al., 2006). Myocd expression in gut mesenchyme is regulated by Hh signaling (Zacharias et al., 2011). Our transcriptional profiling of the SmoM2 mutant gut also showed increased Myocd expression (Figure 4H), and we showed that the induction of SMA expression by SAG in C3H10T1/2 cells requires Myocd (Figure 5D–5E). In contrast, in both Lats1/2 and YAP5SA mutant embryos, Myocd transcription was significantly decreased (Figure 4H–4I and S2), and in C3H10T1/2 cells, SAG-induced Myocd expression was inhibited by YAP activation (Figure 5J).
Recent studies demonstrated that YAP can function as a transcription repressor through recruitment of the NuRD complex in certain genomic regions (Beyer et al., 2013; Kim et al., 2015). It prompted us to examine whether YAP may directly repress Myocd transcription in the context of smooth muscle cell differentiation. In YAP5SA-expressing C3H10T1/2 cells, transcription of CTGF and Cyr61, but not Myocd and SMA, was up-regulated (Figure 6A), and the ChIP-qPCR analysis found that, in addition to the promoters of CTGF and Cyr61, YAP5SA also occupied the promoter region of Myocd, but not SMA (Figure 6B). Further, we showed that ectopic expression of Myocd was able to induce SMA expression in C3H10T1/2 cells even with YAP5SA expression (Figure 6C), highlighting the central role of Myocd in YAP-mediated inhibition of smooth muscle cell differentiation.
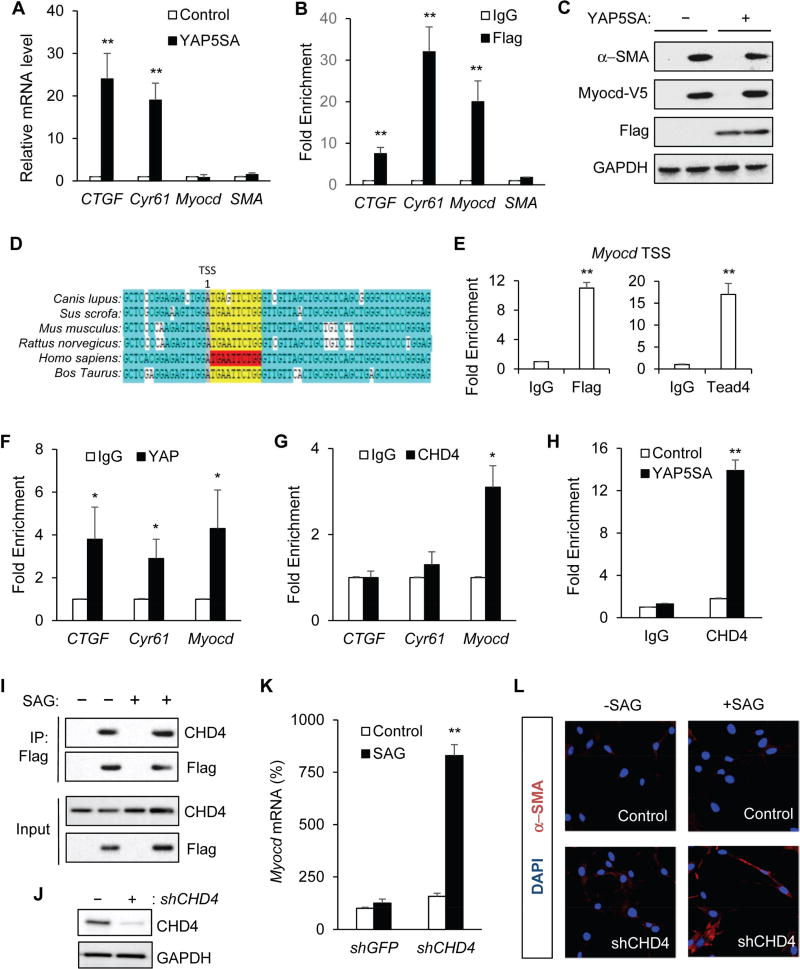
Figure 6. YAP recruits CHD4 to repress Myocardin transcription and smooth muscle cell differentiation.
(A) Real-time PCR analysis of CTGF, Cyr61, Myocd, and αSMA mRNA levels in C3H10T1/2 cells with or without stable expression of YAP5SA. (B) YAP5SA-Flag ChIP-qPCR in C3H10T1/2 cells stably expressing YAP5SA fused with a C-terminal Flag tag. YAP5SA proteins were enriched in the promoter regions of CTGF, Cyr61, Myocd, but not αSMA. Enrichment is calculated based upon qPCR relative to IgG control. (C) Immunoblot analysis of SMA expression in C3H10T1/2 cells with or without Myocd-V5 or YAP5SA-Flag expression. (D) Diagram showing the highly conserved Tead binding site around the transcription starting site (TSS) of the Myocd gene among different species. (E) YAP and Tead bind to the TSS region of the Myocd gene in C3H10T1/2 cells expressing YAP5SA. (F) YAP and (G) CHD4 ChIP-qPCR in embryonic Nkx3.2CreR26YAP5SA stomach at E13.5. Enrichment of YAP and endogenous CHD4 at the promoter regions of CTGF, Cyr61, and Myocd was calculated based upon qPCR relative to IgG control. (H) CHD4 ChIP-qPCR in C3H10T1/2 cells with or without YAP5SA expression. Enrichment of CHD4 at the Myocd promoter was measured by qPCR. (I) YAP5SA binds to endogenous CHD4. In C3H10T1/2 cells with or without SAG treatment, YAP5SA-Flag was immunoprecipitated with an anti-Flag antibody, and immunoblot analysis of endogenous CHD4 was done by an anti-CHD4 antibody. (J) Immunoblot analysis of CHD4 in C3H10T1/2 cells with or without CHD4 knockdown. (K) Real-time PCR analysis of Myocd mRNA level in control (shGFP) or CHD4 knockdown (shCHD4) C3H10T1/2 cells with or without SAG treatment. (L) αSMA immunofluorescence staining in control or SAG-treated C3H10T1/2 cells with or without CHD4 knockdown. Data are mean ± S.D., * = p value ≤0.05, ** = p value ≤0.01.
We also identified a highly conserved Tead binding site around the transcription starting site (TSS) of the Myocd gene (Figure 6D), and showed that both YAP and Tead proteins bound to the site in cells (Figure 6E). More importantly, we found that CHD4, the core component of the NuRD complex, also bound to the Myocd promoter region in YAP5SA-expressing C3H10T1/2 cells (Figure 6H). Furthermore, our ChIP-qPCR analyses in YAP5SA mutant gut mesenchyme revealed that both YAP and CHD4 were highly enriched on the Myocd promoter in vivo (Figure 6F–6G). In addition, we showed that activated YAP interacted with the endogenous CHD4 proteins in C3H10T1/2 cells (Figure 6I), and that the YAP-CHD4 interaction was not affected by SAG treatment (Figure 6I), consistent with our observation that Hh activation could not overcome YAP inhibition of smooth muscle differentiation in vivo and in cells. To further test the idea that YAP-mediated recruitment of CHD4 leads to block Myocd transcription and inhibition of differentiation, we used lentiviral based shRNAs to knockdown CHD4 expression (Figure 6J), and found that, in CHD4 knockdown cells, SAG was able to induce Myocd transcription (Figure 6K) and αSMA expression (Figure 6L) in the cells with YAP5SA ectopic expression. Altogether, these results showed that YAP activation could inhibit smooth muscle cell differentiation through Myocd repression in a NuRD complex-dependent mechanism. Our data, together with two recent reports on possible direct interactions of YAP/Tead and Myocd/SRF proteins (Xie et al., 2012; Liu et al., 2014), suggested a critical functional connection between Hippo/YAP and Myocd in regulation of cell differentiation.
Zone-specific YAP/TAZ down-regulation enables smooth muscle specification by Hedgehog activation
The fact that persistent activation of YAP/TAZ blocks smooth muscle differentiation predicts that YAP/TAZ might have to be down-regulated for proper induction of the smooth muscle lineage in gut mesenchyme. During initial lineage specification from primitive mesenchyme, the αSMA+ smooth muscle progenitors are restricted to a tight ring of cells in the outer half of the mesenchyme. However, it is not clear how this highly organized patterning event is regulated both temporally and spatially. Our IHC analysis revealed that YAP/TAZ are highly expressed throughout the gut mesenchyme prior to smooth muscle differentiation at E11.5 (Figure 7A); however, their expression was significantly lower in this restricted αSMA+ differentiation zone at E13.5 (Figure 7B).
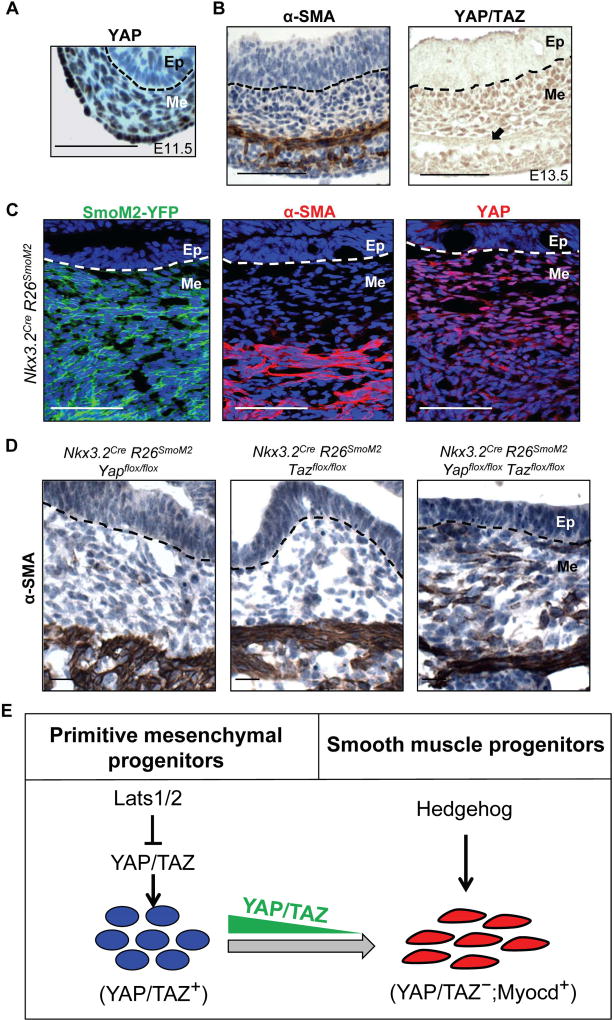
Figure 7. YAP/TAZ downregulation in differentiation zone allows Hedgehog-induced smooth muscle specification in vivo.
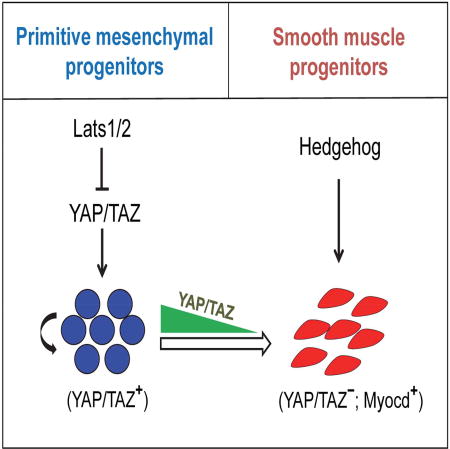
(A) Immunohistochemical staining of YAP in wild-type stomach at E11.5 (B) Immunohistochemical staining of α-SMA and YAP/TAZ in wild-type stomach at E13.5. (C) αSMA and YAP immunoflourescence staining in SmoM2-YFP-expressing gastrointestinal mesenchyme in Nkx3.2CreR26SmoM2 animals at E13.5. (D) Immunohistochemical staining of αSMA in stomach of Nkx3.2CreR26SmoM2Yapflox/flox, Nkx3.2CreR26SmoM2Tazflox/flox, Nkx3.2CreR26SmoM2Yapflox/floxTazflox/flox animals at E13.5. (E) A schematic model showing YAP/TAZ coordination of gut mesenchymal growth and differentiation in concert with Hedgehog signaling. High-level expression of YAP/TAZ in the early mesenchyme is required for expansion of primitive progenitor populations and overall gastrointestinal mesenchymal growth; however, down-regulation of YAP/TAZ in differentiation zone during later development is essential for Hedgehog signaling-induced smooth muscle specification.
Upon close examination of the Nkx3.2Cre;R26SmoM2 mutant embryos, we noticed that, despite the wide-spread expression of SmoM2 in the gastrointestinal mesenchyme (Figure 7C), a mesenchymal domain adjacent to the epithelia, where SmoM2 was not able to drive αSMA expression, exhibiting much higher nuclear YAP expression in cells (Figure 7C). To test whether this high YAP and/or TAZ expression affect smooth muscle differentiation, we generated Nkx3.2Cre R26SmoM2 embryos with concomitant loss of YAP, TAZ or both. We found that, only when both YAP and TAZ were lost, αSMA expression was able to be extended in these mesenchymal cells adjacent to the epithelium (Figure 7D). These results suggested that high YAP/TAZ activity in the sub-epithelial mesenchyme prevents Hh/Smo-induced differentiation therefore providing a positional cure to allow proper spatial induction of the smooth muscle lineage only in the outer layer of gut mesenchyme.
DISCUSSION
Our genetic studies here revealed a compartmental requirement of YAP/TAZ in the anterior lung endodermal epithelia, but not in the posterior gastric and intestinal epithelia, and suggested that this differential requirement is in part due to the higher level of YAP/TAZ expression in the lung epithelia as compared to that of the gastrointestinal tract. It remains to be determined how this expression pattern of YAP/TAZ along the endodermal anterior-posterior axis is established; however it provides a possible molecular explanation as to why YAP/TAZ are dispensable for epithelial development and homeostasis in gastrointestinal tract.
Although YAP/TAZ are not required for normal development and homeostasis in gastrointestinal epithelium, we demonstrated that they are essential for the development of the gastrointestinal mesenchyme; a functional link to the Hippo signaling pathway that was previously unappreciated. Our data place Lats1/2-regulated YAP/TAZ activity at the center stage of mesenchymal development through their pivotal role in coordinating growth and differentiation. We propose a model (Figure 7E) that high YAP/TAZ activity in primitive mesenchyme is critical for expansion of early progenitor populations and overall gut mesenchymal growth; meanwhile, YAP/TAZ also act as the gatekeeper to maintain the primitive progenitor status in these cells to prevent further differentiation into the smooth muscle lineage, a prominent early patterning event during mesenchymal development.
Smooth muscle differentiation in the GI tract is thought to be regulated by several critical developmental pathways, including the Hh pathway. However, it remains largely unknown how this process is coordinated and how the positional cue is provided to establish the highly restricted αSMA+ differentiation domain in the outer mesenchymal layer. Our data identified the essential genetic requirement of the Hippo-YAP pathway in smooth muscle specification, and a molecule mechanism that underlies its tight temporal-spatial control: specific down-regulation of YAP/TAZ in the specific differentiation zone relieves transcriptional repression of Myocd, thereby enabling the induction of the smooth muscle lineage by the pro-differentiation Hh signal (Figure 7E).
The functional interactions between Hippo and Hedgehog signaling were previously reported in medulloblastoma and neural differentiation, where YAP positively regulates Hedgehog pathway (Fernandez et al., 2009; Lin et al., 2012). However, in the context of gastrointestinal development, our data suggested that YAP activation did not directly affect Hedgehog signal transduction in mesenchymal cells; instead it blocked the ability of Hh/Smo to activate transcription of Myocd, the master regulator of smooth muscle differentiation. Consistent with a previous report (Zacharias et al., 2011), our data could not establish the direct Gli binding to the Myocd promoter (Data not shown). It remains possible that the Hh pathway regulates Myocd expression indirectly via downstream signaling such as BMP. The mesenchymal smooth muscle progenitors later give rise to smooth muscle layers, smooth muscle cells inside the villi, and myofibroblasts surrounding the crypts, all of which play critical roles in gastrointestinal homeostasis. It is intriguing to speculate about the possible roles of Hippo-YAP signaling in postnatal gut mesenchyme, and further investigation of the interplays among these pathways may therefore shed light on the mechanisms underlying gastrointestinal regeneration and tumorigenesis.
STAR*MRTHOD
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Junhao Mao (Junhao.mao@umassmed.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
ShhCre (Harfe et al., 2004), VillinCre (Madison et al., 2002), Myh11CreER (Wirth et al., 2008), and R26mT/mG (Muzumdar et al., 2007) mice were obtained from the Jackson laboratory. R26SmoM2 (Mao et al., 2006), Lats1flox and Lats2flox (Yi, et al., 2016) mice were described previously. Nkx3.2Cre (Verzi et al., 2009), Yapflox (Xin et al., 2011) and Tazflox (Xin et al., 2013) mice were kindly provided by Drs. RA Shivdasani, WE Zimmer, and EN Olson. To generate the R26YAP5SA allele, the cDNA fragment encoding YAP5SA (a gift from Kunliang Guan, Addgene plasmid #27371) with the N-terminal Flag and NLS sequences was inserted into the pCN vector with a C-terminal IRES-nuclear lacZ before being cloned into the pROSA targeting vector. The University of Massachusetts Medical School Transgenic Animal Core performed ES cell targeting and blastocyst injection to generate chimeric animals.
To target the endodermal and gastrointestinal epithelia, ShhCre and VillinCre animals were bred with Yapflox and Tazflox mice. To target gastrointestinal mesenchyme and smooth muscle cells, Nkx3.2Cre and Myh11CreER mice were bred with Yapflox, Tazflox, R26YAP5SA, R26SmoM2, R26mT/mG, Lats1flox and Lats2flox mice. Cre recombination in Myh11CreER R26YAP5SA R26SmoM2 embryos was induced with intraperitoneal 200 mg/kg tamoxifen injection in pregnant females at 12.5 dpc and embryos harvested at 15.5 dpc. All mouse experiments were conducted according to the University of Massachusetts Medical School IACUC guidelines.
METHOD DETAILS
Tissue Collection and Histology
Following euthanasia, tissue was dissected from adult or embryonic mice and fixed in 10% Neutral Buffered Formalin (NBF) at 4°C overnigh t. For paraffin sections, tissue was dehydrated, embedded in paraffin, and sectioned at 6 µm. For frozen sections, tissue was dehydrated in 30% sucrose overnight at 4°C, embedded in OCT, and sectioned at 12 µm. Paraffin sections were stained using standard hematoxylin & eosin reagents. For intestinal epithelium and mesenchyme isolation, mouse small intestinal tissues at different postnatal stages are dissected and washed in cold PBS, before transferring to PBS containing 3mM EDTA for rotation at 4°C for 30 mins. After vigorous shaking for 2 mins, the epithelial tissues are collected in the supernatant, while the remaining mesenchymal tissues are washed and incubated with the digestion buffer containing Collagenase XI and Dispase at 37°C for 3 0min, and the samples are then subjected to western blot analysis.
Immunohistochemistry, Immunofluorescence and β-galactosidase staining
For immunohistochemistry (IHC), sections were deparaffinized and rehydrated before undergoing heat-induced antigen retrieval in 10mM sodium citrate buffer (pH 6.0) for 30 minutes. Slides were blocked for endogenous peroxidase for 20 minutes, then blocked for 1 hour in 5% BSA, 1% goat serum, 0.1% Tween-20 buffer in PBS, and incubated overnight at 4°C in primary antibody diluted in blo cking buffer or SignalStain® Antibody Diluent (Cell Signaling). Slides were incubated in biotinylated secondary antibodies for 1 hour at room temperature and signal was detected using the Vectastain Elite ABC kit (Vector Laboratories). For β-galactosidase staining, frozen sections were cut at 12µm intervals and subjected to standard β-galactosidase staining. For immunofluorescence (IF), cells or tissue sections were fixed by 4% paraformaldehyde for 5 minutes, blocked for 1 hour and incubated overnight at 4°C in primar y antibody diluted in blocking buffer. Slides were then incubated for 1 hour at room temperature in Alexa Fluor-conjugated secondary antibodies (Invitrogen) at 1:500 dilution in blocking buffer and mounted using mounting media with DAPI (EMS).
Primary antibodies used for IHC/IF were: YAP (1:200, Cell Signaling), YAP/TAZ (1:200, Cell Signaling), Sox2 (1:2000, Seven Hills Bioreagents), Ki67 (1:10,000, Abcam), phosphor-histone H3 (1:300, Cell Signaling), H/K-ATPase (1:500, MBL), Lysozyme (1:1000, Novus Biologicals), β-catenin (1:500, BD Biosciences), CD44 (1:400, eBioscience), Sox9 (1:200, Abcam), Desmin (1:400, ThermoFisher), α-smooth muscle actin (αSMA) (1:5,000, Abcam), β-tubulin III (1:800, Covance), CD31 (1:200, BD PharMingen), cleaved caspase 3 (1:400, Cell Signaling) and PDGFRα (1:400, BD Biosciences). Image quantification of Ki67 immunofluorescence staining in the gastrointestinal mesenchyme were performed using Fiji software (Schindelin et al., 2012).
Cell culture, lentiviral infection and Luciferase reporter assay
HEK293T and C3H10T1/2 cells were obtained from ATCC, and cultured in DMEM supplemented with 10% FBS, and C3H10T1/2 cells were cultured with Eagle's Basal medium supplemented with 2 mM L-glutamine, 1.5 g/L sodium bicarbonate and Earle's BSS and 10% FBS. For lentiviral infection, pGIPZ based constructs expressing YAP5SA or shRNAs against mouse CHD4 (ACGCTTCTACCGCTATGGAATA and CGCGGACACAGTTATTATATAT), or Myocd (AAGTTCCGATCAGTCTTACAG and TTGTTTCCCAAGGAGATTC) were transfected along with the packing plasmids into growing HEK293T cells. Viral supernatants were collected 48 hours after transfection, and C3H10T1/2 cells were infected and underwent selection with puromycin for 3–4 days. C3H10T1/2 cells were cultured at 30–50% confluency to prevent differentiation. To induce smooth muscle cell differentiation, cells were switched to differentiation media (DMEM with 0.5%FBS) for 12 hr, followed by addition of the Smoothed agonist SAG (25nM, EMD Millipore) in culture for 36 hr. For Gli-BS-Luciferase reporter assay, C3H10T1/2 cells were transfected with the luciferase reporter construct, Gli-BS-Luc (gift of Dr. H. Kondoh, Osaka University), pGIPZ-YAP5SA, pBABE-TAZ4SA (gift of Dr. Kunliang Guan, UCSD), and Renila luciferase expression vector, and treated with or without SAG (25nM). Luciferase assays were conducted 72 hours after transfection using the dual-luciferase reporter kit (Promega). Assays were conducted in triplicates.
RNAseq and Affymetrix gene chip analysis
Embryonic stomachs were dissected from E13.5 control and Nkx3.2CreR26YAP5SA embryos, and RNA was extracted for subsequent RNAseq analysis, using independent biological quadruplicates. Differential expressed genes were determined by cufflinks. Transcriptional profiling of E13.5 Nkx3.2CreR26SmoM2 gastrointestinal using Affymetrix Mouse Gene 1.0ST chips was described previously (Huang et al., 2013). The heatmap was generated using pheatmap (v 1.0.7) package.
Chromatin immunoprecipitation-qPCR analysis
ChIP assays were performed according to the manufacturer’s instructions (Active Motif, CA). Briefly, 2×107 cells were fixed with 1% formaldehyde, washed with cold PBS and lysed in lysis buffer. After sonication, protein-DNA complexes were incubated with Flag or CHD4 () antibodies-coupled protein G beads at 4°C overnight. After elution and reverse cross-link, DNA was purified for subsequent PCR analysis. The primers used for real-time PCR of the promoter regions were described in Supplemental Table 2.
Protein immunoprecipitation and western blotting
C3H10T1/2 with YAP5SA stable transfection were washed with PBS and subsequently lysed in RIPA lysis buffer. Cell lysate was incubated with indicated antibody overnight. The immunoprecipitates were washed five times with RIPA buffer, before subjecting to immunoblot analysis. For tissue samples from perinatal and adult intestine were dissected and lysed. Protein lysates were probed with the following primary antibodies: Taz (1:1,000, BD Pharmingen), YAP (1:1,000, Cell Signaling), Flag (1:5,000, Sigma), V5 (1:1,000, Cell Signaling), CHD4 (1:1,000, Abcam), and GAPDH (1:1,000, Bethyl). HRP-conjugated Secondary antibodies were obtained from Jackson Laboratories.
Quantitative Real-Time PCR
RNA of animal tissues or cultured cells was isolated using Trizol reagent (Invitrogen), followed by reverse-transcription using Superscript IV Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed using Sybr Mastermix (Kapa Bioscience). The primers used for real-time PCR were described in Supplemental Table 3. All qPCR experiments were conducted in biological triplicates, error bars represent mean ± standard deviation, and Student’s t-test was used to generate p-values (* = p value ≤0.05; ** = p value ≤0.01).
Supplementary Material
Table 1.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Yap | Cell Signaling | 14074 |
| Rabbit anti-YAP/TAZ | Cell Signaling | 8414 |
| Rabbit anti-Sox2 | Seven Hills Bioreagents | WRAB-1236 |
| Rabbit anti-Ki67 | Abcam | ab15580 |
| Rabbit anti-Phospho-Histone H3 | Cell Signaling | 9710 |
| Rabbit anti-Proton Pump (H, K-ATPase) | MBL | D032-3 |
| Mouse anti-Lysozyme | Novus Biologicals | NBP1-95509 |
| Mouse anti-bCatenin | BD Biosciences | 610153 |
| Rat anti-CD44 | eBiosciences | 67044-82 |
| Rabbit anti-Sox9 | Abcam | ab185230 |
| Rabbit anti-Desmin | Thermo Fisher | RB-9014-P0 |
| Rabbit anti-aSMA | Abcam | ab5694 |
| Mouse anti-β-tubulin III | Convance | MMS-453P |
| Mouse anti-CD31 | BD Biosciences | 550274 |
| Rabbit anti-Cleaved caspase 3 | Cell Signaling | 9664 |
| Rat anti-PDGFRa | BD Biosciences | 558774 |
| Mouse anti-TAZ | BD Biosciences | 560235 |
| Rabbit anti-CHD4 | Abcam | ab72418 |
| Mouse anti-Flag M2 | Sigma | SLBF6631 |
| Rabbit anti-V5 | Cell Signaling | 13202 |
| Rabbit anti-GAPDH | Bethyl | A300-641A |
| HRP conjugated donkey anti-rabbit | Jackson Immunoresearch | 711-035-152 |
| HRP conjugated donkey anti-mouse | Jackson Immunoresearch | 115-035-003 |
| Alexa Fluor 568, goat anti-rabbit | Invitrogen | A-11036 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SAG, Smoothened Agonist | EMD Millipore | 566660 |
| Critical Commercial Assays | ||
| Vectastain Elite ABC kit | Vector Laboratories | PK-6100 |
| Dual-Luciferase® Reporter Assay System | Promega | E1910 |
| ChIP-IT® Express ChIP Kit | Active Motif | 53008 |
| SuperScript IV Reverse Transcriptase | Invitrogen | 18091060 |
| SYBR® FAST qPCR Kit Master Mix | Kapa Biosystems | Kk4600 |
| Deposited Data | ||
| SmoM2 Microarray | Huang et al., 2013 | Submission to GEO |
| YAP5SA RNAseq | This study | Submission to GEO |
| Experimental Models: Cell Lines | ||
| C3H/10T1/2 | ATCC | CCL-226 |
| Experimental Models: Organisms/Strains | ||
| Mouse: R26YAP5SA | This study | N/A |
| Mouse: R26SmoM2 | Mao et al., 2006 | JAX # 005130 |
| Mouse: R26mT/mG | Muzumdar et al., 2007 | JAX # 007676 |
| Mouse: ShhCre | Harfe et al., 2004 | JAX # 005622 |
| Mouse: VillinCre | Madison et al., 2002 | JAX # 024842 |
| Mouse: Myh11CreER | Wirth et al., 2008 | JAX # 019079 |
| Mouse: Nkx3.2Cre | Verzi et al., 2009 | N/A |
| Mouse: Yapflox | Xin et al., 2011 | N/A |
| Mouse: Tazflox | Xin et al., 2013 | N/A |
| Mouse: Lats1flox | Yi et al., 2016 | JAX# 024941 |
| Mouse: Lats2flox | Yi et al., 2016 | JAX# 025428 |
| Oligonucleotides | ||
| Primers for qRT-PCR | This study | Table S2 |
| Primers for qCHIP-PCR | This study | Table S2 |
| shCHD4 sequence #1 | This study | ACGCTTCTACCGCTATGGAAT |
| shCHD4 sequence #2 | This study | CGCGGACACAGTTATTATATAT |
| shMyocd sequence #1 | This study | AAGTTCCGATCAGTCTTACAG |
| shMyocd sequence #2 | This study | TTGTTTCCCAAGGAGATTC |
| Recombinant DNA | ||
| pCMV-YAP5SA | Gift of Dr. Kun-liang Guan | Addgene #27371 |
| pBABE-TAZ4SA | Gift of Dr. Kun-liang Guan | N/A |
| Gli-BS-Luciferase reporter | Gift of Dr. Dr. H. Kondoh | N/A |
| pGIPZ-YAP5SA | This study | N/A |
| pGIPZ-Myocd | This study | N/A |
| Software and Algorithms | ||
| Fiji (Image J) | Schindelin et al., 2012 | http://fiji.sc |
Highlights.
YAP/TAZ coordinate growth and patterning in gut mesenchyme.
Lats1/2 kinases suppress expansion of primitive gut mesenchyme.
YAP blocks smooth muscle (SM) cell differentiation via Myocardin repression.
Zone-specific YAP/TAZ down-regulation allows Hedgehog-induced SM differentiation.
Acknowledgments
This work was supported by a grant from NIH/NIDDK (5R01DK099510). The authors thank Annie Pang for technical assistance, and members of the Mao lab for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Table S1. (Related to Figure 4 and 5). RNAseq analysis of E13.5 wild type and Nkx3.2-Cre;R26-YAP5SA stomach
AUTHOR CONTRIBUTIONS
J.L.C. and J. M. designed research; J.L.C, Q.L., L.M., and J. W. performed most of the mouse experiments and in vitro cell-based assays. J.L.C designed and generated the R26YAP5SA mice. Y.T.I and R. L. J provided the key resources, including the Lats1/2 mutant mice used in this study and analyzed the data. J. P. and J.L.C performed the RNAseq experiments. J.O. and L.J.Z. provides the Bioinformatics analysis of the RNAseq data. J.L.C and J.M. wrote the manuscript.
References
- Barry ER, Morikawa T, Butler BL, Shrestha K, De La Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, Wrana JL. Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 2013;5:1611–1624. doi: 10.1016/j.celrep.2013.11.021. [DOI] [PubMed] [Google Scholar]
- Cai J, Maitra A, Anders RA, Taketo MM, Pan D. beta-Catenin destruction complex-independent regulation of Hippo-YAP signaling by APC in intestinal tumorigenesis. Genes Dev. 2015;29:1493–1506. doi: 10.1101/gad.264515.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhang N, Zheng Y, De Wilde RF, Maitra A, Pan D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LA, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, Taylor MD, Kenney AM. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Camargo FD. The hippo tumor suppressor network: from organ size control to stem cells and cancer. Cancer Res. 2013;73:6389–6392. doi: 10.1158/0008-5472.CAN-13-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Huang H, Cotton JL, Wang Y, Rajurkar M, Zhu LJ, Lewis BC, Mao J. Specific requirement of Gli transcription factors in Hedgehog-mediated intestinal development. J Biol Chem. 2013;288:17589–17596. doi: 10.1074/jbc.M113.467498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imajo M, Ebisuya M, Nishida E. Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat Cell Biol. 2015;17:7–19. doi: 10.1038/ncb3084. [DOI] [PubMed] [Google Scholar]
- Kedinger M, Duluc I, Fritsch C, Lorentz O, Plateroti M, Freund JN. Intestinal epithelial-mesenchymal cell interactions. Ann N Y Acad Sci. 1998;859:1–17. doi: 10.1111/j.1749-6632.1998.tb11107.x. [DOI] [PubMed] [Google Scholar]
- Kim M, Kim T, Johnson RL, Lim DS. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 2015;11:270–282. doi: 10.1016/j.celrep.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Kolterud A, Grosse AS, Zacharias WJ, Walton KD, Kretovich KE, Madison BB, Waghray M, Ferris JE, Hu C, Merchant JL, et al. Paracrine Hedgehog signaling in stomach and intestine: new roles for hedgehog in gastrointestinal patterning. Gastroenterology. 2009;137:618–628. doi: 10.1053/j.gastro.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715–728. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- Lin YT, Ding JY, Li MY, Yeh TS, Wang TW, Yu JY. YAP regulates neuronal differentiation through Sonic hedgehog signaling pathway. Exp Cell Res. 2012;318:1877–1888. doi: 10.1016/j.yexcr.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Liu F, Wang X, Hu G, Wang Y, Zhou J. The transcription factor TEAD1 represses smooth muscle-specific gene expression by abolishing myocardin function. J Biol Chem. 2014;289:3308–3316. doi: 10.1074/jbc.M113.515817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- Mahoney JE, Mori M, Szymaniak AD, Varelas X, Cardoso WV. The hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev Cell. 2014;30:137–150. doi: 10.1016/j.devcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Kim BM, Rajurkar M, Shivdasani RA, McMahon AP. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development. 2010;137:1721–9. doi: 10.1242/dev.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, McMahon AP. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Wu LW, Grivennikov SI, De Jong PR, Lian I, Yu FX, Wang K, Ho SB, Boland BS, Chang JT, et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014;141:1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- Varelas X, Wrana JL. Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 2012;22:88–96. doi: 10.1016/j.tcb.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Stanfel MN, Moses KA, Kim BM, Zhang Y, Schwartz RJ, Shivdasani RA, Zimmer WE. Role of the homeodomain transcription factor Bapx1 in mouse distal stomach development. Gastroenterology. 2009;136:1701–1710. doi: 10.1053/j.gastro.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horvath B, Maser-Gluth C, Greiner E, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- Xie C, Guo Y, Zhu T, Zhang J, Ma PX, Chen YE. Yap1 protein regulates vascular smooth muscle cell phenotypic switch by interaction with myocardin. J Biol Chem. 2012;287:14598–14605. doi: 10.1074/jbc.M111.329268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci USA. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Lu L, Yanger K, Wang W, Sohn BH, Stanger BZ, Zhang M, Martin JF, Ajani JA, Chen J, et al. Large tumor suppressor homologs 1 and 2 regulate mouse liver progenitor cell proliferation and maturation through antagonism of the coactivators YAP and TAZ. Hepatology. 2016;64:1756–1762. doi: 10.1002/hep.28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias WJ, Madison BB, Kretovich KE, Walton KD, Richards N, Udager AM, Li X, Gumucio DL. Hedgehog signaling controls homeostasis of adult intestinal smooth muscle. Dev Biol. 2011;355:152–162. doi: 10.1016/j.ydbio.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.