Abstract
The MHC region encodes HLA genes and is the most complex region in the human genome. The extensively polymorphic nature of the HLA hinders accurate localization and functional assessment of disease risk loci within this region. Using targeted capture sequencing and constructing individualized genomes for transcriptome alignment, we identified 908 novel transcripts within the human MHC region. These include 593 novel isoforms of known genes, 137 antisense strand RNAs, 119 novel long intergenic noncoding RNAs, and 5 transcripts of 3 novel putative protein-coding human endogenous retrovirus genes. We revealed allele-dependent expression imbalance involving 88% of all heterozygous transcribed single nucleotide polymorphisms throughout the MHC transcriptome. Among these variants, the genetic variant associated with Behçet’s disease in the HLA-B/MICA region, which tags HLA-B*51, is within novel long intergenic noncoding RNA transcripts that are exclusively expressed from the haplotype with the protective but not the disease risk allele. Further, the transcriptome within the MHC region can be defined by 14 distinct coexpression clusters, with evidence of coregulation by unique transcription factors in at least 9 of these clusters. Our data suggest a very complex regulatory map of the human MHC, and can help uncover functional consequences of disease risk loci in this region.
Keywords: MHC, HLA, transcriptome, polymorphisms, allele-specific expression, lncRNA, retroviral elements
Introduction
The human major histocompatibility complex (MHC) is a highly complex polymorphic genomic region containing many important immune-related genes. This region includes the human leukocyte antigen (HLA) genes involved in both self-tolerance and antigen presentation. Polymorphisms within HLA genes have been associated with over 100 autoimmune diseases and cancers, and allelotyping of translated genes has been the focus of extensive research (1–3). Intergenic variants within the MHC region, which may serve a role in gene regulation, have also been associated with several immune-related diseases (4, 5). However, the role of these intergenic variants is often not clear because regulation within the MHC is incompletely understood. The MHC contains a complex regulatory network including cis-acting and trans-acting regulation bridging inside and outside the MHC region (6, 7). Due to both the high rate of polymorphism and the complex regulatory networks within the MHC, the functional effects of specific disease-associated variants are difficult to elucidate.
Long intergenic noncoding RNAs (lincRNAs) have been extensively implicated in transcriptional regulation by recruitment of regulatory proteins. These proteins proceed to regulate gene expression by epigenetic modification, such as DNA methylation and chromatin modification (8, 9). Recruitment of transcription factors by lincRNAs has also been described (10). However, many lincRNAs are weakly expressed, and therefore may not be detected by RNA sequencing that spans the entire transcriptome.
Sequence-specific enrichment by magnetic bead pulldown has previously been used to sequence HLA genes for allelotyping, and to elucidate regulatory regions of individual genes (2, 4). We performed targeted enrichment of the entire MHC region in primary human monocytes using sequence-specific capture probes, followed by high throughput sequencing of DNA and RNA (CaptureSeq) (11), to allow for deep sequencing coverage of the MHC region. We targeted the entire MHC, including both intergenic regions and known splice variants. We identified genetic variants, then constructed personalized genomes to accurately align RNA sequences. After enrichment and alignment to personalized genomes, we were able to detect lowly expressed transcripts, and by including all genomic regions, we were able to identify novel intergenic transcripts. We also comprehensively assessed allelic expression imbalance and revealed extensive allele-specific expression throughout the entire MHC, indicating that polymorphism is a mechanism of complex transcriptional regulation in this region.
Materials and Methods
Probe Design
Sequence-specific capture probes were designed to target the complete reference genomic sequence of the MHC region (chr6:28.5 Mb-33.5 Mb, hg19), as well as splice sites for known transcripts the region contains. By including intergenic and intronic genomic regions, sequences that overlap with previously unannotated regions could be captured and subsequently sequenced; moreover, the same set of probes were designed to enable us to capture both DNA and RNA. This pool of 75 base long capture probes was designed to target 35,895 sequences throughout the region. For the main reference allele, probes directly overlapped 75.9% of the genome, with 88% estimated total sequence coverage. However, because the MHC region is highly polymorphic, the seven alternative reference haplotypes for the MHC were used in addition to the reference allele to design probes targeting all reference genomic sequences in this region. In total, this region, including all alternative haplotypes, was 65.4% covered by the probes, and had a 75.7% estimated net coverage. Of the total target region, including alternate haplotypes, 10% was not covered due to shared homology with other parts of the genome, while 14.2% was not covered because of incomplete sequence information in the alternative haplotypes.
Isolation of Primary Monocyte DNA and RNA
Peripheral blood mononuclear cells (PBMCs) isolated from 12 healthy individuals were initially collected by density gradient centrifugation and immediately stored in liquid nitrogen. Cells were thawed, treated with 25 U/mL benzonase, and incubated at 37°C in RPMI/10% heat inactivated fetal bovine serum for 90 minutes. Thawed PBMCs had a minimum viability of 90%, with an average viability of 98.1% ± 3.6%, measured by tryphan blue staining. Primary monocytes were then isolated from thawed peripheral blood mononuclear cells via negative selection using the Pan Monocyte Isolation Kit, following the manufacturer’s instructions (Miltenyi Biotec Inc., San Diego California). The remaining monocyte-depleted PBMCs were flushed from the magnetic column, and DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen, Hilden Germany). RNA was isolated from primary monocytes using the Direct-zol RNA Isolation Kit (Zymo Research, Irvine California), and then DNase treated using the TURBO DNA-free kit (Invitrogen, Carlsbad California). The purity of the isolated monocytes was measured by flow cytometry using the iCyt Synergy SY3200 cell sorter (Sony Biotechnology, Inc, San Jose California), staining with APC/Cy7 anti-CD14 (BioLegend, San Diego California). Monocyte purity was found to be greater than 90%.
DNA and RNA Sequencing
RNA integrity and concentration was verified using the Agilent Bioanalyzer (RIN> 8) (Agilent Technologies, Santa Clara California). A minimum of 500ng RNA was processed per sample. RNA was ribo-depleted using the NEBNext rRNA Depletion Kit (Human/Mouse/Rat) (New England Biolabs, Ipswich Massechusetts), and sequencing libraries were prepared for every DNA and RNA sample. Sequence-specific magnetic bead capture was performed on DNA and RNA libraries according to the manufacturer’s instructions, using the custom-designed probes (SeqCap EZ Choice XL Library System, Roche Nimblegen, Inc, Madison Wisconsin). Samples were multiplexed, four samples per capture reaction. All post-capture gDNA libraries were sequenced in one lane, while all post-capture cDNA libraries were sequenced in another. All samples were sequenced with the Illumina HiSeq 2500, with paired, 125 bp reads.
Developing Individualized Genotypes
DNA reads were quality trimmed using Trimmomatic, then aligned to the hg19 reference sequence using the Burrows-Wheeler aligner (BWA MEM) (12). Duplicate sequences were then removed using Picard, and indels were processed using GATK (13–15). From these aligned reads, SAMtools was used to generate an mpileup file, then VarScan mpileup2snp was used to identify SNPs (16, 17). For each individual, SNPs were called based on variation from the reference genome (hg19), and all called SNPs have a total read depth of at least 8 and a maximum variant calling p value of 0.01. For all heterozygous SNPs, each allele also has a minimum variant-supporting read depth of 2, a minimum average variant-supporting read base quality of 20, and a minimum allele frequency of 0.2. From these identified and quality filtered SNPs, individualized lists of variants were created for each sample. On average, 23,575 heterozygous variants were identified in the MHC region per individual. The average read depth on heterozygous variants identified in all samples was 417 ± 95.2 with an average variant-supporting read base quality of 230.6 ± 11.7.
RNA Alignment and Assembly
RNA sequencing reads were quality trimmed using Cutadapt, then aligned to the human reference genome (hg19, chromosome 6, RefSeq transcriptome annotation) using GSNAP (18, 19). Alternate haplotypes for chromosome 6 in the reference genome were not used for alignment, to prevent errors from multi-mapped reads. RNA reads were aligned in a SNP-tolerant manner, meaning that variants that were called from the DNA sequencing alignment were not included in the mismatch penalty scores for RNA reads. Reads that successfully aligned to the target region were assembled into transcripts using StringTie, guided by the Ensembl transcriptome annotation (20).
Identification of Novel Transcripts
During RNA assembly, transcripts were annotated using an Ensembl reference. To identify novel transcripts, we followed the following workflow (Figure S1). All transcripts that were successfully annotated using the Ensembl reference during alignment were excluded. Using CuffCompare, the remaining transcripts were annotated using Gencode Comprehensive v25 (hg19) as a reference (21, 22). All transcripts were assigned class codes based on their relation to transcripts in the reference. All transcripts that were assigned the class codes I (intronic), J (novel isoform), U (intergenic), and X (antisense) were identified as novel, while transcripts containing all other class codes were defined as not novel. The remaining novel transcripts were annotated with CuffCompare using the human lincRNA catalog (23). The transcripts that were found to be novel using all three references were next filtered to include only transcripts with an FPKM of 0.1 in two or more samples.
The coding potential of each novel transcript was analyzed using the Coding Potential Analysis Tool (CPAT) (24). The sequence of each novel transcript was determined using genomic coordinates determined by StringTie and the sequence of the reference genome, and these sequences were used to determine coding potential for each transcript. Transcripts with a coding probability of 0.364 or greater were defined as putatively coding, while all others were defined as noncoding. All novel transcripts were categorized based on their Gencode annotation class codes and by these coding predictions.
Predicted Function of Coding Genes
Of the five putative coding transcripts that did not share exons with known genes, structure and function were predicted using IntFOLD3. Using the open reading frame predicted by CPAT, the putative amino acid sequence was determined from the transcript sequences using A plasmid Editor (ApE). Using the IntFOLD3 pipeline, we predicted the tertiary structure of the novel peptides, guided by sequence homology with known proteins (25). In addition, putative ligand binding domains and gene ontology term annotation were predicted using the FunFOLD pipeline, which is integrated into IntFOLD3.
Retroviral Element Sequence Alignment
The five novel putative coding intergenic transcripts described were categorized based on their alignment to retroviral sequences. The predicted open reading frames of each of the five transcripts were translated into a protein sequence. Two transcripts that were isoforms of the same gene shared an open reading frame, so four protein sequences were generated. These sequences were aligned to the human proteome using protein-protein BLAST with the non-redundant protein sequences database(26). Each of these four sequences aligned to known retroviral elements (E Value < 1 × 10−10). Sequence alignments were visualized using MView(27).
Allele Specific Expression
Allele specific expression of aligned transcript and genomic reads at each heterozygous SNP was assessed using GATK ASEReadCounter under the default settings, which includes a read downsampling step (13, 14). Only alignments with base quality and mapping quality no lower than 20 were used. The read counts for each transcript were then normalized to genomic allele-specific read counts derived from DNA reads using GATK ASEReadCounter under the default settings. The DNA allelic imbalance (AI) ratio was first calculated for both the reference and alternate allele of each variant as follows: . Read counts for both alleles of each variant was then calculated from the following formula: .
SNPs containing allelic imbalance were defined by a chi-squared test p value less than 0.05, calculated based on the normalized read counts. Relative allelic imbalance for all expressed heterozygous SNPs was calculated as the reference SNP expression: total expression ratio. Heterozygous SNPs and relative reference allele expression for all individuals were merged based on SNP position and reference allele. Allele specific expression at rs76546355 was also confirmed using the program bam-readcount (28).
Co-expression Analysis
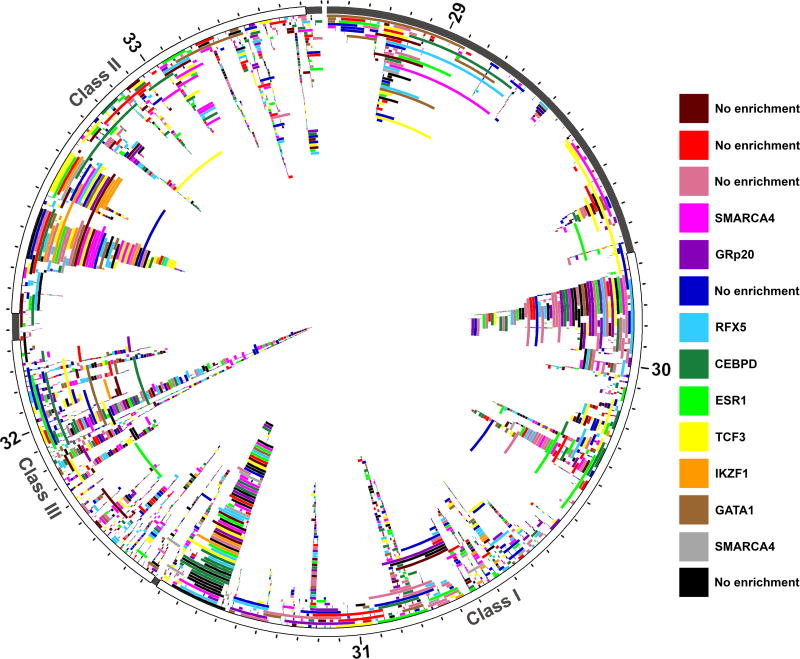
Co-expression networks were assigned using the R package WGCNA(29). This package clusters every sequenced transcript based on the normalized read counts (FPKM) in all twelve samples, using a weighted correlation network analysis. For initial quality filtering, all transcripts that were missing from more than one half of all samples were removed from analysis; 320 transcripts were removed. Samples were then clustered according to transcription patterns to remove any outlier samples; however, no outlier samples were observed. From this filtered set of 2753 genes, a co-expression network was created, with a soft-thresholding power of 7, a dendrogram cut height of 0.25, and a minimum cluster size of 30 transcripts. All transcripts were categorized within an expression dendrogram, then successfully assigned to a co-expression cluster. A total of fourteen clusters were defined. The genomic localization of each cluster was visualized using Circos(30).
Transcription Factor Binding Site Enrichment Analysis
Transcription factor binding site enrichment analysis was performed for each of the fourteen co-expression clusters using GenomeRunner Web(31), which compares the genomic coordinates of each transcript to the genomic positions of known transcription factor binding sites, using a database that includes the non-cell specific binding patterns of 161 transcription factors, measured via transcription factor ChIP-seq distributed by ENCODE. The coordinates for the promoter region of each gene in each coexpression cluster was used as input, defined as the 1500bp preceding transcriptional start sites. As a background for enrichment analysis, we included the promoter region of every gene within the MHC region, as annotated by the UCSC known genes list, and also included the novel genes that we have described. The UCSC known gene list contains an aggregation of gene annotations from across the RefSeq, GenBank, CCDS, Rfam, and tRNAscan-SE databases. Transcription factor enrichment was calculated for each co-expression cluster individually, and a cluster was called enriched for a specific transcription factor when an increased frequency of the target was observed in the cluster compared to the background (OR>1, chi-square p<0.05). Nine of the fourteen co-expression clusters were enriched for specific transcription factors.
HLA Typing
HLA allelotypes for each sample were determined using BWAkit. This pipeline calls types by aligning reads to each HLA gene using the BWA-MEM algorithm, and comparing the exons of each gene to alleles defined by IMGT/HLA. The called types (Table S1) are defined as the alleles that have minimal exonic mismatch with the individual’s sequence.
Sanger Sequencing
Allele-specific expression was validated by Sanger sequencing for the target variant rs76546355. RNA was saved from each individual before sequence-specific capture and was converted into cDNA using the Verso cDNA Synthesis Kit (Thermo Fisher Scientific Inc, Waltham, Massachusetts). This cDNA was then amplified via PCR, using primers that flank the target SNP (forward primer: TGCTTGCCTGTTGTGAGATG, reverse primer: AAGCAACAGTAATTTGGATCTTCC). The proportion of each allele represented in this PCR product was estimated using a Sanger sequencing trace file for each sample.
Results
Targeted genome and transcriptome sequencing in the human MHC region
We performed deep targeted genome and transcriptome sequencing of the human MHC region (Chromosome 6 (hg19): 28.5 Mb-33.5 Mb) in primary human monocytes. Constructing individualized genomes for aligning RNA sequencing reads generated by deep targeted transcriptome sequencing improved transcript alignment and characterization in is this complex polymorphic region. DNA sequence reads aligned against the reference genome human MHC region with a mean read depth of 334.8 ± 84.3 in all samples (Figure S2). Genetic polymorphisms in each sample were identified and an individualized MHC genome in each sample was constructed. A total of 65,289 genetic variants relative to the reference genome were identified, including 62,449 genetic variants that are heterozygous in at least one sample.
Targeted RNA sequencing was performed following ribosomal RNA depletion, allowing for high density coverage with an average 36.3 million alignments to the MHC region, and a mean read depth of 594.5 ± 87.2 per gene in this region in all samples. RNA sequence alignment was performed in an individualized SNP-tolerant mode using DNA sequencing data from each sample to allow alignment to polymorphic loci identified in each corresponding sample. This strategy significantly enhanced successful alignment of transcript reads to the polymorphic MHC region, which coupled with highly dense targeted RNA sequencing, allowed for accurate identification of known and novel transcripts in the MHC region, including transcripts with low expression levels.
Identification and classification of novel transcripts within the MHC region
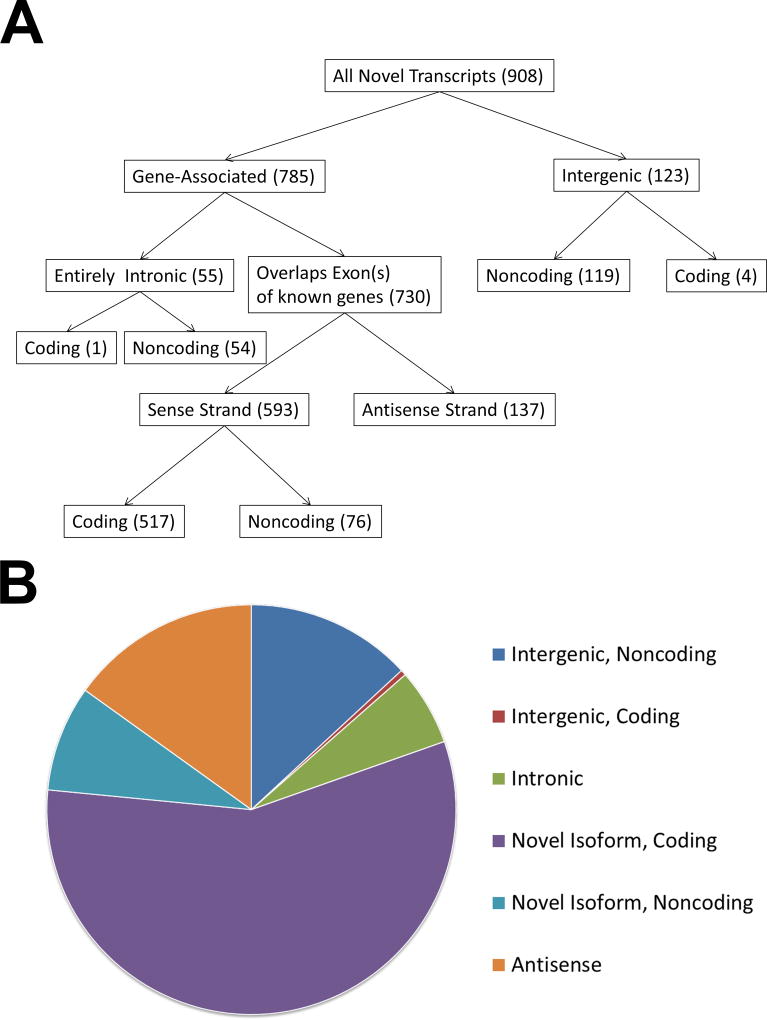
We identified a total of 3,072 transcripts aligned to the human MHC region in human primary monocytes. Of these, 908 transcripts were identified as novel transcripts that were present in at least 2 independent samples (Table S2). This includes 517 and 76 novel coding and noncoding transcript isoforms of known genes, respectively. In addition, we identified 137 novel anti-sense strand transcripts, 119 lincRNA transcripts, 54 intronic noncoding RNA transcripts, and 5 transcripts of three novel coding genes (Figure 1).
Figure 1.
A flow chart (A) and pie chart (B) depicting and summarizing the filtering categories used to classify novel transcripts identified in this study. The categories for intronic, novel isoforms, antisense, and intergenic transcripts were defined via a CuffCompare annotation using the Comprehensive Gencode Resease 25 annotation (hg19) reference transcriptome. Coding potential of novel transcripts was predicted using CPAT.
Evidence for extensive cis allele-specific expression within the human MHC
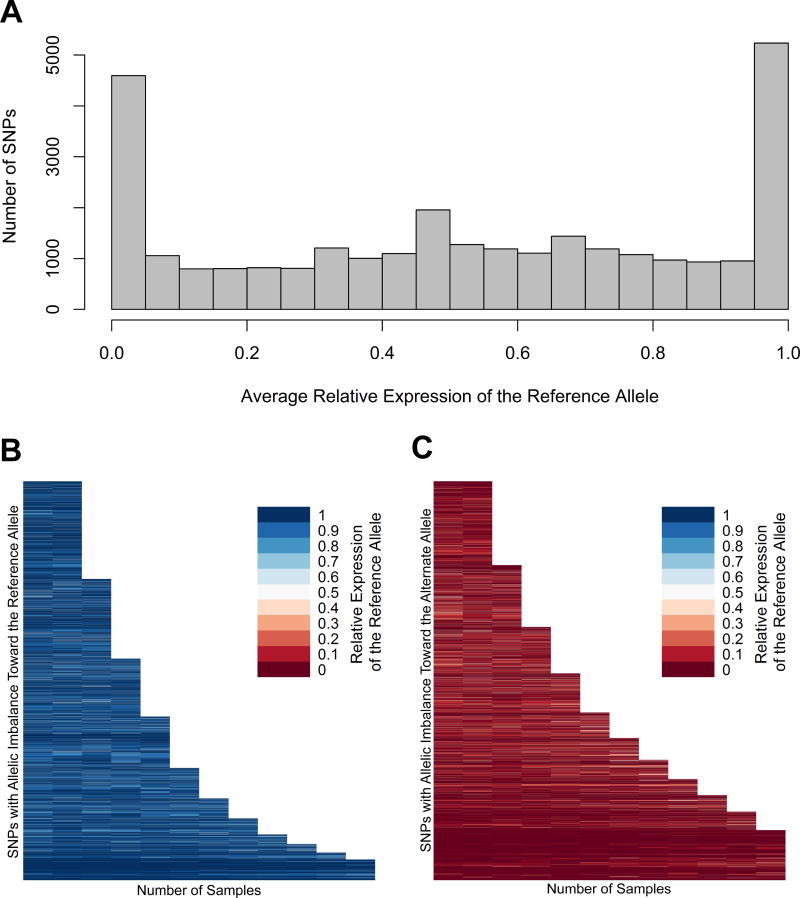
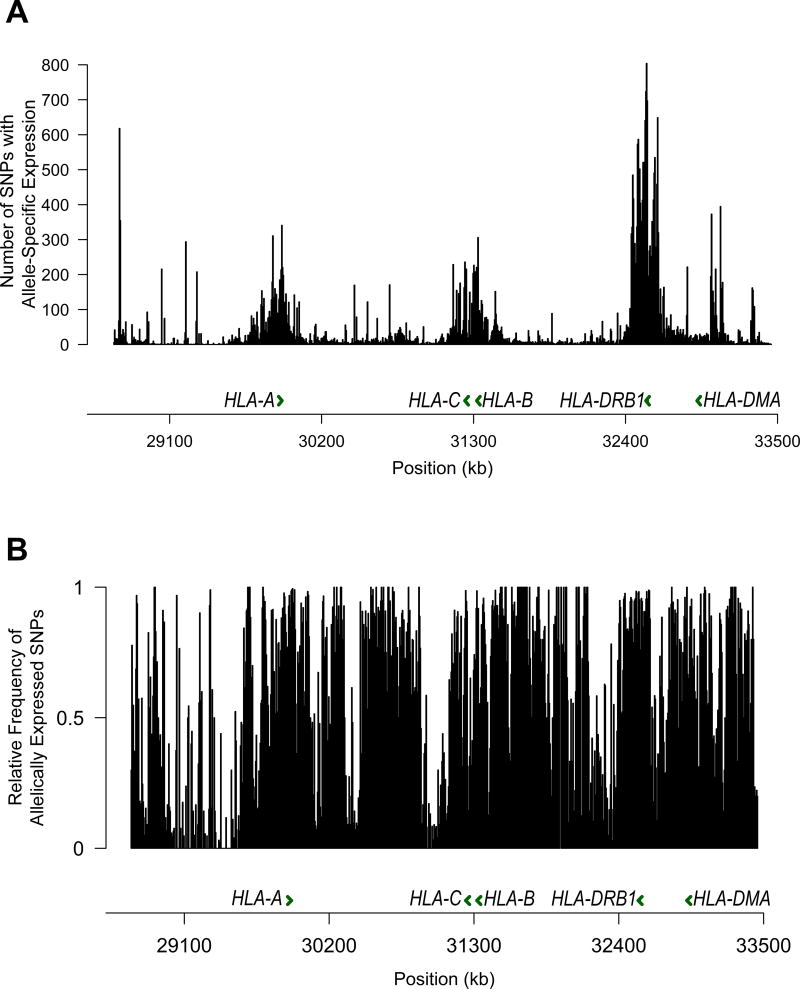
Next, we evaluated the extent of allele-specific expression imbalance in MHC region transcripts that overlap with heterozygous single nucleotide polymorphisms identified using DNA sequencing. We show that 88% of heterozygous transcribed SNPs within the MHC region are associated with significant allele-dependent transcriptional imbalance, with 43% demonstrating extreme allele-dependent expression (>95% expression on either the reference or alternative allele) (Figure 2A). Indeed, allelic imbalance is observed in over 69% of all heterozygous SNPs identified in our study within the MHC region (Table S3 and S4). This remarkably extensive allele-specific expression pattern was non-stochastic and consistent across independent samples in heterozygous SNPs that are present in two or more samples (Figure 2B and 2C). While the overall number of heterozygous SNPs with evidence of allelic expression imbalance was highest in the HLA class II gene region within the MHC, the frequency of transcribed SNPs with allelic imbalance relative to all transcribed SNPs was consistent throughout the HLA regions within the MHC (Figure 3 and Table S5).
Figure 2.
(A) Frequency distribution histogram of instances of allele-specific expression. The average relative expression (proportion of reads containing the reference allele) was calculated for all transcribed heterozygous SNPs identified in our study. Each bin spans a relative expression range of 0.05. (B) Variants in which the average relative expression of the reference allele is greater than 0.5, and in which the average relative expression is less than 0.5 (C). The relative expression of the reference allele in each SNP with allelic imbalance (binomial p<0.05) in two or more samples is represented on the Y axis. The reference allele is defined by the genotype of the reference genome, which is consistent across all samples. Relative expression ranges from 0 (red) to 1 (blue). The allelic imbalance of specific SNPs is shown to be highly consistent across individuals.
Figure 3.
Histogram depicting the number of SNPs with allelic expression imbalance (A), and frequency of SNPs with allelic expression imbalance relative to all heterozygous SNPs detected in the MHC region (B). Each bin spans 5,000 base pairs.
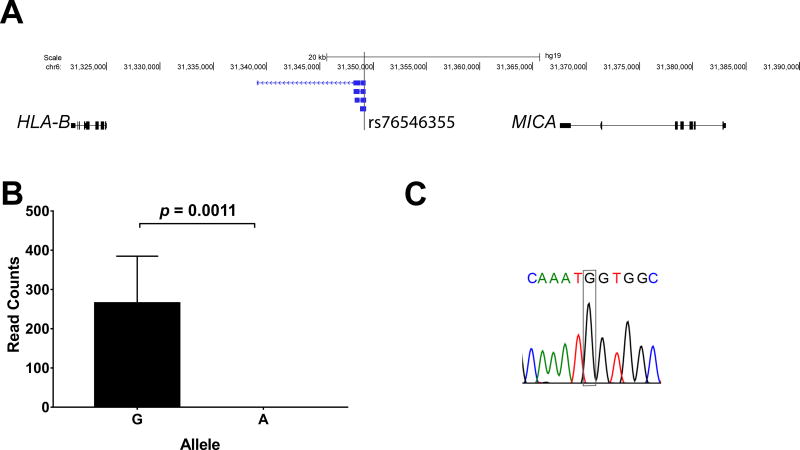
To demonstrate allelic imbalance in a disease relevant locus in the MHC region, we examined the expression of novel transcripts that overlap with and include the SNP rs76546355 (rs116799036) localized between HLA-B and MICA. This genetic variant tags the most robust genetic association in Behçet’s disease (5). We show that rs76546355, previously thought to be intergenic, is expressed within four lincRNA transcripts we identified between HLA-B and MICA. Importantly, these four transcripts are exclusively expressed from the haplotype with the disease protective allele in this SNP. There was no expression of these transcripts from the haplotype with the disease risk allele in heterozygous individuals (Figure 4). These data suggest evidence for haploinsufficiency involving the expression of novel lincRNAs, induced by a disease risk variant within the MHC region in a complex polygenic disease.
Figure 4.
(A) The genetic variant rs76546355 (rs116799036) which explains the most robust genetic association for Behçet’s disease and previously thought to be in a non-transcribed genetic region is expressed within four lincRNA transcripts between HLA-B and MICA. (B) RNA sequencing revealed that these lincRNA transcripts are exclusively expressed from the DNA strand with the disease protective allele (allele G), and no expression was detected from the disease risk allele (allele A). RT PCR followed by Sanger sequencing confirmed expression of the novel lincRNA transcripts in this locus, and allele expression imbalance in rs76546355 (a representative chromatogram of seven heterozygous samples is shown) (C).
Co-expression patterns and transcription factor binding analyses identify transcriptional clusters in the human MHC transcriptome
We characterized the expression patterns of the transcripts within the MHC using a co-expression network analysis. We defined a co-expression network including all aligned transcripts, based on the normalized read counts across all twelve samples, using a weighted correlation network analysis (WGCNA) (29). Based on this network, the transcripts were grouped into fourteen co-expression clusters, which do not localize to specific genomic regions within the MHC (Figure 5). Nevertheless, co-expression remains highly aggregated within individual clusters, and there is a high degree of separation between each cluster within the network (Figure S3).
Figure 5.
All unique transcripts plotted according to genomic position within the MHC region (chr6:28.7Mb-33.5Mb (hg19)). Chromosome six position (labelled in Mb) is plotted on the outer ideogram, and each MHC class is marked by color. The MHC class I is shown in red, MHC class II is green, and MHC class III is blue. Each aligned transcript, including novel transcripts, were grouped into co-expression clusters using the normalized read counts from each sequenced individual (n=12). Every transcript is plotted according to position, and colored according to cluster identity (red, dark red, orange, yellow, lime green, green, light blue, dark blue, purple, magenta, pink, black, grey, and brown). Multiple isoforms of the same gene can be found in the same co-expression cluster, but this is not a requirement and is never the case across all isoforms of a gene. There is no localization of transcripts within individual co-expression clusters based on genomic position. Nine of the fourteen clusters, however, were enriched for specific transcription factors, the most significantly enriched transcription factor for each cluster is listed.
We further described transcription factor binding site enrichment in each cluster (Table S6). Of the fourteen co-expression clusters, nine were enriched for specific transcription factors (OR>1, p<0.05). For these clusters, the transcription factor binding sites most significantly enriched were TCF3 (OR: 2.17, p: 8.06 × 10−7), ESR1 (OR: 2.72, p: 2.21 × 10−5), RFX5 (OR: 1.68, p: 4.27 × 10−5), SMARCA4 (OR: 2.56, p: 2.10 × 10−4), GATA1 (OR:2.08, p: 2.25 × 10−4), IKZF1 (OR: 4.80, p: 6.92 × 10−4), GRp20 (OR: 2.57, p: 2.39 × 10−3), CEBPD (OR: 1.52, p: 5.44 × 10−3), and SMARCA4 (OR: 3.68, p: 6.74 × 10−3), respectively. The enrichment of these specific transcription factor binding sites suggests that these nine clusters may show co-expression due to transcription factor-dependent co-regulation.
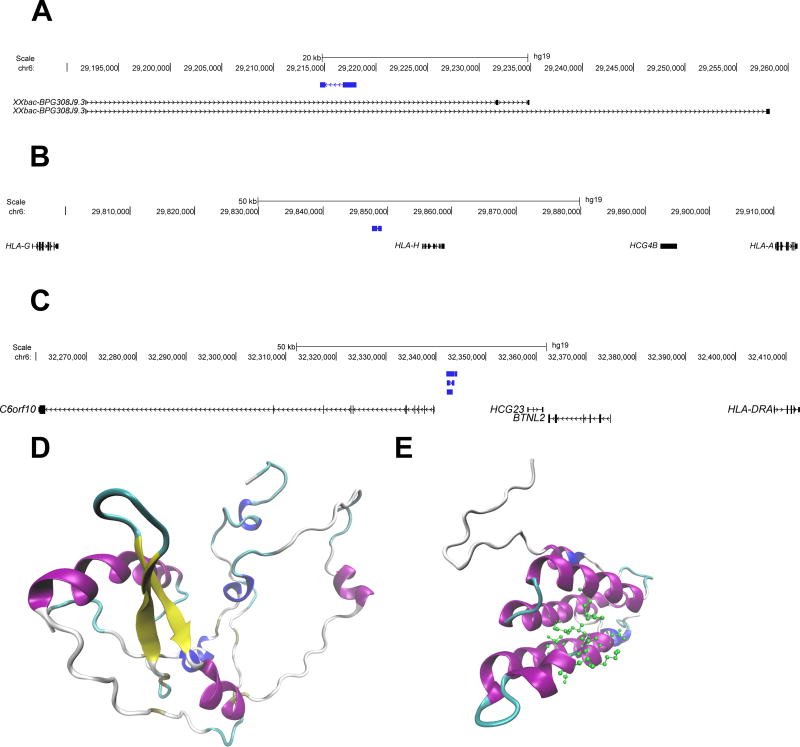
Identification of novel retroviral genes with the human MCH
We identified 3 novel genes with an open reading frame that are predicted to be protein coding within the human MHC region, and demonstrate gene expression at the mRNA level. Using protein function and structure prediction algorithms, two of the three coding genes we identified are predicted with very high and moderate certainty to be novel endogenous retroviral pol and gag genes, respectively (Figure 6). The structure and function of the third gene could not be predicted. The predicted amino acid sequences of all three genes were aligned to the human proteome using protein-protein Blast(26). Based on the homology between each novel sequence and the HERVs to which it is aligned, we predict that these genes are retroviral pol, gag, and gag proteins, respectively (Table S7).
Figure 6.
Genomic position (hg19) and predicted protein structure of the three novel protein-coding genes identified in this study. (A) One protein-coding novel transcript (blue) contained within the intronic region of the gene XXbac-BPG308J9.3. (B) and (C) depicts novel protein coding transcripts (blue) in intergenic regions, near HLA-A and HLA-DRA, respectively. Each of these transcripts shares homology with endogenous retroviral elements. (D) The Predicted protein structure of transcript A (prediction p value= 1.17 × 10−4). This structure shares homology with an endogenous retroviral pol protein, and no predicted ligands are available. (E) The predicted protein structure of transcript C (prediction p value= 0.037). This structure shares homology with a retroviral gag protein, and is predicted to bind to a leucine residue (predicted active site amino acids are shown in green). In both D and E coloring is based on secondary structure: alpha helices are purple, 3–10 helices are blue, beta sheets are yellow, beta bridges are tan, turns are cyan, and coils are white.
Discussion
Variation within the MHC contributes to genetic risk of immune and inflammatory disease. However, this region is characterized by complex variation patterns that complicate identifying causal variants and their direct effects on disease etiology (32). Moreover, these complex variation patterns play a role in the complex alternative splicing and gene regulation networks that have been described in this region (7, 33). Quantification of MHC transcription by RNA sequencing has been limited by both the high rate of polymorphism and the high rate of splice variants, resulting in limitations in RNA sequence alignment (18). Using individualized genomes to map RNA sequencing reads, we accurately measured gene and splice variant expression within the MHC, which can be used to further elucidate the functional effects of variations relevant to disease.
Sequence variation can affect the expression of transcripts by interfering with cis-regulation, such as altering promoter or enhancer activity, altering DNA methylation patterns, or altering the sequence of regulatory RNAs. Variants linked to these cis-effects (cis-eQTL) affect expression in an allele-specific manner. Haplotype-specific gene expression within the HLA, and allelic imbalance linked to cis eQTLs in autoimmunity have been previously described (4, 34). Our findings suggest extensive allele-specific expression throughout the MHC, which involves 88% of all transcribed SNPs in this region.
Many lincRNA transcripts are expressed at low levels, rendering them undetectable without sequence enrichment. By targeting the MHC region using sequence-specific capture probes, we identified novel noncoding transcripts throughout the region. As lincRNAs have been implicated in transcriptional regulation, this suggests a far more complex regulatory network within the MHC than what has been previously described. Variation within the MHC further affects the patterns of transcription regulation, due to allelic imbalance as we demonstrate.
The genetic association of polygenetic diseases within the HLA is complex, and often the identification of causal genetic variants is complicated by the extensive linkage disequilibrium within this region. While specific amino acid residues and classical HLA allelotypes have been considered to contribute to disease pathogenesis in several immune-mediated diseases, our data highlight the importance of including regulatory effects of these disease-associated polymorphisms to better understand the functional role of genetic variants within the HLA.
When we compare the expression patterns of all transcripts across all twelve sequenced individuals, a pattern of co-expression develops. While the co-expression of genes does not intrinsically imply co-regulation, regulation by the same transcription factors is one mechanism by which co-expression can occur. After quantifying the enrichment of the transcription factors binding to the promoter regions of the transcripts in each cluster, we found that nine of the fourteen clusters were enriched for specific transcription factors. This suggests that regulation by these transcription factors may play a role in the expression patterns of the transcripts in each cluster. Some of the enriched transcription factors identified play a role in specific immunological processes. For example, one of these co-expression clusters was found to be enriched for RFX5, a transcription factor that activates MHC class II expression by enhancing CIITA activity(35). Another transcription factor, enriched in a different cluster, CEBPD is directly involved in promoting macrophage activation, M1 macrophage polarization, and pro-inflammatory cytokine production in macrophages(36). The transcription factor GATA1 is involved in dendritic cell differentiation and survival(37). These enriched transcription factors each have a unique role in monocyte differentiation, suggesting that they have a role not only in determining co-expression patterns of transcripts, but in downstream determination of cellular phenotypes.
We found five novel putative coding transcripts, identifying three novel human endogenous retroviral elements (ERVs). ERVs comprise 8% of the human genome(38). Though mutations have silenced the expression of the majority of these elements, approximately 7% of all known ERVs are transcriptionally active(39). Moreover, mutations in these elements have been linked to diseases, including cancer(33) and multiple sclerosis (40). Translated ERVs have been shown to play a role in lymphocyte activation, and transcribed ERVs play a role in transcriptional regulation (41). The exact function of these novel ERVs, and their precise effects on transcription and immune function, has yet to be fully elucidated.
In summary, we performed deep sequencing of both the genome and the transcriptome, targeting the MHC region with sequence-specific capture probes in human monocytes. We accurately identified and quantified the expression of 908 novel transcripts in this region, including 123 transcripts aligning to regions previously thought to be intergenic. In addition, we uncovered extensive allele-specific expression imbalance within the MHC region, which appears to be non-stochastic, suggesting complex cis-acting transcriptional regulation throughout the human MHC. This allelic imbalance can have functional consequences upon disease risk loci within the MHC region.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Institute Of Arthritis And Musculoskeletal And Skin Diseases (NIAMS) of the National Institutes of Health under Award Number R01AR070148.
Footnotes
Competing Interests
None of the authors have any conflicts of interest to disclose.
Author contributions
E.G. contributed to experimental design, performing the experiments and analyzing the data, and drafting the manuscript; W.W. contributed to performing data analysis, and editing the manuscript; A.H.S. conceived the study, contributed to experimental design, data analysis, and drafting the manuscript.
References
- 1.Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hosomichi K, Shiina T, Tajima A, Inoue I. The impact of next-generation sequencing technologies on HLA research. J Hum Genet. 2015;60:665–673. doi: 10.1038/jhg.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deitiker P, Atassi MZ. MHC Genes Linked to Autoimmune Disease. Critical reviews in immunology. 2015;35:203–251. doi: 10.1615/critrevimmunol.2015014510. [DOI] [PubMed] [Google Scholar]
- 4.Raj P, Rai E, Song R, Khan S, Wakeland BE, Viswanathan K, Arana C, Liang C, Zhang B, Dozmorov I, Carr-Johnson F, Mitrovic M, Wiley GB, Kelly JA, Lauwerys BR, Olsen NJ, Cotsapas C, Garcia CK, Wise CA, Harley JB, Nath SK, James JA, Jacob CO, Tsao BP, Pasare C, Karp DR, Li QZ, Gaffney PM, Wakeland EK. Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. Elife. 2016;5 doi: 10.7554/eLife.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes T, Coit P, Adler A, Yilmaz V, Aksu K, Duzgun N, Keser G, Cefle A, Yazici A, Ergen A, Alpsoy E, Salvarani C, Casali B, Kotter I, Gutierrez-Achury J, Wijmenga C, Direskeneli H, Saruhan-Direskeneli G, Sawalha AH. Identification of multiple independent susceptibility loci in the HLA region in Behcet's disease. Nature genetics. 2013;45:319–324. doi: 10.1038/ng.2551. [DOI] [PubMed] [Google Scholar]
- 6.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, Ellis P, Langford C, Vannberg FO, Knight JC. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJ, Smolonska A, Weersma RK, Hofstra RM, Buurman WA, Rensen S, Wolfs MG, Platteel M, Zhernakova A, Elbers CC, Festen EM, Trynka G, Hofker MH, Saris CG, Ophoff RA, van den Berg LH, van Heel DA, Wijmenga C, Te Meerman GJ, Franke L. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015;21:2007–2022. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, Sun H, Wang H. Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci. 2016;6:45. doi: 10.1186/s13578-016-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell. 2012;45:470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer TR, Clark MB, Crawford J, Brunck ME, Gerhardt DJ, Taft RJ, Nielsen LK, Dinger ME, Mattick JS. Targeted sequencing for gene discovery and quantification using RNA CaptureSeq. Nat Protoc. 2014;9:989–1009. doi: 10.1038/nprot.2014.058. [DOI] [PubMed] [Google Scholar]
- 12.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43 doi: 10.1002/0471250953.bi1110s43. 11 10 11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R S. Genome Project Data Processing. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 20.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature biotechnology. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic acids research. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGuffin LJ, Atkins JD, Salehe BR, Shuid AN, Roche DB. IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic acids research. 2015;43:W169–173. doi: 10.1093/nar/gkv236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown NP, Leroy C, Sander C. MView: a web-compatible database search or multiple alignment viewer. Bioinformatics. 1998;14:380–381. doi: 10.1093/bioinformatics/14.4.380. [DOI] [PubMed] [Google Scholar]
- 28.Koboldt DC, Larson DE, Wilson RK. Using VarScan 2 for Germline Variant Calling and Somatic Mutation Detection. Current protocols in bioinformatics. 2013;44 doi: 10.1002/0471250953.bi1504s44. 15 14 11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dozmorov MG, Cara LR, Giles CB, Wren JD. GenomeRunner web server: regulatory similarity and differences define the functional impact of SNP sets. Bioinformatics. 2016;32:2256–2263. doi: 10.1093/bioinformatics/btw169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, Forbes S, Gilbert JG, Halls K, Harrow JL, Hart E, Howe K, Jackson DK, Palmer S, Roberts AN, Sims S, Stewart CA, Traherne JA, Trevanion S, Wilming L, Rogers J, de Jong PJ, Elliott JF, Sawcer S, Todd JA, Trowsdale J, Beck S. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goering W, Schmitt K, Dostert M, Schaal H, Deenen R, Mayer J, Schulz WA. Human endogenous retrovirus HERV-K(HML-2) activity in prostate cancer is dominated by a few loci. Prostate. 2015;75:1958–1971. doi: 10.1002/pros.23095. [DOI] [PubMed] [Google Scholar]
- 34.Vandiedonck C, Taylor MS, Lockstone HE, Plant K, Taylor JM, Durrant C, Broxholme J, Fairfax BP, Knight JC. Pervasive haplotypic variation in the spliceo-transcriptome of the human major histocompatibility complex. Genome Res. 2011;21:1042–1054. doi: 10.1101/gr.116681.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reith W, LeibundGut-Landmann S, Waldburger JM. Regulation of MHC class II gene expression by the class II transactivator. Nat Rev Immunol. 2005;5:793–806. doi: 10.1038/nri1708. [DOI] [PubMed] [Google Scholar]
- 36.Ko CY, Chang WC, Wang JM. Biological roles of CCAAT/Enhancer-binding protein delta during inflammation. J Biomed Sci. 2015;22:6. doi: 10.1186/s12929-014-0110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutierrez L, Nikolic T, van Dijk TB, Hammad H, Vos N, Willart M, Grosveld F, Philipsen S, Lambrecht BN. Gata1 regulates dendritic-cell development and survival. Blood. 2007;110:1933–1941. doi: 10.1182/blood-2007-05-091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J C. International Human Genome Sequencing. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 39.Oja M, Peltonen J, Blomberg J, Kaski S. Methods for estimating human endogenous retrovirus activities from EST databases. BMC Bioinformatics. 2007;8(Suppl 2):S11. doi: 10.1186/1471-2105-8-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brutting C, Emmer A, Kornhuber M, Staege MS. A survey of endogenous retrovirus (ERV) sequences in the vicinity of multiple sclerosis (MS)-associated single nucleotide polymorphisms (SNPs) Mol Biol Rep. 2016;43:827–836. doi: 10.1007/s11033-016-4004-0. [DOI] [PubMed] [Google Scholar]
- 41.Suntsova M, Garazha A, Ivanova A, Kaminsky D, Zhavoronkov A, Buzdin A. Molecular functions of human endogenous retroviruses in health and disease. Cell Mol Life Sci. 2015;72:3653–3675. doi: 10.1007/s00018-015-1947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.