Preface
How can we treat cancer more effectively? Traditionally, tumours from the same anatomical site are treated as one tumour entity. This concept has been challenged by recent breakthroughs in cancer genomics and translational research enabling molecular tumour profiling. The identification and validation of cancer drivers, which are shared between different tumour types, spurred the new paradigm to target driver pathways across anatomical sites by off-label drug use, or within so called “basket or umbrella trials”, which are designed to test whether molecular alterations in one tumour entity can be extrapolated to all others. However, recent clinical and preclinical studies suggest that there are tissue- and cell type-specific differences in tumourigenesis and the organization of oncogenic signalling pathways. In this Opinion article, we focus on the molecular, cellular, systemic and environmental determinants of organ-specific tumourigenesis and mechanisms of context-specific oncogenic signalling outputs. Investigation, recognition and in-depth biological understanding of these differences will be vital for the design of next-generation clinical trials and the implementation of molecularly-guided cancer therapies in the future.
Introduction
In the past three decades, molecular tumour profiling and functional studies have led to the identification and validation of critical genes and pathways, which are dysregulated or mutated in specific tumour types. In parallel, a continuously increasing toolbox of rationally targeted drugs has been developed, which block some of these cancer drivers with high efficacy. In biomarker guided early clinical trials several targeted drugs showed unparalleled activity and became the gold standard of care for patients with the matching molecular tumour profile1. Examples for successfully targeted cancer drivers include ERBB2 overexpression in breast cancer, BRAF mutations in melanoma, ABL1 rearrangements in chronic myeloid leukaemia (CML), KIT mutations in gastrointestinal stroma tumours (GIST), and anaplastic lymphoma kinase (ALK) rearrangements or epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC)2–9. They clearly demonstrate the feasibility and power of biomarker driven precision medicine.
Traditionally, treatment decisions in cancer care are made by organ-specific multi-disciplinary tumour boards, in which experts in different disciplines, such as medical oncology, radiation oncology and pathology review and discuss the medical condition and treatment options of a patient together with experts in the respective cancer entity, e.g. abdominal surgical oncology, gastroenterology, gynaecology, or urology. Recommendations are based on histology, practical treatment options and evidence-based medicine, in which adequately powered phase 3 randomized controlled trials (RCT) provide robust data regarding treatment efficacy and safety. In the last decade, technologies for large scale genomic profiling rapidly evolved enabling the comprehensive analysis of cancer genomes across different tumour entities1. This revealed that similar cancer driver mutations initially discovered in a single tumour entity are present also in tumours of other anatomical sites. For example, oncogenic BRAF mutations occur in 100% of hairy cell leukaemias, ~50% of melanomas, ~50% of papillary thyroid cancers, ~10% of brain tumours, ~10% of colorectal cancers (CRCs) and with lower frequency in a variety of other cancer types10, 11.
Because cancer drivers are shared between different tumour types, it has been proposed that the traditional classifications of cancers based on the tissue or organ of origin should be replaced by a new classification according to the molecular phenotype based on molecular alterations shared by tumours across different tissue types12–14. They might represent common targetable vulnerabilities irrespective of the cell or tissue of origin. This led to the idea to extrapolate and generalize the use of targeted drugs across anatomically distinct cancer types after initial proof of efficacy in one tumour type. Consequently, oncologists are increasingly using molecularly targeted drugs off-label and patients are included into so called “basket or umbrella trials”, which enrol patients with anatomically different cancer types that share a specific molecular alteration that is thought to be responsive to a specific drug15–20. A prominent example of an “umbrella trial” is the recently launched National Cancer Institute - Molecular Analysis for Therapy Choice (NCI-MATCH) study, which will examine tumour biopsy specimens from as many as 5,000 patients to identify potentially druggable targets. This trial will provide important insights regarding the feasibility and effectiveness of such novel clinical trial designs21, 22. Such trials are an important step forward to identify subgroups of patients that respond to molecularly targeted therapies and discover associated biomarkers of response, as well as elucidating mechanisms of primary treatment resistance19, 23. In addition, such trials will generate a rich resource needed to direct future basic and translational cancer research to generate data-driven human cancer models and investigate mechanistically the biological implications of co-occurring and mutually exclusive genetic alterations 19, 23.
However, recent basket trials provide evidence that the response to a molecular alteration-specific anti-cancer drug often depends on the anatomical cancer type; in addition, off-label use of targeted therapies across different tumour entities may not be superior to standard of care12, 14, 17, 18. For example, drugs targeting the same oncogenic BRAFV600E mutation showed unprecedented efficacy in melanoma, NSCLC and hairy cell leukaemia, but failed in BRAFV600E mutated CRC7, 12, 24. Therefore, the assumption that a driver mutation behaves similarly across different tumour entities may not be generally valid. In line with this, there is compelling experimental evidence that oncogenic drivers and the organization of oncogenic signalling pathways are tissue-specific and are therefore important determinants of treatment response and resistance.
In this Opinion article we summarize molecular, cellular and systemic determinants of tissue-specific tumour development. We show that the signalling output of an oncogenic driver can differ substantially between tissue types, and provide selected examples that describe how environmental factors shape the tissue-specific signalling of cancer genes. We discuss the implications of tissue context for the design of molecularly targeted therapies and advocate the importance of studying the biology and targeting of cancer in a multi-dimensional way. It is our opinion that therapy should account for various aspects, including the organ type, environmental context and genetic confounders, such as co-occurring mutations. Because our understanding of tissue specific oncogenic signalling is still in its infancy, we stress the need for investigating the biology of each potential cancer driver in its tissue context. This has the potential to inform and guide clinical trial design, to enrol the right patient to the right ‘basket’. Combining the molecular tumour profile with the tissue type will increase treatment efficacy and pave the way towards molecularly precise therapies.
Tissue-specific tumourigenesis
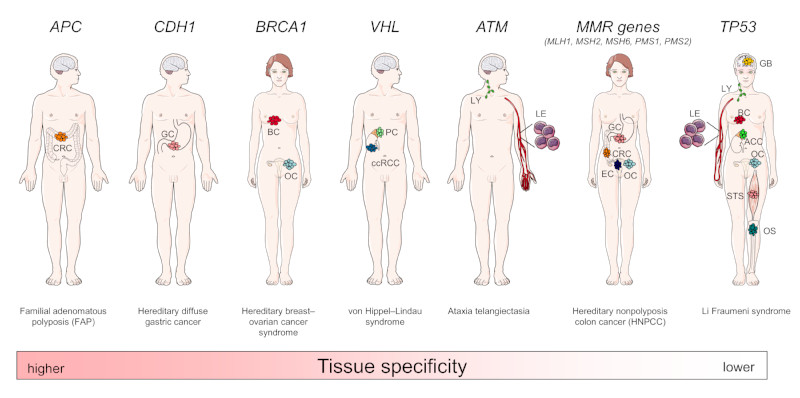
Different cell and tissue types show profound differences in their response to oncogenic driver mutations25, 26. Such differences are most obvious in hereditary cancer predisposition syndromes such as familial adenomatous polyposis (FAP) caused by adenomatous polyposis coli (APC) germline mutations27, 28. Affected persons have a nearly 100% lifetime risk to develop CRC, whereas other tumour types are rare. Similar associations are true for other germline mutations, such as those affecting BRCA1 and BRCA2 (which cause hereditary breast and ovarian cancer syndrome), cadherin 1 (CDH1) (also known as E-cadherin; which cause hereditary diffuse gastric cancer syndrome), RB1 (Retinoblastoma), von Hippel-Lindau tumour suppressor (VHL, which causes von Hippel–Lindau Syndrome and clear cell renal cell cancer (ccRCC) predisposition) and KIT (which cause gastrointestinal stromal tumour predisposition)27–29 (Figure 1). These well known examples demonstrate that tissues differ substantially in their susceptibility towards specific oncogenic events and that barriers to tumour formation are highly tissue-specific. However, other genetic alterations, such as TP53 germline mutations, which cause Li-Fraumeni syndrome, are associated with a much broader spectrum of cancer types and are therefore considered a general cancer gene alteration.
Figure 1. Hereditary cancer predisposition syndromes and tissue-specific tumourigenesis.
Gene defects underlying hereditary cancer predisposition syndromes such as alterations in adenomatous polyposis coli (APC), cadherin 1 (CDH1), BRCA1, von Hippel-Lindau tumour suppressor (VHL) and ataxia telangiectasia mutated (ATM) are associated with a high risk to develop tissue-specific cancer types, whereas others, such as DNA mismatch repair (MMR) genes (MLH1, MSH2, MSH6, PMS1, PMS2) or TP53 are associated with cancers from many different tissues of origin. For each syndrome, associated cancer entities with an at least 4-fold increased risk are indicated. CRC, colo-rectal cancer; GC, gastric cancer; BC, breast cancer; OC, ovarian cancer; PC, pheochromocytoma (adrenal gland tumour); ccRCC, clear cell renal cell carcinoma; LY, lymphoid malignancies; LE, leukaemia; EC, endometrial cancer; STS, soft tissue sarcoma; OS, osteosarcoma; ACC, adrenal cortical carcinoma; GB, glioblastoma.
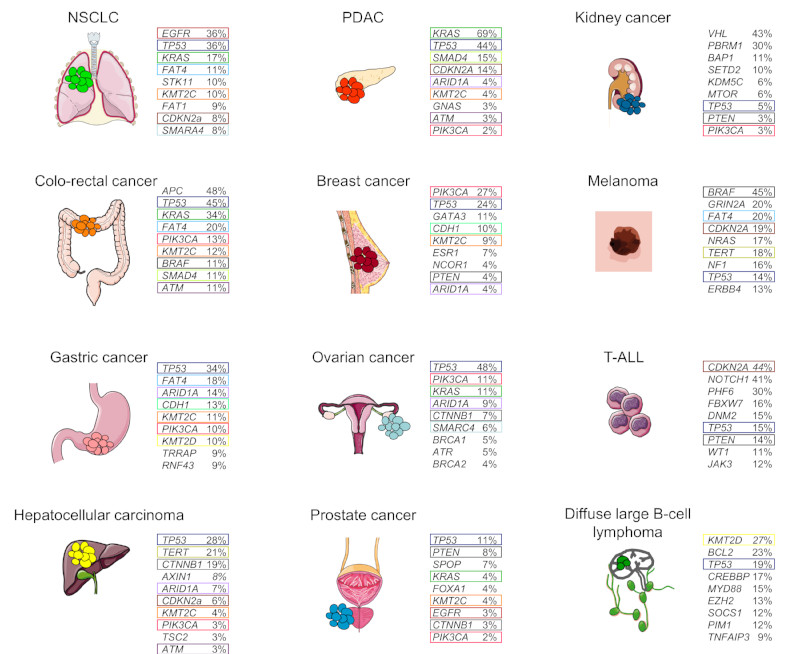
In line with this, a wide variety of sporadic tumours display a predominance of distinct cancer genes based on their site of origin, whereas TP53 mutations are present in many different cancer types (Figure 2). For example, VHL is inactivated in sporadic ccRCC, whereas it is only rarely mutated in other tumour entities30. Other examples are BCR-ABL translocations in CML, APC mutations in CRC, or mutations in the RB1 tumour suppressor gene in small cell lung cancer (SCLC)31–33. These examples of tissue-specific genetic events raise the question about the underlying molecular and cellular mechanisms, allowing or preventing cancer development at different anatomical locations.
Figure 2. Somatic mutation frequencies (single-nucleotides, small insertions or deletions (indels)) in common cancers from the Catalogue of Somatic Mutations in Cancer (COSMIC).
The top 9 mutations occurring in the different depicted common tumour-types are shown. Mutations shared across tumour entities are depicted by coloured boxes. Mutation data were obtained from the COSMIC release version 77 at Welcome Trust Sanger Institute (http://cancer.sanger.ac.uk/cosmic). Please note: only the frequency of somatic mutations (single-nucleotides or indels), but not larger deletions, amplifications or rearrangements are depicted in the figure. NSCLC, non-small cell lung cancer; PDAC, pancreatic ductal adenocarcinoma; T-ALL, T-cell acute lymphoblastic leukaemia. APC, adenomatous polyposis coli, ARID1A, AT-rich interactive domain 1A, ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3-related; BAP1, BRCA1-associated protein 1; CDH1, cadherin 1; CDKN2A, cyclin-dependent kinase inhibitor 2A; CREBBP, CREB-binding protein; CTNNB1, encoding β-catenin; DMN2, dynamin 2; EGFR, epidermal growth factor receptor; ESR1, oestrogen receptor 1; EZH2, enhancer of zeste homologue 2; FAT, atypical cadherin; FBXW7, F-box and WD repeat domain containing 7; GATA3, GATA binding protein 3; GNAS, encoding G protein, GαS; GRIN2A, glutamate receptor ionotropic, NMDA 2A; JAK3, Janus kinase 3; KDM5C, lysine-specific demethylase 5C; KMT2, histone-lysine N-methyltransferase 2; NCOR1, nuclear receptor co-repressor 1; MYD88, myeloid differentiation primary response 88; NF1, neurofibromatosis type 1; PBRM1, polybromo 1; SETD2, SET domain containing 2; PHF6, PHD finger protein 6; SMARCA4, SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily A, member 4; SOCS1, suppressor of cytokine signalling 1; SPOP, speckle-type POZ protein; STK11, serine-threonine kinase 11; TERT, telomerase reverse transcriptase; TNFAIP3, tumour necrosis factor α (TNFα)-induced protein 3; TSC2, tuberous sclerosis 2; TRRAP, transformation/transcription domain associated protein; VHL, von Hippel-Lindau tumour suppressor; WT1, Wilms tumour 1.
The most obvious reason for tissue-specific cancer development could be that expression of the cancer driver is limited to the tissue in which the tumour develops. However, most cancer genes are expressed in a wide variety of tissues and are not restricted to tissues from which the cancer originates26, 34. This suggests the presence and combinatorial action of other factors, such as the tissue-specific oncogenic function of a cancer driver, characteristics of the cell of origin (e.g. stress response, connectivity of signalling pathways, signalling output and compensatory mechanisms) as well as pre-existing or acquired genetic and epigenetic changes25. In addition, cell extrinsic factors such as cell-cell signalling, mosaicism, cooperation and competition between distinct cell types in the context of the respective tumour micro- and macroenvironment, as well as further environmental factors may contribute to the tissue-specificity of individual cancer drivers. Examples of potential molecular and cellular mechanisms that determine which cancer genes are selected during tumour evolution in specific tissue types are discussed below.
Cell of origin, cellular plasticity and transdifferentiation
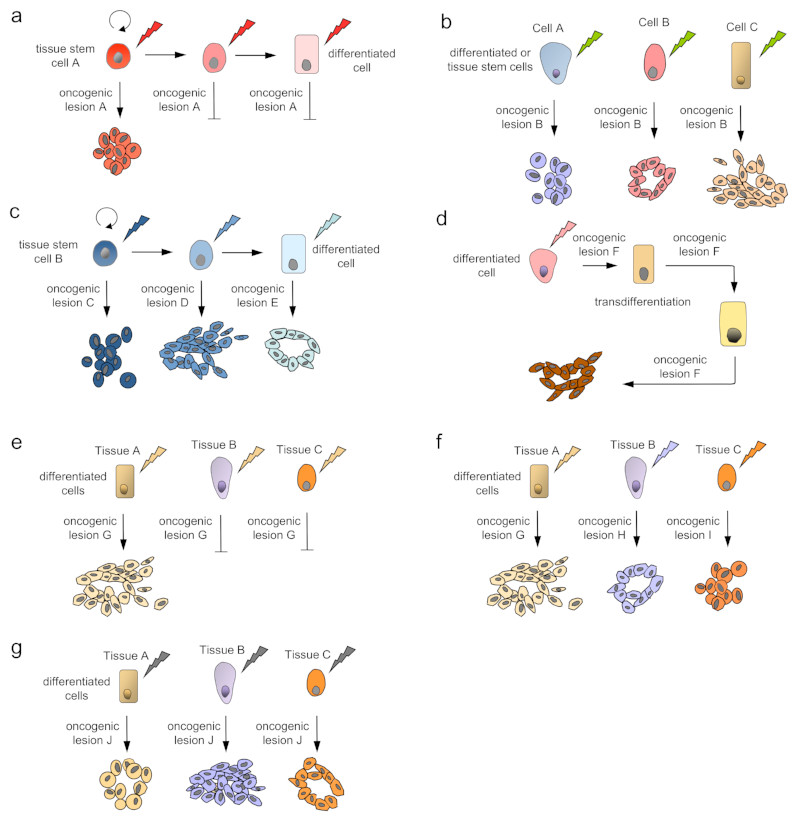
The cell of origin and its differentiation status are important determinants of its susceptibility to oncogenic transformation by a cancer gene35, 36 (Figure 3). Several cancer drivers have context-specific effects on stem and progenitor cell self-renewal, maintenance and lineage commitment (Figure 3a). For example, APC is a critical regulator of the WNT signalling pathway which is essential for intestinal stem cell homeostasis37. Ablation of WNT signalling in mice results in a complete loss of intestinal epithelial cells38, whereas constitutive WNT activation in intestinal stem cells by Apc deletion leads to stem cell-driven intestinal tumourigenesis39. In contrast, Apc inactivation fails to transform differentiated intestinal epithelial cells39.
Figure 3. Models of context-specific carcinogenesis and intertumour heterogeneity based on the cell of origin and its differentiation status.
Genetic and phenotypic variations are observed between individuals with the same tumour-type as well as between tumours of different tissue and cell types. The phenotype of the tumour depends on the specific oncogenic lesion (indicated by a blizzard) as well as the cell of origin (e.g. self-renewing tissue stem cells indicated by curved arrows) in all models. (a) Tumour development in the stem cell compartment. Only stem cells are susceptible to a specific oncogenic event, such as loss of the adenomatous polyposis coli (APC) tumour suppressor in colo-rectal cancer. (b) The same mutation or lesion induces very different tumour phenotypes in different cells of origin of a given tissue type, such as PIK3CA mutations in breast cancer. (c) Different cell populations in the lineage hierarchy, from stem cells to fully differentiated cells, can serve as cells of cancer origin if they acquire the right set of specific mutations, as shown for T-cell acute lymphoblastic leukaemia (T-ALL). (d) Cellular plasticity and transdifferentiation is a driver of tumour development in some tumour types, such as pancreatic ductal adenocarcinoma (PDAC). Here, a specific oncogenic lesion induces the transdifferentiation of acinar cells to a duct-like phenotype and leads to tumourigenesis, which depends on the continuous expression of the oncogenic lesion. (e) Only differentiated cells in a specific tissue type, but not in most others, are susceptible to transformation by a cancer driver, such as von Hippel–Lindau tumour suppressor (VHL) gene mutations in proximal renal tubular epithelial cells that give rise to clear cell renal cell carcinoma (ccRCC). (f) Cells in other tissue types require the right combination of specific molecular alterations to serve as cells of origin of a tumour, such as TP53 and BRCA mutations in serous ovarian cancers that originate from the distal fallopian tube. (g) The same driver lesion induces different tumour types with distinct molecular and phenotypic features, depending on the tissue, in which the oncogene is expressed, such as oncogenic BRAF mutations in melanoma, papillary thyroid cancer and NSCLC.
Other cancer drivers generate divergent phenotypes in different cell types (Figure 3b). The PIK3CA gene (which encodes PI3K catalytic subunit-α) for example, is one of the most frequently mutated genes in human breast cancer and is associated with distinct molecular breast cancer subtypes. Depending on the cell of origin in which the oncogene is initially expressed within the mouse mammary gland, identical Pik3ca mutations induce fundamentally different tumour types with different morphology, growth patterns, invasiveness and aggressiveness40, 41. Multiple differentiated cell types also give rise to KrasG12D-induced NSCLC and the cell of origin influences the NSCLC spectrum and its histopathological phenotype42, 43.
The concept that cancer drivers can have distinct functions in different cell types or at different stages of differentiation has been validated impressively in an unbiased forward genetic transposon-based mutagenesis screen in the lymphoid differentiation lineage36 (Figure 3c). Although transposon mobilization induced T-cell acute lymphoblastic leukaemia (T-ALL) in all cases, there was a remarkable difference in the genes which were activated or inactivated in the different cell types36. These data support the idea that a particular cell of origin may require a unique set of distinct mutations to be able to serve as the cell of origin of a tumour subtype44.
Cellular plasticity is critically involved in tissue-specific tumour development at distinct anatomical sites, such as the pancreas and the skin35, 45–48 (Figure 3d). Acinar to ductal metaplasia (ADM), a first step towards pancreatic ductal adenocarcinoma (PDAC) development, is characterized by the transdifferentiation of acinar cells, which are reprogrammed by oncogenic KRAS to a duct-like phenotype35, 46, 47. The reactivation of embryonic signalling pathways, such as Hedgehog and Notch contributes to cellular plasticity and tumourigenesis47. In basal cell cancer, inducible expression of a constitutively active smoothened mutant in the adult mouse epidermis reprogrammed differentiated epidermal cells into embryonic hair follicle progenitor-like cells and induced cancer formation45. In contrast, the same smoothened mutant was not able to induce ADM or tumour development in the pancreas49 (Figure 3e). These examples support two important conclusions. First, distinct oncogenic drivers are necessary to induce cellular reprogramming and tumour development at specific anatomical sites. Second, reprogramming occurs only in a specific permissive cellular context, explaining the tissue tropism of certain oncogenes and tumour suppressors.
Tumour suppressor barriers
Tumour suppressor barriers are tissue- and context-specific and depend on a variety of interacting signalling molecules50–52. Such signalling networks were demonstrated to play important roles during tumourigenesis, because they trigger tumour suppressor barriers in response to oncogenic stress. For example, PTEN is a negative regulator of the PI3K pathway and a tumour suppressor that is deleted in several cancer types, such as prostate cancer53. In contrast, ablation of Pten in mice suppresses pre-B-ALL development owing to activation of AKT and the tumour suppressor p5354. During normal B cell development, immature B cells bind auto-antigens (self-antigens) through their B cell receptor (BCR), leading to autoreactive BCR signalling and elimination (negative selection)55. In pre-B ALL cells, the absence of PTEN causes strong PI3K/AKT signals, which mimic the negative selection of autoreactive B cells and induce tumour cell death via AKT-mediated activation of the p53 cell cycle checkpoint54. This context is lost in more mature B cell lymphomas, as strong BCR signals lead to proliferation and not elimination of mature B cells56.
Tissue-specific effects have also been described for classical oncogenes, such as mutated RAS family members. Oncogenic KRAS induced cancer development occurs only in specific mouse tissues, such as the lung and the pancreas, whereas most other tissue types resist oncogenic transformation57–60. Importantly, these findings are also reflected in the respective human cancer types (Figure 2). Mechanistically, distinct tumour suppressive pathways are induced by KRAS that contribute to its context-specific oncogenic potency. As an example, oncogenic KRAS triggers the tumour suppressive p19ARF pathway extensively in mesenchymal tissues, such as the musculature, but not in epithelial cells of the lung60. Thereby, KRAS-induced p19ARF expression prevents muscle-derived sarcoma formation in mice60. In line with this, specific deletion of p19 (which is encoded together with p16Ink4a by the cyclin-dependent kinase inhibitor 2a (Cdkn2a) gene) in mice induces a shift of the KRAS-induced tumour spectrum towards sarcomas60. Mechanistically, cell type-specific epigenetic gene regulation of the Cdkn2a locus and thus context-specific expression of its gene products p19ARF and p16INK4A is the critical determinant of this tissue-specific cellular response. Polycomb-group proteins repress p19ARF expression in the lung, whereas the SWI/SNF chromatin-remodelling complex member SNF5 creates a permissive environment for KRAS-induced p19ARF expression in mesenchymal tissues60. This supports the idea that distinct tumour suppressor barriers or signalling thresholds are operative in different tissue types, even in the presence of the same initial oncogenic lesion.
Chromatin organization, replication timing and regulatory elements
Next-generation cancer genome sequencing and functional assays revealed that the rate of somatic mutations varies considerably across the genome of different cell types due to context-specific differences in chromatin organization, DNA accessibility, replication timing and transcription initiation61–66. These effects contribute to the quantitatively and qualitatively different mutation burden between different tumour types. For example, nucleotide excision repair (NER) activity is impaired at active transcription factor binding sites (TFBS). This leads to an increased rate of context-specific DNA mutations in active gene promoter regions of distinct tumours, such as melanoma and lung cancer, that depend on NER for repair of e.g. ultraviolet- or smoke-induced DNA lesions, respectively64, 65. In contrast, cancers that do not rely heavily on NER, such as CRC, show no such enrichment in TFBS mutations65. In adult stem cells of the colon, which are the cells of origin of CRC, context-specific mutation signatures correspond to spontaneous deamination of methylated cytosine (C) residues into thymine (T) at CpG sites, which are strongly associated with replication timing and might reflect the high division rate of colonic stem cells67. Because many point-mutations in CRC driver genes, such as APC, CTNNB1 (encoding β-catenin), TP53 and SMAD family member 4 (SMAD4) are similarly C:G to T:A transitions at CpG dinucleotides67, deamination-induced C to T mutagenesis might be a relevant tissue-specific cancer driver and determinant of point-mutation load in this tumour entity. Consequently, chromatin and epigenomic features of the cell of origin are the best predictors of local somatic mutation densities in a cancer cell and the cell of origin of a cancer can be determined based on the distribution of mutations along its genome62.
The molecular basis of cell-type specific mutations in different RAS family proteins has been uncovered using mouse genetics. KRAS, HRAS and NRAS mutations occur at varying frequencies across different tumour types (Figure 2)68. KRAS is the signature mutation in NSCLC, whereas HRAS mutations are common in skin cancer68. Using an elegant knock-in strategy to express wild-type HRAS from the endogenous Kras locus in mice, it was shown that Hras codon 61 mutations occurred in NSCLC only in the Hras knock-in allele expressed from the Kras locus, but never from the endogenous Hras locus69. These data show that the tissue-specific mechanisms underlying Kras mutations in NSCLC, and Hras mutations in skin cancer involve tissue-specific gene regulatory elements rather than differences in the function of the encoded proteins.
Super-enhancers are another possible determinant of tissue-specific tumourigenesis. They are clusters of regulatory elements that control the transcription of genes and they have been implicated in cell identity70–72. Super-enhancers are bound and controlled by cell-type specific master transcription factors, for example transcriptional effectors of the WNT, transforming growth factor-β (TGFβ), and leukaemia inhibitory factor (LIF) signalling pathways, which are therefore key drivers of cell state and cell-type specific biology73. Cancer cells can acquire super-enhancers at oncogenic drivers through mutation, focal amplification, chromosomal translocation, or overexpression of an oncogenic transcription factor or an epigenetic regulator that controls enhancer activity71, 72, 74. These processes are highly tissue- and context-specific, generating tumour type-specific super-enhancers, which drive cancer development and progression75. For example, in T-ALL, a small mono-allelic insertion that creates a binding site for the haematopoietic transcription factor MYB nucleates the formation of a novel context-specific oncogenic super-enhancer upstream of the T cell acute lymphocytic leukaemia 1 (TAL1) oncogene, thereby driving its aberrant expression76. Additionally, a polymorphism within a super-enhancer element in the first intron of the LIM domain only 1 (LMO1) gene influences specifically neuroblastoma susceptibility and oncogenic addiction to LMO1 through direct modulation of LMO1 expression77. This super-enhancer is not present in cancer cells from other non-neuroblastoma tumour types, such as T-ALL, despite expression of LMO1 in these cells. These examples demonstrate that the cell-type specificity of super-enhancers is widely preserved in cancer and contributes significantly to context-specific tumour formation.
Genetic road to cancer
Genome sequencing studies have identified different classes of complex genomic rearrangements that seem to derive from a catastrophic event, such as chromothripsis, chromoanasynthesis, and chromoplexy78–81. Thus genomes can acquire multiple complex aberrations by a single event rather than by sequential multistep carcinogenesis. The frequency of such catastrophic events varies substantially between different tumour entities, ranging from 0% in head and neck cancer and 1.3% in multiple myeloma, to 32% in oesophageal adenocarcinoma and 38% in glioblastoma82, 83. Some tumour subtypes, such as sonic hedgehog-driven medulloblastoma with mutant TP53, always display chromothripsis83. Chromothripsis can be triggered by various mechanisms, such as high energy ionising radiation, double-strand breaks generated by exogenous agents and/or toxins or replicative stress, aborted apoptosis, or trapped chromosomes within a micronucleus resulting in defective DNA replication83, 84. Such complex rearrangements of chromosomes can cause disruption of tumour suppressors, gene fusions, and amplification of oncogenes85. It is reasonable to assume that differences in the genetic road to cancer have context-specific implications on tumour-driving pathways as well as on therapeutic responses and the development of therapy resistance83, 86. Indeed, chromothripsis has been associated with poor prognosis in melanoma, neuroblastoma and multiple myeloma83. However, whether chromothripsis is indeed a catastrophic single event that drives tumorigenesis and differs substantially from the multi-step process of carcinogenesis remains to be proven experimentally. Another example which demonstrates that distinct roads to cancer may have important therapeutic implications are microsatellite instable (MSI) hypermutated cancers. They are significantly associated with the mismatch repair (MMR) gene deficient CRC subtype (see below) and may be particularly sensitive to immunotherapeutic strategies87.
DNA damage and repair and tolerance of oncogenic stress
DNA repair pathways are significantly associated with distinct cancer types88. For example, CRC has the highest number of DNA MMR gene defects26, 88. MMR seems to be an important mechanism to prevent accumulation of mutations during DNA replication in intestinal stem cells and CRCs arise from rapidly dividing stem cells. In contrast, breast and lung cancers have the highest proportion of altered double-strand break repair (DSBR) genes26, 88. Estrogen and tobacco smoke can induce DNA DSBs under certain conditions, selecting for the accumulation of DSBR gene defects in breast and lung cancer, respectively26.
Hormones such as estrogen play an essential role in maintaining cellular identity, and can also drive proliferation of tissues that express the cognate receptor89. Estrogen exposure is an important risk factor for breast cancer development; estrogen receptors are over-expressed in around 70% of breast cancers and blocking estrogen receptor α (ERα) activity greatly reduces breast cancer risk89. Thus estrogens are clearly drivers of ERα+ breast cancer development. Mechanistically, estrogen-induced DNA DSBs can be mediated by DNA topoisomerase IIβ (TOP2β), which is recruited together with ERα to regulatory sites of target genes90, 91. In addition, conversion of estrogens to genotoxic metabolites is an alternative ERα independent mechanism for estrogen-induced DSBs92.
Recent work suggests that estrogens also play a fundamental role in tolerating otherwise lethal mutations in cancer genes such as BRCA1. Mammary epithelial cells as well as breast cancer cells survive BRCA1 loss due to an estrogen-induced pathway that protects them from reactive oxygen species (ROS)-induced cell death93. In contrast, other tissue types, which do not respond to estrogen, cannot tolerate BRCA1 deficiency. Considering that BRCA1 mutations promote tumour formation almost exclusively in hormone-responsive tissues such as breast and ovary27, 28, the link between estrogens and the survival of BRCA1-deficient cells provides important mechanistic insights into the tissue tropism of BRCA1-deficient cancers.
Oncogenic signalling in context
In addition to the tissue-specific oncogenic function of cancer genes described above, the context-specific organization of oncogenic signalling pathways adds another layer of complexity, which is poorly understood, but has important therapeutic implications. In the following paragraphs, we discuss selected examples that describe molecular and cellular mechanisms that shape oncogenic signalling pathways in tissue context.
Context-specific organization of oncogenic pathways
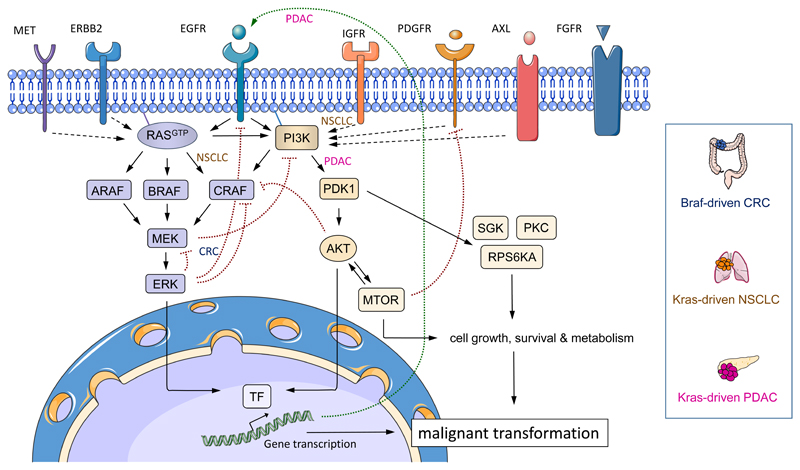
Cellular responses and cell fate decisions are controlled by a limited number of signal transduction pathways. These pathways do not only transmit, but also encode, process and integrate external and internal signals94. Thereby, they fine tune the level of signal propagation and output strength, providing a specific and appropriate response to external stimuli95. Recently, it has become evident that distinct signalling thresholds exist within a cell, which result in diverse and sometimes opposite cellular responses52, 95, 96. The efficacy of drugs targeting distinct cancer driver pathways varies significantly between different cancer entities7, 12, 24. Experimental evidence in genetically engineered mouse models suggests that this is due to tissue-specific signalling outputs (Figure 4). For example, different downstream pathways are engaged by oncogenic KRAS in mouse models of PDAC and NSCLC; signalling via 3-phosphoinositide dependent protein kinase 1 (PDK1) is essential for PDAC development, whereas CRAF is crucial for NSCLC formation59, 97, 98.
Figure 4. Determinants of context-specific oncogenic signalling networks.
Schematic overview of RAS–RAF–MEK–ERK- and PI3K–3-phosphoinositide dependent protein kinase 1 (PDK1)–AKT–MTOR-regulated signalling networks in cancer. Receptor tyrosine kinases (RTK) and RAS, RAF or PI3K oncoproteins signal through both pathways in a context-specific fashion to drive cancer initiation, progression and maintenance. As examples for context-specific oncogenic signalling mechanisms, BRAF-driven colorectal cancer (CRC), KRAS-driven non-small cell lung cancer (NSCLC), and KRAS-driven pancreatic ductal adenocarcinoma (PDAC) specific signalling pathways are highlighted. Signalling output is enhanced by the tissue-specific positive feedback activation of RTKs (e.g. epidermal growth factor receptor (EGFR); long dotted arrow) or other RTKs that are engaged by autocrine and paracrine stimuli. Tissue- and cell-type specific negative feedback loops and inhibitory as well as activating cross-signalling exist at various levels. Activating pro-tumourigenic signalling connections of the canonical signalling pathways are depicted as arrows, inhibitory anti-tumourigenic pathways are shown as dotted lines headed by a perpendicular line. Dotted arrows depict activating pro-tumourigenic signalling loops regulated by the tissue-specific engagement of RTKs. FGFR, fibroblast growth factor receptor; IGFR, insulin-like growth factor receptor; PDGFR, platelet-derived growth factor receptor; PKC, protein kinase C; RPS6KA, ribosomal protein S6 kinase α; SGK, serum and glucocorticoid-regulated kinase; TF, transcription factors.
Importantly, amplification of a signal to a certain level, that is the signalling threshold, seems to be necessary to induce cancer. This threshold differs with tissue type, cell differentiation stage and stage of tumourigenesis, and is often mediated by context-specific engagement of protein kinases and their downstream effector pathways99.
Increasing signal output is sometimes achieved by amplification of the driving oncogene in cancer types such as NSCLC100, but can also be achieved by engagement of autocrine feed-forward loops or upstream signals from receptor tyrosine kinases. Context-specific auto- and paracrine signalling loops are critical amplifiers of tumour-driving pathways in several tumour types, such as breast and ovarian cancer, NSCLC, PDAC and CRC99, 101–106. In the pancreas, oncogenic KRAS induces an autocrine feed-forward loop that activates EGFR, which is necessary to amplify KRAS signalling output to reach a critical threshold necessary for tissue transformation106–109. However, deletion of Egfr in mouse models of KRAS-driven NSCLC or CRC fails to prevent tumorigenesis, unlike KRAS-driven PDAC development that requires EGFR signalling106. In line with this, KRAS mutations are predictors of primary resistance towards EGFR inhibition in patients with NSCLC and CRC, but not PDAC 110–113. Indeed, KRAS-driven NSCLC depends on the coordinate input from oncogenic KRAS as well as insulin-like growth factor receptor 1 (IGFR1), but not EGFR105 (Figure 4).
These results demonstrate that the cellular output of KRAS signalling is highly tissue-specific. When considering therapeutic responses, such findings indicate that treatment efficacy cannot be extrapolated from one KRAS-driven tumour entity to another. For example, treatment of some Kras-mutant lung and pancreatic tumour models with the MEK inhibitor Trametinib uncovered a cell autonomous fibroblast growth factor receptor 1 (FGFR1)-dependent survival pathway, which is not present in Kras-mutant CRC cells114. Furthermore, depending on the cellular context, fibroblast growth factor (FGF) signalling can act via autocrine feed forward loops (i.e. in lung cancer), or as a paracrine mediator of stromal–epithelial interactions via secretion of the ligand from the tumour microenvironment (i.e. in prostate cancer)115–117. The outcome of paracrine FGF signalling is also tissue specific; the pathway drives tumour development in the prostate due to the paracrine upregulation of androgen receptor signalling and AKT activation115–117, whereas it blocks tumour progression in a patched 1 (Ptch1) mutant mouse model of medulloblastoma via inhibition of oncogenic sonic hedgehog signalling118.
All of these findings are supported by recent data showing that receptors often act as cell-type specific mediators and amplifiers of signalling pathways101. Thus, different levels of signalling pathway dysregulation and output exist between cancer types of distinct tissue origin, which impact on signalling organization, such as downstream signalling, signalling crosstalk and signalling loops.
Inhibitory crosstalk between different pathways as well as negative feedback loops affecting the same pathway play a central role in health and disease119, 120. Such inhibitory signalling-circuits fine-tune signalling output under physiological conditions to provide an appropriate response to external stimuli, e.g. growth factors95, 96, 119, 121, 122. Interestingly, persistence of feedback inhibition is often preserved in cancer and specific to the tumour cell of origin122. In other cancers, downregulation of modulators of these negative feedback programs or additional mutational hits that bypass negative feedback, such as inactivation of phosphatases (i.e. PTEN), may also occur during tumourigenesis 123. The context-specific preservation of negative feedback provides an explanation for oncogene addiction to maintain a certain level of signalling output, which counteracts intrinsic feedback inhibition122, 124. This has significant consequences for the design of molecularly-targeted therapies because the blockade of oncoproteins or their downstream effector pathways might impact on negative feedback loops and inversely increase signal output in a tissue-specific manner24, 125. For example, in CRC, pharmacological blockade of the BRAF oncogene decreases a negative feedback loop that would otherwise block EGFR signalling. EGFR signalling is subsequently activated allowing CRC cells to proliferate via an EGFR-induced PI3K–AKT-pathway24 (Figure 4). These findings contrast with BRAF-driven melanoma, where BRAF blockade inhibits MAPK activation without impacting on EGFR signalling7, 24. Therefore, tissue-specific EGFR and PI3K signalling seems to bypass BRAF inhibition and mediate primary treatment resistance in BRAF mutant CRC24, 126, 127. Consequently, blocking EGFR or PI3K–AKT signalling together with BRAF inhibition is an effective treatment strategy in preclinical models of CRC24, 126. These data show that tumours of distinct tissue origin are driven by complex nonlinear signalling dynamics even if they engage an identical activating driver mutation, such as BRAFV600E. Taken together, negative feedback is an important tissue-specific mechanism in cancer used to fine-tune oncogenic signalling output and thereby increase the fidelity of information transmission. This tight control of pathway activation contributes to a permissive window of context-specific tumourigenesis25. For most cancer types and oncogenes, context-specific signalling loops, feedback mechanisms and the signalling crosstalk of cancer driving pathways remain largely unknown. Understanding these mechanisms will be vital for the development of more efficient therapies in the future.
Additivity, epistasis and historical contingency of cancer genes
Tissue-context specific relationships between molecular alterations, such as co-occurrence or mutual exclusivity of mutations, have been observed in many cancer types, but in most cases the underlying biological principles and the therapeutic consequences are to date unclear. As described above, KRAS mutations have a different impact on oncogenic signalling organization in distinct tumour types such as CRC, PDAC and NSCLC59, 98, 106, 128–130. The phenotype of these KRAS-driven tumour types is further modified by tissue-specific co-occurring mutations in other cancer genes, such as loss-of-function mutations in the serine-threonine kinase 11 (STK11; also known as LKB1) in NSCLC131. The co-occurrence of mutant KRAS and STK11 determines distinct biological features in KRAS-driven NSCLC, such as differences in pathway activation and immunogenicity, as well as therapeutic vulnerabilities131.
Co-occurring mutations may act additively or epistatically during the course of cancer development132–134. They are considered additive when the genes do not interact and their biological consequences are the sum of the single effects. However, additive effects are relatively rare in cancer132, 134. Most genes exhibit at least some level of epistatic interaction, having greater or weaker consequences in combination than expected from their individual effects132, 134–136. Thereby, epistatic interactions shape signalling pathways in cancer by permitting some and blocking others. The quantitation of epistatic interactions in vivo in complex situations, such as cancer, which is characterized by dynamically changing environmental conditions, remains a major challenge, and novel approaches to accurately measure such effects are urgently needed.
Using data from > 3,000 cancers, it has been shown that mutual exclusivity and co-occurrence of cancer drivers is frequent and tissue-specific134. More than 90% of cancer driver interactions have only been detected in a single tumour type and at least half of the cancer gene interactions differ in their interaction strength in different tumour types134. Examples include context-specific co-occurring oncogenic mutations in KRAS and PIK3CA in CRC, EGFR amplification and O6-methylguanine-DNA methyltransferase (MGMT) methylation in NSCLC, and mutual exclusivity of EGFR amplification and isocitrate dehydrogenase 1 (IDH1) or TP53 mutation in glioblastoma134. Importantly, these data challenge in part the established concept that genetic alterations impinging on a given molecular pathway tend to be mutually exclusive. Therefore, epistasis may account for the tissue specific accumulation of multiple genetic alterations that act synergistically in a given pathway during tumorigenesis134, 135. Examples include amplifications of MAPK3 (also known as ERK1) in KRAS mutant PDAC (synergistic impingement on the MAPK pathway), co-occurring genetic alterations of KRAS, PIK3CA and PTEN in uterine corpus endometrial carcinoma (synergistic impingement on the PI3K signalling pathway), or EGFR mutations and PTEN deletions in glioblastoma (synergistic impingement on the PI3K pathway) (http://www.cbioportal.org). It will be important to test experimentally, whether epistatic interactions that act synergistically on a given pathway, can be exploited therapeutically by a pathway-focused multiple targeting approach.
Recent work also provides evidence that tissue-specific differences in oncogenic signalling networks, such as feedback loops or signalling cross-talk as well as differences in the cellular environment, affect epistatic interactions133–135. In line with this, epistatic interactions are not only associated with the cell of origin of a tumour, but also affect the context-specific biological functions of the epistatically linked genes and the survival of a cancer patient132. The cell type specificity of epistatic interactions has not only important implications for personalized treatment regimens, but also affects synthetic lethal interactions, where the combination of alterations in two genes leads to cell death, whereas mutation in either gene alone has no effect. The exploitation of synthetic lethality to develop cancer therapies has to consider cell- and tissue-specific epistatic interactions134 as it is predicted that synthetic lethality will be efficient only in a subset of tumours carrying the targeted vulnerability134, 135. This is consistent with data from large-scale screens and trials showing that synthetic lethality strategies that are efficient in a specific cell- or tumour-type fail in others24, 135.
The order in which alterations in cancer genes occur might also be an important determinant of signalling organization and output, as has been shown in a model of BRAFV600E-driven serrated intestinal cancer126. This specific CRC subtype is characterized by a serrated histopathological morphology and progresses through a hyperplasia - serrated adenoma - serrated carcinoma sequence, giving rise to microsatellite-instable sessile cancers. In contrast to the classical CRC progression model described by Vogelstein and colleagues137, which is often initiated by APC mutation and subsequent WNT pathway activation, followed by additional genetic alterations, such as RAS- and MAPK-pathway activation (http://www.cbioportal.org)138, BRAF-driven serrated CRC is initiated by MAPK-signalling amplification, followed by WNT pathway activation during tumour progression126. This “inverted” sequence of MAPK–WNT pathway activation has not only important consequences for oncogenic signalling outputs, such as the specific activation of the p16INK4A and/or p19ARF tumour suppressors in BRAF-driven serrated intestinal carcinogenesis, but might also influence the pathomorphologic (serrated histology), genetic (MSI and DNA hypermethylation of CpG islands) and clinical (poor prognosis) characteristics of this CRC subtype139.
The phenomenon that distinct genetic alterations are beneficial or viable only if other alterations have occurred first, known as historical contingency140–142, differs substantially between different cancer types, such as chronic lymphocytic leukaemia (CLL) and myelodysplastic syndromes (MDS)143, 144. For example, in MDS, a genetic “predestination” exist, in which activation of early cancer drivers, such as genes involved in the RNA splicing machinery, dictate future genetic events and routes of tumour evolution with distinct clinical features and prognostic outcomes143. Mechanistically, historical contingency is exemplified by the tissue-specific interplay between genetic alterations in MYC and BCL-2145–147. The anti-apoptotic proto-oncogene BCL-2 is activated by translocation in a variety of B-cell lymphomas147–149. MYC hyperactivation induces apoptosis of B-lineage cells, but BCL2 overexpression represents one possibility to block this effect and permit oncogenic MYC to drive the tumour147, 150, 151. In a different tissue type, the epidermis, MYC activation is well tolerated by keratinocytes. Here, MYC triggers proliferation, hyperplasia and tumourigenesis, but only very low levels of apoptosis152, 153. Presence of paracrine survival signals in the skin might be the determinant of this distinct tissue-specific apoptotic threshold153.
In human CRC and PDAC, TP53 is commonly inactivated at the transition from high-grade intraepithelial neoplasia to carcinoma, rather than at an earlier stage137, 154. This contrasts with breast and liver cancer, where mutations in TP53 occur early155. This observation points to distinct functions of TP53 in controlling invasiveness of tumour cells in CRC and PDAC126, and potentially cell cycle arrest in breast cancer135, 156, 157. Feedback, signalling crosstalk, environmental factors and complex nonlinear signalling dynamics might dictate the tissue-specificity and timing of such sequential processes. These examples illustrate that exploiting the occurrence of cancer drivers for improved therapies needs to consider the evolution of the cell-type specific signalling networks that act during tumorigenesis.
Environmental factors
Tissue-specific tumourigenesis and context-specific oncogenic signalling pathways are also influenced by non-cell autonomous factors, such as the tumour micro- and macroenvironment, metabolism, the microbiota, acute and chronic inflammatory processes, infection and immunity, as well as on environmental chemicals and toxins26, 158.
The tumour microenvironment
The tumour microenvironment (TME) is an important mediator and modulator of oncogenic signalling pathways159–162 and there are clearly tissue-specific differences in the microenvironment of epithelial cells163–166. One of the best examples of TME heterogeneity is the presence of myofibroblast-like stellate cells specifically in the liver and pancreas164. These cells are an important pathogenic driver of tissue fibrosis in both organs164, 167, 168. After activation by tissue damage or specific oncogenic insults, stellate cells secrete extracellular matrix (ECM) components, proteases, cytokines and growth factors which induce tissue repair, but can also drive tumour formation and cancer desmoplasia164, 167–169. Thus, the prominent desmoplastic stroma reaction can distinguish some tumour entities, such as PDAC from other tumour types, such as sarcomas, even if they are driven by the same oncogene59, 170–173. It remains to be determined if tissue-specific differences exist in the composition of (myo)fibroblast subtypes and lineages, which might promote or restrain tumour development and influence TME heterogeneity160, 166, 174–178. However, this seems likely, as different types of fibroblast have been identified in the skin, which respond differentially to paracrine signals, such as sonic hedgehog or TGFβ175, 179.
A context-specific role for TGFβ signalling in stromal fibroblasts has been shown during tumour initiation180, 181. Global inactivation of the Tgfβ receptor 2 (Tgfbr2) in all fibroblasts of the mouse induced tumourigenesis only in the prostate and forestomach180. In this case, increased hepatocyte growth factor (HGF) secretion by Tgfbr2 knock-out fibroblasts seems to be the underlying mechanism that activates MET in epithelial cells to initiate carcinogenesis specifically in the prostate and forestomach180. Therefore, fibroblasts can increase the oncogenic potential of adjacent epithelial cells in specific tissue types. Because cancer associated fibroblasts can secrete high amounts of HGF in human tumours166, these findings support the view that the successful outgrowth of transformed cells is not only dependent on their molecular alterations, but also on the advantage a given microenvironment confers.
Most adult tissues are continuously being renewed from stem cells165. Dependent on the tissue type, resident stem cells can either receive self-renewal factors from specific cells in their local microenvironment or can generate those signals by themselves165. This might have important consequence for tissue-specific tumorigenesis. In the intestine, mesenchymal cells supply WNT proteins to maintain intestinal stem cells, indicating that in the gut, stem and niche cells are functionally paired165. In contrast, epidermal interfollicular stem cells in the skin produce their own WNT ligands, which are required for self-renewal165. In line with this, stemness of CRC depends on secreted factors from activated myofibroblasts, that overactivate the WNT pathway to drive tumour progression162. This suggests that targeting the TME might be an attractive tissue-specific therapeutic option specifically in CRC by switching off the supply of self-renewing factors to cancer cells. Taken together, these data support the idea that TME heterogeneity is an important functional determinant that drives tissue-specific tumourigenesis and represents an attractive target for therapeutic interventions in specific tumour types.
However, there is still a lack of mechanistic understanding of the complex interplay between tumours and their micro- and macroenvironment and how this interplay affects treatment response and resistance. This is in part due to a lack of appropriate methods to target and analyse specific cell types in vivo. Recent developments, such as the generation of dual-recombinase based systems, allowing highly controlled independent genetic manipulation of specific cell types in whole animals182, 183, provide a means of investigating the co-operation and competition between different cell (sub)populations and the role of mosaicism in cancer183–185. With the use of such models and other systems184, it will also be possible to investigate tissue and cell type-specific cell-cell communication and the context-dependent relationship of gene dose to signalling response to determine signalling outputs more quantitatively.
The tumour macroenvironment and metabolism
Overweight, obesity and type 1 or type 2 diabetes increase cancer risk and death from specific cancer types, such as colon and pancreas, suggesting that distinct organ-specific mechanisms are operative186–190. In normal pancreas, obesity promotes steatosis, inflammation and fibrosis189, 191. In mouse models of PDAC, obesity shapes a specific microenvironment, which is characterized by the accumulation of hypertrophic adipocytes that secrete high amounts of cytokines, such as IL-1β189. This accelerates tumourigenesis, tumour growth and treatment resistance due to stellate cell activation, increased desmoplasia, neutrophil-infiltration and inflammation, which can be blocked by IL-1β inhibition189. This contrasts with the normal intestine and intestinal tumourigenesis, where obesity has distinct effects on stem cell function, but not obesity-related inflammation192, 193. Here, obesity increases the number of intestinal stem cells via peroxisome proliferator-activated receptor δ (PPARδ)-mediated WNT activation, reduces the niche dependency of intestinal stem cells , and induces non-stem cells to form tumours in mice following Apc loss193. Other examples of tissue-specific macroenvironmental cancer-drivers are sex hormones, which are involved in cancers of reproductive tissues, such as the breast and prostate102, 194. Conditional overexpression of the androgen receptor induces oncogenic transformation of the mouse prostate195 and androgen stimulation induces the creation and overexpression of oncogenic ETS fusion genes specifically in human prostate cancer, which cooperates with PI3K pathway activation to drive cancer progression102, 196–200.
Tissue- and context-specific metabolic requirements of cancer cells have been identified recently201, 202. Interestingly, tumour metabolism depends on both the genetic lesions and the tissue of origin202. Cell-type specific metabolic alterations can make tumour cells selectively dependent upon certain nutrients and metabolic pathways, leading to tissue of origin specific therapeutic vulnerabilities. KRAS-driven NSCLCs incorporate circulating branched-chain amino acids (BCAAs) to satisfy their metabolic requirements. This contrasts KRAS-driven PDAC tumours that have a decreased BCAA uptake. Consequently, interfering with this particular metabolic pathway could provide a therapeutic opportunity for NSCLC, but not PDAC203. In addition, metabolic changes in cancers are likely to influence oncogenic signalling organization and output. Targeting metabolic pathways for cancer therapy should therefore account for context-specific metabolic changes in different tumour types.
Infection, inflammation, the microbiome and other environmental factors
Chronic infections due to Helicobacter pylori, hepatitis B and C, Epstein-Barr, or human papilloma viruses as well as chronic inflammatory diseases such as hepatitis, pancreatitis and colitis have been linked to greater risk for cancer of the respective inflamed or infected organs (e.g. oropharynx, stomach, colon, anus, cervix, pancreas, liver)158, 204, 205. Persistent infections and inflammation drive epithelial cell proliferation206, 207. In addition, activated immune cells produce highly reactive molecules containing oxygen and nitrogen, which can damage DNA207, 208. Recent work demonstrates that simultaneous DNA damage and cell division during inflammation leads to cancer because dividing cells are more vulnerable to mutations caused by DNA damage207.
Other environmental factors such as ultraviolet (UV) light exposure or toxins (e.g. smoke) can induce an extraordinary high rate of mutations resulting in the activation of diverse cancer drivers209. Although the developing tumours are subsequently genetically very heterogeneous, they do however, harbour mutational signatures that are associated with the cancer aetiology81. Treatment response to immune checkpoint inhibitors such as cytotoxic T lymphocyte associated antigen 4 (CTLA4), programmed cell death protein 1 (PD-1) and PD1 ligand 1 (PD-L1) antibodies strongly correlates with mutational load, which is the highest in UV-induced melanomas and smoke-induced NSCLC81, 210–213. In addition, MMR-deficient MSI CRC harbour a hypermutation signature and tend to be sensitive to immune checkpoint inhibitors, as discussed above. However, there are also examples of tumours with lower mutation rates, which respond well to immunotherapy. Here, tissue-specific neoantigens, high PD-L1 expression levels, or the context-specific immunobiology of the tumour stroma might influence therapeutic efficacy211, 213–215. A recent study revealed that the cytolytic activity of the local immune cell infiltrate in the tumour microenvironment varies substantially across 18 different tumour types216. It correlates with specific oncogenic signalling pathways, such as WNT or PI3K, neoantigen load and the presence of exogenous or endogenous viruses, as well as with sensitivity to immunotherapy and overall survival211, 216–218. Taken together, these examples show that the underlying pathogenic mechanisms of tissue-specific tumour development often have important therapeutic implications in the clinic.
The role of the microbiota in tumour initiation and maintenance and its influence on anti-tumour immuno-surveillance is an area of active research158, 219. It has been shown that distinct patterns of the intestinal microbiome drives or restrains intestinal cancer formation as well as treatment response and resistance219–223. In addition, there is an increasing body of evidence that differences in the 'estrobolome', the aggregate of enteric bacterial proteins capable of metabolizing estrogens, can substantially affect ER-positive breast cancer development224. Therefore, a role for the microbiome in tissue-specific cancer formation and oncogenic signalling output is an important possibility that remains to be addressed experimentally.
Perspectives
As outlined in this article, multiple factors and their dynamic interactions determine tumour driver selection and oncogenic signalling organization in a highly tissue- and context-specific manner. This indicates that therapeutic success of personalized cancer care cannot always be or extrapolated from efficacy in other tumour types harbouring the same molecular alteration. Rather, there is accumulating evidence that the biology of each potential cancer driver should be investigated in its tissue context to provide the scientific rationale to use molecular tumour profiling to guide drug development, patient stratification and treatment decisions.
How can we address this need? It is clear that we cannot explore the tissue- and context-specific vulnerability of each cancer driver in clinical trials. First, often only small numbers of patients harbour the respective driver lesion especially in rare diseases making adequately powered clinical studies impossible; second, we do not have appropriate clinically approved drugs to target all of the various cancer drivers identified so far in patients; third, we are not able to mechanistically investigate and understand the biology of a molecular alteration in its tissue-specific context in a cancer patient.
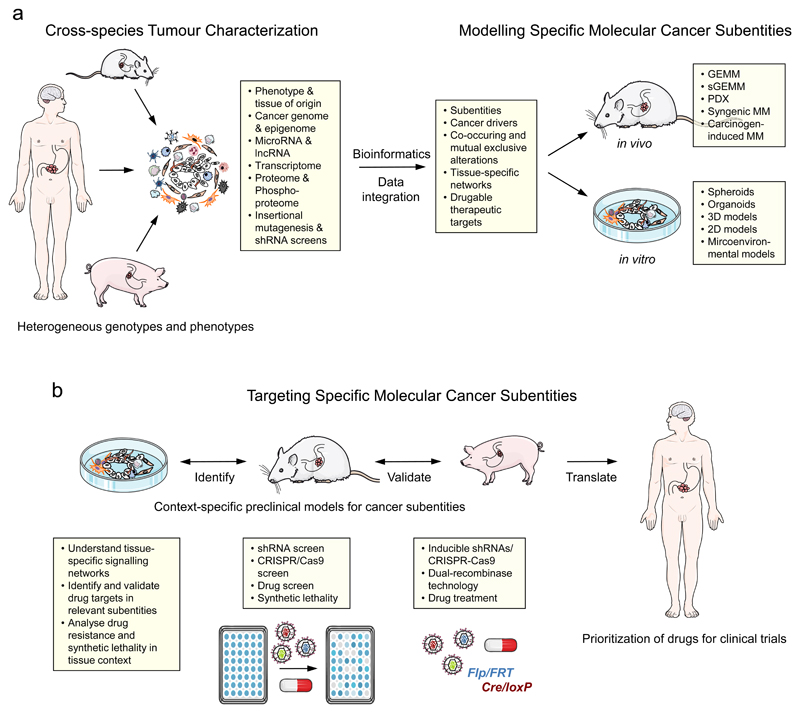
Here pre-clinical model systems are needed that closely reflect the biology of specific molecular subtypes of human cancer225–230. This includes (i) cell-based in vitro models (organoids, spheroids) derived from human, porcine and murine tumours and normal tissues that have been transformed by distinct cancer drivers225, , 226, 230 (ii) in vitro 3D cancer models that mimic the microenvironment of human tumours229, (iii) patient-derived xenografts (PDX) and orthotopic syngenic engraftment models225, 226, 228, and (iv) genetically engineered and carcinogen-induced autochthonous small and large animal models226–228, 231 (Figure 5a). Each model has its own advantages and disadvantages, depending on the scientific question to be addressed. Cancer modelling in pigs is now a rapidly evolving new field, addressing the unmet need for genetically defined human-scale preclinical cancer models231–233.
Figure 5. Approaches to identify, analyse and validate cancer drivers in context.
(a) Cross-species comparative characterization of distinct heterogeneous tumour entities to improve the signal-to-noise ratio and identify substantially altered pathways, transcriptional regulators, missense mutations or copy number changes that likely drive the tumour and are potential therapeutic targets. The comparison of human cancers with corresponding genetically engineered or carcinogen-induced mouse and pig tumour models serves as a filter to identify shared alterations. Pig cancer modelling is a rapidly evolving field, which is driven by the need of more humanized tumour models for pre-clinical studies. Systems biology and data integration is used to define relevant molecular subtypes of a cancer type, based on putative driver mutations, co-occurring alterations, and druggable targets. Such bioinformatic analyzes will help to model these molecular cancer subtypes, e.g. by genetic engineering in the mouse germline (genetically engineered mouse model (GEMM)), or by (multiplexed) somatic gene engineering of an appropriate target cell in vitro (e.g. in organoids) or in vivo (somatic GEMM (sGEMM)) using CRISPR-Cas9, short hairpin RNAs (shRNAs) or overexpression systems. Thereby, it is possible to reproduce most of the features that are unique for the particular context-specific tumour subtype. In addition, patient-derived xenograft models (PDXs), orthotopic syngeneic engraftment models, and carcinogen-induced models, which may recapitulate important features of a specific tumour subtype (e.g. hypermutation in carcinogen-induced models), can be employed. (b) These subtype specific cancer models can then be used to understand tissue-specific signalling networks of molecular alterations or to identify context-specific targets by retroviral shRNA or CRISPR-Cas9-based library, drug or synthetic lethality screens. These models can also be used to analyse drug resistance or validate therapeutic targets preclinically by inducible shRNA or CRISPR-Cas9 systems, dual-recombinase technology, or drug treatment studies. Genetically defined pig cancer models can be used to perform more representative molecularly-guided context-specific treatment trials at human scale. Knowledge gained from this approach can then be exploited to prioritize drugs and treatment trials with stratified patients in the clinic. lncRNA; long non-coding RNA.
The characterization and validation of cancer drivers and their epistatic interactions in distinct tissue types is clearly a great challenge in the context of the many genomic aberrations and their frequent co-occurrence in human cancers. Cross-species comparative tumour analyses and computational approaches defining co-occurring genetic alterations and synergistic epistatic interactions, as well as mutual exclusivity will help us to model these specific molecular cancer subtypes in tissue context134, 226. A strategy might be to introduce a number of mutations into the appropriate target cell in vitro or in vivo in order to reproduce most of the features that are unique for the particular tumour subtype (Figure 5a). This permits us to answer the question how individual mutations contribute to the tumour phenotype and how they influence intervention strategies. The advent of CRISPR–Cas9 mediated gene editing enables us for the first time to introduce combinatorial genetic alterations into specific somatic cells with high efficacy230, 234–238. Multiplexed somatic CRISPR–Cas9-based genome editing has been shown to be a powerful tool to model relevant human cancer subtypes more closely with the potential to better understand the context-specific biological basis of cancer drivers in vitro and in vivo230, 238–242. In addition, new ways to genetically validate cancer genes as appropriate therapeutic targets in their specific tissue-context as well as evaluate primary and secondary therapy resistances have been developed recently (Figure 5b). These include short hairpin RNA (shRNA) or dual-recombinase strategies that allow for the inducible inactivation of cancer genes in autochthonous tumours182, 183, 243, 244. In addition, dual-recombinase based sequential genetic manipulation enables the timing and stages of tumorigenesis to be modelled183. Cancer-subtype specific shRNA, CRISPR–Cas9 or pharmacological screens are another attractive option to identify context-specific therapeutic vulnerabilities245, 246.
Over the past few decades, the efficacy of treatment regimens established in pre-clinical cancer models has often not been translated into the clinic, indicating that conventional models are poor predictors of clinical efficacy228. This is probably due to limitations of xenotransplantation models using established human cancer cell lines and tumour models that do not adequately recapitulate the matching human tumour subtype. However, in the last decade a number of next-generation models have been developed, which faithfully recapitulate human cancer, and predict therapeutic response more accurately227, 228. Therefore, we propose that pipelines of well-characterized model systems for relevant cancer subtypes should be established in order to perform pre-clinically guided and prioritized clinical trials. This will increase success rates and save costs and resources.
Comprehensive molecular tumour profiling will greatly aid clinical decision-making and improve cancer care. Together with an increasing arsenal of targeted drugs, this information will provide us with the tools to match the right patient to the right drug. Effective sharing and dissemination of patient molecular tumour profiles and response data will be essential to achieve this goal. However, tissue-context has to be considered and empirically tested as it is an important determinant of treatment response and resistance.
We believe that considering both the molecular tumour profile and the anatomical site is likely to be superior to either of these in isolation in predicting drug response in the patient. Therefore, a thorough understanding of the molecular mechanisms involved in tissue-specific tumour biology as well as drug responses is essentially needed for the design of novel next-generation targeted treatment regimens.
Glossary Box.
Mosaicism: Presence of two or more genetically distinct cell populations.
Transdifferentiation: A cell fate switch (metaplasia) where a differentiated adult somatic cell transforms into another mature somatic cell type.
Super-enhancers: A genomic regulatory region of multiple enhancers with very strong enrichment of transcriptional coactivator binding that drives gene transcription.
Chromothripsis: The massive catastrophic shattering and reassembly of one or a few chromosomes, which results in the simultaneous acquisition of multiple genetic alterations in a cell.
Chromoanasynthesis: Local gene rearrangements leading to multiple copy number alterations including deletions, duplications, triplications, as well as extensive translocations and inversions that result from template switching during locally defective DNA replication.
Chromoplexy: Complex chained DNA rearrangements that affect multiple chromosomes resulting from several broken DNA strands.
Desmoplasia: A dense fibrous connective tissue reaction, usually to malignant epithelial tumours in the stroma of a carcinoma, due to the proliferation of fibroblasts and increased deposition of extracellular matrix components.
Steatosis: The abnormal retention and accumulation of lipid droplets within cells resulting in fatty changes or degeneration of a solid organ.
Acknowledgements
We apologize to all colleagues whose work was not included in this Opinion article due to space constraints or the need to focus on specific examples. G.S. is supported by grants from the Deutsche Krebshilfe [grant number: 110908], DFG [SFB824/C9], Wilhelm-Sander Stiftung [grant number: 2016.004.1], and a Joint Funding project of the German Cancer Consortium (DKTK), R.R. is supported by grants from the DKTK and the Helmholtz-Alliance Preclinical Comprehensive Cancer Center (PCCC), and D.S. is supported by grants from the European Research Council [ERC CoG No. 648521], DKTK, DFG [SA 1374/4-2 and SFB824/C9] and the Helmholtz-Alliance PCCC.
Biographies
Author Biographies
Günter Schneider studied medicine and received his MD from the Julius-Maximilians-University Würzburg, Germany. He trained as a physician scientist in gastrointestinal oncology at University of Ulm and Technical University of Munich (TUM). Since 2002 he is an independent PI at the Department of Gastroenterology at the University Hospital Klinikum rechts der Isar, TUM School of Medicine, focusing on pancreatic and gastrointestinal cancer subtypes. He is interested how oncogenic signalling is integrated by transcriptional mechanisms and how such understanding can translate into novel therapies.
Marc Schmidt-Supprian studied chemistry, biochemistry and genetics in Germany and Ireland. He received his PhD in immunology from Cologne University and conducted postdoctoral and junior investigator training at Harvard Medical School in Boston. He started his independent research group at the Max Planck Institute of Biochemistry in Martinsried and then moved to the department of Haematology and Oncology of the Technical University of Munich, where he serves as Tenure Track Professor and head of research. Marc’s research interest focuses on signal transduction and post-transcriptional gene regulation in immunopathology and cancer.
Roland Rad is a professor of translational gastrointestinal oncology and experimental cancer genetics at the Technical University of Munich (TUM) School of Medicine and the German Cancer Research Center Heidelberg. He conducted clinical and postdoctoral research training at TUM and the Wellcome Trust Sanger Institute in Cambridge. His research interest is focused on gene discovery, functional gene annotation and signalling in cancer. His group is developing tools for high-throughput forward and reverse genetics in mice, including transposon-based genome-wide screening technologies and in vivo CRISPR-Cas9 genome engineering approaches.
Dieter Saur is a consultant Gastroenterologist & Oncologist and Professor at TUM School of Medicine. He has a long-standing expertise in diagnosing and treating cancer patients in the clinic. Research of his group focuses on a deeper understanding of tissue specific oncogenic signalling pathways in cancer, with a special interest in the role of the tumour microenvironment. He is developing advanced genetically engineered models for cancer subtypes allowing highly controlled independent genetic manipulation of specific cell types in whole animals. Dieter’s translational research interests focus on new endoscopic imaging procedures for early detection of gastrointestinal tumours and the development of novel therapeutic strategies for gastrointestinal cancer subtypes.
Laboratory homepage: http://www.med2.mri.tum.de/en/research/ag-saur.php
Footnotes
Subject Categories
Biological sciences / Cancer / Cancer therapy / Targeted therapies [URI /631/67/1059/602];
Biological sciences / Cancer / Cancer genetics [URI /631/67/68;
Biological sciences / Cancer / Cancer therapy [URI /631/67/1059];
Biological sciences / Cancer / Oncogenes [URI /631/67/395];
Biological sciences / Cancer / Cancer models [URI /631/67/70]
Table of Contents Summary
In this Opinion article, Schneider et al. outline tissue- and cell type-specific differences in tumorigenesis and the organization of oncogenic signalling pathways, and discuss the implications of our understanding of tissue context on molecularly targeted therapy and clinical trial design.
Competing interests statement
The authors declare no competing interests.
References
- 1.Dancey JE, Bedard PL, Onetto N, Hudson TJ. The genetic basis for cancer treatment decisions. Cell. 2012;148:409–420. doi: 10.1016/j.cell.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 3.Demetri GD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 4.Rosell R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 5.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw AT, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 7.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Druker BJ, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 9.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 10.Hall RD, Kudchadkar RR. BRAF mutations: signaling, epidemiology, and clinical experience in multiple malignancies. Cancer Control. 2014;21:221–230. doi: 10.1177/107327481402100307. [DOI] [PubMed] [Google Scholar]
- 11.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 12.Hyman DM, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med. 2015;373:726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter DJ, D'Agostino RB. Let's Not Put All Our Eggs in One Basket. N Engl J Med. 2015;373:691–693. doi: 10.1056/NEJMp1508144. [DOI] [PubMed] [Google Scholar]
- 14.Le Tourneau C, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 15.Krzyzanowska MK. Off-label use of cancer drugs: a benchmark is established. J Clin Oncol. 2013;31:1125–1127. doi: 10.1200/JCO.2012.46.9460. [DOI] [PubMed] [Google Scholar]
- 16.Conti RM, et al. Prevalence of off-label use and spending in 2010 among patent-protected chemotherapies in a population-based cohort of medical oncologists. J Clin Oncol. 2013;31:1134–1139. doi: 10.1200/JCO.2012.42.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullard A. Off-label targeted cancer drugs fail in first randomized trial. Nat Rev Drug Discov. 2015;14:669. doi: 10.1038/nrd4758. [DOI] [PubMed] [Google Scholar]
- 18.Mullard A. Use of personalized cancer drugs runs ahead of the science. Nature. 2015 doi: 10.1038/nature.2015.18389. [DOI] [Google Scholar]
- 19.Redig AJ, Jänne PA. Basket Trials and the Evolution of Clinical Trial Design in an Era of Genomic Medicine. Journal of Clinical Oncology. 2015;33:975–977. doi: 10.1200/JCO.2014.59.8433. [DOI] [PubMed] [Google Scholar]
- 20.Stenzinger A, Weichert W, Lennerz JK, Klauschen F. Basket Trials: Just the End of the First Quarter. Journal of Clinical Oncology. 2015;33:2823–2824. doi: 10.1200/JCO.2015.62.1516. [DOI] [PubMed] [Google Scholar]
- 21.Abrams J, et al. National Cancer Institute's Precision Medicine Initiatives for the new National Clinical Trials Network. Am Soc Clin Oncol Educ Book. 2014:71–76. doi: 10.14694/EdBook_AM.2014.34.71. [DOI] [PubMed] [Google Scholar]
- 22.McNeil C. NCI-MATCH launch highlights new trial design in precision-medicine era. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv193. [DOI] [PubMed] [Google Scholar]
- 23.Mullard A. NCI-MATCH trial pushes cancer umbrella trial paradigm. Nat Rev Drug Discov. 2015;14:513–515. doi: 10.1038/nrd4694. [DOI] [PubMed] [Google Scholar]
- 24.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 25.Sieber OM, Tomlinson SR, Tomlinson IPM. Tissue, cell and stage specificity of (epi)mutations in cancers. Nat Rev Cancer. 2005;5:649–655. doi: 10.1038/nrc1674. [DOI] [PubMed] [Google Scholar]
- 26.Schaefer MH, Serrano L. Cell type-specific properties and environment shape tissue specificity of cancer genes. Sci Rep. 2016;6 doi: 10.1038/srep20707. 20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene. 2004;23:6445–6470. doi: 10.1038/sj.onc.1207714. [DOI] [PubMed] [Google Scholar]
- 28.Garber JE, Offit K. Hereditary cancer predisposition syndromes. J Clin Oncol. 2005;23:276–292. doi: 10.1200/JCO.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 29.Ricci R. Syndromic gastrointestinal stromal tumors. Hered Cancer Clin Pract. 2016;14:15. doi: 10.1186/s13053-016-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore LE, et al. Von Hippel-Lindau (VHL) inactivation in sporadic clear cell renal cancer: associations with germline VHL polymorphisms and etiologic risk factors. PLoS Genet. 2011;7:e1002312. doi: 10.1371/journal.pgen.1002312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447–1462. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heisterkamp N, Stam K, Groffen J, de Klein A, Grosveld G. Structural organization of the bcr gene and its role in the Ph' translocation. Nature. 1985;315:758–761. doi: 10.1038/315758a0. [DOI] [PubMed] [Google Scholar]
- 34.Lage K, et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Natl Acad Sci U S A. 2008;105:20870–20875. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 36.Berquam-Vrieze KE, et al. Cell of origin strongly influences genetic selection in a mouse model of T-ALL. Blood. 2011;118:4646–4656. doi: 10.1182/blood-2011-03-343947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 38.Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 40.Koren S, et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature. 2015;525:114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- 41.Van Keymeulen A, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 42.Rowbotham SP, Kim CF. Diverse cells at the origin of lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111:4745–4746. doi: 10.1073/pnas.1401955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutherland KD, et al. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014;111:4952–4957. doi: 10.1073/pnas.1319963111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon M-c, Berns A. Mouse models for lung cancer. Mol Oncol. 2013;7:165–177. doi: 10.1016/j.molonc.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Youssef KK, et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat Cell Biol. 2012;14:1282–1294. doi: 10.1038/ncb2628. [DOI] [PubMed] [Google Scholar]
- 46.Morris JPt, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy N, Hebrok M. Regulation of Cellular Identity in Cancer. Dev Cell. 2015;35:674–684. doi: 10.1016/j.devcel.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian H, et al. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc Natl Acad Sci U S A. 2009;106:4254–4259. doi: 10.1073/pnas.0813203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchert M, et al. Genetic dissection of differential signaling threshold requirements for the Wnt/beta-catenin pathway in vivo. PLoS Genet. 2010;6:e1000816. doi: 10.1371/journal.pgen.1000816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zandstra PW, Lauffenburger DA, Eaves CJ. A ligand-receptor signaling threshold model of stem cell differentiation control: a biologically conserved mechanism applicable to hematopoiesis. Blood. 2000;96:1215–1222. [PubMed] [Google Scholar]
- 52.Fu G, et al. Fine-tuning T cell receptor signaling to control T cell development. Trends Immunol. 2014;35:311–318. doi: 10.1016/j.it.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abeshouse A, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shojaee S, et al. PTEN opposes negative selection and enables oncogenic transformation of pre-B cells. Nat Med. 2016;22:379–387. doi: 10.1038/nm.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Werner M, Hobeika E, Jumaa H. Role of PI3K in the generation and survival of B cells. Immunol Rev. 2010;237:55–71. doi: 10.1111/j.1600-065X.2010.00934.x. [DOI] [PubMed] [Google Scholar]
- 56.Küppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 57.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 58.Guerra C, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 59.Eser S, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23:406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 60.Young NP, Jacks T. Tissue-specific p19Arf regulation dictates the response to oncogenic K-ras. Proc Natl Acad Sci U S A. 2010;107:10184–10189. doi: 10.1073/pnas.1004796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuster-Böckler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 62.Polak P, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, Lopez-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. 2016;532:264–267. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- 65.Perera D, et al. Differential DNA repair underlies mutation hotspots at active promoters in cancer genomes. Nature. 2016;532:259–263. doi: 10.1038/nature17437. [DOI] [PubMed] [Google Scholar]
- 66.Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berns A. Kras and Hras--what is the difference? Nat Genet. 2008;40:1149–1150. doi: 10.1038/ng1008-1149. [DOI] [PubMed] [Google Scholar]
- 69.To MD, et al. Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat Genet. 2008;40:1240–1244. doi: 10.1038/ng.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whyte WA, et al. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016 doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
- 73.Hnisz D, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loven J, et al. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang X, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mansour MR, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oldridge DA, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418–421. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu P, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C-Z, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rode A, Maass KK, Willmund KV, Lichter P, Ernst A. Chromothripsis in cancer cells: An update. Int J Cancer. 2016;138:2322–2333. doi: 10.1002/ijc.29888. [DOI] [PubMed] [Google Scholar]
- 84.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 85.Notta F, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–382. doi: 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kass EM, Moynahan ME, Jasin M. When Genome Maintenance Goes Badly Awry. Mol Cell. 2016;62:777–787. doi: 10.1016/j.molcel.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le DT, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30:326–339. doi: 10.1016/j.tig.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 89.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 90.Haffner MC, De Marzo AM, Meeker AK, Nelson WG, Yegnasubramanian S. Transcription-induced DNA double strand breaks: both oncogenic force and potential therapeutic target? Clin Cancer Res. 2011;17:3858–3864. doi: 10.1158/1078-0432.CCR-10-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Williamson LM, Lees-Miller SP. Estrogen receptor alpha-mediated transcription induces cell cycle-dependent DNA double-strand breaks. Carcinogenesis. 2011;32:279–285. doi: 10.1093/carcin/bgq255. [DOI] [PubMed] [Google Scholar]
- 92.Savage KI, et al. BRCA1 deficiency exacerbates estrogen-induced DNA damage and genomic instability. Cancer Res. 2014;74:2773–2784. doi: 10.1158/0008-5472.CAN-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gorrini C, et al. Estrogen controls the survival of BRCA1-deficient cells via a PI3K-NRF2-regulated pathway. Proc Natl Acad Sci U S A. 2014;111:4472–4477. doi: 10.1073/pnas.1324136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blüthgen N. Signaling output: it's all about timing and feedbacks. Mol Syst Biol. 2015;11:843. doi: 10.15252/msb.20156642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karreth FA, Frese KK, DeNicola GM, Baccarini M, Tuveson DA. C-Raf is required for the initiation of lung cancer by K-Ras(G12D) Cancer Discov. 2011;1:128–136. doi: 10.1158/2159-8290.CD-10-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blasco RB, et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell. 2011;19:652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fleuren EDG, Zhang L, Wu J, Daly RJ. The kinome 'at large' in cancer. Nat Rev Cancer. 2016;16:83–98. doi: 10.1038/nrc.2015.18. [DOI] [PubMed] [Google Scholar]
- 100.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531:110–113. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schaefer MH, Yang J-S, Serrano L, Kiel C. Protein conservation and variation suggest mechanisms of cell type-specific modulation of signaling pathways. PLoS Comput Biol. 2014;10:e1003659. doi: 10.1371/journal.pcbi.1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mills IG. Maintaining and reprogramming genomic androgen receptor activity in prostate cancer. Nat Rev Cancer. 2014;14:187–198. doi: 10.1038/nrc3678. [DOI] [PubMed] [Google Scholar]
- 103.Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS, Downward J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov. 2013;3:548–563. doi: 10.1158/2159-8290.CD-12-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Navas C, et al. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:318–330. doi: 10.1016/j.ccr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Diersch S, et al. Kras(G12D) induces EGFR-MYC cross signaling in murine primary pancreatic ductal epithelial cells. Oncogene. 2016;35:3880–3886. doi: 10.1038/onc.2015.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ardito CM, et al. EGF Receptor Is Required for KRAS-Induced Pancreatic Tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schneeweis C, Wirth M, Saur D, Reichert M, Schneider G. Oncogenic KRAS and the EGFR loop in pancreatic carcinogenesis - a connection to licensing nodes. Small GTPases. 2016 doi: 10.1080/21541248.2016.1262935. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eberhard DA, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 111.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tol J, et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med. 2009;360:563–572. doi: 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]
- 113.da Cunha Santos G, et al. Molecular predictors of outcome in a phase 3 study of gemcitabine and erlotinib therapy in patients with advanced pancreatic cancer: National Cancer Institute of Canada Clinical Trials Group Study PA.3. Cancer. 2010;116:5599–5607. doi: 10.1002/cncr.25393. [DOI] [PubMed] [Google Scholar]
- 114.Manchado E, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 116.Memarzadeh S, et al. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell. 2007;12:572–585. doi: 10.1016/j.ccr.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437:199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 118.Fogarty MP, Emmenegger BA, Grasfeder LL, Oliver TG, Wechsler-Reya RJ. Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells. Proc Natl Acad Sci U S A. 2007;104:2973–2978. doi: 10.1073/pnas.0605770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015;160:816–827. doi: 10.1016/j.cell.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Torday JS. Homeostasis as the Mechanism of Evolution. Biology (Basel) 2015;4:573–590. doi: 10.3390/biology4030573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu RC, et al. Negative feedback that improves information transmission in yeast signalling. Nature. 2008;456:755–761. doi: 10.1038/nature07513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov. 2012;2:311–319. doi: 10.1158/2159-8290.CD-12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stebbing J, et al. The regulatory roles of phosphatases in cancer. Oncogene. 2014;33:939–953. doi: 10.1038/onc.2013.80. [DOI] [PubMed] [Google Scholar]
- 124.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yaktapour N, et al. BRAF inhibitor-associated ERK activation drives development of chronic lymphocytic leukemia. J Clin Invest. 2014;124:5074–5084. doi: 10.1172/JCI76539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rad R, et al. A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer Cell. 2013;24:15–29. doi: 10.1016/j.ccr.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Custodio A, Feliu J. Prognostic and predictive biomarkers for epidermal growth factor receptor-targeted therapy in colorectal cancer: beyond KRAS mutations. Crit Rev Oncol Hematol. 2013;85:45–81. doi: 10.1016/j.critrevonc.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 129.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. 2014;111:817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cohen RL, Settleman J. From cancer genomics to precision oncology--tissue's still an issue. Cell. 2014;157:1509–1514. doi: 10.1016/j.cell.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 131.Skoulidis F, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang X, Fu AQ, McNerney ME, White KP. Widespread genetic epistasis among cancer genes. Nat Commun. 2014;5:4828. doi: 10.1038/ncomms5828. [DOI] [PubMed] [Google Scholar]
- 133.Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet. 2013;14:168–178. doi: 10.1038/nrg3404. [DOI] [PubMed] [Google Scholar]
- 134.Park S, Lehner B. Cancer type-dependent genetic interactions between cancer driver alterations indicate plasticity of epistasis across cell types. Mol Syst Biol. 2015;11:824. doi: 10.15252/msb.20156102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 136.Lehner B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 2011;27:323–331. doi: 10.1016/j.tig.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 137.Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 138.Giannakis M, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rustgi AK. BRAF: a driver of the serrated pathway in colon cancer. Cancer Cell. 2013;24:1–2. doi: 10.1016/j.ccr.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 140.Fisher R, et al. Development of synchronous VHL syndrome tumors reveals contingencies and constraints to tumor evolution. Genome Biol. 2014;15:433. doi: 10.1186/s13059-014-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Harms MJ, Thornton JW. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature. 2014;512:203–207. doi: 10.1038/nature13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.New AM, Lehner B. Systems biology: Network evolution hinges on history. Nature. 2015;523:297–298. doi: 10.1038/nature14537. [DOI] [PubMed] [Google Scholar]
- 143.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang J, et al. Tumor evolutionary directed graphs and the history of chronic lymphocytic leukemia. Elife. 2014;3 doi: 10.7554/eLife.02869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fanidi A, Harrington EA, Evan GI. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature. 1992;359:554–556. doi: 10.1038/359554a0. [DOI] [PubMed] [Google Scholar]
- 146.Bissonnette RP, Echeverri F, Mahboubi A, Green DR. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature. 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- 147.Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- 148.Aukema SM, et al. Biological characterization of adult MYC-translocation-positive mature B-cell lymphomas other than molecular Burkitt lymphoma. Haematologica. 2014;99:726–735. doi: 10.3324/haematol.2013.091827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Horn H, et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121:2253–2263. doi: 10.1182/blood-2012-06-435842. [DOI] [PubMed] [Google Scholar]
- 150.Eischen CM, Woo D, Roussel MF, Cleveland JL. Apoptosis triggered by Myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol Cell Biol. 2001;21:5063–5070. doi: 10.1128/MCB.21.15.5063-5070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Delbridge ARD, Strasser A. The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ. 2015;22:1071–1080. doi: 10.1038/cdd.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 153.Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010;20:91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2140–2141. doi: 10.1056/NEJMc1412266. [DOI] [PubMed] [Google Scholar]
- 155.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Connor F, et al. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 158.Elinav E, et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 159.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 161.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 162.Vermeulen L, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 163.Polyak K, Kalluri R. The role of the microenvironment in mammary gland development and cancer. Cold Spring Harb Perspect Biol. 2010;2:a003244. doi: 10.1101/cshperspect.a003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Omary MB, Lugea A, Lowe AW, Pandol SJ. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J Clin Invest. 2007;117:50–59. doi: 10.1172/JCI30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 166.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 167.Apte MV, Wilson JS, Lugea A, Pandol SJ. A starring role for stellate cells in the pancreatic cancer microenvironment. Gastroenterology. 2013;144:1210–1219. doi: 10.1053/j.gastro.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Erkan M, et al. StellaTUM: current consensus and discussion on pancreatic stellate cell research. Gut. 2012;61:172–178. doi: 10.1136/gutjnl-2011-301220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sherman MH, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tomlinson J, et al. Different patterns of angiogenesis in sarcomas and carcinomas. Clin Cancer Res. 1999;5:3516–3522. [PubMed] [Google Scholar]
- 171.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 172.Kirsch DG, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–997. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 173.Erkan M, et al. The role of stroma in pancreatic cancer: diagnostic and therapeutic implications. Nat Rev Gastroenterol Hepatol. 2012;9:454–467. doi: 10.1038/nrgastro.2012.115. [DOI] [PubMed] [Google Scholar]
- 174.Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Driskell RR, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wagner EF. Cancer: Fibroblasts for all seasons. Nature. 2016;530:42–43. doi: 10.1038/530042a. [DOI] [PubMed] [Google Scholar]
- 177.Koliaraki V, Pasparakis M, Kollias G. IKKb in intestinal mesenchymal cells promotes initiation of colitis-associated cancer. J Exp Med. 2015;212:2235–2251. doi: 10.1084/jem.20150542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pallangyo CK, Ziegler PK, Greten FR. IKKb acts as a tumor suppressor in cancer-associated fibroblasts during intestinal tumorigenesis. J Exp Med. 2015;212:2253–2266. doi: 10.1084/jem.20150576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lichtenberger BM, Mastrogiannaki M, Watt FM. Epidermal beta-catenin activation remodels the dermis via paracrine signalling to distinct fibroblast lineages. Nat Commun. 2016;7:10537. doi: 10.1038/ncomms10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bhowmick NA, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 181.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 2011;71:4040–4047. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schönhuber N, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med. 2014;20:1340–1347. doi: 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Muzumdar MD, et al. Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers. Nat Commun. 2016;7:12685. doi: 10.1038/ncomms12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fernandez LC, Torres M, Real FX. Somatic mosaicism: on the road to cancer. Nat Rev Cancer. 2016;16:43–55. doi: 10.1038/nrc.2015.1. [DOI] [PubMed] [Google Scholar]
- 186.Giovannucci E, et al. Diabetes and cancer: a consensus report. CA Cancer J Clin. 2010;60:207–221. doi: 10.3322/caac.20078. [DOI] [PubMed] [Google Scholar]
- 187.Harding JL, Shaw JE, Peeters A, Cartensen B, Magliano DJ. Cancer risk among people with type 1 and type 2 diabetes: disentangling true associations, detection bias, and reverse causation. Diabetes Care. 2015;38:264–270. doi: 10.2337/dc14-1996. [DOI] [PubMed] [Google Scholar]
- 188.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 189.Incio J, et al. Obesity-Induced Inflammation and Desmoplasia Promote Pancreatic Cancer Progression and Resistance to Chemotherapy. Cancer Discov. 2016;6:852–869. doi: 10.1158/2159-8290.CD-15-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lauby-Secretan Ba, et al. Body Fatness and Cancer--Viewpoint of the IARC Working Group. N Engl J Med. 2016;375:794–798. doi: 10.1056/NEJMsr1606602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Khasawneh J, et al. Inflammation and mitochondrial fatty acid beta-oxidation link obesity to early tumor promotion. Proc Natl Acad Sci U S A. 2009;106:3354–3359. doi: 10.1073/pnas.0802864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Johnson AMF, et al. High fat diet causes depletion of intestinal eosinophils associated with intestinal permeability. PLoS One. 2015;10:e0122195. doi: 10.1371/journal.pone.0122195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Beyaz S, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Clocchiatti A, Cora E, Zhang Y, Dotto GP. Sexual dimorphism in cancer. Nat Rev Cancer. 2016;16:330–339. doi: 10.1038/nrc.2016.30. [DOI] [PubMed] [Google Scholar]
- 195.Zhu C, et al. Conditional expression of the androgen receptor induces oncogenic transformation of the mouse prostate. J Biol Chem. 2011;286:33478–33488. doi: 10.1074/jbc.M111.269894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mani R-S, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Haffner MC, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Weischenfeldt J, et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23:159–170. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 200.King JC, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hensley CT, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yuneva MO, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mayers JR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353:1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12:584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 206.Guerra C, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kiraly O, Gong G, Olipitz W, Muthupalani S, Engelward BP. Inflammation-induced cell proliferation potentiates DNA damage-induced mutations in vivo. PLoS Genet. 2015;11:e1004901. doi: 10.1371/journal.pgen.1004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Akbani R, et al. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic b-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 218.Peng W, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and Anticancer Immunosurveillance. Cell. 2016;165:276–287. doi: 10.1016/j.cell.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 220.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Vetizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kwa M, Plottel CS, Blaser MJ, Adams S. The Intestinal Microbiome and Estrogen Receptor-Positive Female Breast Cancer. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15:747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Eifert C, Powers RS. From cancer genomes to oncogenic drivers, tumour dependencies and therapeutic targets. Nat Rev Cancer. 2012;12:572–578. doi: 10.1038/nrc3299. [DOI] [PubMed] [Google Scholar]
- 227.Tuveson D, Hanahan D. Translational medicine: Cancer lessons from mice to humans. Nature. 2011;471:316–317. doi: 10.1038/471316a. [DOI] [PubMed] [Google Scholar]
- 228.Herter-Sprie GS, Kung AL, Wong K-K. New cast for a new era: preclinical cancer drug development revisited. J Clin Invest. 2013;123:3639–3645. doi: 10.1172/JCI68340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Gu L, Mooney DJ. Biomaterials and emerging anticancer therapeutics: engineering the microenvironment. Nat Rev Cancer. 2016;16:56–66. doi: 10.1038/nrc.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 231.Flisikowska T, Kind A, Schnieke A. Pigs as models of human cancers. Theriogenology. 2016;86:433–437. doi: 10.1016/j.theriogenology.2016.04.058. [DOI] [PubMed] [Google Scholar]
- 232.Flisikowska T, et al. A porcine model of familial adenomatous polyposis. Gastroenterology. 2012;143:1173–1175.e1171–1177. doi: 10.1053/j.gastro.2012.07.110. [DOI] [PubMed] [Google Scholar]
- 233.Saalfrank A, et al. A porcine model of osteosarcoma. Oncogenesis. 2016;5:e210. doi: 10.1038/oncsis.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sanchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sanchez-Rivera FJ, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zuckermann M, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Weber J, et al. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1512392112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Maresch R, et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun. 2016;7:10770. doi: 10.1038/ncomms10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Guernet A, et al. CRISPR-Barcoding for Intratumor Genetic Heterogeneity Modeling and Functional Analysis of Oncogenic Driver Mutations. Mol Cell. 2016;63:526–538. doi: 10.1016/j.molcel.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Walton J, et al. CRISPR/Cas9-Mediated Trp53 and Brca2 Knockout to Generate Improved Murine Models of Ovarian High-Grade Serous Carcinoma. Cancer Res. 2016;76:6118–6129. doi: 10.1158/0008-5472.CAN-16-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Annunziato S, et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 2016;30:1470–1480. doi: 10.1101/gad.279190.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Dow LE, Lowe SW. Life in the fast lane: mammalian disease models in the genomics era. Cell. 2012;148:1099–1109. doi: 10.1016/j.cell.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Fellmann C, Lowe SW. Stable RNA interference rules for silencing. Nat Cell Biol. 2014;16:10–18. doi: 10.1038/ncb2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Christensen CL, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell. 2014;26:909–922. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]