Abstract
Hysteresis in equilibrium protein folding titrations is an experimental barrier that must be overcome to extract meaningful thermodynamic quantities. Traditional approaches to solving this problem involve testing a spectrum of solution conditions to find ones that achieve path independence. Through this procedure, a specific pH of 3.8 was required to achieve path-independence for the water-to-bilayer equilibrium folding of outer membrane protein OmpLA. We hypothesized that the neutralization of negatively charged side chains (Asp and Glu) at pH 3.8 could be the physical basis for path-independent folding at this pH. To test this idea, we engineered variants of OmpLA with Asp→Asn and Glu→Gln mutations to neutralize the negative charges within various regions of the protein and tested for reversible folding at neutral pH. Although not fully resolved, our results show that these mutations in the periplasmic turns and extracellular loops are responsible for 60% of the hysteresis in WT folding. Overall, our study suggests that negative charges impact the folding hysteresis in outer membrane proteins and their neutralization may aid in protein engineering applications.
Keywords: membrane protein folding, membrane protein stability, lipid bilayers
Table of Contents
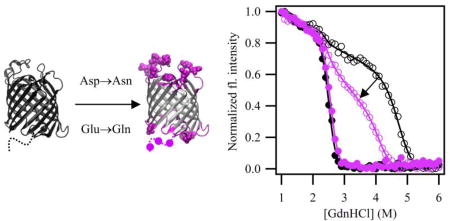
Introduction
Thermodynamic investigations of membrane proteins are essential for quantifying the forces that stabilize native structures in membranes. Such measurements allow for the interrogation of membrane protein energetics, such as the interactions with lipids,1, 2 the effects of bilayer thickness,3 assessment of thermodynamic coupling in side chain interactions,1, 2 and the interrogation of folding mechanisms.4 To date there are few measurements of membrane protein folding free energies; in our experience this is primarily due to aggregation or precipitation of the unfolded state ensemble.5–7 Even if conditions can be found to fold (or re-fold) membrane proteins reversibly, it can still be the case that equilibrium titrations conducted in both the folding and unfolding directions fail to coincide, indicating a lack of path-independence. Closing this hysteresis is a significant technical challenge that must be overcome as a prerequisite for the extraction of thermodynamic quantities for a greater number of membrane proteins.6, 8, 9
In a previous study,10 the solution condition of pH 3.8 closed the hysteresis gap for three outer membrane proteins (OMPs) from E. coli: Outer membrane Phospholipase A (OmpLA), PagP and OmpW. This pH was the only parameter that worked, even after an exhaustive search different lipid bilayer compositions, varying temperatures, and different denaturants2, 6. The physical basis underlying why this unusual pH provides path-independence in these thermodynamic experiments is unknown. Interestingly, the solution condition of pH 3.8 could not resolve hysteresis in four other E. coli OMPs (OmpT, FadL, OmpX or full-length OmpA) even though these proteins all have theoretical pI values that are similar. These data also demonstrated that path-independence is not determined by size of the barrel: OmpW and PagP are 8-stranded and path-independent, but OmpX and OmpA are also 8-stranded and showed hysteresis. Additionally, the extent of hysteresis (area in between titration curves) could not be correlated to either the change in accessible surface area (ΔASA) of folding, or the ratio of water exposed mass to mass buried in lipid, or the total protein charge at pH 3.8. A direct comparison with other in vitro folding systems yields no further insight because those studies either measure different endpoint transitions or employ different denaturants4 or use different lipid compositions or geometries.3, 7 The fact that pH 3.8 does not solve the hysteresis challenge for other E. coli OMPs suggests that this pH is not conferring a bulk effect on properties of the bilayer. In proteins, the source could be the ionizable aspartic and glutamic acid side chains, which have model compound pKa values equal to 3.71 and 4.15, respectively.11 Partial or full neutralization of some of these side chains at pH 3.8 could reduce the activation barrier to folding or unfolding or both.
Statistical surveys have shown that acidic side chains are found distributed across the structures of OMPs including within the interiors of the beta-barrels as well as in both the intracellular periplasmic turns and the extracellular loops of OMPs12. Because folding and unfolding both require at least one of these regions to pass through the membrane, any ionized groups in the extra-membranous region of a membrane protein must cross the dehydrated, hydrophobic bilayer center, an energetically costly process. This affects equilibrium folding titrations because slow rates of folding or unfolding or both can prevent equilibrium from being attained.8, 9 We hypothesize that the pH 3.8 solution conditions reduce this energetic penalty by neutralization of negatively charged residues thus resulting in an acceleration of the kinetics of either the folding or unfolding reaction or both. This could collapse the hysteresis. To test this idea and potentially enable thermodynamic measurements at neutral pH, we engineered a number of Asp→Asn and Glu→Gln mutations in distinct domains of OmpLA and tested for path-independence at pH 8.0. For efficiency, we engineered many mutations at once to identify regions of OmpLA that would show a response to this strategy. Although the absolute free energies of folding in these engineered constructs would not be easily relatable to the native sequence, constructs displaying path-independent equilibrium folding will still serve as valid reference sequences for subsequent investigations of cooperative pair-wise interaction energies3 between side chains as well as protein-lipid interactions.1, 13 Our results show a partial reduction of the hysteresis suggesting that this strategy has potential as an engineering tool for rendering membrane proteins more amenable to thermodynamic analysis.
Materials and Methods
Engineering and Expression negative charge neutralization constructs
WT OmpLA was cloned as previously described.14 Variant mutations were chosen in different regions of the barrel; sites are shown in Table S1. Plasmids were cloned into the pET28b vector using gene synthesis by Genewiz with codon optimization; sequences of these variants are shown in Table S2. Initial expression and purification of these variants resulted in low yields. To increase yields, genes were transferred from pET28b to pET11a using the In-fusion kit (Clonetech). Variants and WT protein were transformed into hms174(DE3) cells, purified from inclusion bodies as previously described15 and stored at −20 °C before use. Inclusion body pellets were dissolved in 8 M GdnHCl (Fisher Scientific), filtered with a 0.22 µm filter, and the protein stock concentrations were diluted to 100 µM.
SDS-PAGE folding assays
Samples were folded under the same conditions as folding titrations as previously described6, 16. Final folded samples contained 1 M GdnHCl, 20 mM tris(hydroxymethyl)aminomethane (TRIS), 2 mM ethylenediaminetetraacetic acid (EDTA), 0.4 µM protein and a 2000:1 1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) ratio. Because GdnHCl precipitates in sodium dodecyl sulfate (SDS), samples were dialyzed in Slide-A-Lyzer cassettes with a 3.5 kDa molecular weight cut-off (Pierce) into 20 mM TRIS, 2 mM EDTA overnight at room temperature. Samples were then concentrated with 0.5 mL with PES spin concentrators with a 10 kDa molecular weight cutoff (Pierce) to a final concentration of 4 µM protein. SDS-PAGE was performed as in17 using 12% Mini-Protean TGX pre-cast gels (Bio-Rad). The ladder used was PageRuler pre-stained 10 to 180 kDa (Thermo).
Reversible folding titrations and quantitation of hysteresis
Reversible folding titrations were carried out as previously described2, 6. Experiments at pH 3.8 were conducted in 100 mM citrate, 2 mM EDTA; experiments at pH 8 were conducted in 20 mM TRIS and 2 mM EDTA. The wavelength of maximum emission, λmax, was determined by fitting two averaged fluorescence wavelength spectra to the spectral log normal equation16. Normalized intensities at 330 nm titrations were fitted with sigmoid equations modified with equations of lines in the baselines using Igor Pro to enable quantitation of hysteresis at pH 8. One sigmoid (Eq 1) was used to describe folding data while a sum of two sigmoid (Eq 2) functions was used to fit the unfolding data, which displayed an intermediate state. The single sigmoid equation used as a function of denaturant was:
| (Eq 1) |
where, x1 is the midpoint of the transition; r1 is a parameter that describes the steepness of the transition; I1 and m1 are the intercept and slope of the folded baseline, respectively; and I2 and m2 are the intercept and slope of the unfolded baseline, respectively. The equation with two sigmoid used as a function of denaturant was:
| (Eq 2) |
Where xi and ri describe the steepness of the transitions as indicated; Ii and mi describe the intercept and slopes of the two baselines for each sigmoid. Areas under the curves were calculated using trapezoidal integration using Igor Pro to y=0 with an x-range of 1–6 M GdnHCl. The area between any two curves was determined by subtraction of these quantities. This analysis was performed with two independent experiments, and data were normalized to WT levels.
Results
Variants with mutations in the loops and turns fold to 100% efficiencies in LUVs
Five different constructs were created to ascertain if there were regions of OmpLA responsible for the hysteresis in folding/unfold titrations under equilibrium conditions. Three of these constructs were engineered with Asp→Asn and Glu→Gln mutations within the soluble loop and turn regions of the transmembrane β-barrel. Shown in Figure 1a are variants “P” (turn region), “E” (loop region), and “EP” (sum of loop and turn regions). Specific sites of mutations are listed in Table S1, and the sequences are in Table S2. Because microbial OMPs show a heat modifiable migration between the unfolded and folded forms on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)14, we first used this assay to monitor the folded fraction of variants under the same conditions as in GdnHCl titrations. Figure 1b shows that these three variants all fold to 100%, similar to the WT protein.
Figure 1.
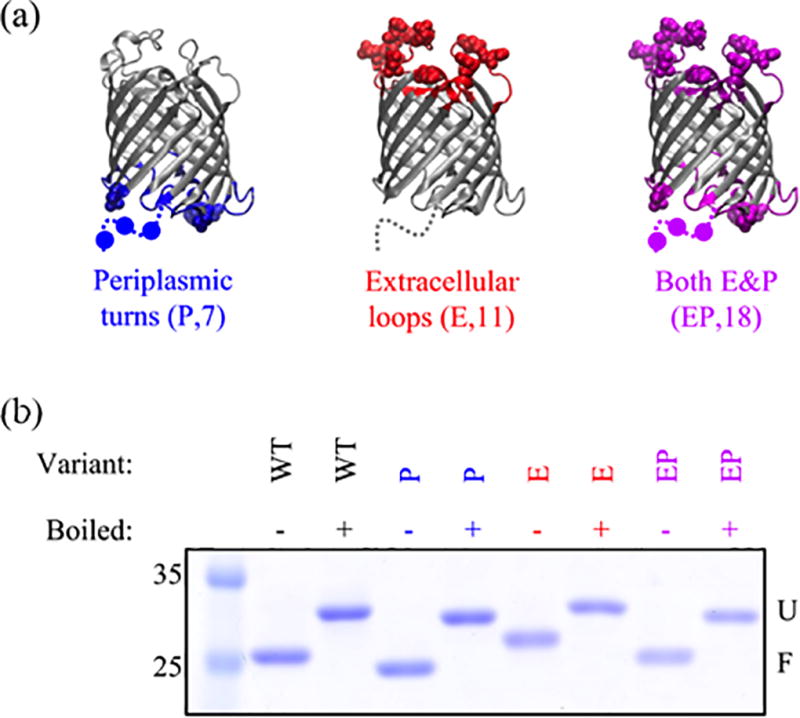
OmpLA variants with neutralizing mutations in the loops and turns adopt a folded state. (a) Illustration of constructs used in this study with mutations highlighted as spheres. Disordered regions in the crystal structure 1qd521 are shown as dashed lines and mutations as circles. Below each variant is the name; in parentheses are the name abbreviations and total number of mutations. The sequences are given in Supplementary Table 2. These images were created with VMD22. (b) SDS-PAGE gel showing differential migration for folded (not boiled) and unfolded (boiled) states. All samples were folded under solution conditions of 1 M GdnHCl, 20 mM TRIS (pH 8), 2 mM EDTA and at a 2000:1 DLPC to protein ratio. Although the folded form of the E variant is shifted to a higher molecular weight relative to WT, its pattern upon boiling is consistent with a protein that can fold, and we speculate may indicate differential binding of negatively charged SDS.
All loop and turn variants display path-independent folding at pH 3.8
We next assessed folding by conducting equilibrium denaturant titrations at pH 3.8 to test if the engineered constructs maintained reversible folding. The reversible folding of each variant at pH 3.8 was monitored using fluorescence spectroscopy. Figure S1 shows these data in which three parameters were used to monitor folding and unfolding titrations: (i) wavelength of maximum emission (λmax); (ii) Rayleigh-Gans-Debye (RGD) scattering; and (iii) fluorescence intensity at 330 nm as previously described.6, 16 Figure S1 shows there is excellent agreement between all parameters for all variants demonstrating path-independent folding. Upon fitting the intensity at 330 nm in Figure S1, we found the folding free energies of the P, E, and EP variants are all within error of WT. This analysis indicates these mutations do not perturb the reversible folding behavior at pH 3.8.
Neutralizing variants in the soluble loop and turn regions fold and unfolded to similar conformations as the WT protein and result in a reduction in hysteresis at pH 8.0
We next tested these engineered variants for path-independent folding at pH 8.0. We first examined the fluorescence spectra in the folded and unfolded baseline regions. These indicate that the endpoint conformations of the loop and turn constructs are very similar to that of WT OmpLA, which means that the overall folding reactions are comparable. The unfolded states of variants at pH 8 were off the membrane (Figure S2) and the wavelength of maximum emission in this denatured state ensemble was within error of WT (Figure S3). This is an interesting finding because point mutations to charged residues have been shown to perturb the denatured states of a small number of soluble proteins when using urea as the denaturant.18 In contrast, the denatured states of the variants in this study do not appear largely altered. We attribute this to the ionic nature of the denaturant GdnHCl, which is a salt and could therefore screen the charges in the denatured state ensemble. Under folding conditions, as shown in Figure S4, the fluorescence spectra of the variant folded states are also very similar to each other and to WT except for a small shift to longer wavelengths in the E and EP variants. This might be due to mutations affecting local environment of tryptophan side chains (OmpLA has 9 in total). For example, there is a tryptophan in the extracellular loops where many of the mutations may affect the conformational dynamics, as illustrated in Figure S4C.
Despite having similar endpoint conformations of the WT protein, hysteresis is still observed for all constructs at pH 8.0 (Figure 2 and S5). This is demonstrated by a lack of complete overlay of the two titrations by fluorescence (both λmax and intensity at 330 nm). This hysteresis persists in these equilibrium experiments even though the reactions are reversible in the sense that the folded and unfolded baselines overlay (e.g. left and right extremes of each plot). Although the mid-regions of folding and unfolding titrations do not coincide, there is still an improvement as indicated by a reduction in the area between titrations for some variants as compared to WT. To quantify the hysteresis reduction, these data were fitted to sigmoid functions, and the areas between curves were determined using trapezoidal integration. Typical titrations are shown in Figure 2a with the shaded area illustrating the area in between curves. In Figure 2b the reduction is shown as percent hysteresis relative to the WT signal. The P, E, and EP variants show a reduction of hysteresis by 17, 54, and 61% respectively.
Figure 2.
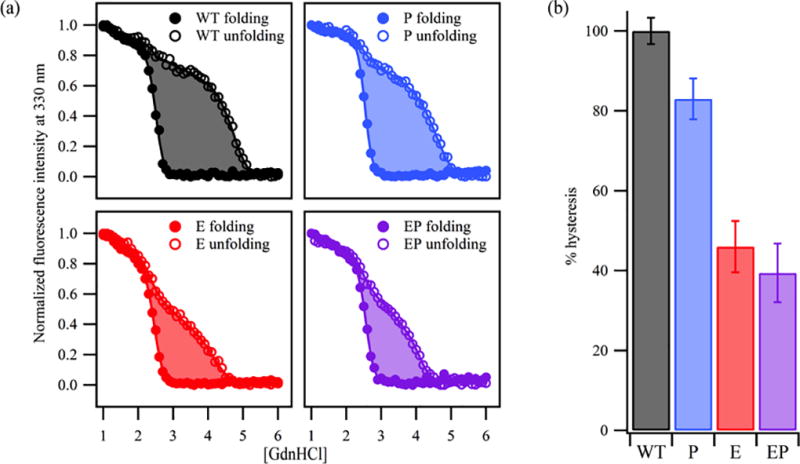
Negative charge neutralization in the loops and turns reduces hysteresis by 60% at pH 8 relative to WT. (a) Representative titrations illustrating the area in between sigmoidal fits to the data. Hysteresis is reduced further as the number of mutations increase. (b) The area between curves was quantitated from two titrations and standard errors are shown. These data have been normalized by the WT area; numbers are given in Table S3.
Reduction of the hysteresis appears to be due to a collapse of the unfolding titration towards lower denaturant concentrations; by comparison the folding titrations of the engineered variants are minimally perturbed and are similar to that of the WT. The unfolding titrations at pH 8.0 also displayed a pronounced intermediate that is reminiscent of the three-state equilibrium intermediate observed at pH 3.8. Intriguingly, mutations introducing polar or ionizable side chains at position 210 enhance the population of the equilibrium intermediate at pH 3.8 whereas neutralizing mutations in this study appear to reduce its appearance.2 A more rigorous investigation of this curious finding must await the achievement of path-independence at pH 8.0.
Variants neutralizing acidic groups in the β-barrel interior do not fold
Two additional constructs shown in Figure S6a were engineered with negative charge neutralization mutations within the barrel (“B”) or with all (“A”) of the negative charges mutated. Neither of these latter variants folded as indicated by the absence of a gel-shift by SDS-PAGE (Figure S6b) and by their fluorescence spectra. As shown in Figure S6c, the fluorescence spectra under folding conditions (low [GdnHCl]) either in the presence or absence of LUVs are the same for the B and A variants, while they are different for the WT protein. To confirm this finding, we performed GdnHCl titrations without LUVs, which detect the fluorescence changes due to aggregation. These experiments confirmed that the B and A variants are aggregating in titrations and the negative charges that point inside the barrel are required for folding.
Discussion
Using a protein engineering approach we demonstrated that Asp→Asn and Glu→Gln mutations can produce a significant reduction in the hysteresis gap present in equilibrium folding titrations of the membrane protein OmpLA. Mutations in the soluble extracellular loop regions showed the largest reduction in hysteresis, consistent with the idea that those soluble domains of the protein must cross the bilayer during equilibrium titrations as they do in the biological context. A further decrease in the hysteresis was observed when acidic side chains in the periplasmic turn region were neutralized by mutation. This result is less straightforward to explain because these regions of the protein are not expected to cross the bilayer, however one possibility is that they may have non-native interactions at some rate-limiting step during the folding or unfolding reaction. Nevertheless, because path-independent equilibrium titrations are still achieved for both of these constructs at pH 3.8 but not neutral pH, we conclude that the physical basis of this reduction in hysteresis is due to the neutralization of one or more negative charges in the extra-membranous loops and/or turns of OmpLA.
Mutations neutralizing acidic residues located within the transmembrane β-barrel region were inconclusive because the large number we introduced all at once (13 in total) resulted in constructs that did not fold. We hypothesize that this result may reflect the disruption of specific side chain interactions within the barrel. In particular, removing ionizable acidic side chains in this region creates the potential for unsatisfied ionizable basic side chains in the water-filled interior of the barrel. A more fine-grained approach will therefore be required to fully address the contributions of this region to equilibrium hysteresis. It should be recognized that the combinatorial nature of mutating the acidic residues within the barrel region in OmpLA quickly results in a large number of engineered variants. However, based on the promising results with the extra-membranous regions of OmpLA, such mutational efforts may warrant further investigation. Because side chain pKa values have been shown to be altered significantly by the local environment19, a solution to this challenging biophysical problem would also lead to an improved understanding of side chain pKa determinants in membrane proteins and how they contribute to protein folding and stability.
Interestingly, for the constructs showing a reduction in hysteresis, we found distinct differences in the folding and unfolding titration responses to the engineered mutations. The folding titrations are largely unaffected by mutations and thus not sensitive to the charge neutralization changes on acidic side chains. In contrast, the unfolding titrations moved to lower GdnHCl concentrations, reducing the equilibrium hysteresis. One speculative explanation for this result is that the folding titrations are not rate limiting in this context because outer membrane β-barrel sequences have evolved to solve the problem of moving charges across the membrane during folding. But because they do not unfold in the biological setting, the sequences have not experienced evolutionary pressure to develop mechanisms to unfold. Thus, the unfolding half of the equilibrium titration experiment may be more amenable to sequence changes that accelerate its rate. It is worth noting that a lack of unfolding capability is not necessarily a problem in the cellular environment because bacteria are increasing in membrane area as they grow and divide. In addition, mini-vesicles containing outer membrane components are released from gram-negative bacteria,20 further diluting OMP numbers in cells. These growth characteristics in outer membranes may explain the absence of an evolutionary pressure for fast OMP unfolding reactions.
Supplementary Material
Acknowledgments
We thank Dr. Nathan Zaccai for help with the design of the variants in this study.
FUNDING SOURCES: This work was funded by grants NIH R01 GM079440 and T32 GM008403.
ABBREVIATIONS
- ΔASA
Change in accessible surface area
- DLPC
1,2-dilauroyl-sn-glycero-3-phosphocholine
- EDTA
Ethylenediaminetetraacetic acid
- E
Extracellular loop variant
- EP
Extracellular loop and periplasmic turn variant
- GdnHCl
Guanidine hydrochloride
- LUVs
Large unilamellar vesicles
- OMPs
Outer membrane proteins
- OmpA
Outer membrane protein A
- OmpLA
Outer membrane phospholipase A
- P
Periplasmic turn variant
- RGD
Rayleigh-Gans-Debye
- SDS
Sodium dodecyl sulfate
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TRIS
Tris(hydroxymethyl)aminomethane
- λmax
wavelength of maximum emission
- WT
Wild-type
Footnotes
ASSOCIATED CONTENT
The following files are available free of charge.
Combined SI with text and figures (PDF)
AUTHOR CONTRIBUTIONS
SKM and KGF designed experiments. SKM conducted all experiments. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Literature Cited
- 1.Hong H, Park S, Jimenez RH, Rinehart D, Tamm LK. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J. Am. Chem. Soc. 2007;129:8320–8327. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- 2.Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc Natl Acad Sci U S A. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong H, Tamm LK. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. 2004;101:4065–4070. doi: 10.1073/pnas.0400358101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huysmans GH, Baldwin SA, Brockwell DJ, Radford SE. The transition state for folding of an outer membrane protein. Proc Natl Acad Sci U S A. 2010;107:4099–4104. doi: 10.1073/pnas.0911904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanley AM, Fleming KG. The process of folding proteins into membranes: challenges and progress. Archives of biochemistry and biophysics. 2008;469:46–66. doi: 10.1016/j.abb.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 6.Moon CP, Kwon S, Fleming KG. Overcoming Hysteresis to Attain Reversible Equilibrium Folding for Outer Membrane Phospholipase A in Phospholipid Bilayers. J. Mol. Biol. 2011;413:484–494. doi: 10.1016/j.jmb.2011.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming KG. Energetics of membrane protein folding. Annu Rev Biophys. 2014;43:233–255. doi: 10.1146/annurev-biophys-051013-022926. [DOI] [PubMed] [Google Scholar]
- 8.Andrews BT, Capraro DT, Sulkowska JI, Onuchic JN, Jennings PA. Hysteresis as a Marker for Complex, Overlapping Landscapes in Proteins. J Phys Chem Lett. 2013;4:180–188. doi: 10.1021/jz301893w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Street TO, Courtemanche N, Barrick D. Protein folding and stability using denaturants. Methods Cell Biol. 2008;84:295–325. doi: 10.1016/S0091-679X(07)84011-8. [DOI] [PubMed] [Google Scholar]
- 10.Moon CP, Zaccai NR, Fleming PJ, Gessmann D, Fleming KG. Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proc. Natl. Acad. Sci. 2013;110:4285–4290. doi: 10.1073/pnas.1212527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes WM. CRC Handbook of Chemistry and Physics. 92. CRC Press; Hoboken: 2011. 92th ed. [Google Scholar]
- 12.Slusky JS, Dunbrack RL., Jr Charge asymmetry in the proteins of the outer membrane. Bioinformatics. 2013;29:2122–2128. doi: 10.1093/bioinformatics/btt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald SK, Fleming KG. Aromatic Side Chain Water-to-Lipid Transfer Free Energies Show a Depth Dependence across the Membrane Normal. J Am Chem Soc. 2016;138:7946–7950. doi: 10.1021/jacs.6b03460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgess NK, Dao TP, Stanley AM, Fleming KG. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J. Biol. Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plummer AM, Gessmann D, Fleming KG. The Role of a Destabilized Membrane for OMP Insertion. Methods Mol Biol. 2015;1329:57–65. doi: 10.1007/978-1-4939-2871-2_5. [DOI] [PubMed] [Google Scholar]
- 16.Moon CP, Fleming KG. Using Tryptophan Fluorescence to Measure the Stability of Membrane Proteins Folded in Liposomes. Method Enzymol. 2011;492:189–211. doi: 10.1016/B978-0-12-381268-1.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook MRGaJ. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 18.Cho JH, Meng WL, Sato S, Kim EY, Schindelin H, Raleigh DP. Energetically significant networks of coupled interactions within an unfolded protein. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12079–12084. doi: 10.1073/pnas.1402054111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castaneda CA, Fitch CA, Majumdar A, Khangulov V, Schlessman JL, Garcia-Moreno BE. Molecular determinants of the pKa values of Asp and Glu residues in staphylococcal nuclease. Proteins. 2009;77:570–588. doi: 10.1002/prot.22470. [DOI] [PubMed] [Google Scholar]
- 20.Henry T, Pommier S, Journet L, Bernadac A, Gorvel JP, Lloubes R. Improved methods for producing outer membrane vesicles in Gram-negative bacteria. Res Microbiol. 2004;155:437–446. doi: 10.1016/j.resmic.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, Dijkstra BW. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 22.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


