Abstract
SNCA missense mutations are a rare cause of autosomal dominant Parkinson’s disease (PD). To date, six missense mutations in SNCA have been nominated as causal. Here, we assess the frequency of these six mutations in public population databases and PD case-control datasets in order to determine their true pathogenicity. We found that one of the six reported SNCA mutations, His50Gln, was consistently identified in large population databases and no enrichment was evident in PD cases compared to controls. These results suggest that His50Gln is probably not a pathogenic variant. This information is important to provide counseling for His50Gln carriers and has implications for the interpretation of His50Gln α-synuclein functional investigations.
Introduction
Parkinson’s disease (PD) disease is one of the most common neurodegenerative disorders. The pathological hallmark of PD are Lewy bodies, which are neuronal cytoplasmic inclusions consisting of misfolded α-synuclein encoded by the SNCA gene (Spillantini et al., 1997). To date, six missense mutations in the SNCA gene have been reported to cause PD: three well established mutations (Ala30Pro, Glu46Lys and Ala53Thr) and three more recently described mutations (His50Gln, Gly51Asp and Ala53Glu) (Figure 1 and Table 1). Whilst atypical presentations and a later onset have been reported, SNCA mutation carriers typically develop autosomal dominant early-onset PD characterized by a severe, rapidly progressive course and cognitive decline that commonly progresses to Lewy body dementia (Papadimitriou et al., 2016; Trinh et al., 2014). A fuller understanding of exactly which mutations are truly causal for PD will help direct research on the pathophysiology of PD driven by SNCA mutations, and is of crucial importance for counseling of mutation carriers and their family members. Here, we explore the frequency and spectrum of these different SNCA mutations in several large public datasets, and then examine their presence in several large PD case-control datasets.
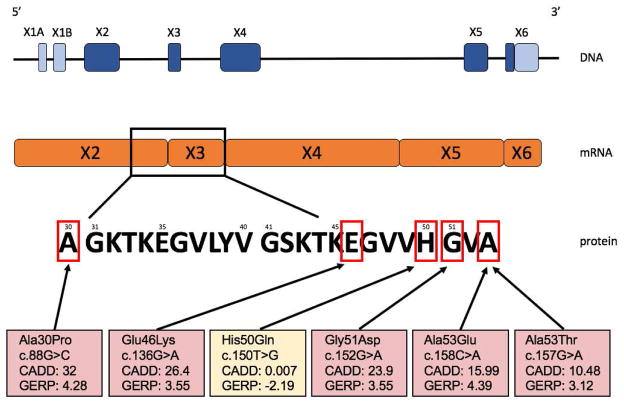
Figure 1. Overview of the SNCA gene on DNA, mRNA and protein level.
The SNCA gene has six exons of which several are non-coding (light-blue). NB the SNCA gene is located on the antisense strand of the human genome. On mRNA level five exons are left totaling 423 nucleotides (NM_000345) which result in a 141-amino acid protein. All six missense mutations are located in exon two and three in relatively close proximity. When comparing pathogenicity algorithm scores (CADD (Quang et al., 2015), GERP (Davydov et al., 2010)) between the six missense variants, His50Gln scores very poor compared to the other five missense mutations.
Table 1. Prevalence of six reported pathogenic SNCA mutations and LRRK2 Gly2019Ser in large population databases and pathogenicity algorithm scores.
Twenty-two individuals in gnomAD (3 Africans and 19 Europeans, n=138,587) and seven individuals in HRC (all Europeans, n=32,488) were heterozygous for SNCA His50Gln (Lek et al., 2016; McCarthy et al., 2016). The LRRK2 Gly2019Ser mutation was found in 136 individuals in gnomAD (3 African, 8 Latino, 118 European [including Ashkenazi Jewish] and 7 other) and 13 individuals in the HRC (all Europeans). gnomAD = Genome Aggregation Database, HRC = Haplotype Reference Consortium, D = damaging, T = tolerant, B = benign.
| Gene: amino acid change | rs number | chr:bp (hg19) | gnomAD (frequency) | HRC (frequency) | SIFT (score) | Polyphen2 (score) | CADD phred | DANN score | GERP++ RS |
|---|---|---|---|---|---|---|---|---|---|
| SNCA: Ala30Pro (Kruger et al., 1998) | rs104893878 | 4:90756731 | 0 | 0 | D (0.001) | D (0.996) | 32 | 0.998 | 4.28 |
| SNCA: Glu46Lys (Zarranz et al., 2004) | rs104893875 | 4:90749321 | 0 | 0 | D (0.006) | B (0.426) | 26.4 | 0.998 | 3.55 |
| SCNA: His50Gln (Appel-Cresswell et al., 2013; Proukakis et al., 2013) | rs201106962 | 4:90749307 | 22 (0.0079%) | 7 (0.011%) | T (1) | B (0.012) | 0.007 | 0.425 | −2.19 |
| SNCA: Gly51Asp (Kiely et al., 2013; Lesage et al., 2013) | rs431905511 | 4:90749305 | 0 | 0 | D (0.004) | D (0.999) | 23.9 | 0.996 | 3.55 |
| SNCA: Ala53Glu (Pasanen et al., 2014) | n/a | 4:90749299 | 0 | 0 | D (0.007) | B (0.015) | 15.99 | 0.952 | 4.39 |
| SNCA: Ala53Thr (Polymeropoulos et al., 1997) | rs104893877 | 4:90749300 | 0 | 0 | T (1) | B (0) | 10.48 | 0.203 | 3.12 |
| LRRK2: Gly2019Ser (Gilks et al., 2005) | rs34637584 | 12:40734202 | 136 (0.049%) | 13 (0.020%) | D (0) | D (1) | 35 | 0.998 | 5.69 |
Results
The only SNCA missense mutation identified in the population databases was His50Gln (Table 1). To assess whether the His50Gln mutation is found in PD cases, we accessed several PD case/control datasets, which cumulatively totaled 11,095 PD cases and 12,615 controls. From these data, we identified two controls and one case carrying the SNCA His50Gln mutation. Additionally, two PD cases carrying Ala53Thr and a single PD case with Gly51Asp mutation were found (Table 2 and Supplementary data). We next assessed pathogenicity prediction algorithms for all six SNCA mutations and overall SNCA His50Gln scored poorly compared to the other five SNCA mutations (Figure 1, Table 1 and Supplementary data). For comparison, we used the known pathogenic LRRK2 Gly2019Ser mutation as it is also present in the general population (Table 2). Unlike for SNCA His50Gln, analysis in PD case-control datasets revealed a consistent increase in the frequency of LRRK2 Gly2019Ser mutation carriers amongst PD cases than controls (Table 2).
Table 2.
Frequency of SNCA His50Gln and LRRK2 Gly2019Ser in large PD case/control datasets.
| Dataset | Population description | No. controls | No. cases | His50Gln alleles in controls (frequency) | His50Gln alleles in cases (frequency) | Gly2019Ser alleles in controls (frequency) | Gly2019Ser alleles in cases (frequency) | Average sequencing coverage at SNCA locus |
|---|---|---|---|---|---|---|---|---|
| IPDGC whole-exome sequencing | Mainly European ancestry | 5,774 | 2,440 | 1 (0.0087%) | 0 | 4 (0.0346%) | 25 (0.51%) | >35x |
| IPDGC resequencing data | Mainly European ancestry | 2,391 | 3,481 | 1 (0.021%) | 0 | 0 | 56 (0.80%) | >60x |
| McGill resequencing data1 | Mainly European and Jewish ancestry | 2,460 | 2,175 | 0 | 0 | 11 (0.22%) | 125 (2.9%) | >300x |
| COURAGE-PD resequencing data2 | Both Asian and European ancestry | 1,490 | 1,490 | 0 | 0 | 0 | 6 (0.20%) | >400x |
| French PDG group resequencing data | Mainly of French European origin | 500 | 1,509 | 0 | 2 (1 case in HMZ state) (0.07%) | 0 | 42 (1 HMZ + 40 HTZ) (1.39%) | >260x |
| Total | 12,615 | 11,095 | 2 (0.0079%) | 0 (0.0090%) | 15 (0.059%) | 254 (1.1%) |
HMZ = homozygous and HTZ = Heterozygous.
Samples included cohorts from Quebec, France, Israel and Columbia University NY (Alcalay et al., 2015).
Allele counts in the COURAGE-PD dataset were called from sequencing of DNA pools representing 10 cases or 10 controls. Average coverage is given per sample pool. For subsets of COURAGE-PD participants with available data on LRRK2 Gly2019Ser status, known mutation carriers have been excluded from the resequencing study.
Discussion
Here, we examined the presence of reported pathogenic SNCA missense mutations in large population control databases and identified that His50Gln is relatively frequent in both the European and African population. In contrast, the other five reported pathogenic mutations were not observed in these control databases. Follow-up analysis in large PD case-control cohorts identified two additional control individuals carrying this variant, representing a similar frequency to the public population databases. We identified the His50Gln mutation in a homozygous state in one sporadic early-onset PD case with an age at onset at 32 years and two heterozygous controls with last known ages of 62 and 89. Notably, two other SNCA mutations, Ala53Thr and Gly51Asp, were found twice and once respectively in cases, demonstrating the power of our large dataset to detect rare mutations in the SNCA gene. The SNCA His50Gln case presented with a classic PD phenotype and was free of dementia and cognitive decline after almost 10 years of disease, which is unusual for patients with pathogenic SNCA missense mutations (Papadimitriou et al., 2016). Currently, with the lack of other homozygous cases or controls there is insufficient evidence to conclude that the His50Gln mutation is pathogenic in a homozygous state. One possibility is that the His50Gln mutation has reduced penetrance, as has been reported for other PD mutations, such as LRRK2 Gly2019Ser (Healy et al., 2008; Latourelle et al., 2008). However, the lack of enrichment of SNCA His50Gln in PD cases versus controls argues against this and this contrasts with the enrichment observed for the LRRK2 Gly2019Ser mutation, Table 2. Assuming a life-time risk of 1.3–2% (depending on sex) to be diagnosed with PD (Elbaz et al., 2002), one would expect ~2,600 individuals from gnomAD to develop PD. If the SNCA His50Gln mutation is indeed pathogenic, fully penetrant and inherited in an autosomal dominant fashion, the 22 carriers of this allele would represent around ~1% of all PD cases. Similarly, we would expect to identify over 100 PD patients in our PD case cohorts. This was not observed and argues against pathogenicity of this vairant. In general, segregation data, case-control enrichments, absence in population databases and pathogenicity prediction algorithms are considered important criteria for establishing the causality of sequence variants for genetic disease (MacArthur et al., 2014; Richards et al., 2015), however the SNCA His50Gln mutation does not fulfill any of these criteria (Table 1). In conclusion, while it is tempting to speculate about the pathogenicity of SNCA His50Gln, especially given limited in vitro evidence indicating an increased propensity to form α-synuclein fibrils (Rutherford et al., 2014), we conclude that insufficient evidence exists to nominate the His50Gln mutation as a causative mutation or high risk mutation. This finding has important implications for the interpretation of functional investigations of His50Gln mutated α-synuclein isoforms as well as for future study design. Furthermore, when identifying the SNCA His50Gln mutation in either a patient or an asymptomatic individual, caution should be used by clinicians and genetic counselors, as genetic evidence suggests this is a rare benign variant.
Supplementary Material
Highlights.
Publicly available data show that SNCA His50Gln has a frequency of ~0.01% in Europeans
SNCA His50Gln was reported to cause Parkinson’s disease, but it is too common to do so
There is no evidence of variant enrichment in Parkinson’s disease cases compared to controls
Acknowledgments
The authors would like to thank all members of the IPDGC (http://pdgenetics.org/partners) and COURAGE-PD consortia for proving data, support and comments. This work was supported (in part) by the Intramural Research Program of the National Institutes of Health (National Institute of Neurological Disorders and Stroke, National Institute on Aging; projects 1ZIA-NS003154-2 and Z01-AG000949) and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. Data used in the preparation of this article were obtained from the Parkinson’s Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data). For up-to-date information on the study, visit www.ppmi-info.org. PPMI - a public-private partnership - is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, including Abbvie, Avid, Biogen, Bristol-Myers Squibb, Covance, GE Healthcare, Genentech, GlaxoSmithKline, Lilly, Lundbeek, Merck, Meso Scale Discovery, Pfizer, Piramal, Roche, Servier, Teva, UCB, and Golub Capital. The study on the cohort from McGill University was financially supported by the Michael J. Fox Foundation and by the Canadian Consortium on Neurodegeneration in Aging (CCNA). The study on the cohort from France was performed by the French Parkinson’s disease genetics study group (PDG) and supported by the France Parkinson’s association. Also we thank dr. Thibaud Lebouvier as the neurologist who examined the homozygous His50Gln SNCA patient. Columbia University Spot cohort was funded by the NIH (K02NS080915) and the Parkinson’s Disease Foundation. We thank Guy Rouleau, Jennifer Ruskey, Sandra B Laurent, Pascale Hince, Dan Spiegelman, Alexandre Dionne-Laporte, Helene Catoire, Cynthia Bourassa, Pierre Provencher, Cathy Mirarchi and Vessela Zaharieva for their assistance. We thank the Quebec Parkinson’s Network and its members (http://rpq-qpn.ca/) for their collaboration. COURAGE-PD (COmprehensive Unbiased Risk factor Assessment for Genetics and Environment in Parkinson’s Disease) is a transnational project funded by the EU Joint Programme - Neurodegenerative Disease Research (JPND). transnational project funded by the EU Joint Programme - Neurodegenerative Disease Research (JPND).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcalay RN, Levy OA, Waters CC, Fahn S, Ford B, Kuo SH, Mazzoni P, Pauciulo MW, Nichols WC, Gan-Or Z, Rouleau GA, Chung WK, Wolf P, Oliva P, Keutzer J, Marder K, Zhang X. Glucocerebrosidase activity in Parkinson’s disease with and without GBA mutations. Brain. 2015;138(Pt 9):2648–2658. doi: 10.1093/brain/awv179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel-Cresswell S, Vilarino-Guell C, Encarnacion M, Sherman H, Yu I, Shah B, Weir D, Thompson C, Szu-Tu C, Trinh J, Aasly JO, Rajput A, Rajput AH, Jon Stoessl A, Farrer MJ. Alpha-synuclein p.H50Q, a novel pathogenic mutation for Parkinson’s disease. Mov Disord. 2013;28(6):811–813. doi: 10.1002/mds.25421. [DOI] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz A, Bower JH, Maraganore DM, McDonnell SK, Peterson BJ, Ahlskog JE, Schaid DJ, Rocca WA. Risk tables for parkinsonism and Parkinson’s disease. J Clin Epidemiol. 2002;55(1):25–31. doi: 10.1016/s0895-4356(01)00425-5. [DOI] [PubMed] [Google Scholar]
- Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, Lynch J, Healy DG, Holton JL, Revesz T, Wood NW. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet. 2005;365(9457):415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- Healy DG, Falchi M, O’Sullivan SS, Bonifati V, Durr A, Bressman S, Brice A, Aasly J, Zabetian CP, Goldwurm S, Ferreira JJ, Tolosa E, Kay DM, Klein C, Williams DR, Marras C, Lang AE, Wszolek ZK, Berciano J, Schapira AH, Lynch T, Bhatia KP, Gasser T, Lees AJ, Wood NW, International LC. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol. 2008;7(7):583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiely AP, Asi YT, Kara E, Limousin P, Ling H, Lewis P, Proukakis C, Quinn N, Lees AJ, Hardy J, Revesz T, Houlden H, Holton JL. alpha-Synucleinopathy associated with G51D SNCA mutation: a link between Parkinson’s disease and multiple system atrophy? Acta Neuropathol. 2013;125(5):753–769. doi: 10.1007/s00401-013-1096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18(2):106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- Latourelle JC, Sun M, Lew MF, Suchowersky O, Klein C, Golbe LI, Mark MH, Growdon JH, Wooten GF, Watts RL, Guttman M, Racette BA, Perlmutter JS, Ahmed A, Shill HA, Singer C, Goldwurm S, Pezzoli G, Zini M, Saint-Hilaire MH, Hendricks AE, Williamson S, Nagle MW, Wilk JB, Massood T, Huskey KW, Laramie JM, DeStefano AL, Baker KB, Itin I, Litvan I, Nicholson G, Corbett A, Nance M, Drasby E, Isaacson S, Burn DJ, Chinnery PF, Pramstaller PP, Al-hinti J, Moller AT, Ostergaard K, Sherman SJ, Roxburgh R, Snow B, Slevin JT, Cambi F, Gusella JF, Myers RH. The Gly2019Ser mutation in LRRK2 is not fully penetrant in familial Parkinson’s disease: the GenePD study. BMC Med. 2008;6:32. doi: 10.1186/1741-7015-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation C. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Anheim M, Letournel F, Bousset L, Honore A, Rozas N, Pieri L, Madiona K, Durr A, Melki R, Verny C, Brice A French Parkinson’s Disease Genetics Study G. G51D alpha-synuclein mutation causes a novel parkinsonian-pyramidal syndrome. Ann Neurol. 2013;73(4):459–471. doi: 10.1002/ana.23894. [DOI] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, Conrad DF, Cooper GM, Cox NJ, Daly MJ, Gerstein MB, Goldstein DB, Hirschhorn JN, Leal SM, Pennacchio LA, Stamatoyannopoulos JA, Sunyaev SR, Valle D, Voight BF, Winckler W, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508(7497):469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki AE, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PI, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R Haplotype Reference C. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou D, Antonelou R, Miligkos M, Maniati M, Papagiannakis N, Bostantjopoulou S, Leonardos A, Koros C, Simitsi A, Papageorgiou SG, Kapaki E, Alcalay RN, Papadimitriou A, Athanassiadou A, Stamelou M, Stefanis L. Motor and Nonmotor Features of Carriers of the p. A53T Alpha-Synuclein Mutation: A Longitudinal Study. Mov Disord. 2016;31(8):1226–1230. doi: 10.1002/mds.26615. [DOI] [PubMed] [Google Scholar]
- Pasanen P, Myllykangas L, Siitonen M, Raunio A, Kaakkola S, Lyytinen J, Tienari PJ, Poyhonen M, Paetau A. Novel alpha-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson’s disease-type pathology. Neurobiol Aging. 2014;35(9):2180e2181–2185. doi: 10.1016/j.neurobiolaging.2014.03.024. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Proukakis C, Dudzik CG, Brier T, MacKay DS, Cooper JM, Millhauser GL, Houlden H, Schapira AH. A novel alpha-synuclein missense mutation in Parkinson disease. Neurology. 2013;80(11):1062–1064. doi: 10.1212/WNL.0b013e31828727ba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang D, Chen Y, Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31(5):761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Moore BD, Golde TE, Giasson BI. Divergent effects of the H50Q and G51D SNCA mutations on the aggregation of alpha-synuclein. J Neurochem. 2014;131(6):859–867. doi: 10.1111/jnc.12806. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Trinh J, Guella I, Farrer MJ. Disease penetrance of late-onset parkinsonism: a meta-analysis. JAMA Neurol. 2014;71(12):1535–1539. doi: 10.1001/jamaneurol.2014.1909. [DOI] [PubMed] [Google Scholar]
- Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, Llorens V, Gomez Tortosa E, del Ser T, Munoz DG, de Yebenes JG. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55(2):164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.