Abstract
Dengue is the most common mosquito-borne viral infection in tropical and sub-tropical countries. In recent years, India has reported increased incidences of concurrent infection with multiple serotypes of dengue viruses (DENV). In the present study, we have characterized DENV circulating during a single season of 2016 in Pune, India. A total of 64 serum samples from NS1 ELISA positive dengue patients were used for PCR amplification of CprM region of the viral genome and sequencing. Phylogenetic analysis documented circulation of all the four DENV serotypes with predominance of DENV-2 (40.6%). DENV genotyping classified DENV-1 to Genotype V, DENV-2 to Genotype IV, DENV-3 to Genotype III and DENV-4 to Genotype I. Further analysis revealed emergence of a novel clade (D) of genotype I of DENV-4. Subsequent isolation of three DENV-4 viruses in cell culture followed by complete genome sequence analysis confirmed this observation. Additionally, a new genotype within serotype-4 with >6.7% sequence variation from other genotypes was identified. This first report of significant co-circulation of all the four serotypes in a single outbreak in Pune reconfirms need for molecular monitoring of DENV.
Introduction
An estimated 40% of the global population (~3.9 billion) is at risk of dengue virus (DENV) infection [1, 2]. About 2.5% of people affected with severe dengue die each year [3]. The disease is endemic in more than 125 countries and the spread to newer areas is mainly attributed to returning travelers from endemic countries [4, 5]. There are four serotypes of DENV (DENV-1 to -4) and all of them can cause dengue fever (DF), a self-limiting febrile illness. A variable proportion of patients progress to life threatening dengue hemorrhagic fever (DHF) characterized by thrombocytopenia and hemorrhage, and dengue shock syndrome (DSS) due to excessive plasma leakage [6, 7].DENV has been in circulation in the Indian subcontinent since 1950s [8]. The first virologically proven epidemic of DF occurred in Kolkata in 1963–1964 and at present the virus has spread to 35 states and union territories in the country (NVBDCP, http://nvbdcp.gov.in/den-cd.html) [9].
On account of sequence variability, dengue serotypes are further classified into distinct genotypes that differ >6% within a single serotype [10–12].Emergence of new serotype or lineage \ clade shifts in circulating DENV genotypes led to enhanced severity during dengue outbreaks [13–17]. A lineage shift in DENV-3 was reported to cause severe disease in Sri Lanka [13, 18]. Emergence of genotype III of DENV-3in 2005 resulted in dengue outbreak in Northern India [19]. Recently, emergence of Asian or genotype I of DENV-1 also caused large outbreak of dengue with 12,000 cases in Tamil Nadu, South India [20].
All the four serotypes of DENV have circulated in India at different times, but generally one serotype dominates a given outbreak. Dengue outbreak in 1996 in Delhi was caused by genotype IV of DENV-2 replacing genotype V isolates of 1957 and 1967[21]and virus remained in circulation till 2002. Second outbreak in 2003 in Delhi was due to emergence of DENV-3 which remained as dominant serotype till 2006 [19]. Over a period from 2007–2009, DENV-1 became the predominant serotype in Delhi by replacing DENV-2 and DENV-3 [22]. Earlier dengue outbreaks were attributed to sudden emergence of serotype or genotype that co-circulate along with existing genotype for some time before getting replaced by others in subsequent years. In recent years, co-circulation of multiple serotypes has been reported from different parts of India [23]. High percentage of co-infection with more than one serotype was also observed with increased disease severity [24–26]. In 2017, co-circulation of all four DENV serotypes in single outbreak was reported from Odisha [27] and Hyderabad [28].
Pune city, western India with a population of 112 million (census 2011) is endemic for dengue [29]. In view of the possibility of introduction of dengue vaccine in near future, it is essential to understand the type and proportion of circulating DENV strains. The present study reports molecular characterization of dengue viruses circulating in Pune during the 2016-dengue season.
Methods
Sample collection
Patients presenting with dengue like symptoms for < 4days to the Medicine and Pediatric OPDs of the Bharati hospital, a tertiary care hospital from Pune were included in the study. To avoid second prick, consent for the use of blood sample for dengue molecular studies was obtained from all the suspected patients. This included written informed consent from the parents (subjects below 7 years of age), written informed assent and consent (subjects and their parents respectively, age group 7–17 years) and written informed consent (subjects above 17 years of age). NS1 positive (Dengue Early ELISA, Panbio, Windsor, Qld, Australia), leftover serum samples (n = 120) were collected from the diagnostic laboratory of the hospital. The study was approved by Institutional Ethics Committee, Bharati Vidyapeeth Deemed University, Pune with approval number IEC/2017/04.
Virus isolation
One day prior to infection, 1 x 104 Vero cells were seeded in each well of 96-well plate and incubated at 37°C in 5% CO2 incubator. 100μl of 10-fold serially diluted patient’s serum in quadruplate wells in 96-well plate was used to infect Vero cells grown in Minimum Essential Medium (MEM, Gibco, Thermo Scientific) containing 2% fetal bovine serum, 1% penicillin and streptomycin. 7 days post infection, culture supernatant was harvested from NS1 positive wells and aliquots were stored at -80°C.
Viral RNA extraction
Total RNA was extracted from 140μl of human serum or cell culture isolates using a QIAmp viral RNA kit (QIAGEN, INC, Valencia, CA), as per manufacturer’s protocol. RNA was eluted in 50μl of AVE buffer provided with the kit. For a conventional gel-based PCR, a minimum of one negative control every four samples with no presence of target RNA was included as a part of the extraction procedure.
cDNA synthesis
Dengue specific viral RNA was reverse transcribed and amplified for CprM region of the viral genome as reported by Chien et al (2006) [30]. For this, single-stranded cDNA was synthesized from total RNA using the high capacity cDNA reverse transcription kit (Invitrogen). Briefly, 10μl of the extracted RNA was added to the 2 X RT master mix consisting of 2μl of 10X RT buffer, 0.8μl of 100mM dNTP mix, 2μl of reverse primer D2 (TTGCACCAACAGTCAATGTCTTCAGGTTC-616) and 1μl of MultiScribe reverse transcriptase. The reaction was then subjected to reverse transcription at 25°C for 1min, 37°C for 120min, 85°C for 5min. The prepared cDNA was immediately used or stored at -20°C until use.
PCR and sequencing
CprM region was PCR amplified using AmpliTaq polymerase kit (Invitrogen). 5μl of the synthesized cDNA was then added to the PCR mix containing 10μl of PCR buffer, 10μl of MgCl2, 5μl of primers mD1 (134-TCAATATGCTGAAACGCGAGAGAAACCG) and D2 each, 0.5μl dNTPs, 1μl of polymerase. The reaction mixture was then subjected to 35 cycles of denaturation at 94°C for 1min, annealing at 55°C for 1min, and extension at 72°C for 1min. The products were then visualized for 511bp by ethidium bromide agarose gel staining [30]. Amplified products were then extracted from the gels using Qiaquick Gel extraction kit (QIAGEN, INC, Valencia, Calif) and both strands were sequenced by using a Big Dye Terminator Cycle Sequencing kit (Applied Biosystems). The CprM sequences were confirmed by BLAST (www.ncbi.nlm.nih.gov/BLAST). The forward and reverse sequences were aligned and manually edited using Codon Code aligner v.7.0.1 software to obtain the consensus sequence. New partial CprM sequences were submitted to GenBank at www.ncbi.nlm.nih.gov/genbank (accession number MG053110-MG053173).
Complete genome sequencing
Full genome sequencing of viral genomes was done using Ion Proton system (Life technologies, USA). Briefly, products were purified, size selected, amplified and quantified. Clonal amplification was carried out by emulsion PCR and the Ion sphere particles were deposited on to Ion PI chip. All proton quality-approved, trimmed and filtered (against human genome) data were exported as BAM files for bioinformatics analysis. Unmapped reads were quality filtered with mean quality score > = 20, minimum length 20 and trimmed using PrinSeq-Lite program. Resulting high quality reads were assembled using MIRA v4.0.2 assembler and contigs were annotated using BLAST against NCBI database. Complete genome sequences were submitted to GenBank at www.ncbi.nlm.nih.gov (accession number MG272272-MG272274).
Phylogenetic analysis
The sequences obtained in the present study and other sequences retrieved from GenBank were aligned using MAFFT online alignment tool [31]. Phylogenetic trees were constructed using Maximum Likelihood method based on Tamura Nei model in MEGA 6.06 software [32]. Genetic distances were calculated using the p-distance model of nucleotide and amino acid substitution. The robustness of the resulting tree was assessed with 1000 bootstrap replicates.
Results
Patient characteristics
During 2016-dengue season, serum samples from 109 NS1 positive patients were subjected to RT-PCR and 53(48.6%) scored positive for DENV-RNA. Further, 11 cell culture-grown DENV isolates obtained from additional NS1 positive patients were subjected to RT-PCR. Of the 64 patients, age of the patients ranged from 5 months to 65 years with median age of 28.6 years. Male (n = 35) to female (n = 29) ratio was 1: 0.8. Based on WHO 2009 guidelines, 63patients were categorized as dengue illness of which 59 without warning signs and 4 with warning signs. One patient was classified as severe dengue. Details are provided in Table 1.
Table 1. Demographics and clinical parameters of patients infected with DENV with different serotypes.
| Sr. No. | Sample No. | Sample type | NS1 Detection | Age | Gender | Clinical Manifestation | GenBank Accession No. |
|---|---|---|---|---|---|---|---|
| Dengue Serotype 1 and genotype V | |||||||
| 1 | S44 | Serum | pos | 36 | M | DwoWS | MG053110 |
| 2 | S58 | Serum | pos | 32 | F | DwoWS | MG053111 |
| 3 | S59 | Serum | pos | 18 | M | DwoWS | MG053112 |
| 4 | S105 | Vero isolates | pos | 27 | F | DwoWS | MG053113 |
| 5 | S5 | Serum | pos | 10 | F | DwoWS | MG053114 |
| 6 | S19 | Serum | pos | 24 | M | DwoWS | MG053115 |
| 7 | S16 | Serum | pos | 10 | M | DwoWS | MG053116 |
| 8 | S10 | Serum | pos | 4 | M | DwoWS | MG053117 |
| 9 | S51 | Serum | pos | 47 | M | DwoWS | MG053118 |
| Dengue Serotype 2 and genotype IV (Cosmopolitan) | |||||||
| 10 | 141 | Serum | pos | 30 | M | DwoWS | MG053119 |
| 11 | 812 | Serum | pos | 26 | F | DwoWS | MG053120 |
| 12 | 815 | Serum | pos | 22 | M | DwoWS | MG053121 |
| 13 | 892 | Serum | pos | 60 | F | DwoWS | MG053122 |
| 14 | 1008 | Serum | neg | 21 | F | DwoWS | MG053123 |
| 15 | 1053 | Serum | pos | 20 | M | DwoWS | MG053124 |
| 16 | 1571 | Vero isolates | pos | 25 | F | DwoWS | MG053125 |
| 17 | S107 | Vero isolates | pos | 16 | M | DwoWS | MG053126 |
| 18 | S47 | Serum | pos | 25 | M | DwoWS | MG053127 |
| 19 | S57 | Serum | pos | 24 | M | DwoWS | MG053128 |
| 20 | S77 | Serum | pos | 5 month | M | DWS | MG053129 |
| 21 | S85 | Serum | pos | 11 | F | DWS | MG053130 |
| 22 | S94 | Serum | pos | 27 | M | DwoWS | MG053131 |
| 23 | S87 | Serum | pos | 35 | M | DwoWS | MG053132 |
| 24 | S25 | Serum | pos | 25 | F | DwoWS | MG053133 |
| 25 | S26 | Serum | pos | 26 | M | DwoWS | MG053134 |
| 26 | S52 | Serum | pos | 34 | F | DwoWS | MG053135 |
| 27 | S53 | Serum | pos | 50 | M | DwoWS | MG053136 |
| 28 | S66 | Vero isolates | pos | 41 | M | DwoWS | MG053137 |
| 29 | S67 | Vero isolates | pos | 38 | F | DwoWS | MG053138 |
| 30 | S97 | Vero isolates | pos | 22 | M | DwoWS | MG053139 |
| 31 | S100 | Vero isolates | pos | 18 | M | DwoWS | MG053140 |
| 32 | S4 | Serum | pos | 16 | F | DwoWS | MG053141 |
| 33 | S15 | Serum | pos | 12 | F | DwoWS | MG053142 |
| 34 | S9 | Serum | pos | 34 | F | DwoWS | MG053143 |
| 35 | S78 | Serum | neg | 21 | F | DwoWS | MG053144 |
| Dengue Serotype 3 and genotype III | |||||||
| 36 | 1389 | Serum | pos | 19 | F | DwoWS | MG053145 |
| 37 | 984 | Serum | pos | 18 | F | DwoWS | MG053146 |
| 38 | S108 | Serum | pos | 24 | M | DwoWS | MG053147 |
| 39 | S73 | Serum | pos | 41 | F | DwoWS | MG053148 |
| 40 | S45 | Vero isolates | pos | 26 | F | DwoWS | MG053149 |
| 41 | S33 | Serum | pos | 25 | F | DwoWS | MG053150 |
| 42 | S111 | Vero isolates | pos | 30 | F | DwoWS | MG053151 |
| 43 | S56 | Serum | neg | 42 | M | DwoWS | MG053152 |
| 44 | S54 | Serum | pos | 39 | M | DwoWS | MG053153 |
| 45 | S50 | Vero isolates | pos | 13 | M | DwoWS | MG053154 |
| 46 | S74 | Serum | pos | 20 | M | DwoWS | MG053155 |
| 47 | S112 | Vero isolates | pos | 20 | M | DwoWS | MG053156 |
| 48 | S1 | Serum | pos | 26 | F | DwoWS | MG053157 |
| 49 | S76 | Vero isolates | pos | 8 | M | DwoWS | MG053158 |
| 50 | S81 | Serum | pos | 10 | M | DwoWS | MG053159 |
| 51 | S82 | Serum | pos | 41 | M | DWS | MG053160 |
| 52 | S65 | Serum | pos | 18 | F | DwoWS | MG053161 |
| Dengue Serotype 4 and genotype I | |||||||
| 53 | S28 | Serum | pos | 55 | M | DwoWS | MG053162 |
| 54 | S30 | Serum | pos | 42 | M | DwoWS | MG053163 |
| 55 | S46 | Serum | pos | 37 | M | DWS | MG053164 |
| 56 | S80 | Serum | pos | 21 | M | DwoWS | MG053165 |
| 57 | S2 | Serum | pos | 16 | F | DwoWS | MG053166 |
| 58 | 36 | Serum | pos | 6 | F | SD | MG053167 |
| 59 | 294 | Serum | pos | 65 | F | DwoWS | MG053168 |
| 60 | 1018 | Serum | pos | 2 | M | DwoWS | MG053169 |
| 61 | 1021 | Serum | pos | 20 | F | DwoWS | MG053170 |
| 62 | 1028 | Serum | pos | 17 | M | DwoWS | MG053171 |
| 63 | S49 | Serum | pos | 47 | F | DwoWS | MG053172 |
| 64 | S41 | Serum | pos | 23 | F | DwoWS | MG053173 |
DwoWS: Dengue illness without warning signs
DWS: Dengue illness with warning signs
SD: Severe dengue
DENV serotype distribution
Fig 1 depicts CprM gene phylogeny-based serotyping of 64 DENV sequences obtained during this study. Clearly, all the four serotypes were circulating in Pune during the 2016 season. Of these, DENV-1 was detected in 9 (14.1%, Pune-2016-DENV1), DENV-2 in 26 (40.6%, Pune-2016-DENV2), DENV-3 in 17 (26.6%, Pune-2016-DENV3) and DENV-4 in 12 (18.7%, Pune-2016-DENV4) samples. Thus, DENV-2 was found to be the predominant serotype and a substantial proportion of patients were infected with other serotypes as well. As far as serotypic distribution among different clinical forms is considered, the only severe dengue patient was infected with serotype4, patients with dengue illness without warning signs were infected with either of 4 serotypes and those with warning signs were infected with serotypes 2, 3 or 4.
Fig 1. Phylogenetic analyses of CprM gene sequences from 64 DENV positive cases for serotype determination.
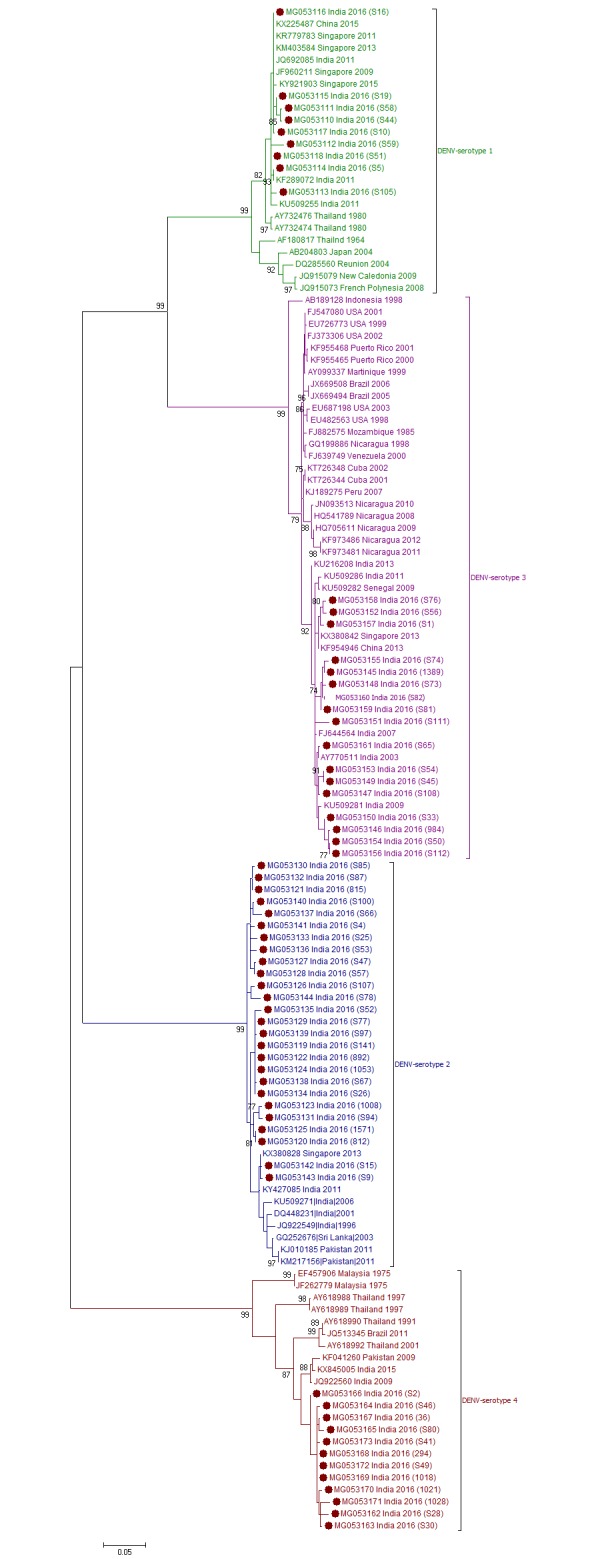
Each strain is identified by Genbank accession number followed by country and year of isolation. Numbers at the nodes are support values for the major branches (bootstrap; 1000 replicates). The sequences obtained in this study are marked in filled colored circles. Scale bar indicates number of base substitutions per site.
DENV genotype distribution
To determine the genotype distribution of DENV within each serotype, CprM gene sequences obtained during this study and sequences from different geographical locations across the globe were retrieved from NCBI database and used for phylogenetic analyses (Figs 2–5).
Fig 2. Genotyping analyses of CprM gene sequences of DENV-1 serotype isolates (n = 9) from Pune.
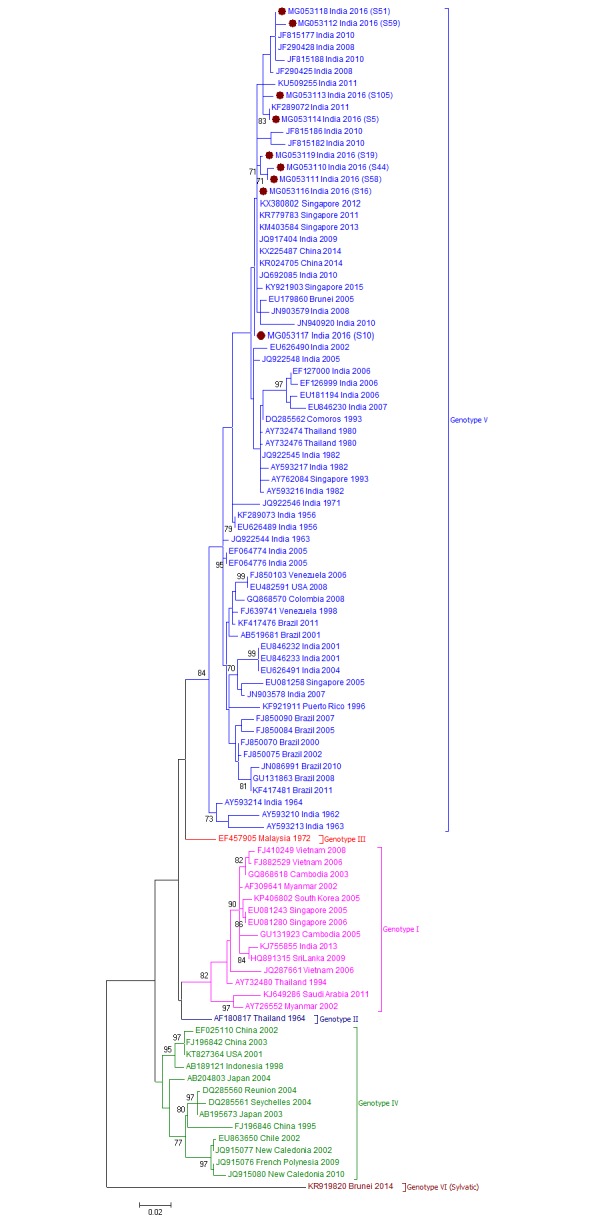
Each strain is indicated by Genbank accession number followed by country and year of isolation. Numbers at the nodes are support values for the major branches (bootstrap; 1000 replicates). The sequences obtained in this study are marked in filled colored circles. Scale bar indicates number of base substitutions per site.
Fig 5. Genotyping analyses of CprM gene sequences of DENV-4 serotype isolates (n = 12) from Pune.
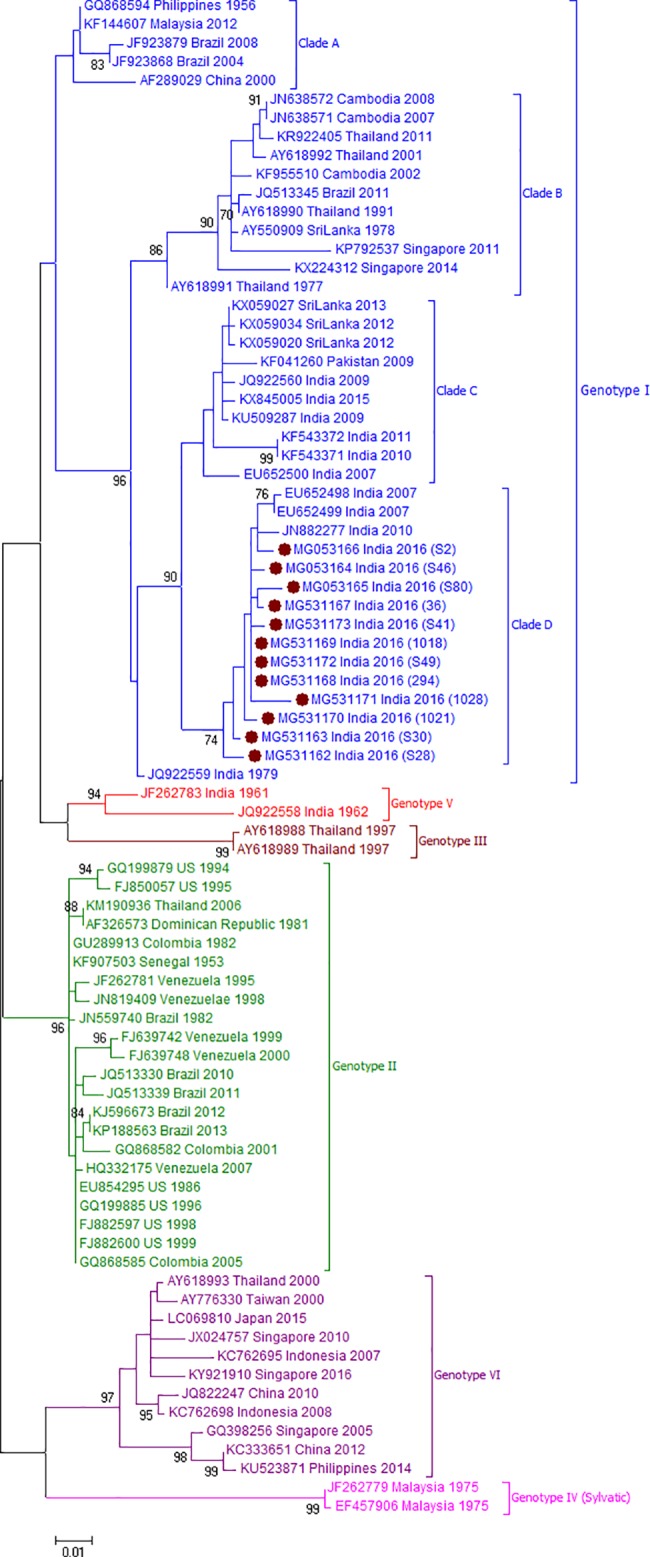
Each strain is indicated by Genbank accession number followed by country and year of isolation. Numbers at the nodes are support values for the major branches (bootstrap; 1000 replicates). The sequences obtained in this study are marked in filled colored circles. Scale bar indicates number of base substitutions per site.
For DENV-1 strains, phylogenetic tree revealed clustering of DENV-1 sequences into six genotypes. The Pune-2016-DENV1isolates (n = 9) grouped into American/African (AM/AF) or genotype V together with other Indian isolates from 1962 to 2011(Fig 2). As reported earlier (Cecilia et al, 2017), one isolate from Kerala, 2013 (KJ755855) belonged to Asian or genotype I. The Pune-2016-DENV1 sequences were similar with 99.8 ± 0.3% nucleotide identities. The current sequences clustered with isolates from India (2008–2016), Singapore (2011–15), China (2014) and Brunei (2005).
As evident from Fig 3, DENV-2 sequences were classified into six genotypes. Pune-2016 sequences (n = 26) grouped together in Cosmopolitan or genotype IVand exhibited 99.3 ± 0.3% nucleotide similarity. This genotype is divided into two geographically distinct lineages, lineage A (isolates from Southeast Asia, China and Oceania) and lineage B (isolates mostly from Indian subcontinent). Pune-2016 sequences belonged to lineage B and clustered with strains from India (2008–12), Pakistan (2008–13), China (1999), Singapore (2013) and Sri Lanka (2003).
Fig 3. Genotyping analyses of CprM gene sequences of DENV-2 serotype isolates (n = 26) from Pune.
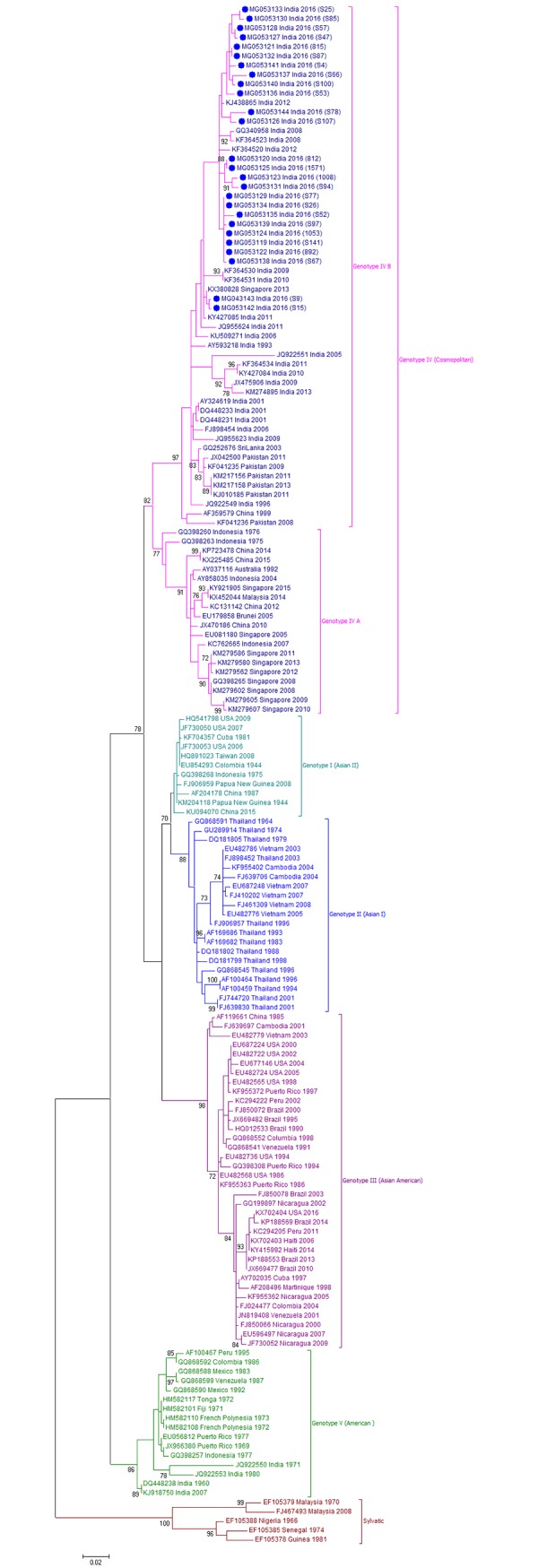
Each strain is indicated by Genbank accession number followed by country and year of isolation. Numbers at the nodes are support values for the major branches (bootstrap; 1000 replicates). The sequences obtained in this study are marked in filled colored circles. Scale bar indicates number of base substitutions per site.
Phylogenetic analysis classified Pune-2016-DENV3 (n = 17) CprM sequences in genotype III (Fig 4) with nucleotide sequence similarity of 99.3 ± 0.3%. Genotype III strains exhibit wide geographic distribution from Asia, Caribbean, Americas and Europe.Pune-2016 sequences were closely related to the other isolates from India (2004–2016), China (2009, 2013), Singapore (2009), Pakistan (2008–09) and a single isolate from Senegal (2009) with 99.4 ± 0.3% nucleotide similarities. Among the other India isolates, 1984-isolate (KF955477) grouped into Genotype II while a single isolate from northern India (KC787098, 2009) was assigned to genotype IV and was found to be closely related to DENV-3 prototype strain of Philippines, 1956 (M93130).
Fig 4. Genotyping analyses of CprM gene sequences of DENV-3 serotype isolates (n = 17) from Pune.
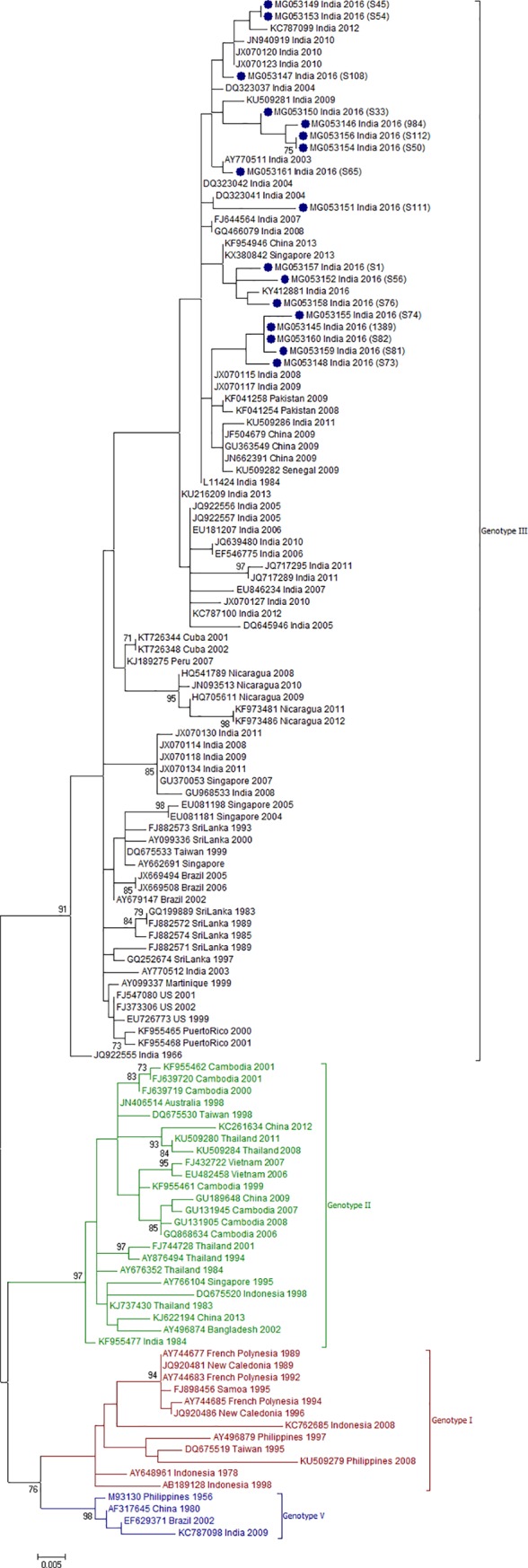
Each strain is indicated by Genbank accession number followed by country and year of isolation. Numbers at the nodes are support values for the major branches (bootstrap; 1000 replicates). The sequences obtained in this study are marked in filled colored circles. Scale bar indicates number of base substitutions per site.
DENV-4, the rare serotype in India was previously reported in 2003 from Delhi, and 2007 from Hyderabad and 2010 from Kerala [33–35]. In Maharashtra state, last report of DENV-4 cases was in 1975 from Amalner district and later detected in Pune in 2009 after a gap of 30 years [36]. Phylogenetic analysis revealed that DENV-4 sequences have been grouped into 5 genotypes with an inter-genotypic sequence divergence of > 6% [10–12, 37, 38]. Our data documented that (1) the Pune-2016 viruses (n = 12) formed a distinct cluster within genotype I and (2) 11 sequences earlier classified elsewhere [39, 40] as genotype II constituted a separate cluster (Fig 5) that included isolates mostly from East and Southeast Asian countries such as Japan, China, Taiwan, Indonesia, Singapore and Philippines.
We further compared percent nucleotide divergence in CprM region among different clusters within genotype I and different clusters constituting genotypes within serotype IV (Table 2). The novel cluster including current Pune strains was 3.0±0.6% to 5.6±0.8% divergent when compared to the other clusters/clades within genotype I and tentatively designated as clade D. DENV-4 viruses isolated in 2007 (EU652498-EU652499) and 2010 (JN882277) from two southern Indian states together with Pune-2016 sequences belonged to clade D. The distinct cluster of sequences earlier classified as genotype II was 6.8% - 10.2% different from the known genotypes I–V (Table 2). These results suggested that this cluster may represent a novel genotype VI.
Table 2. Nucleotide diversity in CprM region between (A) clades within genotype I and (B) across genotypes of DENV-4 viruses.
| (A) | Clade A | Clade B | Clade C | ||
| Clade A | -- | -- | -- | ||
| Clade B | 5.5 ± 0.9 | -- | -- | ||
| Clade C | 5.0 ± 0.9 | 5.8 ± 0.9 | -- | ||
| Clade D | 4.6 ± 0.9 | 5.6 ± 0.8 | 3.0 ± 0.6 | ||
| (B) | GT* I | GT II | GT III | GT IV (sylvatic) | GT V |
| GT I | -- | -- | -- | -- | -- |
| GT II | 6.9±0.9 | -- | -- | -- | -- |
| GT III | 7.8±1.0 | 7.3±1.0 | -- | -- | -- |
| GT IV (sylvatic) | 10.9±1.2 | 9.3±1.3 | 10.7±1.3 | -- | -- |
| GT V | 7.4±0.9 | 6.2±0.9 | 7.3±1.0 | 11.5±1.2 | -- |
| GT VI | 8.6±1.0 | 6.8±1.0 | 8.9±1.1 | 10.2±1.2 | 8.0±0.9 |
* GT—abbreviation for genotype
--indicates blank spaces
For confirmation of these observations, full genome sequence analysis was done. We obtained complete genome sequences of three DENV-4 strains, isolated in Vero cells (accession number MG272272-MG272274). The complete genome lengths of Pune-2016 isolates are 10653 nucleotides (nt). The length of 5’ and 3’ untranslated regions are 103 nt and 386 nt respectively with an ORF of 10164 nt coding for 3388 amino acids. As evident from Fig 6, phylogenetic analysis on complete genome sequences confirmed emergence of a novel clade “D” within genotype I that differed by 3.3 to 5.9% (nt) and 1.2 to 1.9% (aa) from the other clades (Table 3). Pune-2016 complete genome sequences showed nucleotide similarity of 99.2 ± 0.1% among themselves and diversity of 3.3 ± 0.1% when compared with Pune, 2009 isolate (JQ922560). Genomic diversity within genotype I was highest of 4.4% as compared to other genotypes of DENV-4 viruses. The existence of an additional genotype VI was confirmed by the full genome analysis with nucleotide divergence of 6.7% - 13.5% across genotypes I to V (Table 3). Inter-genotypic divergence for genotype I to VI ranged from 6.7 to 13.7% in nucleotide and 2.2 to 5.2% in amino acid sequences (Table 3).
Fig 6. Genotyping analyses of complete gene sequences of DENV-4 serotype isolates (n = 3) from Pune.
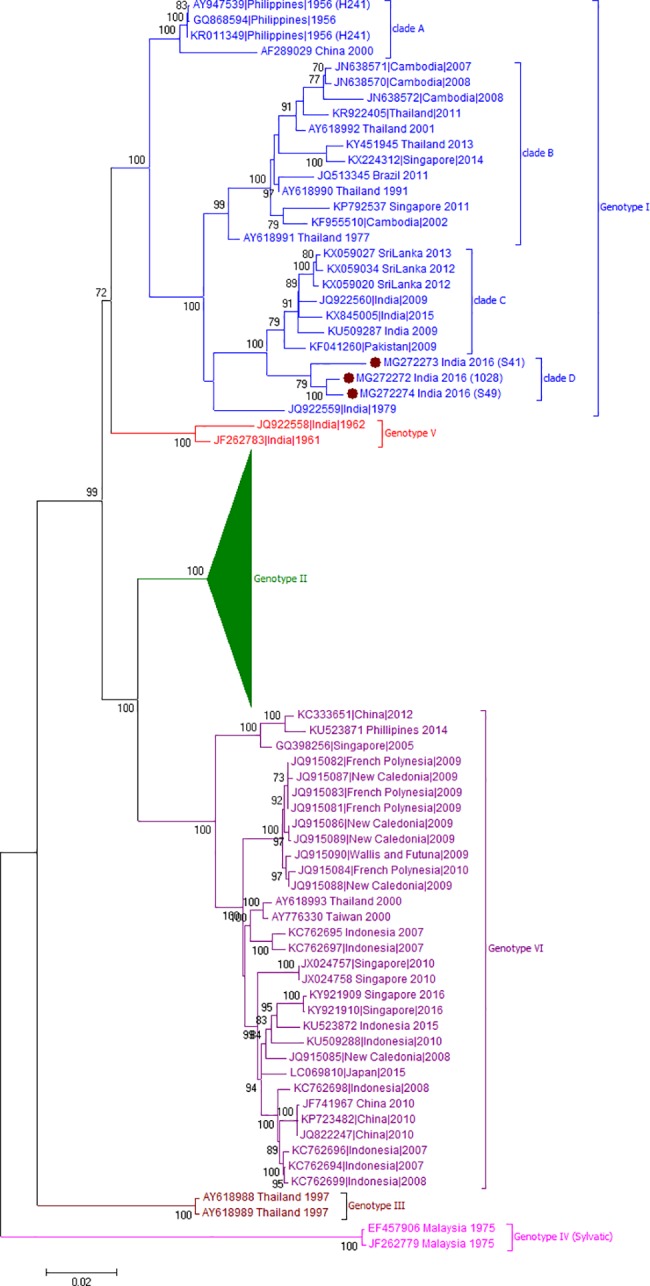
Each strain is indicated by Genbank accession number followed by country and year of isolation. 141 Genotype II sequences obtained from Genbank are shown here as compressed tree (S1 Text provides the accession numbers). Numbers at the nodes are support values for the major branches (bootstrap; 1000 replicates). The sequences obtained in this study are marked in filled colored circles. Scale bar indicates number of base substitutions per site.
Table 3. Nucleotide and amino acid diversity in complete genome between (A) clades within genotype I and (B) across genotypes of DENV-4 viruses.
Pairwise distances and standard errors of nucleotide and amino acid diversity are displayed in lower-left and upper-right matrix respectively.
| (A) | Clade A | Clade B | Clade C | Clade D | ||
| Clade A | -- | 1.9±0.2 | 1.8±0.2 | 1.9±0.2 | ||
| Clade B | 5.4±0.2 | -- | 1.7±0.2 | 1.7±0.2 | ||
| Clade C | 5.3±0.2 | 5.4±0.2 | -- | 1.2±0.2 | ||
| Clade D | 5.7±0.2 | 5.9±0.1 | 3.3±0.1 | -- | ||
| (B) | GT* I | GT II | GT III | GT IV (sylvatic) | GT V | GT VI |
| GT I | -- | 2.7±0.2 | 2.9±0.3 | 4.8±0.4 | 3.0±0.2 | 3.1±0.3 |
| GT II | 7.9±0.2 | -- | 2.8±0.3 | 4.5±0.4 | 3.0±0.3 | 2.2±0.2 |
| GT III | 9.2±0.2 | 9.2±0.3 | -- | 4.4±0.4 | 3.2±0.3 | 3.1±0.3 |
| GT IV (sylvatic) | 13.4±0.3 | 13.3±0.3 | 13.7±0.3 | -- | 5.2±0.4 | 4.7±0.4 |
| GT V | 7.3±0.2 | 7.1±0.2 | 8.5±0.3 | 13.4±0.3 | -- | 3.2±0.2 |
| GT VI | 8.2±0.2 | 6.7±0.2 | 9.4±0.3 | 13.5±0.3 | 7.6±0.2 | -- |
* GT—abbreviation for genotype
--indicates blank spaces
To understand the mutation sites associated with the divergence of Pune-2016 DENV-4 isolates to “clade D”, amino acid sequence comparison with different clades of genotype I and Indian isolates of genotype V was carried out (Table 4). Unique substitutions in coding region of clade D was identified in comparison to reference strain H241 isolated in Philippines, 1956 (AY947539, clade A).A total of 7 amino acid changes in polyprotein (M271I, I411V, K479T, N645S, F945L, V1262Aand C1310R) were found to be specific to clade D. In fact, none of the other genotype I clades exhibited these amino acid substitutions. Individual protein analysis revealed that amino acid substitutions specific to Clade D were confined to membrane glycoprotein precursor (n = 1), envelope (n = 3), NS1 (n = 1), and NS2A (n = 2) regions (Table 4). Envelope region showed two and one amino acid substitutions in domain II (I132V, K200T) and domain III (N366S) respectively. Domain III (residues 300–495) is responsible for receptor binding and contains virus neutralizing epitopes. Two amino acid substitutions in NS2A region (V136A and C184R) might affect virus replication. As compared to the reference strain, clades C and D shared identical substitutions at only one amino acid position in NS3 (M605V) region. Three amino acid substitutions specific to clade C at positions 130, 202 in envelope and at position 383 in NS3wasreversed back to the original amino acids of reference strain in clade D.
Table 4. Comparative analyses of amino acid substitutions among 4 clades of genotype I, genotype V and Indian isolate, 1979 to corresponding residues in the reference strain, H241 (AY947539, Philippines 1956) of clade A.
| Sr No. | Genomic region | Polyprotein position | Gene position | Reference strain (H241) | Genotype I | JQ922559(India, 1979) | Genotype V(India, 1961–62) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Clade A | Clade B | Clade C | Clade D(Pune 2016) | |||||||
| 1 | prM (114–279,166aa) | 269 | 156 | V | * | I | I | I | I | * |
| 2 | 271 | 158 | M | * | * | * | I | * | * | |
| 3 | Env (280–774, 495aa) | 409 | 130 | V | * | * | I | * | * | * |
| 4 | 411 | 132 | I | * | * | * | V | * | * | |
| 5 | 479 | 200 | K | * | * | * | T | * | * | |
| 6 | 481 | 202 | K | * | * | N | * | * | * | |
| 7 | 512 | 233 | Y | * | H | H | H | * | * | |
| 8 | 645 | 366 | N | * | * | * | S | * | * | |
| 9 | NS1 (775–1126, 352aa) | 776 | 2 | T | * | M | M | M | M | M |
| 10 | 903 | 129 | K | * | R | R | R | R | R | |
| 11 | 945 | 171 | F | * | * | * | L | * | * | |
| 12 | NS2A (1127–1344, 218aa) | 1219 | 93 | R | * | * | * | K! | * | * |
| 13 | 1262 | 136 | V | * | * | * | A | * | * | |
| 14 | 1281 | 155 | R | * | K | K | K | K | * | |
| 15 | 1310 | 184 | C | * | * | * | R | * | * | |
| 16 | NS2B (1345–1474, 130aa) | 1433 | 89 | I | * | V | V | V | V | V |
| 17 | NS3(1475–2092, 618aa) | 1536 | 62 | T | * | S | S | S | S | * |
| 18 | 1645 | 171 | T | * | I | I | I | I | I | |
| 19 | 1795 | 321 | A | * | T | T | T | T | * | |
| 20 | 1857 | 383 | I | * | * | V | * | * | * | |
| 21 | 1954 | 480 | K | * | R | R | R | R | * | |
| 22 | 2079 | 605 | M | * | * | V | V | * | * | |
| 23 | NS4A (2093–2219, 127aa) | 2209 | 117 | V | * | * | A | A!! | * | A |
| 24 | 2K peptide (2220–2242, 23aa) | 2240 | 21 | I | * | V | V | V | V | * |
| 25 | NS4B(2243–2487, 245aa) | 2440 | 198 | V | * | I | I | I | I | I |
| 26 | NS5(2488–3387, 900aa) | 2733 | 246 | R | * | * | * | K | K | * |
| 27 | 2734 | 247 | H | * | * | Y∉ | Y | * | * | |
| 28 | 2743 | 256 | V | * | A | A | A | A | * | |
| 29 | 2762 | 275 | T | * | * | A∉ | A | * | * | |
*—sequence similar to reference strain Philippines, 1956
Bold–amino acid substitution unique to clade D
!—amino acid substitution unique to clade D except MG272272 (1028)
!! - amino acid substitution unique to clade D except MG272273 (S41)
Italics—amino acid substitution unique to clade C
∉—amino acid substitution unique to clade C except KU509287
Discussion
This study documents co-circulation of all the four serotypes of DENV during a single season at Pune, India. Interestingly, though DENV-2 was the most prevalent serotype (40.6%), 18.7% isolates belonged to serotype IV. This is especially important since this serotype was introduced in this city in 2009 after a gap of 30 years [36]. Since 2005, DENV-1, 2 and 3 were shown to co-circulate. However, each year was dominated by a single serotype; DENV-1 in 2005 and 2007, DENV-2 in 2008 and DENV-3 in 2009. In 2010, both DENV-2 and DENV-3 were co-dominant [41]. 2016 witnessed prevalence of all the four serotypes to an appreciable extent and presents possible risk of secondary infection with serotype 4 leading probably to severe disease. Though one patient infected with serotype 4 led to severe disease, no conclusions can be made because of small numbers.
On account of highest mutation rate among the Flavivirus group, DENV serotypes are divided into different genotypes and further into lineages or clades [10–12]. Infection of populations with a new genotype not exposed to earlier or with a virus with lineage shift within genotype has been attributed to severe form of disease [19, 42–44]. In the light of these observations, it is important to note that Pune-2016 DENV-4 isolates formed a distinct clade-D within genotype I that were 3.0%- 5.6% (CprM) and 3.3%-5.9% (complete genome) divergent from clades A, B and C. Interestingly, viruses isolated earlier (2007, 2010) from two southern states also belonged to this clade. Identification of novel clade within genotype I emphasizes high rate of genomic diversity in this continually evolving genotype of DENV-4 viruses. It would be desirable to assess the role of this clade in disease severity when presenting with primary or secondary infection. Further monitoring is essential to identify emergence of novel DENV-4 viruses, especially, as introduction of dengue vaccine remains a distinct possibility in endemic areas including India. As far as serotypes I, II and III are concerned, similar to earlier reports from India [25, 45–48], persistent circulation of genotype V of DENV-1, genotype IV of DENV-2 and genotype III of DENV-3 was noted.
Another significant observation of this study is the identification of an additional genotype (VI) within serotype-4. This genotype includes viruses from East Asia, Southeast Asia and Oceania countries, isolated during 2000-2016that were earlier grouped in genotype II [39–40, 49–50]. Genotype VI is proposed since > 6% divergence from other genotypes was observed as evidenced by complete genome based phylogenetic analysis (Table 3, Fig 6). Further study is required to correlate disease profile of patients infected with different genotypes of DENV-4 viruses. Different regions of dengue genome like Envelope, E-NS1 and C-prM have been largely utilized for genotyping. CprM gene based genotyping is faster and economical due to usage of single set of primer pair for both amplification and sequencing [30, 51]. Our study emphasizes the utility of this region for genotyping as the CprM based observations of emergence of a new clade in genotype I or a distinct cluster within genotype II were confirmed by complete genome based analysis.
Chances of co-infection with more than one serotype are likely to be much higher when multiple dengue serotypes co-circulate in a population. Co-infection with multiple serotypes poses risk of emergence of recombinant virus strains that could have distinct properties. Co-circulation of all the four serotypes in a single outbreak has been reported earlier with 42.9% and 45.4% cases of co-infection in Karnataka and Hyderabad respectively [28, 52]. Significant co-infection (15% -43%) has been reported from northern [24, 26], and eastern India[25] without detecting all the 4 serotypes. These regions with circulation of more than one serotype simultaneously are of high significance as they are more prone to severe dengue infection [28, 52]. However, we did not find evidence of co-infection among the patients studied.
In summary, in contrast to the predominance of a single serotype observed earlier, we provide recent evidence of significant co-circulation of all the four serotypes in Pune and emergence of a novel clade in genotype I of DENV-4 viruses. In view of the role of novel strains in increased severity and vaccine availability in near future, a comprehensive molecular surveillance programme for DENV is urgently needed.
Supporting information
(DOCX)
Acknowledgments
The authors thank Dr. Ruta Kulkarni and Mrunal Gosavi for testing clinical samples for NS1 ELISA. Special thanks are due to Mr. Tushar Bhosale and Mr. Mandar Bhutkar for collecting samples and clinical information used in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is supported by Indian Council of Medical Research (ICMR) grant number: ECD/NTF/8/2016-17 received by ACM. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013. April 25;496(7446):504–7. doi: 10.1038/nature12060 Epub 2013 Apr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman MG, Gubler DJ, Izquierdo A, Martinez E, Halstead SB. Dengue infection. Nat Rev Dis Primers. 2016. August 18;2:16055 doi: 10.1038/nrdp.2016.55 Review. [DOI] [PubMed] [Google Scholar]
- 3.WHO report. Global Strategy for dengue prevention and control, 2012–2020, http://apps.who.int/iris/bitstream/10665/75303/1/9789241504034_eng.pdf (Accessed 11 November 2017)
- 4.Schwartz E, Weld LH, Wilder-Smith A, von Sonnenburg F, Keystone JS, Kain KC, et al. Seasonality, annual trends, and characteristics of dengue among ill returned travelers, 1997–2006. Emerg Infect Dis. 2008. July;14(7):1081–8. doi: 10.3201/eid1407.071412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shihada S, Emmerich P, Thomé-Bolduan C, Jansen S, Günther S, Frank C, et al. Genetic Diversity and New Lineages of Dengue Virus Serotypes 3 and 4 in Returning Travelers, Germany, 2006–2015. Emerg Infect Dis. 2017. February;23(2):272–275. doi: 10.3201/eid2302.160751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubler DJ, Clark GG. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerg Infect Dis. 1995. Apr-Jun;1(2):55–7. doi: 10.3201/eid0102.952004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halstead SB, Cohen SN. Dengue Hemorrhagic Fever at 60 Years: Early Evolution of Concepts of Causation and Treatment. Microbiol Mol Biol Rev. 2015. September;79(3):281–91. doi: 10.1128/MMBR.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakravarti A, Arora R, Luxemburger C. Fifty years of dengue in India. Trans R Soc Trop Med Hyg. 2012. May;106(5):273–82. doi: 10.1016/j.trstmh.2011.12.007 Epub 2012 Feb 21. Review. [DOI] [PubMed] [Google Scholar]
- 9.National Vector Borne Disease Control Programme (NVBDCP), Government of India. Dengue cases and deaths in the Country since 2010, http://nvbdcp.gov.in/den-cd.html (Accessed 11 November 2017)
- 10.Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–41. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol. 2009. July;9(4):523–40. doi: 10.1016/j.meegid.2009.02.003 Epub 2009 Feb 13. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen R, Vasilakis N. Dengue—quo tu et quo vadis? Viruses. 2011. September;3(9):1562–608. doi: 10.3390/v3091562 Epub 2011 Sep 1. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003. July;9(7):800–9. doi: 10.3201/eid0907.030038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kukreti H, Mittal V, Chaudhary A, Rautela RS, Kumar M, Chauhan S, et al. Continued persistence of a single genotype of dengue virus type-3 (DENV-3) in Delhi, India since its re-emergence over the last decade. J Microbiol Immunol Infect. 2010. February;43(1):53–61. doi: 10.1016/S1684-1182(10)60008-4 Epub 2010 Mar 29. [DOI] [PubMed] [Google Scholar]
- 15.Hapuarachchi HC, Koo C, Rajarethinam J, Chong CS, Lin C, Yap G, et al. Epidemic resurgence of dengue fever in Singapore in 2013–2014: A virological and entomological perspective. BMC Infect Dis. 2016. June 17;16:300 doi: 10.1186/s12879-016-1606-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha K, Ghosh M, Firdaus R, Biswas A, Seth B, Bhattacharya D, et al. Changing pattern of dengue virus serotypes circulating during 2008–2012 and reappearance of dengue serotype 3 may cause outbreak in Kolkata, India. J Med Virol. 2016. October;88(10):1697–702. doi: 10.1002/jmv.24529 Epub 2016 Mar 29. [DOI] [PubMed] [Google Scholar]
- 17.Choudhary MC, Gupta E, Sharma S, Hasnain N, Agarwala P. Genetic signatures coupled with lineage shift characterise endemic evolution of Dengue virus serotype 2 during 2015 outbreak in Delhi, India. Trop Med Int Health. 2017. July;22(7):871–880. doi: 10.1111/tmi.12898 Epub 2017 Jun 19. [DOI] [PubMed] [Google Scholar]
- 18.Ospina MC, Diaz FJ, Osorio JE. Prolonged co-circulation of two distinct Dengue virus Type 3 lineages in the hyperendemic area of Medellin, Colombia. Am J Trop Med Hyg. 2010. September;83(3):672–8. doi: 10.4269/ajtmh.2010.09-0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dash PK, Parida MM, Saxena P, Abhyankar A, Singh CP, Tewari KN, et al. Reemergence of dengue virus type-3 (subtype-III) in India: implications for increased incidence of DHF & DSS. Virol J. 2006. July 6;3:55 doi: 10.1186/1743-422X-3-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cecilia D, Patil JA, Kakade MB, Walimbe A, Alagarasu K, Anukumar B, et al. Emergence of the Asian genotype of DENV-1 in South India. Virology. 2017. October;510:40–45. doi: 10.1016/j.virol.2017.07.004 Epub 2017 Jul 10. [DOI] [PubMed] [Google Scholar]
- 21.Singh UB, Maitra A, Broor S, Rai A, Pasha ST, Seth P. Partial nucleotide sequencing and molecular evolution of epidemic causing Dengue 2 strains. J Infect Dis. 1999. October;180(4):959–65. doi: 10.1086/315043 [DOI] [PubMed] [Google Scholar]
- 22.Chakravarti A, Kumar A, Matlani M. Displacement of dengue virus type 3 and type 2 by dengue virus type 1 in Delhi during 2008. Indian J Med Microbiol. 2010. Oct-Dec;28(4):412 doi: 10.4103/0255-0857.71806 [DOI] [PubMed] [Google Scholar]
- 23.Reddy MN, Dungdung R, Valliyott L, Pilankatta R. Occurrence of concurrent infections with multiple serotypes of dengue viruses during 2013–2015 in northern Kerala, India. PeerJ. 2017. March 14;5:e2970 doi: 10.7717/peerj.2970 eCollection 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bharaj P, Chahar HS, Pandey A, Diddi K, Dar L, Guleria R, et al. Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi, India. Virol J. 2008. January 9;5:1 doi: 10.1186/1743-422X-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das B, Das M, Dwibedi B, Kar SK, Hazra RK. Molecular investigations of dengue virus during outbreaks in Orissa state, Eastern India from 2010 to 2011. Infect Genet Evol. 2013. June;16:401–10. doi: 10.1016/j.meegid.2013.03.016 Epub 2013 Mar 22. [DOI] [PubMed] [Google Scholar]
- 26.Tazeen A, Afreen N, Abdullah M, Deeba F, Haider SH, Kazim SN, et al. Occurrence of co-infection with dengue viruses during 2014 in New Delhi, India. Epidemiol Infect. 2017. January;145(1):67–77. Epub 2016 Sep 13. doi: 10.1017/S0950268816001990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra B, Turuk J, Sahu SJ, Khajuria A, Kumar S, Dey A, et al. Co-circulation of all four dengue virus serotypes: First report from Odisha. Indian J Med Microbiol. 2017. Apr-Jun;35(2):293–295. doi: 10.4103/ijmm.IJMM_15_536 [DOI] [PubMed] [Google Scholar]
- 28.Vaddadi K, Gandikota C, Jain PK, Prasad VSV, Venkataramana M. Co-circulation and co-infections of all dengue virus serotypes in Hyderabad, India 2014. Epidemiol Infect. 2017. September;145(12):2563–2574. doi: 10.1017/S0950268817001479 Epub 2017 Jul 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cecilia D. Current status of dengue and chikungunya in India. WHO South East Asia J Public Health. 2014. Jan-Mar;3(1):22–26. doi: 10.4103/2224-3151.206879 Review. [DOI] [PubMed] [Google Scholar]
- 30.Chien LJ, Liao TL, Shu PY, Huang JH, Gubler DJ, Chang GJ. Development of real-time reverse transcriptase PCR assays to detect and serotype dengue viruses. J Clin Microbiol. 2006. April;44(4):1295–304. doi: 10.1128/JCM.44.4.1295-1304.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013. April;30(4):772–80. doi: 10.1093/molbev/mst010 Epub 2013 Jan 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013. December;30(12):2725–9. doi: 10.1093/molbev/mst197 Epub 2013 Oct 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dar L, Gupta E, Narang P, Broor S. Cocirculation of dengue serotypes, Delhi, India, 2003. Emerg Infect Dis. 2006. February;12(2):352–3. doi: 10.3201/eid1202.050767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dash PK, Sharma S, Srivastava A, Santhosh SR, Parida MM, Neeraja M, et al. Emergence of dengue virus type 4 (genotype I) in India. Epidemiol Infect. 2011. June;139(6):857–61. doi: 10.1017/S0950268810001706 Epub 2010 Jul 30. [DOI] [PubMed] [Google Scholar]
- 35.Kumar NP, Jayakumar PR, George K, Kamaraj T, Krishnamoorthy K, Sabesan S, et al. Genetic characterization of dengue viruses prevalent in Kerala State, India. J Med Microbiol. 2013. April;62(Pt 4):545–52. doi: 10.1099/jmm.0.052696-0 Epub 2013 Jan 3. [DOI] [PubMed] [Google Scholar]
- 36.Cecilia D, Kakade MB, Bhagat AB, Vallentyne J, Singh A, Patil JA, et al. Detection of dengue-4 virus in pune, western india after an absence of 30 years—its association with two severe cases. Virol J. 2011. February 1;8:46 doi: 10.1186/1743-422X-8-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patil JA, Cherian S, Walimbe AM, Bhagat A, Vallentyne J, Kakade M, et al. Influence of evolutionary events on the Indian subcontinent on the phylogeography of dengue type 3 and 4 viruses. Infect Genet Evol. 2012. December;12(8):1759–69. doi: 10.1016/j.meegid.2012.07.009 Epub 2012 Aug 6. [DOI] [PubMed] [Google Scholar]
- 38.Vaddadi K, Gandikota C, Venkataramana M. Complete genome characterization and evolutionary analysis of serotype-4 associated with severe dengue. Epidemiol Infect. 2017. May;145(7):1443–1450. doi: 10.1017/S0950268817000243 Epub 2017 Feb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao H, Yu XD, Zhang XY, Jiang T, Hong WX, Yu M, et al. Complete genome sequence of a dengue virus serotype 4 strain isolated in Guangdong, China. J Virol. 2012. June;86(12):7021–2. doi: 10.1128/JVI.00858-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasmono RT, Wahid I, Trimarsanto H, Yohan B, Wahyuni S, Hertanto M, et al. Genomic analysis and growth characteristic of dengue viruses from Makassar, Indonesia. Infect Genet Evol. 2015. June;32:165–77. doi: 10.1016/j.meegid.2015.03.006 Epub 2015 Mar 14. [DOI] [PubMed] [Google Scholar]
- 41.Cecilia D, Shah PS, Alagarasu K. Dengue: Achievements in the last decade. In: NIV Golden to Diamond Jubilee: The Glorious Decade. Eds. Arankalle VA, Cecilia D. 2012. pp. 141–162. [Google Scholar]
- 42.Zhang FC, Zhao H, Li LH, Jiang T, Hong WX, Wang J, et al. Severe dengue outbreak in Yunnan, China, 2013. Int J Infect Dis. 2014. October;27:4–6. doi: 10.1016/j.ijid.2014.03.1392 Epub 2014 Aug 11. [DOI] [PubMed] [Google Scholar]
- 43.Heringer M, Nogueira RM, de Filippis AM, Lima MR, Faria NR, Nunes PC, et al. Impact of the emergence and re-emergence of different dengue viruses' serotypes in Rio de Janeiro, Brazil, 2010 to 2012. Trans R Soc Trop Med Hyg. 2015. April;109(4):268–74. doi: 10.1093/trstmh/trv006 Epub 2015 Jan 28. [DOI] [PubMed] [Google Scholar]
- 44.Heringer M, Souza TMA, Lima MDRQ, Nunes PCG, Faria NRDC, de Bruycker-Nogueira F, et al. Dengue type 4 in Rio de Janeiro, Brazil: case characterization following its introduction in an endemic region. BMC Infect Dis. 2017. June 9;17(1):410 doi: 10.1186/s12879-017-2488-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma S, Dash PK, Agarwal S, Shukla J, Parida MM, Rao PV. Comparative complete genome analysis of dengue virus type 3 circulating in India between 2003 and 2008. J Gen Virol. 2011. July;92(Pt 7):1595–600. doi: 10.1099/vir.0.030437-0 Epub 2011 Mar 16. [DOI] [PubMed] [Google Scholar]
- 46.Dash PK, Sharma S, Soni M, Agarwal A, Sahni AK, Parida M. Complete genome sequencing and evolutionary phylogeography analysis of Indian isolates of Dengue virus type 1. Virus Res. 2015. January 2;195:124–34. doi: 10.1016/j.virusres.2014.08.018 Epub 2014 Sep 6. [DOI] [PubMed] [Google Scholar]
- 47.Afreen N, Naqvi IH, Broor S, Ahmed A, Parveen S. Phylogenetic and Molecular Clock Analysis of Dengue Serotype 1 and 3 from New Delhi, India. PLoS One. 2015. November 4;10(11):e0141628 doi: 10.1371/journal.pone.0141628 eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afreen N, Naqvi IH, Broor S, Ahmed A, Kazim SN, Dohare R, et al. Evolutionary Analysis of Dengue Serotype 2 Viruses Using Phylogenetic and Bayesian Methods from New Delhi, India. PLoS Negl Trop Dis. 2016. March 15;10(3):e0004511 doi: 10.1371/journal.pntd.0004511 eCollection 2016 Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao-Lormeau VM, Roche C, Aubry M, Teissier A, Lastere S, Daudens E, et al. Recent emergence of dengue virus serotype 4 in French Polynesia results from multiple introductions from other South Pacific Islands. PLoS One. 2011;6(12):e29555 doi: 10.1371/journal.pone.0029555 Epub 2011 Dec 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai Z, Liu Q, Jiang LY, Liu LC, Cao YM, Xu Y, et al. Complete genome sequence of dengue virus serotype 4 from guangzhou, china. Genome Announc. 2013. May 30;1(3). pii: e00299-13. doi: 10.1128/genomeA.00299-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992. March;30(3):545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vinodkumar CS, Kalapannavar NK, Basavarajappa KG, Sanjay D, Gowli C, Nadig NG, et al. Episode of coexisting infections with multiple dengue virus serotypes in central Karnataka, India. J Infect Public Health. 2013. August;6(4):302–6. doi: 10.1016/j.jiph.2013.01.004 Epub 2013 Apr 3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


