Summary
Few studies have quantitatively probed how adaptive mutations result in increased fitness. Even in simple microbial evolution experiments, with full knowledge of the underlying mutations and specific growth conditions, it is challenging to determine where within a growth-saturation cycle those fitness gains occur. A common implicit assumption is that most benefits derive from an increased exponential growth rate. Here we instead show that in batch serial transfer experiments, adaptive mutants’ fitness gains can be dominated by benefits that are accrued in one growth cycle but not realized until the next growth cycle. For thousands of evolved clones (most with only a single mutation) we systematically varied the lengths of fermentation, respiration, and stationary phases to assess how their fitness, as measured by barcode sequencing, depends on these phases of the growth-saturation-dilution cycles. These data revealed that while all adaptive lineages gained similar and modest benefits from fermentation, most of the benefits for the highest fitness mutants came instead from the time spent in respiration. From monoculture and high-resolution pairwise fitness competition experiments for a dozen of these clones, we determined that that the benefits accrued during respiration are only realized later as a shorter duration of lag phase in the following growth cycle. These results reveal hidden complexities of the adaptive process even under ostensibly simple evolutionary conditions, in which fitness gains can accrue during time spent in a growth phase with little cell division, and that the memory of those gains can be realized in the subsequent growth cycle.
Keywords: Yeast, adaptation, experimental evolution, fitness, trade-offs, serial-dilution, antagonistic pleiotropy, high-throughput, Ras/PKA, diploidy
The eTOC Blurb
Quantitative details of how adaptive genotypes gain fitness have rarely been studied. By quantifying fitness benefits of thousands of yeast clones in different parts of the growth-saturation cycle, Li et al. identify distinct adaptive strategies, with some strategies showing clear trade-offs between growth and survival.
Introduction
Experimental microbial evolution combined with genomics has succeeded in delineating the molecular basis and population dynamics of adaptation for multiple species and under diverse conditions [1–14]. However, this stands in sharp contrast to the difficulty of quantitatively understanding how these genetic events lead to fitness benefits [15,16]. While the beneficial effects of some mutations are obvious, e.g. amplification of a gene encoding a transporter of the limiting nutrient, there are many cases where it is far from clear. Furthermore, beneficial mutations may have multiple phenotypic effects at the organismic level. Whether the net effect of these will be beneficial, and if so by how much, likely depends on the subtleties of the specific environmental conditions. While some detailed analyses of observed fitness gains in experimentally evolved microbes have been carried out [17,18], these approaches were low-throughput and required detailed knowledge of the fitness-related phenotypic changes.
For serial batch culture experiments with distinct physiological growth phases one possible approach is to study where within the growth cycle adaptive clones acquire their fitness benefits. A challenge of this approach is that the growth cycle phase where fitness gains might accrue may be distinct from the phase where reproduction occurs, even in a “simple” system such as asexually growing single cells. An example from metazoans provides a useful analogy: in some animal species, such as Richardson’s ground squirrels (Spermophilus richardsonii), females that gain more body mass outside of the reproductive season have a higher fecundity the following spring when they do reproduce [19]. Thus, understanding fitness by focusing on the parts of the growth cycle that are associated with a faster rate of cell division might miss the importance of the part of the growth cycle where the physiological gains actually accrue.
We previously isolated yeast clones containing single adaptive mutations from a glucose-limited serial transfer evolution [6,12]; in the evolutionary condition, the clones experienced lag, fermentation and respiration phases. Here, we have quantitatively investigated where in the growth cycle these clones accrue and realize their fitness benefits. We first studied realized fitness by generating detailed cell number measurements throughout the growth cycle under the EC using both monocultures and high-resolution pairwise competitions, for a dozen representative clones. Cell number measurements capture where within the growth cycle fitness benefits are realized, but not necessarily where those benefits are accrued.
Next, using barcode-based fitness measurements, we quantified where in the growth cycle fitness was accrued by measuring how per-cycle fitness of ~1,600 adaptive clones [12] scaled with the systematically varied lengths of fermentation, respiration, and stationary phases. We found that the most fit adaptive clones primarily accrue their fitness during respiration, but realize that fitness only later, by having a shorter lag phase in the next growth cycle; such clones often also experienced a significant fitness loss during stationary phase, suggesting an intrinsic trade-off. In summary, combining cell-number based approaches with high throughput quantitative fitness measurements enabled us to infer the adaptive strategies for all 1,600 adaptive clones.
RESULTS
We previously isolated 4,800 clones from generation 88 of the Levy et al. [6] evolution experiments, measured each clone’s fitness, and sequenced the whole genomes of several hundred adaptive clones to characterize the spectrum of beneficial mutations [12]. We divided the 4,800 lineages into four classes: 1) putatively neutral haploids, whose fitness was indistinguishable from that of the ancestor; 2) adaptive haploids; 3) “pure diploids” that had self-diploidized, but had no additional beneficial mutations; and 4) “high-fitness diploids” with additional beneficial mutations. Here, we sought to understand the adaptive strategies selected for in the original evolutionary condition (EC) by studying “realized” and “accrued” fitness benefits from different phases of the growth cycle under the EC and how adaptive clones tradeoff in stationary phase, which was not part of the EC.
Monoculture and pairwise competition measurements
Monoculture measurements
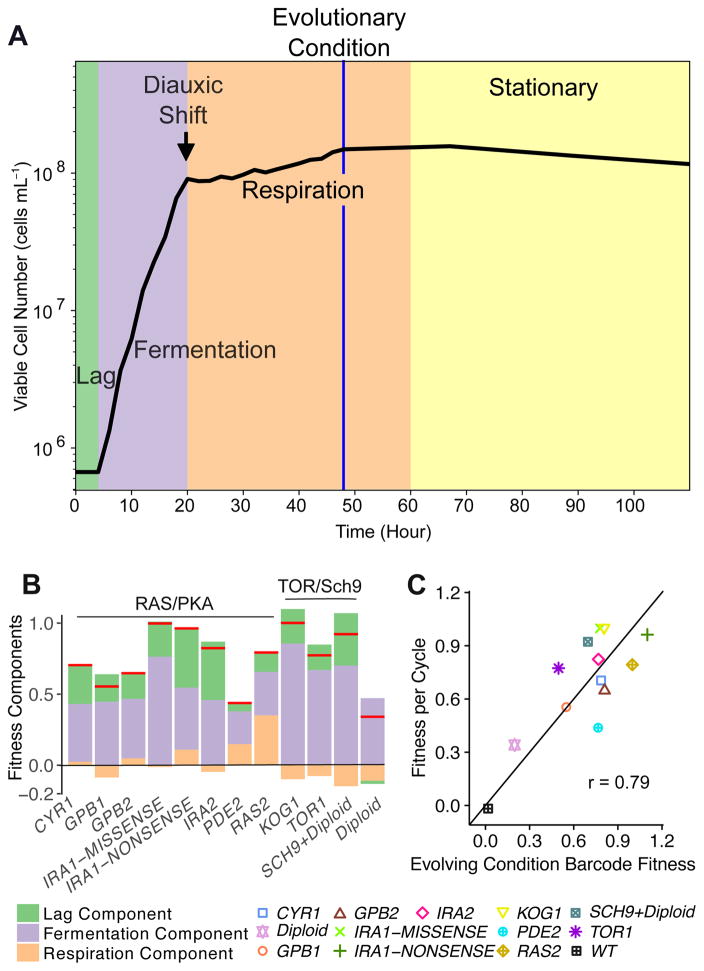
We characterized monoculture growth of twelve clones with known adaptive mutations, plus a wild-type control (STAR Methods; Table S1), using the same culture conditions as the EC, in which cells spend 48h per growth cycle with approximately 4 hours in lag phase, 16 hours in fermentation, and 28 hours in respiration (Figure 1A). Cell number, bio-volume, glucose concentration, ethanol concentration and cellular trehalose were measured during the 48h growth cycle (Figure S1; Data S1). We used these measurements to calculate each adaptive clone’s fitness change during lag (lag component), fermentation (fermentation component) and respiration (respiration component) in the EC.
Figure 1. Yeast growth curve under the evolutionary condition and beyond, and fitness components of adaptive clones in monoculture.
(A) Example of number of viable yeast cells during the growth cycle. Lag phase was extrapolated from cell number measurements at early time-points (STAR Methods). These data are from adaptive clone PDE2 in replicate 1.
(B) Lag, fermentation, and respiration components of fitness of mutant clones estimated from differences in the monoculture cell number relative to WT. Red lines indicate clones’ per-cycle fitness. Negative numbers imply a fitness decrease in that phase.
(C) Per cycle fitness estimated from monoculture cell number measurements vs. high-throughput barcode fitness measurements. The adaptive clones’ fitnesses were estimated by combining the lag, fermentation and respiration components estimated from the monoculture cell number measurements. The black line represents y=x.
All adaptive clones gained fitness during fermentation and most (except pure diploids) also gained fitness in lag phase relative to the ancestral control (Figure 1B). The fitness gains of all clones during fermentation are consistent with their faster consumption of glucose and faster production of ethanol during fermentation (Figures S1D and S1E). By contrast, only the pde2 and RAS2 mutants clearly exhibit a positive respiration component and several adaptive mutants even have negative respiration components (Figure 1B). The total fitness gains across the entire EC growth cycle, as estimated by the monoculture growth curves, are consistent with our prior estimates using barcode sequencing (Figure 1C, Pearson’s r = 0.79, 95% CI [0.19,0.94]) [12]. The mean fitness difference between these estimates is 0.16 per growth cycle, which corresponds to less than one fifth of a cell division per growth cycle (e0.16-1 ≈ 0.17 divisions).
However, there are issues with estimating EC fitness from monoculture measurements (STAR Methods). Most importantly, the EC fitness was measured – indeed defined – in a pool in which the ancestor was the dominant type and set the dynamics of glucose, ethanol, and other chemical concentrations in the cell culture. The absence of the ancestor in monoculture will likely affect the nutrient composition in the culture and thus the growth and measured fitness of the mutants.
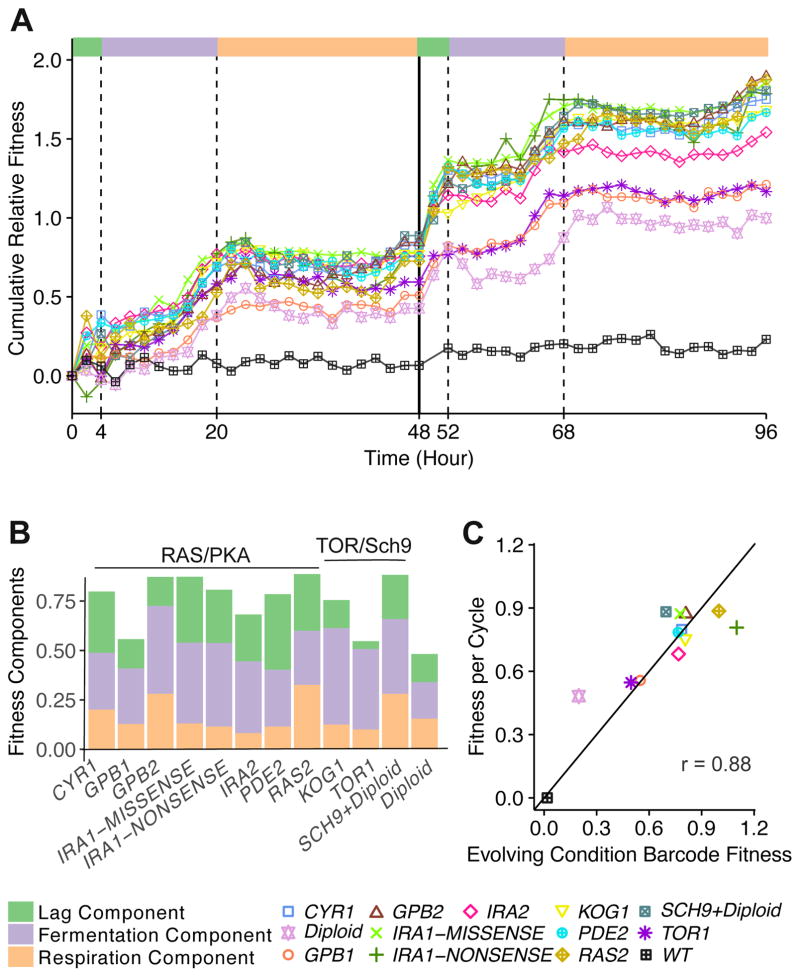
Pairwise competition
To measure fitness in the EC relative to the ancestor, we conducted high-resolution pairwise competitions for the same 12 adaptive clones (plus a WT control), each grown in competition with a YFP-tagged ancestral strain (STAR Methods). From the relative frequency of the adaptive clones in each pairwise competition, we estimated the cumulative fitness relative to the ancestor compared over two consecutive growth cycles (STAR Methods; Data S1). The fitness trajectories have high curvature, are somewhat non-monotonic, and sometimes have significant fitness gains in short periods of time, most notably at the start of the cycle close to lag phase (Figure 2A). This complex behavior is in contrast to the common assumption implicit in the analysis of most serial transfer evolution experiments: fitness is usually reported per generation, as if the fitness benefits all derive from a steady growth advantage during exponential phase. To parameterize the components of fitness from lag, fermentation and respiration phases respectively, we used cumulative fitness differences from 0 to 4 hours, from 4 to 20 hours and from 20 to 48 hours. All of the adaptive lineages showed a clear fitness increase during all three phases (Figure 2B); interestingly, for some clones this is in contrast to the monoculture data estimates (compare Figure 1B to Figure 2B). The total per-cycle fitness estimated from 0 to 48 hours is strongly correlated to EC fitness (Figure 2C; r = 0.88, 95% CI [0.57, 0.97]) [12], with an average difference of only 0.1 per cycle. This fitness difference is comparable to the smallest barcode-measured fitness differences between mutants in the same gene and the variations in the barcode-measured diploid fitnesses.
Figure 2. Frequency dynamics of mutant clones in competitive assays with wild type under the evolutionary condition, and the resulting mutants’ fitness components.
(A) Mutant lineages’ cumulative relative fitness in pairwise competition with a YFP marked ancestor. The first growth cycle ends at 48h. Color bars at the top of the figure indicate growth phases, consistent with the color scheme in Figure 1A.
(B) Lag, fermentation and respiration components estimated from the pairwise competition assays.
(C) Per-cycle fitness estimated from pairwise competition assays vs. high-throughput barcode fitness measurements. The black line represents y=x.
High-throughput barcode fitness measurements
While the monoculture and pairwise competition assays provided insight into the growth-phase dependent fitness effects of our adaptive mutations, both assays are limited in throughput. Furthermore, they only measured cell number change in a given growth phase (“realized” fitness gains), and thus do not reflect where within the growth cycle those benefits might be “accrued”.
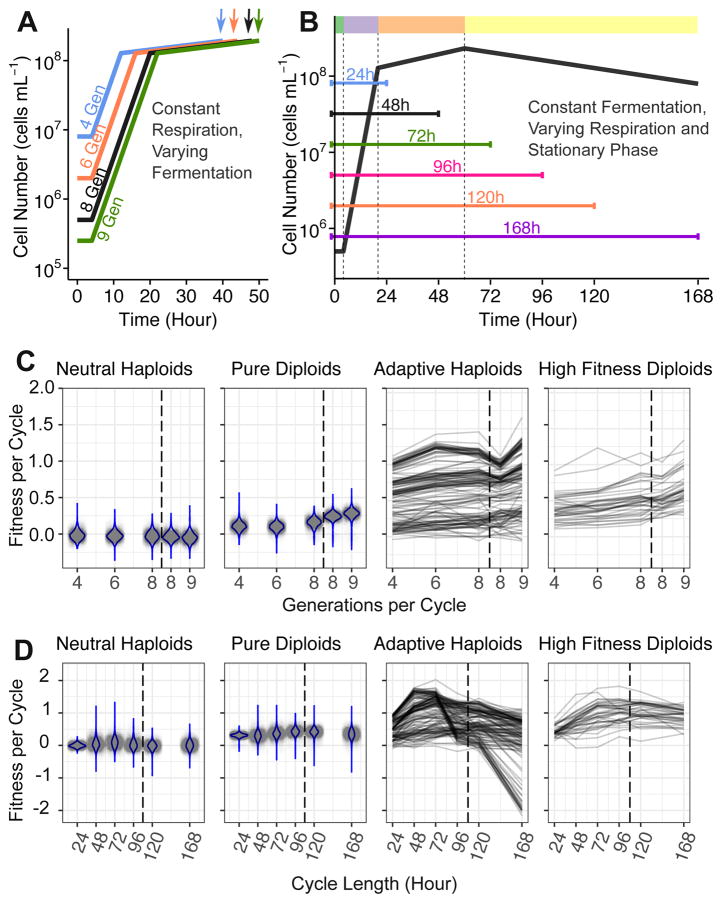
To identify where the fitness gains accrue, we used a high throughput fitness measurement approach to determine how the full-cycle fitness of the ~4,800 evolved clones [12] changed under a series of conditions in which we systematically varied the lengths of fermentation and respiration, and also varied the duration of stationary phase to explore fitness trade-offs (Figures 3A and 3B; Table S2). A subset of these clones were removed from further analysis due to either poor quality fitness measurements or to an ambiguous ploidy designation [12] (STAR Methods). This resulted in 3,048 high-quality lineages: 1) 1,464 neutral haploids, 2) 1,400 pure diploids, 3) 144 adaptive haploids, and 4) 40 high fitness diploids.
Figure 3. High-throughput barcode fitness measurements of 4,800 evolved clones under varying growth conditions.
(A) Schematic of conditions with varying dilution rate to control the number of generations in the fermentation phase. Arrows indicate the end of the growth cycle; cycle length was adjusted to maintain a constant respiration phase length.
(B) Schematic of conditions with varying cycle length to change the amount of time spent in the respiration (24h and 48h cycles) and in stationary phases (72h, 96h, 120h and 168h cycles), with the respiration phase ending around 60h. Dashed vertical lines indicate lag, fermentation, respiration and stationary phase; colors in color bar at the top of the figure are consistent with the color scheme in Figure 1A.
(C) Fitness measurements under conditions with varying number of generations in the fermentation phase.
(D) Fitness measurements under conditions with varying time in the respiration phase and the stationary phase. Respiration ended around 60h.
Note, fitness scales in (C) differ from (D) as the fitness changes are larger in (D). Violin plots are shown for neutral haploids and pure diploids. Fitness trajectories across this series of conditions are shown for adaptive haploids and high-fitness diploids (which carry a beneficial mutation). The vertical black dashed line separates the experiments in different batches (Table S2).
Experiments to determine the fermentation fitness component were conducted in two separate batches, with experiments using 4, 6, or 8 generations of growth in batch A, and 8 or 9 generations in batch B (Table S2). Pure diploids, adaptive haploids, and high fitness diploids increased their per-cycle fitness as they spent more generations in fermentative growth (Figure 3C). While there are clear batch effects, analysis of the data from batch A alone yields the same quantitative trends as when combining data from both batches (Figures S3B and S3C); batch B alone has too small a dilution range to be useful.
The pure diploids’ fitness did not change significantly across conditions with increased time in either respiration or stationary phases (Figure 3D). By contrast, the fitness changes of adaptive haploids and high fitness diploids were both non-linear and non-monotonic with cycle length (Figure 3D). The majority of these lineages increased their fitness as they spent more time in the respiratory growth phase (20–60h) but then decreased their fitness as they spent more time in stationary phase (Figure 3D), suggesting the presence of trade-offs between different parts of the growth cycle.
Using the fitness measurements across both sets of experiments (varying dilution and varying cycle length), we calculated the fermentation-dependent, respiration-dependent and stationary-dependent components of fitness (STAR Methods). Note, these phase-“dependent” components are conceptually distinct from the fitness components calculated from the monoculture and pairwise competition assays above – they are inferred by perturbing the environment and measuring how overall per-cycle fitness changes, rather than by measuring cell number change within specific portions of the EC growth cycle.
Contributions to Evolutionary Condition fitness from fermentation and respiration
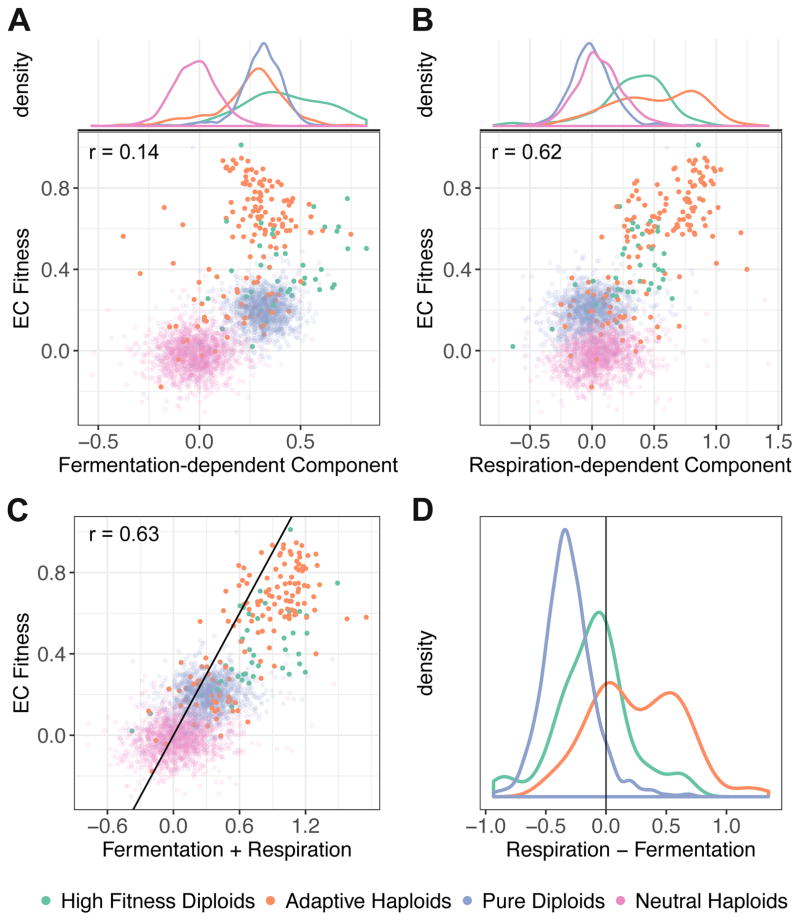
We studied how both accrued fermentation-dependent and respiration-dependent components vary between classes of adaptive lineages, and how these correlated with the fitness of individual clones under the EC [12]. Almost all adaptive lineages accrued fitness during fermentation (Figure 4A, see STAR Methods for analysis), with high-fitness diploids having a significantly higher averaged fermentation-dependent component than pure diploids (one-sided WRS p = 8e-6) and adaptive haploids (one-sided WRS p = 7e-7) (Figure 4A). This suggests that self-diploidization and additional adaptive mutations affect fermentative growth by non-redundant mechanisms. The fermentation-dependent component was weakly correlated with the EC fitness (r = 0.14, 95% CI [0.07, 0.21] for all adaptive lineages) and thus variation in per-cycle EC fitness among adaptive lineages cannot be explained by differences during fermentative growth (Figure 4A). By contrast, we found a strong positive correlation between the respiration-dependent component and EC fitness for all adaptive lineages (r = 0.62 95% CI [0.57,0.66]; Figure 4B), with adaptive haploids (r = 0.70, 95% CI [0.62,0.78]) and high fitness diploids (r = 0.65, 95% CI [0.37,0.80]) being more highly correlated. The pure diploid class has a negligible respiration-dependent component (mean of −0.004, compared to the batch effect of 0.1 per cycle) (Figure 4B). These data suggest that the EC fitness variation between adaptive lineages (other than pure diploids) comes primarily from differences in benefits accrued during respiration and, potentially, changes in the diauxic shift from fermentation to respiration.
Figure 4. Quantification of fermentation-dependent and respiration-dependent components of fitness in the evolutionary condition, inferred from the variable dilution and cycle length measurements of Figure 3.
(A) Fermentation-dependent component and (B) Respiration-dependent component vs. the per-cycle fitness in the evolutionary condition.
(C) Estimated fitness from combining the fermentation-dependent and the respiration-dependent components against their measured EC fitness. The black line represents y=x. For panels A–C, each dot represents one evolved lineage and is colored by ploidy and adaptive class under the EC; Pearson correlation is calculated for all adaptive lineages.
(D) The difference between the respiration-dependent component and the fermentation-dependent component among four groups of evolved clones.
See also Data S3.
In general, adaptive haploids, especially those with the highest fitness, have larger respiration-dependent components than fermentation-dependent components (Figure 4D; one-sided paired WRS p = 5e-12), while high fitness diploids gain slightly more from fermentation (Figure 4D; one-sided paired WRS p = 0.01) and pure diploids gain fitness from the fermentation phase exclusively (Figures 4A, 4B and 4D; z-test p < 2e-16). The sums of these components are strongly correlated with the EC fitness measurements across all classes of adaptive lineages (Figure 4C; r = 0.63, 95% CI [0.59, 0.66] with r = 0.73, 95% CI [0.66,0.79] for adaptive haploids and high fitness diploids). The average fitness difference between the sums of components and the EC fitness is only 0.19 per growth cycle among adaptive clones, suggesting that these two components are sufficient to recapitulate EC fitness.
Within-Cycle Fitness Trade-offs
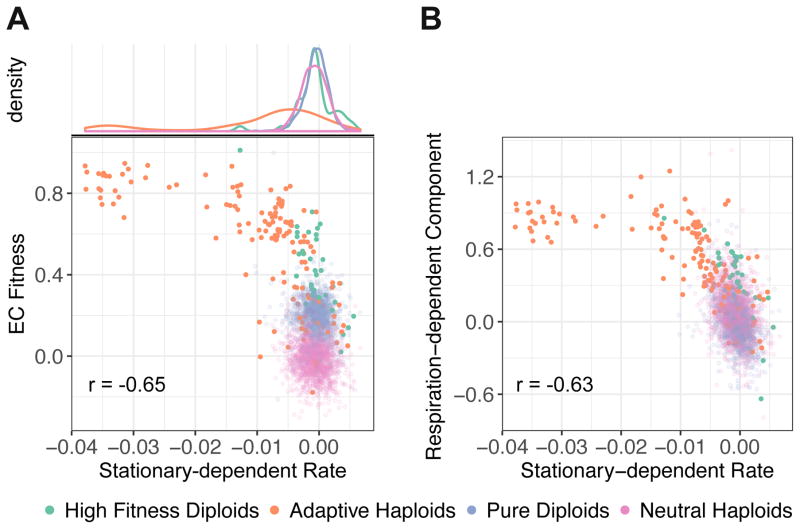
We next examined how fitness changed as a result of cells entering stationary phase, which was not experienced in the EC. We observed a general negative correlation between the EC fitness and the fitness change per hour during stationary phase (the stationary-dependent rate) (Figure 5A; r = −0.65, 95% CI [−0.69, −0.59] for all adaptive lineages, r = −0.69, 95% CI [−0.75, −0.63] for adaptive haploids, and r = −0.77, 95% CI [−0.89, −0.53] for high fitness diploids). Indeed, some of the most fit adaptive haploids showed a dramatic decrease in fitness. Based on the lack of variation in fermentation-dependent component, we expected and observed only a weak correlation between the fermentation-dependent component and the stationary-dependent rate (Figure S3A, r = 0.06, 95% CI [0.01, 0.1] for all adaptive lineages). However, there is a strong anti-correlation between the respiration-dependent component and the stationary-dependent rate (Figure 5B, r = −0.63, 95% CI [−0.66, −0.59] for all adaptive lineages), suggesting the existence of a physiological trade-off between the adaptive benefits accrued in respiration and survival during stationary phase.
Figure 5. Quantification of stationary-dependent rate.
(A) Fitness change per hour during stationary phase (stationary-dependent rate, inferred from variable cycle length barcode measurements) vs. measured EC fitness.
(B) Stationary-dependent rate vs. respiration-dependent component.
Pearson correlation is calculated for all adaptive lineages in A and B.
See also Figure S2 and S3 and Data S3.
We hypothesized that the loss of fitness during stationary phase of many adaptive clones was due to decreased cell viability. We found that the 12 tested adaptive clones exhibited significantly decreased viability over a six-day monoculture growth compared to the ancestral WT clone (Figure S2A; Data S2) and that there was a strong correlation between the stationary-dependent rate and the viability loss rate (Figure S2B, r = 0.92, 95% CI [0.56, 0.98]; R2 = 0.9). In addition, the adaptive clones exhibiting this trade-off had reduced cellular accumulation of trehalose (Data S2), a storage carbohydrate synthesized during respiration and important for cell survival during stationary phase [20–22].
Combining monoculture, pairwise competition assays and barcode fitness measurements
While the pairwise competition assays (Figure 2C) and the barcode based fitness measurements (Figure 4C) are both able to recapitulate the total EC fitness, they measure different things: the first measures realized fitness benefits, while the second measures accrued benefits, that may or may not be realized in the same growth phase in which they accrue. To determine whether there are fitness benefits accrued in one part of the growth cycle, yet realized later, we compared realized and accrued fitness components measured with these two approaches.
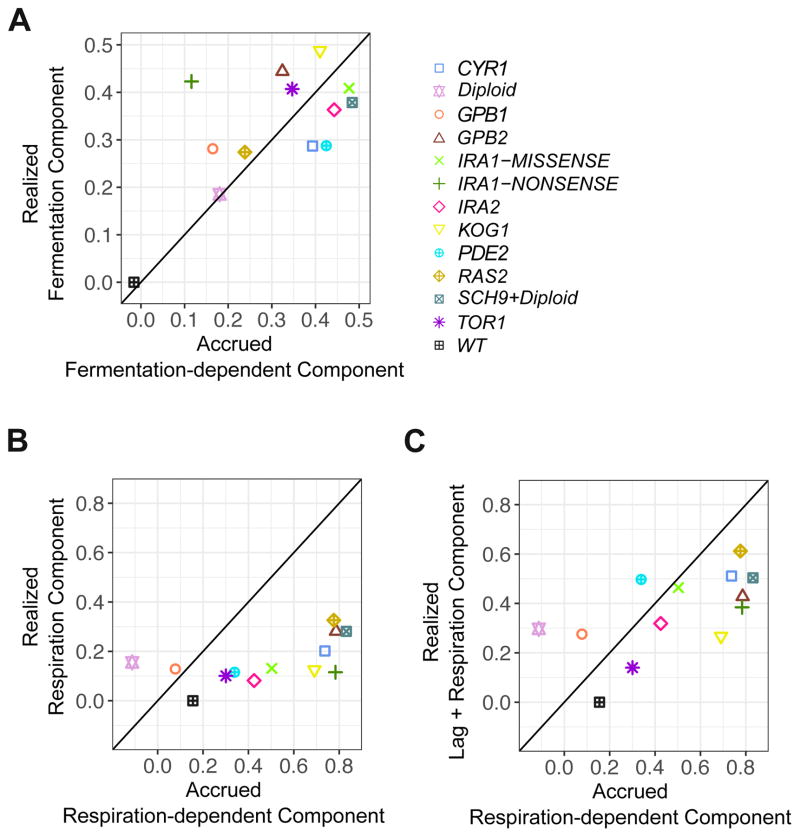
The accrued fermentation-dependent component estimated from our barcode fitness measurements is positively correlated with the realized fermentation component estimated from our pairwise competition assays (Figure 6A, r = 0.80, 95% CI [0.23, 0.94]; note, IRA1-NONSENSE is excluded from correlation analyses in each panel of Figure 6, as it is a significant outlier) with an average difference of only 0.08 (comparable to our observed batch effects). This suggests that fitness benefits accrued during fermentation are realized in the fermentation phase.
Figure 6. Comparison of fitness components estimated from different approaches.
Comparison of the realized fitness components estimated from the pairwise competition assays with the corresponding accrued fitness components estimated from the high-throughput barcode fitness measurements: (A) realized fermentation component from the pairwise competition with the accrued fermentation-dependent component from the barcode fitness measurements; (B) realized respiration component from the pairwise competition with the accrued respiration-dependent component from the barcode fitness measurements; and (C) combined realized fitness from the lag and the respiration phases estimated from the pairwise competition with the accrued respiration-dependent component estimated from the barcode fitness measurements. See also Figure S4.
However, the magnitude of the accrued respiration-dependent component estimated from the barcode fitness measurements is in general larger than that of the realized respiration component measured directly from the pairwise competition assays (Figure 6B). Strikingly, for the high fitness lineages the accrued respiration-dependent component is larger than could possibly be realized during respiration, given that the cells divide very little during respiration (see STAR Methods for quantitative explanation). We therefore suggest that the accrued respiration benefits are not fully realized within the respiration phase itself and thus must be realized in other phases of the growth cycle, either lag phase or fermentation. Since the fermentation components were fully accounted for, we hypothesized that the fitness accrued in respiration is instead largely realized in the subsequent lag phase. We thus combined the realized lag and respiration components from the pairwise competitions and compared this sum to the accrued respiration-dependent component from the barcode fitness measurements. Confirming our hypothesis, the sum of the realized lag and respiration components is not only significantly correlated with (r = 0.62, 95% CI [0.29, 0.87]), but is also comparable in magnitude to, the accrued respiration-dependent component, with an average difference of 0.23 (Figure 6C). Similarly, the sum of the realized lag and respiration components estimated from the monoculture measurements is more comparable to the accrued respiration-dependent component estimated from the barcode fitness measurements than is the realized respiration component alone (Figure S4). These combined data support the notion that benefits accrued during respiration are partially realized in the subsequent lag phase.
Fitness profiles denote genotypes and identify modes of adaptation
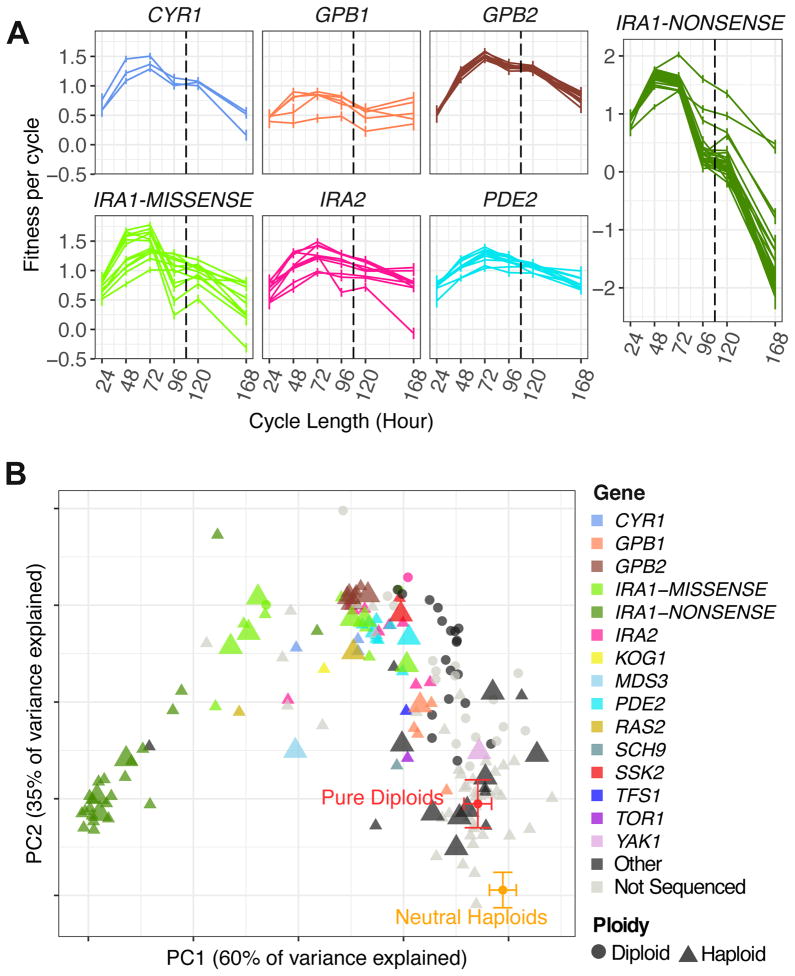
The combined set of barcode fitness measurements, across all nine conditions (Figures 3A and 3B) can be considered as a fitness “profile” for each of our assayed lineages. Visual inspection of the fitness of lineages carrying mutations in the Ras/PKA pathway showed that lineages with a similar genetic basis have similar fitness profiles (Figure 7A). We investigated whether these fitness profiles provide predictive power of their genetic basis of adaptation (within the spectrum of observed adaptive mutants), and/or reveal distinct modes of adaptation. Using principal components analysis (PCA) on the entire set of fitness data for 3,048 clones, we identified two large clusters, corresponding to neutral haploids and pure diploids, and determined that adaptive haploids and high fitness diploids were separated from these two groups (Figure S5); 87% of the variance is accounted for by the first two PCs. We then conducted PCA (Figure 7B) using the fitness data from only the adaptive haploids (144 lineages) and the high fitness diploids (40 lineages) of which 78 adaptive haploids and 22 high fitness diploids had previously been genotyped [12] (small icons in Figure 7B); we also genotyped an additional 24 adaptive haploid clones (large icons in Figure 7B; also see Data S3 and S4). The first PC accounts for 60% of the variance and captures the non-monotonic nature of the fitness trajectories under the varying growth cycle time conditions, while the second PC accounts for 35% of the variance and corresponds to the combination of the fitness changes under both series of conditions (Figure S6A). The 102 sequenced adaptive haploids cluster by the identity of the adaptive mutation (Figure 7B). Furthermore, all lineages with mutations in the same Ras/PKA pathway gene were on average significantly closer (Euclidean distance) in this PC-space than the average for all pairs of lineages (t-test p < 0.001, IRA1, IRA2, GPB1, GPB2, PDE2). This suggests that fitness profiles might be useful not only for predicting mutant genotypes, but also for guiding the selection of a diverse subset of representative strains for further detailed analysis in future studies. Indeed, the newly sequenced clones whose fitness profiles grouped them with previously sequenced Ras/PKA pathway mutants (large icons in Figure 7B) invariably had mutations in the same genes as those previously sequenced clones. By contrast, those newly sequenced clones whose fitness profiles did not group with known signaling pathway mutants, did not have mutations in either the Ras/PKA or TOR/Sch9 pathways.
Figure 7. Fitness profiles of recurrent mutants and principal component analysis (PCA) using adaptive haploids and high fitness diploids.
(A) Fitness profiles, from varying cycle length experiments, for selected adaptive haploids grouped by the mutant gene (and mutant type for IRA1). The vertical dashed line separates Batch C and Batch D experiments (Table S2).
(B) PCA of only adaptive haploids and high fitness diploids using their fitness measurements across all (both variable dilution and variable cycle length) experiments described in Figure 3A and 3B. Each symbol represents one lineage. 15 lineages carrying mutations outside of the nutrient response pathways are labeled as “Other” (see Data S3, S4 for details). Large symbols correspond to clones whole genome sequenced in this study; small symbols are clones sequenced in [12]. The neutral haploids and pure diploids symbols show the averages over the PCs of the neutral haploid population and the pure diploid population (as determined in Figure S5).
See also Figure S5 and S6 and Data S3 and S4.
The fitness profiles also provide a way to capture the variety of possible “modes of adaptation” that are selected for during evolution. Based on the PCA analysis of the fitness profiles, we grouped the adaptive clones into six clusters (Figure S6B), suggesting distinct modes of adaptation (which we define as including the trade-offs that result from the adaptation). Clusters 1 through 4 were enriched in nutrient response pathway mutations (Figures 7B and S6B; Data S3, S4) with each of the clusters showing a different rate of fitness trade-off during stationary phase (Fig S6C). By contrast, clusters 5 and 6 captured other modes of adaptation (Figures S6B–S6D), including those outside of the nutrient response pathways (Data S3, S4).
DISCUSSION
Underlying causes of fitness gain
Three groups of beneficial mutants were characterized under the evolutionary condition (EC): 1) pure diploids caused by self-diploidization; 2) high-fitness diploids carrying additional mutations; and 3) adaptive haploids, with the majority of them containing mutations in Ras/PKA or TOR/Sch9 pathways, which couple nutrient sensing to the decision to undergo cell division.
We observed that all adaptive mutants “accrue” a similar amount of fitness benefit during fermentation regardless of their molecular basis of adaptation. By contrast, adaptive mutants with different underlying genetic bases accrue a highly variable amount of benefit during respiration, with the most adaptive haploids gaining a larger benefit from respiration than from the ~8 generations of fermentation. The ability of S. cerevisiae to efficiently ferment sugars has likely been selected for both during its domestication for beverage production, as well as under laboratory conditions when growing on rich media [23]. It is possible that the fermentation ability of the S. cerevisiae strain used in our experiments is close to the maximum possible and that there is little room to improve it. By contrast, the respiration ability has likely been under less of a selective pressure during this history. This may explain why fermentation benefits across all three groups of adaptive mutants are both minor and similar in magnitude, while the magnitude of respiration benefits are variable over a broad range. The variable capacity of accruing benefits during respiration of different mutants in the Ras/PKA or TOR/Sch9 pathways, may come from the different extent of activation of these pathways, together with different magnitudes of deleterious pleiotropic effects of the mutations.
Based on our comparison of realized fitness components to accrued fitness components, we propose that benefits accrued during respiration can be partially carried over and realized in the subsequent lag phase. Cell size measurements show that the tested adaptive haploids are on average 26% larger than the ancestor by the end of the growth cycle. Adaptive haploids with a larger size may benefit from the nutrient-limited batch culture condition by dividing sooner upon the addition of fresh medium, thus shortening lag phase, though the increased size by itself is insufficient to account for the large respiration-dependent fitness increases. The difference between the respiration-dependent component of the fitness and that realized during respiration can be as large as 0.6, which, under the simple hypothesis that the shortening of lag phase is due to cells being closer to the size needed to divide, would require them to have been almost twice as large as the wild type. Furthermore, pure diploids also have a larger cell size yet do not accrue benefits during respiration. Combined, these data suggest that increased size by itself is insufficient for respiration-dependent fitness increases, and thus that other factors may also be important for shortening the subsequent lag phase of the adaptive haploids. For example, physiological differences in regulation of cell division that occur during diauxic shift and respiration might cause a subsequent short lag phase duration. Identification of the key physiological changes that cause this “memory” effect is a challenge for future research.
Ras/PKA hyperactive mutations
Fitness benefits accrued during respiration are strongly correlated with the amount of fitness lost during stationary phase, implying a trade-off between respiration benefits and survival in stationary phase for the large-effect mutations. The majority of these large-effect mutants are predicted to have up-regulated the Ras/PKA pathway. Mutants with an activated PKA have reduced accumulation of intracellular trehalose [24,25] and glycogen [26], repress autophagy [27], and are extremely sensitive to both carbon and nitrogen starvation [26], and do not arrest as unbudded cells when deprived of nutrients, preventing then from entering stationary phase properly [28]. We observed that adaptive clones with substantial accrued respiration benefits indeed had reduced intracellular trehalose storage, implying that these adaptive clones may sacrifice trehalose synthesis to boost growth during respiration, which would cause loss of survival in stationary phase. However, as the precise physiology underlying accrued respiration benefits is not clear and the causes of stationary survival are complex, further investigation is needed to understand this trade-off fully. It is known that decreased Ras/PKA activity results in a decreased growth rate during both fermentation and respiration [29], and that exogenous addition of cAMP affects both fermentative and respiratory activities [30]. This may explain the selection of likely hyperactive Ras/PKA pathway mutants in serial-dilution experiments with fermentation and prolonged respiration [29 and in this work], as well as in chemostat experiments, where the cells ferment the limiting glucose as fast as possible [4,5,32].
In contrast to well-controlled “simple” laboratory conditions, yeast cells in nature are exposed to complex and changing conditions, so that adaptive mutations observed under laboratory conditions may differ vastly from those selected in natural environments. In particular, starvation is frequently encountered by yeast cells in nature, and hyperactive nutrient pathway mutations will likely be strongly deleterious in such conditions, as observed for many of our mutants. Consistent with this scenario, hyperactive nutrient pathway mutants have been identified at high temperature QTLs in a domesticated palm wine strain, but were not seen in a natural oak bark isolate [32].
Fitness profiles
Phenotypic profiling [34–36], and fitness profiling [37,38] of systematic mutant collections – typically deletion or transposon insertion mutants – under a variety of conditions have greatly contributed to our knowledge of the functional roles of genes. However, we are the first to conduct a high-throughput survey of fitness effects across many conditions using thousands of adaptive clones.
The way in which the conditions were rationally chosen allowed us to use the combined fitness data (fitness “profiles”) to capture the general adaptive strategies among different classes of adaptive clones and to identify different modes of adaptation under the EC, which would be impossible by measuring fitness of only a handful of adaptive clones using traditional methods. Our work shows that having quantitative measurements of fitness across multiple environments not only captures broad similarities (e.g. the clustering of nutrient response pathway mutants), but also resolves the differences between mutations in different genes in the same pathway (and sometimes even between types of mutation in the same gene) with even simple analysis such as PCA. More importantly, fitness profiles improve our understanding of adaptation from a functional perspective, above what is gained solely from the genotypic point of view. Thus, we propose that using a combination of experimental evolution and fitness profiling to study the genetics and functions of a large number of adaptive mutants will result in a greater understanding of the adaptive process. By measuring evolved lineages’ fitness in a wide range of conditions and analyzing the resulting fitness profiles, we can gain a broad picture of the adaptive strategies during adaptation. Importantly, given finite sequencing and phenotyping resources, this approach also enables informed choices as to which adaptive clones to sequence to identify potentially novel mutations, including those in only modestly adaptive clones.
Lessons for experimental evolution
Fitness gains in experimental evolution are often reported per generation [6,39]. This implicitly suggests that the fitness gains will scale linearly with time spent in exponential growth phase. However, in our evolutionary condition the fitness gains for adaptive haploids are not primarily from fermentation (where most growth occurs), but instead, are realized mainly in lag phase due to benefits accrued during respiration. The per-cycle fitness is thus a more appropriate measure of the fitness gain, as it avoids any implicit assumption about where within the growth cycle those gains might occur. This approach has been used for evolution in complex conditions [40], and our data suggest that per-cycle fitness may be more appropriate even for seemingly “simple” conditions such as serial batch culture.
The net fitness effect for any beneficial mutation will be the sum of its positive and negative effects, which will depend on the particular environment. This balance suggests that the net fitness of adaptive lineages can be highly sensitive to modest changes in conditions. Thus qualitative characterizations of the environment — such as “batch culture in rich medium” or “in low glucose” — are insufficient. Detailed quantitative parameterization of the conditions is needed to determine the first-mutants that will dominate the initial evolution and the distribution of fitness effects (DFE) of potential beneficial mutations. For example, if our evolutionary condition had included a substantial stationary phase, then the most fit IRA1-nonsense mutants would have had much lower (perhaps even negative) fitness, and would likely not have been be observed at all. More importantly, even subtle differences in the DFE can lead to large differences in the relative abundance of adaptive clones during evolution since differences in fitness accumulate exponentially with time. Quantitatively, a 0.6 change in the per-cycle fitness of a mutant, which is the value of the accrued respiration-dependent components of some of our mutants, would change its abundance at the time point at which our mutants were chosen by a factor of ~1,000. Furthermore, changes in the DFE lead to different genetic backgrounds for future evolution, which can change evolutionary trajectories through both epistasis and via environmental feedback changing how phenotypic changes are selected.
Much of the sensitivity to conditions we observed appears due to the periodic nature of the environment. All adaptive lineages (except pure diploids) took advantage of the consistent transfer into fresh media via mutations that appear to reduce lag phase. This is underscored by the trade-off in stationary phase: when experiencing conditions that were not part of the EC these lineages were ill-prepared and had severe fitness defects. These sorts of complex tradeoffs may be common in evolution with constant as well as periodic conditions [41,42]. Mutations can be selected for within the narrow range of external pressures felt by the organism, and they may carry unknown and unpredictable costs even in seemingly very similar environments. While beyond the scope of this study, it will be interesting to quantify how adaptive mutations that are selected for in predictable environments differ from those selected in varying conditions. One might predict that varying conditions would select for smaller effect mutations that show fewer trade-offs. Quantifying these differences would in part require understanding the joint distribution of fitness effects of various mutations over different environments – this can now be straightforwardly measured using our approach.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information regarding the manuscript and requests for reagents may be directed to, and will be fulfilled by the lead contact, Gavin Sherlock (gsherloc@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The yeast strains used in this study can be grown and maintained using standard methods (e.g., YPD media in test tubes, glycerol stocks for long term storage at −80°C), but should be propagated in the selection environment (glucose limited minimal media – M3 medium) for optimal phenotypic and fitness measurements. We refer to the 5x Delft media with 4% ammonium sulfate and 1.5% dextrose [6] as M3 medium in this manuscript.
Twelve evolved strains and a WT strain were assayed in both monocultures and pairwise competition assays. Ten out of the twelve evolved strains are haploids and harbor mutations in CYR1, GPB1, GPB2, IRA1, IRA2, KOG1, PDE2, RAS2 and TOR1. Two IRA1 haploid mutants with different mutation types (missense and nonsense) were tested. The other two tested strains were a diploid with no additional mutations (“pure” diploid) and a diploid with an additional mutation in SCH9 (SCH9+Diploid) (see detailed information of tested strains in Table S1). Note that the strains IRA1-NONSENSE, RAS2 and SCH9+Diploid carry additional mutations outside of the genes listed here (see Data S4 for details). The genes in which these additional mutations occur only appear mutated once each across the whole set of sequenced mutant lineages and thus are less likely than those in multiply-hit genes to be adaptive under the evolutionary condition. The fitness profile for this IRA1-NONSENSE lineage is similar to that of other IRA1-NONSENSE lineages supporting this hypothesis.
All high-throughput barcode fitness measurements were conducted using the pool of 4,800 evolved barcoded clones described in [12]. Briefly, this pool was constructed by picking 4,800 individual single colonies that had been plated out from frozen samples of the two replicate evolution experiments of [6] at generation 88, and then pooling these clones into a single culture. The pool culture was mixed with glycerol (17% glycerol in final concentration), aliquoted into 1.5ml Eppendorf tubes and stored at −80°C.
Lineages carrying mutations in Ras/PKA pathway genes or TOR/Sch9 pathway genes are referred to as nutrient response pathway mutants in this work: RAS2, GPB1, GPB2, PDE2, IRA1, CYR1, TFS1, SSK2 and YAK1 gene are involved in Ras/PKA signaling pathway; SCH9, TOR1, KOG1 and MDS3 are involved in TOR/Sch9 signaling pathway. If a mutant harbors more than one mutation, as long as one of the mutations is located in above genes, it is classified as a nutrient response pathway mutant in this work.
METHOD DETAILS
Pre-culture test strains
Strains were streaked out from freezer stocks onto M3 agar plates and grown for 3 days until colonies were visible. A single colony was used to inoculate 3mL of M3 medium, which was grown for 48 hours (30°C roller drum). After saturation, 400μl was used to inoculate evolutionary condition pre-cultures (100mL M3 media in 500mL Delong flasks, 223 RPM 30°C), which were grown for 48 hours.
Monoculture measurements
Monoculture growth measurements over a 48-hour growth cycle were conducted in two independent replicates, which were performed on different days with independent single colony isolates. We assayed 12 evolved strains and a WT strain in both replicates (see EXPERIMENTAL MODEL AND SUBJECT DETAILS for strain information). Cells were precultured as described above, then for each clone, 5e+7 cells (as measured by a Beckman Coulter Z2 particle counter) were transferred into a fresh evolution condition flask (100mL M3 media in 500mL Delong flasks, 223 RPM 30°C) and the culture was tracked throughout the 48 hour growth cycle for various attributes. We sampled ~300μl of cell culture every two hours starting at eight hours of growth (the cell density was too low for accurate measurements prior to eight hours), measured cell number and cell size using the Coulter counter and collected ~200μl supernatant from the sampled cell culture for glucose and ethanol concentration measurements. Bio-volume was calculated by computing the weighted average cell size using the cell size distribution measured by the particle counter and multiplying the weighted average cell size by the total cell number. An additional 1mL of cell culture was sampled at 20, 22, 32 and 48 hours for cellular trehalose concentration measurements. Cellular trehalose concentrations were measured using the Megazyme Trehalose kit (Megazyme K-TREH), while glucose and ethanol concentrations in cell culture were measured using the Megazyme D-Glucose-HK kit (Megazyme K-GLUHK-110A) and Megazyme Ethanol kit (Megazyme K-ETOH), respectively. Except for ethanol measurements, all measurements were conducted in both independent replicates/growth cycles. Due to the technical difficulties in accurately measuring residual ethanol in cell culture, we only measured ethanol concentration in one replicate. Some clones showed a glucose concentration slightly higher than 1.5% at eight hours – 1.5% is the starting glucose concentration of the M3 medium, which could be caused by either measurement errors or variation in medium preparation; this may underlie batch effects we observed in barcode fitness measurements. All raw measurement data are shown in Data S1 (sheet 1 – 11). The batch effects of the cell number, bio-volume and glucose concentration measurements between these two independent replicates are similar to those observed in our previous work [12]. Trehalose per unit of cell volume was calculated by dividing trehalose measurements by median cell size measurements at the same time point. Averaged trehalose per unit of cell volume over two batches was used for analysis. However, due to the unusually slow growth of the wild-type control in replicate 1, only the wild type control in replicate 2 was used when calculating trehalose per unit of cell volume. Median cell size measurements at 46 and 48 hours are averaged and used as the cell size at the end of the growth cycle.
Viability measurement
Twelve evolved strains plus a WT strain (see EXPERIMENTAL MODEL AND SUBJECT DETAILS for strain information) were pre-cultured as described above. For each clone, 5e+7 cells (as measured by a Beckman Coulter Z2 particle counter) were transferred into a fresh evolution condition flask (100mL M3 media in 500mL Delong flasks, 223 RPM 30°C), the exact same condition as the monoculture measurements, and the culture was tracked for a total of six days. We measured viability every 24 hours starting at 48h by sampling the cell culture, diluting the cells by 1e+5 and plating 150μL of diluted cells onto YPD+AT agar plates. Plates were scored for the number of viable colonies after 2 days of growth at 30°C. The expected number of colonies was estimated using the Beckman Coulter Z2 particle counter of the cell culture at the time of plating. The observed colony number was then divided by the expected number, to calculate viability.
Pairwise competition growth curves
Twelve evolved strains plus a WT strain (see EXPERIMENTAL MODEL AND SUBJECT DETAILS for strain information) and a YFP-tagged ancestor [6] were pre-cultured as described above. We then mixed each pre-cultured clone with the YFP-tagged ancestor at a 1:9 ratio and performed high-resolution pairwise competition assays between these evolved strains (Data S1, sheet 13 and 14) plus the WT strain and the YFP-tagged ancestor. The pairwise competition population was sampled every two hours across two successive growth cycles, which was a total of 96 hours. Two independent replicates were conducted as described above, with each replicate being tracked over two successive growth cycles, which gave us a high-resolution frequency trajectory for evolved strains over four total cycles. The relative frequencies of the evolved and YFP strains were estimated as described [6]. Due to errors in the flow cytometry calibration during the first cycle of replicate 1, we had only three, rather than four cycles of usable data. In addition, measurements at 48h in replicate 2 were discarded as they were low quality and were replaced with the 46h measurements.
High-throughput fitness measurements
All high-throughput barcode fitness measurements were conducted using the pool of 4,800 evolved clones (see EXPERIMENTAL MODEL AND SUBJECT DETAILS for detailed information). Fitness measurements were conducted under a number of different growth regimens, each of which was a variant of our previously used “evolutionary condition” (EC) regimen [6]. Briefly, the EC regimen was as follows: 400μL of saturated cell culture (~5e+7 cells) were inoculated into 100mL of M3 (minimal, glucose-limited) medium in 500mL DeLong flasks and grown at 30°C in an incubator shaken at 223 RPM; the population was diluted to 5e+7 cells every 48 hours (2 days) into fresh M3 medium, resulting in ~8 generations of growth every 48 hours. The various differing growth conditions used for the fitness measurements in this study are described in Figure 3. Details of the varying growth conditions are listed in Table S2.
All conditions are variations of the EC regimen (see Results for details), and parameters were identical to the EC except where noted. In preliminary experiments, we determined that each generation during the fermentation growth phase took about two hours under our growth conditions (data not shown), so for conditions in Batch A and Batch B (see Table S2), we modulated the transfer time between growth cycles appropriately to maintain a consistent amount of growth time during respiration. Note, the fitness measurements in the EC itself were previously reported [12]. Before fitness assays, a tube of the 4,800 clone pool was thawed and cultured in the EC condition for two days to acclimate the yeast cells to the experimental condition after recovery from being frozen; cells reached saturation after this two-day growth. All fitness assays were conducted by mixing the saturated culture of the 4,800 clone pool with a pre-cultured ancestral clone (as described above) in a 1:9 ratio (time 0), and growing this mixture for four successive growth/dilution cycles under the appropriate condition (timepoints 1, 2, 3 and 4). The fitness measurement assays were thus sampled five times, at time 0 before the beginning of the assay, and at the end of each of the four growth/dilution cycles. For each sampling, cells were spun down, mixed with sorbitol solution (0.9M sorbitol, 0.1M Tris-HCL pH 7.5, 0.1M EDTA pH 8.0) and stored at −80°C for future genome extraction and barcode region amplification [12]. The frequencies of each of the lineages were tracked by Illumina sequencing of the DNA barcodes, which were then used to estimate their frequencies in the population as a whole [12].
All fitness measurements were performed with 2–3 replicates, and were conducted for 4 growth cycles. Fitness assays were conducted under nine unique test conditions and were divided into four batches (Table S2). Assays in each batch were performed independently with a different aliquot of the 4,800 clone pool and a different ancestral clone picked on different days. All batches of experiments contained one set of measurements in the EC to account for systematic effects between experimental batches, except for the long-cycle length (batch D) measurements, where the EC does not overlap much temporally with the other growth conditions in the batch (Table S2).
Whole-genome sequencing
Library preparation, whole-genome sequencing and analysis were conducted as previously described [12]. Briefly, we generated low-volume Nextera libraries [43], and identified small variants (SNPs and short indels) using the GATK [44–46] and large structural variants using CLC Genomics Workbench.
78 adaptive haploids and 22 high fitness diploids were previously sequenced [12] with beneficial mutations successfully identified in 76 adaptive haploids and 16 high fitness diploids. 25 adaptive haploids were sequenced in this work, with candidate beneficial mutations successfully identified in 24 of them. The sequenced clones with no identified candidate beneficial mutations are labeled as “Other” on the PCA plot and in Data S3. Details can be found in Data S4.
QUANTIFICATION AND STATISTICAL ANALYSIS
Monoculture fitness estimation and challenges
Cell number was measured in monocultures under the original evolutionary condition. Linear models were fit to log(mutant cell number) − log(WT cell number), and the slopes were used to infer the relative growth rates (per hour). Measurements from 10h to 16h were used to infer fermentation growth rates, and measurements from 24h to 48h were used to estimate respiration growth rates. The average of the relative growth rates over two independent replicates (equal to two growth cycles) were used in this manuscript. The relative growth rate error was estimated as the difference of the relative growth rate between the two replicates.
The monoculture cell number measurements were also used to estimate the lag phase fitness advantage. By linearly extrapolating the exponential growth back to the starting cell number, we gained a rough estimate of the time spent in lag. We also estimated the difference in fitness by comparing the log ratio of the cell numbers at an early time-point, given that all cultures were inoculated with the same number of cells. We used data from both growth cycles of the monoculture experiments to make this estimate of the lag fitness. We used the 8h, 10h, and 12h time-points to estimate the cell number at 8h via log-linear extrapolation, then computed the difference in log cell numbers to find the relative effects of the lag phase. The fitness advantages were measured relative to the wild type control in replicate 2. The averaged lag phase fitness estimations over two replicates were used.
In addition to issues noted in the main text itself, the inference of fitness from the monoculture data depends on an implicit model of the population dynamics, for example, the growth data are not strictly log-linear, and there is curvature near both the lag end and the diauxic shift (Figures S1A and S1B), which affects the fit. Even without these fundamental problems, the monoculture measurements are systematically different between replicates (Data S2), causing large uncertainties in the inferred parameters and limiting their utility.
Relative viability estimation
Twelve evolved strains plus a WT strain (see EXPERIMENTAL MODEL AND SUBJECT DETAILS for strain information) were cultured in monoculture for viability measurements. The number of viable cells which formed colonies on plates was divided by the expected number measured by Coulter Counter to calculate viability: the fraction of clones that are viable. Linear models were fit to log(viability of adaptive clones) − log(viability of WT clone) to infer the relative viability loss of the adaptive clones.
Fitness estimation using pairwise competition data
Twelve evolved strains plus a WT strain (see EXPERIMENTAL MODEL AND SUBJECT DETAILS for strain information) were competed pairwise with a YFP-tagged ancestor. We computed the cumulative fitness advantage of our test clones relative to the YFP tagged ancestor via the equation C(t) = log{F(T)/[1−F(T)]} − log{F0/[1−F0]}, where F(T) is a lineage’s frequency at time T and F0 is a lineage’s initial frequency at time 0. The expression for C(t) can be obtained by integrating the logistic growth function df/dt = s(t)(1−f)f, the basic model for competition between individuals with time-dependent fitness difference s(t). Frequency measurements at 48h were regarded as time 0 for the 2nd growth cycle of replicate 1. We used fitness differences from 0 hour to 4 hours, from 4 hours to 20 hours and from 20 hours to 48 hours to estimate the components of fitness from lag phase, fermentation and respiration respectively. We reported the average of the fitness components over all three growth cycles, with error given by the sample standard deviation. We estimated the total fitness change per cycle as log[F46h/(1−F46h)] − log[F0/(1−F0)] for the 1st growth cycle in replicate 2, and by log[F94h/(1−F94h)] − log[F46h/(1−F46h)] for the 2nd growth cycle in replicates 1 and 2. The per-cycle fitness was calculated by averaging fitness change per cycle across all 3 cycles. In addition, to decrease measurement noise, a moving window-based approach was used to estimate per-cycle fitness from pairwise competition assays. In replicate 2, each test clone’s frequency measurements from 20 hours to 46 hours were averaged and used as its start frequency; each clone’s frequency measurements from 68 hours to 96 hours were averaged and used as its end frequency. Per-cycle fitness was estimated using the same equation with the averaged start frequency and end frequency described above. The per-cycle fitness estimates between these two different approaches are similar to each other (mean difference = 0.037 and Pearson’s r = 0.98). The averaged fitness change per-cycle across all 3 cycles was used for our analyses.
High-throughput barcode fitness estimation
High-throughput barcode fitness measurements with the 4,800 clone evolved pool (see EXPERIMENTAL MODEL AND SUBJECT DETAILS for detailed information) were conducted under nine unique test conditions; the nine conditions were divided into four batches (see Table S2). Fitness measurements under each condition were performed with 2–3 replicates, and were conducted for 4 growth cycles. The frequencies of each of the lineages were tracked by Illumina sequencing of the DNA barcodes, which were then used to estimate their frequencies in the population as a whole, as previously described [12].
Fitness under a certain growth regimen was estimated using lineages’ frequency change over growth cycles under this regimen, as previously described [12]. Fitness under conditions with varying dilution rate (batch A and B in Table S2) was estimated using frequency data from time-points 1, 2, 3 and 4. Fitness under conditions with varying cycle length (batch C and D in Table S2) was estimated using frequency data from timepoints 0, 1, 2, and 3. Estimates were combined across the 2–3 replicates for each growth regimen using inverse variance weighting. The source code for computing these fitness estimates can be found at https://github.com/barcoding-bfa/fitness-assay-python.
Removal of lineages from analysis
From our pool of 4,800 clones, a subset of lineages was removed from subsequent analyses due to the following four reasons: 1) all lineages were cultured individually before they were pooled; however, some lineages did not grow and thus were not included in the pool [12]; 2) lineages present in the starting pool but that presumably declined rapidly in frequency and were thus undetectable under any condition were not included; 3) lineages with fitness estimation errors larger than 0.5 per cycle under any test condition were not included; and 4) lineages with an ambiguous ploidy designation (for details about ploidy identification see [12]) were not included. 3,048 lineages remained after these removals and were used for subsequent analyses described in this work.
Classification of strains
The 3,048 lineages with high-quality fitness measurements in the pool were classified into four groups, based on their ploidy and fitness: neutral haploids, adaptive haploids, diploids presumed (or found from sequencing) to have no additional adaptive mutations (“pure” diploids), and diploids with additional adaptive mutations (high fitness diploids). Lineages were grouped first by their ploidy (haploid or diploid; see [12]), then by their fitness assay data to determine which lineages were likely to contain adaptive mutations other than diploidization events. Under our modeling assumptions, lineages’ fitness estimates were assumed to be Gaussian distributed with an estimated standard deviation σ, which were supported by analysis of the neutral haploids and pure diploids [12]. Adaptive haploids were defined as lineages that had low probability (p < 1e-3) of having fitness of 0 or less in at least two conditions. Using this approach, 144 adaptive haploid lineages and 1,464 effectively-neutral haploid lineages were identified. High fitness diploids were defined as diploid lineages with additional adaptive mutations that had low probability (p < 1e-3) of having fitness less than the mean fitness of the diploid class in at least two conditions. The mean fitness of the diploid population was calculated using inverse variance weighting, then 40 high fitness diploid and 1,400 “pure” diploid lineages were identified using the above approach. The pure diploids have measured fitness averaging about 0.2 per cycle, while the high fitness diploids have fitnesses mostly in the range 0.3 to 0.8 (5th to 95th percentile). The vast majority of the adaptive haploids carry mutations in the Ras/PKA or TOR/Sch9 nutrient response pathways with per-cycle fitness ranging from 0.1 to 1.1 (5th to 95th percentile) [12].
Quantification of fitness components
We measured lineages’ fitness under two sets of systematically varied conditions where the length of the fermentation, respiration and stationary phases were changed independently (Figure 3; see Table S2), and used these measurements to quantify the fitness contribution from the three different growth phases. Specifically, we estimated fitness change per generation during the fermentation phase, fitness change per hour during the respiration phase and fitness change per hour during the stationary phase for each individual lineage in the pool. To do this, we fit a number of weighted linear models using the estimated fitness of the lineage under different conditions. All weighted models used inverse variance weighting, as under our modeling assumptions the inferred fitness values are Gaussian distributed. Note: this choice will lead to roughly equal weights for the high fitness lineages, which don’t obey the Gaussian assumption, as their error is dominated by a multiplicative noise term that is consistent across experiments.
We estimated the per generation fitness by fitting a weighted linear model to the per-cycle fitness against the estimated number of generations per growth cycle (log2 of the dilution factor) (fermentation-dependent fitness change rate). Note that during fermentation, each generation equals about two hours. We similarly estimated the rate of fitness change per hour during the respiratory growth phase (respiration-dependent fitness change rate) using fitness measurements from experiments with 24h and 48h growth cycles (as the cells undergo respiratory growth between 20 and 60h), and the fitness change per hour during stationary phase (stationary-dependent fitness change rate, called stationary-dependent rate in the main text) using the fitness measurements from the experiments with 72h, 96h, 120h and 168h growth cycles. We estimated fitness components using the fitness change rate estimates: fermentation-dependent component = fermentation-dependent fitness change rate x 8 generations; respiration-dependent component = respiration-dependent fitness change rate x 28 hours. We made this choice of measurements in order to avoid excess correlation between the respiration and stationary rate measurements; the 72h experiment shares its first time-point with the 96h, 120h, and 168h experiments. If we use only the 96h, 120h, and 168h growth cycles for the stationary phase and include the 72h experiment in the respiratory phase inference (extrapolating back to 60h where respiration actually ends), the stationary fitness change rate between the two estimates with/without 72h growth cycle on average differ 0.001 per hour with a Pearson correlation 0.88; the respiration fitness change rate between the two estimates with/without 72h growth cycle on average differ 0.004 per hour (0.11 per cycle under the EC) with a Pearson correlation 0.8. Despite the small quantitative change, no large qualitative change is observed and all of our results hold (especially comparisons to the EC fitness).
The errors in the stationary-dependent fitness change rate were calculated directly from the weighted linear model. The respiration-dependent fitness change rate was assumed to be the difference of two Gaussian distributed random variables, so its variance was taken to be the sum of the variances of the individual measurements, scaled appropriately. Systematic deviations between the two batches of experiments (Table S2) dominate the errors of the fermentation-dependent rate calculations. These deviations cause enough of a kink in the fermentation fitness profiles that the standard linear regression machinery cannot be used to quantify errors in the estimation. To understand the effects that systematic errors had on adaptive haploids, we plotted the fermentation rate as computed from the full data against the rate computed from the 4, 6, and the 8 generation experiment from the same batch (Figures S3B and S3C). The differences between the two fermentation rates are on the order of 0.01/gen. This suggests that the actual errors are typically on the scale of 0.01/gen to 0.02/gen (0.1–0.15 per cycle in EC), as the partial data consist of about half the full data. To test whether or not adaptive haploids had positive fermentation fitness, we performed a z-test with a significance level of p < 0.05 using the conservative estimate of 0.02/gen for the errors on the fitness. This resulted in 56% of adaptive haploid lineages as having a non-zero fermentation component. We performed the same test on the high fitness diploids, with 80% of them being called as having a non-zero fermentation component. For the pure diploids, since we had a large number of putatively genetically identical individuals, we performed a standard t-test on the set of fermentation components and showed that the component was significantly greater than 0 (p < 2.2e-16).
Accrued vs. realized respiration benefits
The magnitude of the accrued respiration-dependent component estimated from the barcode fitness measurements is, for all but the pure diploid and the GPB1 mutant, higher than the realized respiration component estimated from the pairwise competition assays (Figure 6B). For example, for IRA1-missense, the barcode derived accrued respiration-dependent component is ~0.35 higher than the realized respiration component derived from the pairwise fitness competition. This corresponds to almost half an extra doubling (e0.35 – 1 ≈ 0.42 divisions) compared to the ancestral type in the growth cycle, and it is not even the most extreme case. From the monoculture data, we know that both ancestral and adaptive cells undergo only about half a division during respiration. Thus, adaptive clones are unlikely to gain almost half an extra division and derive the respiration-dependent component entirely through more rapid division during respiration.
Evolutionary Condition fitness
The evolutionary condition fitness (EC fitness) used in this work is from Table S2 in [12], where EC fitness was measured in four independent batches and reported in both per generation units and per-cycle units: fitness per cycle = fitness per generation * 8. Fitness measurements from batch 1 were not used for any fitness component estimation in the current work. Batch 2 corresponds to batch C (respiration-dependent component estimation) in the current work. Batches 3 and 4 correspond to batches A and B respectively (fermentation-dependent component estimation) in the current work. Fitness estimates from monoculture (Figure 1) and pairwise competition (Figure 2) were compared to weighted averaged per-cycle EC fitness from [12] using all four batches of EC fitness measurements. Fitness component estimates from the barcode fitness measurements (Figures 4 and 5) are compared only to EC fitness from batch 1 in the previous work [12], to avoid spurious correlation between the estimated components and the estimated EC fitness in our analyses.
Correlation testing
For comparison of fitness and fitness components measured with different methods, we used Pearson correlation (Pearson’s r) since the units of measurement were the same, and we had many data points. All confidence intervals were computed using bootstrap percentiles due to the lack of an explicit model for most of our data.
Principal Components Analysis and clustering
The prcomp function in the software package R was used for Principal Component Analysis. The hclust and cutree functions in R were used to cluster lineages into 6 different groups.
DATA AND SOFTWARE AVAILABILITY
Data resource
All Illumina sequencing data (for both the whole-genome sequencing and the fitness measurement assays) can be found under NIH BioProject: PRJNA388215.
Software
The software repository for the barcode counting code can be found at https://github.com/sunthedeep/BarcodeCounter.
The source code for computing these fitness estimates can be found at https://github.com/barcoding-bfa/fitness-assay-python.
Supplementary Material
Data S1: Growth characteristics and metabolite measurements of twelve evolved strains in monoculture and frequency measurements in high-resolution pairwise competition assays; Related to Figures 1 and 2
Data S2: Measured properties of the 12 assayed adaptive clones; Related to Figures 1 and 2
This table includes fitness components from monoculture, pairwise competition assay and barcode measurements, estimated viability loss rate per hour, trehalose per unit of cell volume at 48h and the change rate of other metabolites measured in Data S1.
Data S3: Measured properties of the 3,048 clones in the pool; Related to Figures 3, 4, 5 and 7
The table includes each clone’s barcode ID, clone ID, fitness measurements in batch 1 from [S1], fitness measurements and error under all tested conditions, as well as ploidy, class (neutral haploids, pure diploids, adaptive haploids and high fitness diploids), global EC fitness and their fermentation-dependent and respiration-dependent fitness components, and stationary-dependent rate (per hour) estimated from the barcode fitness measurements. This table also contains the cluster information of adaptive haploids and high fitness diploids in Figure S6, their genotypes if they carry mutations in the nutrient response pathways (Clones that carry mutations only in non-nutrient response pathway genes are indicated as “Other” in the genotype column), mutation type and in which batch they are sequenced. Note that SCH9+Diploid clone tested in monoculture and pairwise competition were removed from analysis with barcode fitness measurements because it has high fitness estimation error and thus is not included in the 3,048 clones. In order to facilitate comparison, the measurements and estimates of SCH9+Diploid clone are added back, resulting in 3,049 clones in this table.
Data S4: Detailed genotypes of sequenced clones; Related to Figure 7
Data S5: Primers used to amplify the barcode region; Related to the Key Resources Table
Highlights.
Fitness benefits from different phases of a growth dilution cycle were quantified.
Benefits were “accrued” in respiration but “realized” as a shortened lag phase.
Analysis of high throughput data reveals different adaptive strategies.
Trade-offs exist between accrued respiration benefits and stationary survival.
Acknowledgments
We wish to thank Jennifer C. Ewald for discussions and technical help. We thank all members of the D.S.F, G.S., and D.A.P. labs for useful discussions. We thank the Stanford Shared FACS facility for use of their flow cytometers, and the Stanford Center for Personalized Genomics and Medicine for Illumina sequencing services. Y.L. is supported by Genentech Foundation Predoctoral Fellowship; S.V. by NIH/NHGRI T32 HG000044 and the Stanford Center for Computational, Human and Evolutionary Genomics (CEHG); A.A. by a Stanford Bio-X Bowes Fellowship. The work was supported by NIH grants R01 HG003328 and GM110275 to G.S. and R01 GM115919, GM10036601, and GM097415 to D.A.P, and NSF grants PHY-1305433 and PHY-1607606 to D.S.F.
Footnotes
Author Contributions
Conceptualization: Y.L., S.V., A.A., D.S.F, G.S., and D.A.P.. Methodology: Y.L., S.V. and A.A. Formal Analysis: Y.L., S.V. and A.A. Investigation: Y.L., S.V. and B.D.. Writing – Original Draft: Y.L.. Writing – Review & Editing: Y.L, S.V., A.A., B.D., D.A.P., G.S., and D.S.F.. Supervision: D.A.P, G.S. and D.S.F..
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 2.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The Repertoire and Dynamics of Evolutionary Adaptations to Controlled Nutrient-Limited Environments in Yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong J, Gresham D. Molecular Specificity, Convergence and Constraint Shape Adaptive Evolution in Nutrient-Poor Environments. PLOS Genet. 2014;10:e1004041. doi: 10.1371/journal.pgen.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kvitek DJ, Sherlock G. Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment. PLOS Genet. 2013;9:e1003972. doi: 10.1371/journal.pgen.1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy SF, Blundell JR, Venkataram S, Petrov DA, Fisher DS, Sherlock G. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature. 2015;519:181–186. doi: 10.1038/nature14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE. Repeatability and Contingency in the Evolution of a Key Innovation in Phage Lambda. Science. 2012;335:428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller CR, Nagel AC, Scott L, Settles M, Joyce P, Wichman HA. Love the one you’re with: replicate viral adaptations converge on the same phenotypic change. PeerJ. 2016;4:e2227. doi: 10.7717/peerj.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oz T, Guvenek A, Yildiz S, Karaboga E, Tamer YT, Mumcuyan N, Ozan VB, Senturk GH, Cokol M, Yeh P, et al. Strength of Selection Pressure Is an Important Parameter Contributing to the Complexity of Antibiotic Resistance Evolution. Mol Biol Evol. 2014;31:2387–2401. doi: 10.1093/molbev/msu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payen C, Rienzi SCD, Ong GT, Pogachar JL, Sanchez JC, Sunshine AB, Raghuraman MK, Brewer BJ, Dunham MJ. The Dynamics of Diverse Segmental Amplifications in Populations of Saccharomyces cerevisiae Adapting to Strong Selection. G3 Genes Genomes Genet. 2014;4:399–409. doi: 10.1534/g3.113.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toprak E, Veres A, Michel JB, Chait R, Hartl DL, Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkataram S, Dunn B, Li Y, Agarwala A, Chang J, Ebel ER, Geiler-Samerotte K, Hérissant L, Blundell JR, Levy SF, et al. Development of a Comprehensive Genotype-to-Fitness Map of Adaptation-Driving Mutations in Yeast. Cell. 2016;166:1585–1596. e22. doi: 10.1016/j.cell.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voordeckers K, Kominek J, Das A, Espinosa-Cantú A, Maeyer DD, Arslan A, Pee MV, van der Zande E, Meert W, Yang Y, et al. Adaptation to High Ethanol Reveals Complex Evolutionary Pathways. PLOS Genet. 2015;11:e1005635. doi: 10.1371/journal.pgen.1005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, Dahan O. Chromosomal duplication is a transient evolutionary solution to stress. Proc Natl Acad Sci. 2012;109:21010–21015. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long A, Liti G, Luptak A, Tenaillon O. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat Rev Genet. 2015;16:567–582. doi: 10.1038/nrg3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitti JJ, Grossman SR, Sabeti PC. Detecting Natural Selection in Genomic Data. Annu Rev Genet. 2013;47:97–120. doi: 10.1146/annurev-genet-111212-133526. [DOI] [PubMed] [Google Scholar]
- 17.Gerstein AC, Otto SP. Cryptic Fitness Advantage: Diploids Invade Haploid Populations Despite Lacking Any Apparent Advantage as Measured by Standard Fitness Assays. PLOS ONE. 2011;6:e26599. doi: 10.1371/journal.pone.0026599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasi F, Travisano M, Lenski RE. Long-Term Experimental Evolution in Escherichia coli. II Changes in Life-History Traits During Adaptation to a Seasonal. Environment Am Nat. 1994;144:432–456. [Google Scholar]
- 19.Plourde-Owobi L, Durner S, Goma G, François J. Trehalose reserve in Saccharomyces cerevisiae: phenomenon of transport, accumulation and role in cell viability. Int J Food Microbiol. 2000;55:33–40. doi: 10.1016/s0168-1605(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 20.Shi L, Sutter BM, Ye X, Tu BP. Trehalose Is a Key Determinant of the Quiescent Metabolic State That Fuels Cell Cycle Progression upon Return to Growth. Mol Biol Cell. 2010;21:1982–1990. doi: 10.1091/mbc.E10-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silljé HHW, Paalman JWG, ter Schure EG, Olsthoorn SQB, Verkleij AJ, Boonstra J, Verrips CT. Function of Trehalose and Glycogen in Cell Cycle Progression and Cell Viability in Saccharomyces cerevisiae. J Bacteriol. 1999;181:396–400. doi: 10.1128/jb.181.2.396-400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landry CR, Townsend JP, Hartl DL, Cavalieri D. Ecological and evolutionary genomics of Saccharomyces cerevisiae. Mol Ecol. 2006;15:575–591. doi: 10.1111/j.1365-294X.2006.02778.x. [DOI] [PubMed] [Google Scholar]
- 23.François J, Neves MJ, Hers HG. The control of trehalose biosynthesis in Saccharomyces cerevisiae: evidence for a catabolite inactivation and repression of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase. Yeast Chichester Engl. 1991;7:575–587. doi: 10.1002/yea.320070605. [DOI] [PubMed] [Google Scholar]
- 24.Lillie SH, Pringle JR. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cannon JF, Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1987;7:2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cebollero E, Reggiori F. Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim Biophys Acta BBA - Mol Cell Res. 2009;1793:1413–1421. doi: 10.1016/j.bbamcr.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Chang YW, Howard SC, Budovskaya YV, Rine J, Herman PK. The rye mutants identify a role for Ssn/Srb proteins of the RNA polymerase II holoenzyme during stationary phase entry in Saccharomyces cerevisiae. Genetics. 2001;157:17–26. doi: 10.1093/genetics/157.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breviario D, Hinnebusch A, Cannon J, Tatchell K, Dhar R. Carbon Source Regulation of RAS1 Expression in Saccharomyces cerevisiae and the Phenotypes of ras2 - Cells. Proc Natl Acad Sci U S A. 1986;83:4152–4156. doi: 10.1073/pnas.83.12.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dejean L, Beauvoit B, Alonso AP, Bunoust O, Guérin B, Rigoulet M. cAMP-induced modulation of the growth yield of Saccharomyces cerevisiae during respiratory and respiro-fermentative metabolism. Biochim Biophys Acta BBA - Bioenerg. 2002;1554:159–169. doi: 10.1016/s0005-2728(02)00240-2. [DOI] [PubMed] [Google Scholar]
- 30.Leeuwen J, van Pons C, Mellor JC, Yamaguchi TN, Friesen H, Koschwanez J, Ušaj MM, Pechlaner M, Takar M, Ušaj M, et al. Exploring genetic suppression interactions on a global scale. Science. 2016;354:aag0839. doi: 10.1126/science.aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F. Hunger Artists: Yeast Adapted to Carbon Limitation Show Trade-Offs under Carbon Sufficiency. PLOS Genet. 2011;7:e1002202. doi: 10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parts L, Cubillos FA, Warringer J, Jain K, Salinas F, Bumpstead SJ, Molin M, Zia A, Simpson JT, Quail MA, et al. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 2011;21:1131–1138. doi: 10.1101/gr.116731.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown JA, Sherlock G, Myers CL, Burrows NM, Deng C, Wu HI, McCann KE, Troyanskaya OG, Brown JM. Global analysis of gene function in yeast by quantitative phenotypic profiling. Mol Syst Biol. 2006;2:20060001. doi: 10.1038/msb4100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol. 2005;1:20050001. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 36.Deutschbauer A, Price MN, Wetmore KM, Tarjan DR, Xu Z, Shao W, Leon D, Arkin AP, Skerker JM. Towards an Informative Mutant Phenotype for Every Bacterial Gene. J Bacteriol. 2014;196:3643–3655. doi: 10.1128/JB.01836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The Chemical Genomic Portrait of Yeast: Uncovering a Phenotype for All Genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiser MJ, Ribeck N, Lenski RE. Long-Term Dynamics of Adaptation in Asexual Populations. Science. 2013;342:1364–1367. doi: 10.1126/science.1243357. [DOI] [PubMed] [Google Scholar]
- 39.Poltak SR, Cooper VS. Ecological succession in long-term experimentally evolved biofilms produces synergistic communities. ISME J. 2011;5:369–378. doi: 10.1038/ismej.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bono LM, Smith LB, Pfennig DW, Burch CL. The emergence of performance trade-offs during local adaptation: insights from experimental evolution. Mol Ecol. 2017;26:1720–1733. doi: 10.1111/mec.13979. [DOI] [PubMed] [Google Scholar]
- 41.Schick A, Bailey SF, Kassen R. Evolution of Fitness Trade-Offs in Locally Adapted Populations of Pseudomonas fluorescens. Am Nat. 2015;186:S48–S59. doi: 10.1086/682932. [DOI] [PubMed] [Google Scholar]
- 42.Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science. 2014;344:1519–1522. doi: 10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11.10.1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Growth characteristics and metabolite measurements of twelve evolved strains in monoculture and frequency measurements in high-resolution pairwise competition assays; Related to Figures 1 and 2
Data S2: Measured properties of the 12 assayed adaptive clones; Related to Figures 1 and 2
This table includes fitness components from monoculture, pairwise competition assay and barcode measurements, estimated viability loss rate per hour, trehalose per unit of cell volume at 48h and the change rate of other metabolites measured in Data S1.
Data S3: Measured properties of the 3,048 clones in the pool; Related to Figures 3, 4, 5 and 7
The table includes each clone’s barcode ID, clone ID, fitness measurements in batch 1 from [S1], fitness measurements and error under all tested conditions, as well as ploidy, class (neutral haploids, pure diploids, adaptive haploids and high fitness diploids), global EC fitness and their fermentation-dependent and respiration-dependent fitness components, and stationary-dependent rate (per hour) estimated from the barcode fitness measurements. This table also contains the cluster information of adaptive haploids and high fitness diploids in Figure S6, their genotypes if they carry mutations in the nutrient response pathways (Clones that carry mutations only in non-nutrient response pathway genes are indicated as “Other” in the genotype column), mutation type and in which batch they are sequenced. Note that SCH9+Diploid clone tested in monoculture and pairwise competition were removed from analysis with barcode fitness measurements because it has high fitness estimation error and thus is not included in the 3,048 clones. In order to facilitate comparison, the measurements and estimates of SCH9+Diploid clone are added back, resulting in 3,049 clones in this table.
Data S4: Detailed genotypes of sequenced clones; Related to Figure 7
Data S5: Primers used to amplify the barcode region; Related to the Key Resources Table