Abstract
Background
The MGMT is a key tumor suppressor gene and aberrant promoter methylation has been reported in many cancers. However, the relationship between MGMT promoter methylation and ovarian cancer remains controversial. This meta‐analysis was first conducted to estimate the clinical significance of MGMT promoter methylation in ovarian carcinoma.
Methods
Literature search was performed in the PubMed, Embase, EBSCO and Cochrane Library databases. The pooled odds ratio (OR) and their corresponding 95% confidence interval (95% CI) were summarized.
Results
Final 10 studies with 910 ovarian tissue samples were included in this meta‐analysis. MGMT promoter methylation was significantly higher in ovarian cancer than in normal ovarian tissues (OR = 4.13, 95% CI = 2.32–7.33, p < .001). The MGMT had a similar methylation status in cancer versus benign lesions and low malignant potential (LMP) samples (OR = 2.01, 95% CI = 0.67–6.04, p = .212; OR = 1.42, 95% CI = 0.46–4.40, p = .543; respectively). MGMT promoter methylation was correlated with pathological types in which it was significantly lower in serous cancer than in nonserous cancer (OR = 0.29, 95% CI = 0.14–0.59, p = .001). The methylation of the MGMT promoter was not associated with clinical stage and tumor grade (OR = 1.46, 95% CI = 0.71–3.02, p = .301; OR = 1.13, 95% CI = 0.51–2.46, p = .767; respectively).
Conclusions
MGMT promoter methylation may be correlated with the tumorigenesis of ovarian cancer. It was associated with tumor histotypes, but not correlated with clinical stage and tumor grade. More prospective studies with lager sample sizes are necessary in the future.
Keywords: clinical features, methylation, MGMT, ovarian cancer
1. INTRODUCTION
Ovarian cancer is the second most frequently and the most deadly gynecological malignancy among women (Siegel, Miller, & Jemal, 2016). According to global statistics, approximately 238,700 new cases were clinically diagnosed with ovarian carcinoma, and it killed 151,900 cases in the world in 2008 (Torre et al., 2015). Due to difficulties of early detection, most patients with ovarian cancer are diagnosed with high stages of this disease. Five‐year survival rate at advanced‐stage ovarian carcinoma remains <20% (Heintz et al., 2006; Kaja et al., 2012). Serous histology is the most common ovarian cancer, and other histotypes include mucinous, endometrioid, clear cell tumors, and undifferentiated carcinomas, etc. (Vang, Shih Ie, & Kurman, 2009).
Epigenetic modifications, especially DNA methylation, a commonly observed epigenetic change, have been noted in the initiation and progression of human cancer (Ghavifekr Fakhr, Farshdousti Hagh, Shanehbandi, & Baradaran, 2013; Huang et al., 2015; Ma, Wang, Zhang, & Gazdar, 2013). Aberrant promoter methylation of tumor suppressor genes (TSGs) is the most common methylation in many types of cancers (Smith et al., 2016; Yokoyama et al., 2016). O6‐methylguanine‐DNAmethyl‐transferase (MGMT), located on 10q26, a DNA repair gene, protects cells against the effects of alkylating agent chemotherapy by eliminating the alkylation of the O6 position of guanine (Esteller et al., 2000; Tano, Shiota, Collier, Foote, & Mitra, 1990). MGMT is widely methylated in the promoter region in various carcinomas, including ovarian cancer (Chaudhry, Srinivasan, & Patel, 2009; Hochhauser et al., 2013; Pierini et al., 2014; Schiffgens et al., 2016). Nevertheless, data with regard to the methylation levels of the MGMT promoter were inconsistent in ovarian cancer. No methylation rate of the MGMT promoter was detected in patients with ovarian carcinoma by Agostini et al. (Agostini et al., 2016). However, An et al. reported that MGMT promoter methylation had a high rate in ovarian cancer (An et al., 2010).
On the basis of numerous studies with small sample size and the different methylation frequencies, we carried out this meta‐analysis to better determine the relationship between the methylation of the MGMT promoter and ovarian cancer in the comparison of cancer and different control groups. Additionally, we also analyzed whether MGMT promoter methylation was associated with clinicopathological characteristics.
2. MATERIALS AND METHODS
2.1. Literature search
The PubMed, Embase, EBSCO, and Cochrane Library databases were systemically searched to identify the studies of the eligibility using the following key words and search terms: (O‐6‐methylguanine‐DNA methyltransferase OR MGMT) AND (ovarian OR ovary) AND (cancer OR carcinoma OR tumor OR neoplasm) AND (methylation OR DNA methylation OR hypermethylation OR epigenetic silencing OR epigenetic inactivation). The search strategy was updated prior November 1, 2016. Manual search from reference lists of eligible studies was also conducted to achieve other potential articles.
2.2. Inclusion criteria
The following selection criteria were used for eligible studies: (i) the patients had a diagnosis with ovarian carcinoma using histopathological examination; (ii) studies must have sufficient information with regard to the methylation data to evaluate the correlation between MGMT promoter methylation and ovarian cancer in cancer versus control groups; (iii) studies provided data about the relationships between MGMT promoter methylation and clinicopathological features of patients with ovarian cancer; (iv) if authors published more than one paper using the same sample data, only paper with more information was included in the current meta‐analysis.
2.3. Data extraction
According to the selection criteria, we extracted necessary data from original articles: first author's surname, year of publication, country, ethnicity, type of sample, sample size, detection methodology of methylation, rate of methylation, total number of cases and controls, and clinicopathological parameters such as tumor grade, tumor stage, and pathological types. Control groups included low malignant potential (LMP) tumors, benign lesions and normal samples.
2.4. Statistical analysis
The pooled data were analyzed using the Stata statistical software (version 12.0; Stata Corporation, College Station, TX, USA). The correlation between MGMT promoter methylation and ovarian cancer in cancer versus control groups was calculated by the combined odds ratio (OR) and corresponding 95% confidence interval (95% CI).
In addition, we also assessed the association of MGMT promoter methylation with clinicopathological characteristics in patients with ovarian cancer. The degree of heterogeneity was detected using the Cochran's Q test and I 2 statistic (Coory, 2010). The random‐effects model was used when substantial heterogeneity existed (I 2 ≥ 50% and p < .1), whereas a fixed‐effects model was applied when there was no obvious evidence of heterogeneity (DerSimonian, 1996; Higgins, Thompson, Deeks, & Altman, 2003).
3. RESULTS
3.1. Baseline characteristics of studies
Our initial search yielded a total of 81 potential studies based on the above described search strategy. In the end, 10 case–control studies involving 910 ovarian tissue samples (Agostini et al., 2016; An et al., 2010; Brait et al., 2013; Chmelarova et al., 2012; Furlan et al., 2006; Makarla et al., 2005; Rimel, Huettner, Powell, Mutch, & Goodfellow, 2009; Roh et al., 2011; Shilpa et al., 2014; Wu et al., 2007) were included in the current meta‐analysis (Figure 1). The main characteristics of eligible studies were extracted and shown in Table 1.
Figure 1.
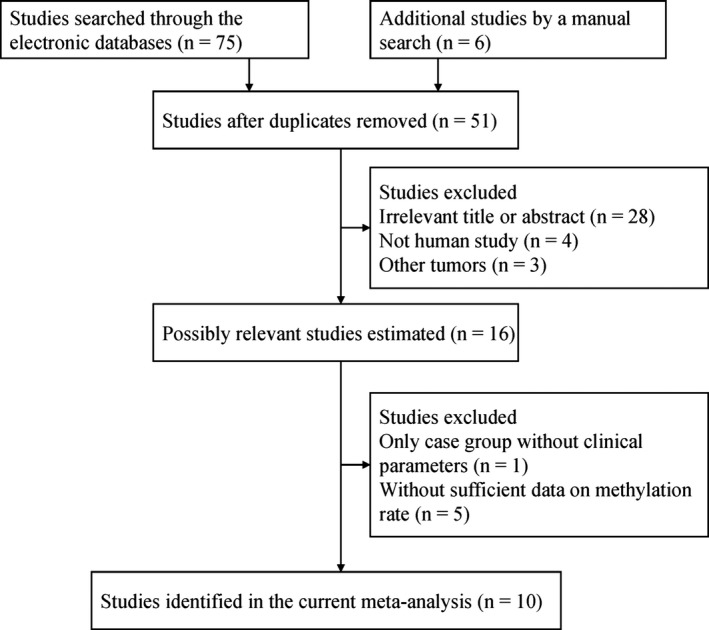
Flowchart of selection of studies
Table 1.
Baseline characteristics of the included studies in this meta‐analysis
| First author | References | Country | Ethnicity | Method | Sample | Case | LMP | Benign | Normal | G3 | G1–2 | Stage 3–4 | Stage 1–2 | Serous | Nonserous |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N (M %) | N (M %) | N (M %) | N (M %) | M/N | M/N | M/N | M/N | M/N | M/N | ||||||
| Makarla | Makarla et al. (2005) | USA | Caucasians | MSP | Tissue | 23 (8.7) | 23 (0) | 23 (0) | 16 (0) | 0/9 | 2/14 | ||||
| Furlan | Furlan et al. (2006) | Italy | Caucasians | MSP | Tissue | 18 (38.9) | 13 (0) | ||||||||
| Wu | Wu et al. (2007) | Norway | Caucasians | MSP | Tissue | 52 (0) | 2 (0) | ||||||||
| Rimel | Rimel et al. (2009) | USA | Caucasians | COBRA | Tissue | 21 (4.8) | 21 (0) | ||||||||
| An | An et al. (2010) | USA | Mix | MSP | Tissue | 101 (32.7) | 100 (14) | 30/94 | 3/7 | ||||||
| Roh | Roh et al. (2011) | Kore | Asians | MSP | Tissue | 34 (14.7) | 10 (0) | 0/4 | 5/30 | 3/17 | 2/17 | 0/16 | 5/18 | ||
| Chmelařová | Chmelarova et al. (2012) | Czech Republic | Caucasians | MS‐MLPA | Tissue | 69 (15.9) | 40 (2.5) | 6/38 | 5/29 | 9/46 | 2/23 | 5/48 | 6/21 | ||
| Brait | Brait et al. (2013) | Brazil | Mix | QMSP | Tissue | 33 (3) | 13 (0) | ||||||||
| Shilpa | Shilpa et al. (2014) | India | Caucasians | MSP | Tissue | 88 (29.5) | 14 (28.6) | 20 (20) | 15 (0) | 18/57 | 6/25 | 21/66 | 5/22 | 11/52 | 15/36 |
| Agostini | Agostini et al. (2016) | Norway | Caucasians | QMSP | Tissue | 126 (0) | 35 (0) | 0/60 | 0/66 |
3.2. Relationship between MGMT promoter methylation and ovarian cancer
Of the 10 available articles, eight studies included 387 ovarian cancer patients and 228 normal ovarian tissue samples, four studies included 289 patients with ovarian cancer and 80 benign lesions, and two studies consisted of 111 ovarian cancer patients and 37 LMP samples. No substantial heterogeneity was found in cancer versus normal ovarian tissues, benign lesions and LMP samples (all I 2 = 0%) (Figure 2). Thus, the fixed‐effects model was applied in this study.
Figure 2.
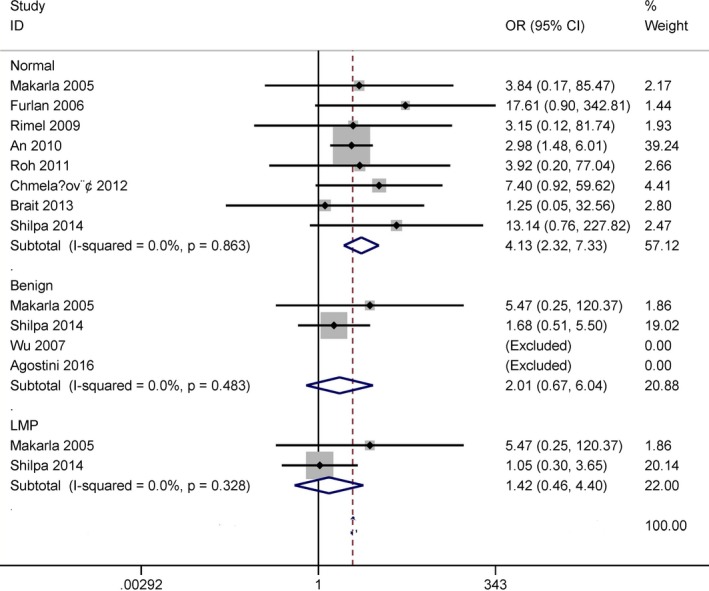
Forest plot for the association between MGMT promoter methylation and ovarian cancer in cancer versus LMP, benign lesions and normal ovarian tissues
The result demonstrated that MGMT promoter methylation was notably correlated with an increased risk of ovarian cancer in the comparison of ovarian cancer patients and normal ovarian tissues (OR = 4.13, 95% CI = 2.32–7.33, p < .001). When cancer was compared with benign lesions and LMP specimens, no significant relationship was found in cancer versus benign lesions and LMP samples (OR = 2.01, 95% CI = 0.67–6.04, p = .212; OR = 1.42, 95% CI = 0.46–4.40, p = .543).
3.3. Subgroup analyses of MGMT promoter methylation in cancer versus normal ovarian tissues
Ethnic population consisted of Caucasians, mixed population and Asians. Testing method included the methylation‐specific polymerase chain reaction (MSP) and non‐MSP.
Subgroup analysis based on ethnicity demonstrated that the OR value was 8.44 (95% CI = 2.51–28.39, p = .001) in Caucasians among five studies, 2.87 (95% CI = 1.44–5.69, p = .003) in mixed population among two studies, and 3.92 (95% CI = 0.20–77.04, p = .369) in Asians among one study (Figure 3).
Figure 3.
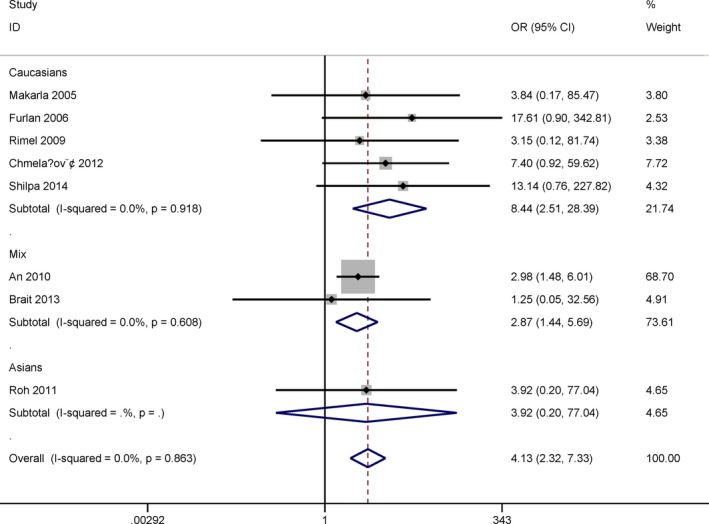
Subgroup analysis by ethnicity for the correlation between MGMT promoter methylation and ovarian cancer in cancer versus normal ovarian tissues
Subgroup analysis by detection method of methylation indicated that the OR of the MSP subgroup was 4.03 (95% CI = 2.17–7.51, p < .001) among five studies, and 4.61 (95% CI = 1.02–20.86, p = .047) in the non‐MSP subgroup among three studies (Figure 4).
Figure 4.
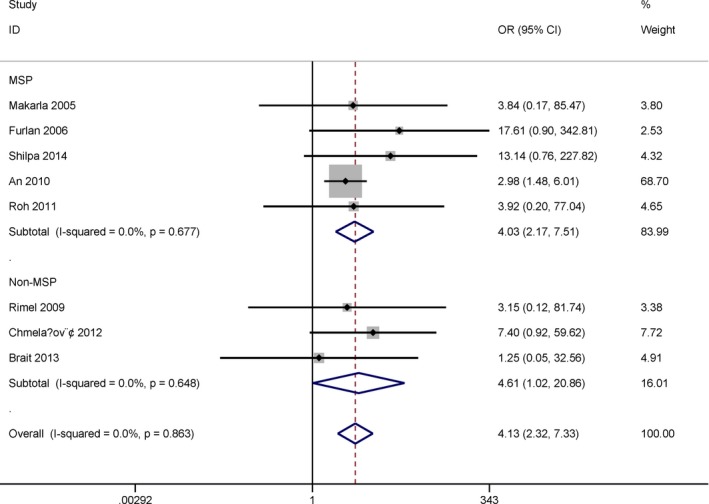
Subgroup analysis by detection method for the relationship between MGMT promoter methylation and ovarian cancer in cancer versus normal ovarian tissues
3.4. Relationship between MGMT promoter methylation and clinicopathological features
No evidence of heterogeneity was detected in relation to pathological types, tumor stage, and grade under the fixed‐effects model (all I 2 = 0%) (Figure 5).
Figure 5.
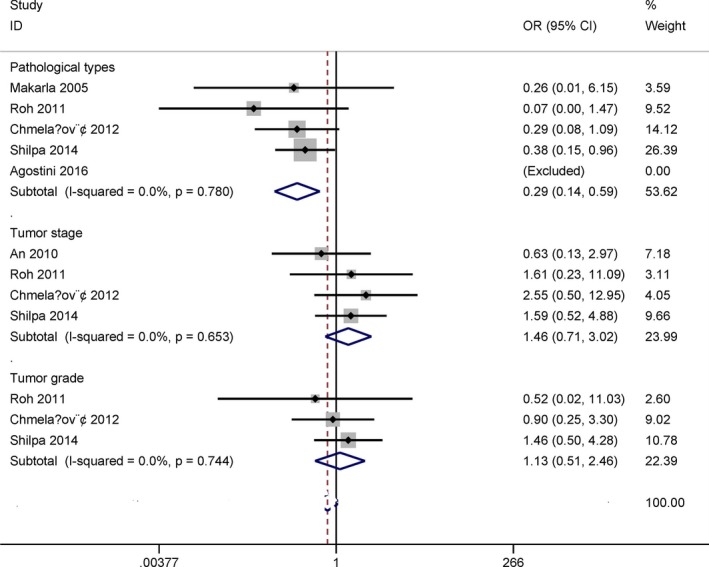
Forest plot for the association between MGMT promoter methylation and clinicopathological features
The pooled OR indicated that MGMT promoter methylation was significantly linked to pathological types in ovarian cancer (OR = 0.29, 95% CI = 0.14–0.59, p = .001), including five studies with 185 serous cancer and 155 nonserous cancer patients.
The pooled OR showed that the methylation of the MGMT promoter was not correlated with clinical stage (OR = 1.46, 95% CI = 0.71–3.02, p = .301), including 223 patients with stage 3–4 and 69 patients with stage 1–2 ovarian carcinoma among four studies.
The pooled OR from three studies involving 99 patients with grade 3 and 84 patients with grade 1–2 ovarian carcinoma indicated that MGMT promoter methylation was not associated with tumor grade (OR = 1.13, 95% CI = 0.51–2.46, p = .767).
4. DISCUSSION
The methylation of DNA of promoter region is a common mechanism for the inactivation of tumor suppressor genes in cancer, which leads to the loss or reduction in gene expression (Herman & Baylin, 2000, 2003). The downregulation of MGMT expression through promoter methylation has been shown in various human cancers, including lung cancer, glioblastoma, gastric carcinoma, and salivary gland carcinoma (Cabrini, Fabbri, Lo Nigro, Dechecchi, & Gambari, 2015; Lee et al., 2008; Pulling et al., 2003; Yousuf et al., 2014). The association between promoter methylation of the MGMT gene and its expression was found in ovarian cancer, with negative MGMT expression (Roh et al., 2011). The methylation frequency of MGMT promoter was found to be different in this study. The methylation rate of MGMT promoter ranged from 0% (Agostini et al., 2016) to 38.9% (Furlan et al., 2006) in ovarian carcinoma. MGMT promoter methylation had a different methylation frequency in LMP samples, with a variation in 0% (Makarla et al., 2005) to 28.6% (Shilpa et al., 2014). The rate of MGMT promoter methylation was different in benign ovarian tissue samples (Agostini et al., 2016; Shilpa et al., 2014). Moreover, An et al. reported that MGMT promoter had a methylation frequency of 14% in normal ovarian tissue specimens (An et al., 2010). Some studies reported that MGMT promoter was unmethylated in normal ovarian tissue specimens (Furlan et al., 2006; Makarla et al., 2005; Rimel et al., 2009). Thus, we performed this study using eligible publications to further identify the relationship between the methylation of MGMT promoter and ovarian carcinoma.
No evidence of heterogeneity existed in ovarian cancer versus LMP, benign lesions and normal ovarian tissues (all I 2 = 0%), suggesting the stability of our results. The result showed that ovarian cancer had a significantly higher methylation status of MGMT promoter than normal ovarian tissues, which suggested that MGMT promoter methylation may be an early event of the carcinogenesis of ovarian cancer. MGMT promoter methylation had s similar frequency in ovarian carcinoma versus LMP and benign lesions. However, the result should be cautious based on fewer studies with small subjects when cancer was compared to LMP and benign lesions.
When cancer was compared to normal ovarian tissue samples, subgroup analyses were conducted to find the correlation in the different subgroups. Subgroup analysis of detection method demonstrated that MGMT promoter methylation was correlated with ovarian cancer in the MSP and non‐MSP subgroups. Subgroup analysis of ethnic population showed that the methylation status of MGMT promoter was associated with ovarian carcinoma in the Caucasian and mixed populations, but not in Asian population, suggesting that the MGMT may be a susceptible gene for Caucasians and mixed population. While the result regard the subgroup analyses should be carefully considered as only small sample size were included, particularly in mixed population and Asians subgroups.
We also determined whether MGMT promoter methylation had a correlation in relation to clinicopathological characteristics, including pathological types, clinical stage, and tumor grade. Our findings indicated that MGMT promoter methylation was significantly correlated with pathological types, and it was significantly lower in serous carcinoma than in nonserous carcinoma (OR = 0.29, p = .001), which suggested that MGMT promoter methylation had a decreased the risk of serous ovarian cancer. No significant association was found in relation to clinical stage and tumor grade.
Several potential limitations should be stated in this study. First, the search strategy was limited to available papers published in English language. Other publications published in other languages were eliminated because of the difficulties in reading. Second, based on small sample size, the results with regard to subgroup analyses should be further done in the future. Third, when cancer was compared to LMP and benign lesions, multiple studies with larger sample size should be performed to clarify our results.
In summary, our findings suggested that MGMT promoter methylation may play a crucial role in the initiation of ovarian cancer. MGMT promoter methylation was linked to pathological types, but not associated with clinical stage and tumor grade. In the future, additional prospective studies remain needed.
CONFLICT OF INTEREST
None declared.
Qiao B, Zhang Z, Li Y. Association of MGMT promoter methylation with tumorigenesis features in patients with ovarian cancer: A systematic meta‐analysis. Mol Genet Genomic Med. 2018;6:69–76. https://doi.org/10.1002/mgg3.349
Contributor Information
Zhenyu Zhang, Email: zhangzhenyu@coga.org.cn.
Yanfang Li, Email: liyanfang8523@126.com.
REFERENCES
- Agostini, A. , Panagopoulos, I. , Davidson, B. , Trope, C. G. , Heim, S. , & Micci, F. (2016). A novel truncated form of hmga2 in tumors of the ovaries. Oncology Letters, 12, 1559–1563. https://doi.org/10.3892/ol.2016.4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, J. , Wei, Q. , Liu, Z. , Lu, K. H. , Cheng, X. , Mills, G. B. , & Wang, L. E. (2010). Messenger rna expression and methylation of candidate tumor‐suppressor genes and risk of ovarian cancer‐a case–control analysis. International Journal of Molecular Epidemiology and Genetics, 1, 1–10. [PMC free article] [PubMed] [Google Scholar]
- Brait, M. , Maldonado, L. , Noordhuis, M. G. , Begum, S. , Loyo, M. , Poeta, M. L. , … Hoque, M. O. (2013). Association of promoter methylation of vgf and pgp9.5 with ovarian cancer progression. PLoS ONE, 8, e70878 https://doi.org/10.1371/journal.pone.0070878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrini, G. , Fabbri, E. , Lo Nigro, C. , Dechecchi, M. C. , & Gambari, R. (2015). Regulation of expression of o6‐methylguanine‐DNA methyltransferase and the treatment of glioblastoma (review). International Journal of Oncology, 47, 417–428. https://doi.org/10.3892/ijo.2015.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry, P. , Srinivasan, R. , & Patel, F. D. (2009). Utility of gene promoter methylation in prediction of response to platinum‐based chemotherapy in epithelial ovarian cancer (eoc). Cancer Investigation, 27, 877–884. https://doi.org/10.1080/07357900902849699 [DOI] [PubMed] [Google Scholar]
- Chmelarova, M. , Krepinska, E. , Spacek, J. , Laco, J. , Nekvindova, J. , & Palicka, V. (2012). Methylation analysis of tumour suppressor genes in ovarian cancer using ms‐mlpa. Folia Biologica, 58, 246–250. [PubMed] [Google Scholar]
- Coory, M. D. (2010). Comment on: Heterogeneity in meta‐analysis should be expected and appropriately quantified. International Journal of Epidemiology, 39, 932; author reply 933. https://doi.org/10.1093/ije/dyp157 [DOI] [PubMed] [Google Scholar]
- DerSimonian, R. (1996). Meta‐analysis in the design and monitoring of clinical trials. Statistics in Medicine, 15, 1237–1248; discussion 1249–1252. https://doi.org/10.1002/(ISSN)1097-0258 [DOI] [PubMed] [Google Scholar]
- Esteller, M. , Garcia‐Foncillas, J. , Andion, E. , Goodman, S. N. , Hidalgo, O. F. , Vanaclocha, V. , … Herman, J. G. (2000). Inactivation of the DNA‐repair gene mgmt and the clinical response of gliomas to alkylating agents. The New England Journal of Medicine, 343, 1350–1354. https://doi.org/10.1056/NEJM200011093431901 [DOI] [PubMed] [Google Scholar]
- Furlan, D. , Carnevali, I. , Marcomini, B. , Cerutti, R. , Dainese, E. , Capella, C. , & Riva, C. (2006). The high frequency of de novo promoter methylation in synchronous primary endometrial and ovarian carcinomas. Clinical Cancer Research, 12, 3329–3336. https://doi.org/10.1158/1078-0432.CCR-05-2679 [DOI] [PubMed] [Google Scholar]
- Ghavifekr Fakhr, M. , Farshdousti Hagh, M. , Shanehbandi, D. , & Baradaran, B. (2013). DNA methylation pattern as important epigenetic criterion in cancer. Genetics Research International, 2013, 317569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz, A. P. , Odicino, F. , Maisonneuve, P. , Quinn, M. A. , Benedet, J. L. , Creasman, W. T. , … Beller, U. (2006). Carcinoma of the ovary. Figo 26th annual report on the results of treatment in gynecological cancer. International Journal of Gynaecology and Obstetrics, 95(Suppl. 1), S161–S192. https://doi.org/10.1016/S0020-7292(06)60033-7 [DOI] [PubMed] [Google Scholar]
- Herman, J. G. , & Baylin, S. B. (2000). Promoter‐region hypermethylation and gene silencing in human cancer. Current Topics in Microbiology and Immunology, 249, 35–54. [DOI] [PubMed] [Google Scholar]
- Herman, J. G. , & Baylin, S. B. (2003). Gene silencing in cancer in association with promoter hypermethylation. The New England Journal of Medicine, 349, 2042–2054. https://doi.org/10.1056/NEJMra023075 [DOI] [PubMed] [Google Scholar]
- Higgins, J. P. , Thompson, S. G. , Deeks, J. J. , & Altman, D. G. (2003). Measuring inconsistency in meta‐analyses. BMJ, 327, 557–560. https://doi.org/10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhauser, D. , Glynne‐Jones, R. , Potter, V. , Gravalos, C. , Doyle, T. J. , Pathiraja, K. , … Sausville, E. A. (2013). A phase ii study of temozolomide in patients with advanced aerodigestive tract and colorectal cancers and methylation of the o6‐methylguanine‐DNA methyltransferase promoter. Molecular Cancer Therapeutics, 12, 809–818. https://doi.org/10.1158/1535-7163.MCT-12-0710 [DOI] [PubMed] [Google Scholar]
- Huang, T. , Chen, X. , Hong, Q. , Deng, Z. , Ma, H. , Xin, Y. , … Duan, S. (2015). Meta‐analyses of gene methylation and smoking behavior in non‐small cell lung cancer patients. Scientific Reports, 5, 8897 https://doi.org/10.1038/srep08897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaja, S. , Hilgenberg, J. D. , Collins, J. L. , Shah, A. A. , Wawro, D. , Zimmerman, S. , … Koulen, P. (2012). Detection of novel biomarkers for ovarian cancer with an optical nanotechnology detection system enabling label‐free diagnostics. Journal of Biomedical Optics, 17, 081412–081411. https://doi.org/10.1117/1.JBO.17.8.081412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. S. , Issa, J. P. , Roberts, D. B. , Williams, M. D. , Weber, R. S. , Kies, M. S. , & El‐Naggar, A. K. (2008). Quantitative promoter hypermethylation analysis of cancer‐related genes in salivary gland carcinomas: Comparison with methylation‐specific pcr technique and clinical significance. Clinical Cancer Research, 14, 2664–2672. https://doi.org/10.1158/1078-0432.CCR-07-1232 [DOI] [PubMed] [Google Scholar]
- Ma, X. , Wang, Y. W. , Zhang, M. Q. , & Gazdar, A. F. (2013). DNA methylation data analysis and its application to cancer research. Epigenomics, 5, 301–316. https://doi.org/10.2217/epi.13.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarla, P. B. , Saboorian, M. H. , Ashfaq, R. , Toyooka, K. O. , Toyooka, S. , Minna, J. D. , … Schorge, J. O. (2005). Promoter hypermethylation profile of ovarian epithelial neoplasms. Clinical Cancer Research, 11, 5365–5369. https://doi.org/10.1158/1078-0432.CCR-04-2455 [DOI] [PubMed] [Google Scholar]
- Pierini, S. , Jordanov, S. H. , Mitkova, A. V. , Chalakov, I. J. , Melnicharov, M. B. , Kunev, K. V. , … Goranova, T. E. (2014). Promoter hypermethylation of cdkn2a, mgmt, mlh1, and dapk genes in laryngeal squamous cell carcinoma and their associations with clinical profiles of the patients. Head and Neck, 36, 1103–1108. https://doi.org/10.1002/pro.v36.8 [DOI] [PubMed] [Google Scholar]
- Pulling, L. C. , Divine, K. K. , Klinge, D. M. , Gilliland, F. D. , Kang, T. , Schwartz, A. G. , … Belinsky, S. A. (2003). Promoter hypermethylation of the o6‐methylguanine‐DNA methyltransferase gene: More common in lung adenocarcinomas from never‐smokers than smokers and associated with tumor progression. Cancer Research, 63, 4842–4848. [PubMed] [Google Scholar]
- Rimel, B. J. , Huettner, P. , Powell, M. A. , Mutch, D. G. , & Goodfellow, P. J. (2009). Absence of mgmt promoter methylation in endometrial cancer. Gynecologic Oncology, 112, 224–228. https://doi.org/10.1016/j.ygyno.2008.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh, H. J. , Suh, D. S. , Choi, K. U. , Yoo, H. J. , Joo, W. D. , & Yoon, M. S. (2011). Inactivation of o(6)‐methyguanine‐DNA methyltransferase by promoter hypermethylation: Association of epithelial ovarian carcinogenesis in specific histological types. The Journal of Obstetrics and Gynaecology Research, 37, 851–860. https://doi.org/10.1111/jog.2011.37.issue-7 [DOI] [PubMed] [Google Scholar]
- Schiffgens, S. , Wilkens, L. , Brandes, A. A. , Meier, T. , Franceschi, E. , Ermani, M. , … Dumitru, C. A. (2016). Sex‐specific clinicopathological significance of novel (frizzled‐7) and established (mgmt, idh1) biomarkers in glioblastoma. Oncotarget, 7, 55169–55180. https://doi.org/10.18632/oncotarget.10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilpa, V. , Bhagat, R. , Premalata, C. S. , Pallavi, V. R. , Ramesh, G. , & Krishnamoorthy, L. (2014). Relationship between promoter methylation & tissue expression of mgmt gene in ovarian cancer. The Indian Journal of Medical Research, 140, 616–623. [PMC free article] [PubMed] [Google Scholar]
- Siegel, R. L. , Miller, K. D. , & Jemal, A. (2016). Cancer statistics, 2016. A Cancer Journal for Clinicians, 66, 7–30. https://doi.org/10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- Smith, B. , Neff, R. , Cohn, D. E. , Backes, F. J. , Suarez, A. A. , Mutch, D. G. , … Goodfellow, P. J. (2016). The mutational spectrum of foxa2 in endometrioid endometrial cancer points to a tumor suppressor role. Gynecologic Oncology, 143, 398–405. https://doi.org/10.1016/j.ygyno.2016.08.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano, K. , Shiota, S. , Collier, J. , Foote, R. S. , & Mitra, S. (1990). Isolation and structural characterization of a cdna clone encoding the human DNA repair protein for o6‐alkylguanine. Proceedings of the National Academy of Sciences of the United States of America, 87, 686–690. https://doi.org/10.1073/pnas.87.2.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre, L. A. , Bray, F. , Siegel, R. L. , Ferlay, J. , Lortet‐Tieulent, J. , & Jemal, A. (2015). Global cancer statistics, 2012. A Cancer Journal for Clinicians, 65, 87–108. https://doi.org/10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- Vang, R. , Shih Ie, M. , & Kurman, R. J. (2009). Ovarian low‐grade and high‐grade serous carcinoma: Pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Advances in Anatomic Pathology, 16, 267–282. https://doi.org/10.1097/PAP.0b013e3181b4fffa [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q. , Lothe, R. A. , Ahlquist, T. , Silins, I. , Trope, C. G. , Micci, F. , … Lind, G. E. (2007). DNA methylation profiling of ovarian carcinomas and their in vitro models identifies hoxa9, hoxb5, scgb3a1, and crabp1 as novel targets. Molecular Cancer, 6, 45 https://doi.org/10.1186/1476-4598-6-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, S. , Higashi, M. , Kitamoto, S. , Oeldorf, M. , Knippschild, U. , Kornmann, M. , … Hollingsworth, M. A. (2016). Aberrant methylation of muc1 and muc4 promoters are potential prognostic biomarkers for pancreatic ductal adenocarcinomas. Oncotarget, 7, 42553–42565. https://doi.org/10.18632/oncotarget.v7i27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousuf, A. , Bhat, M. Y. , Pandith, A. A. , Afroze, D. , Khan, N. P. , Alam, K. , … Mudassar, S. (2014). Mgmt gene silencing by promoter hypermethylation in gastric cancer in a high incidence area. Cellular Oncology, 37, 245–252. https://doi.org/10.1007/s13402-014-0179-3 [DOI] [PubMed] [Google Scholar]


