Abstract
Purpose
As part of the Epilepsy Genetics Initiative, we re-evaluated clinically generated exome sequence data from 54 epilepsy patients and their unaffected parents to identify molecular diagnoses not provided in the initial diagnostic interpretation.
Methods
We compiled and analyzed exome sequence data from 54 genetically undiagnosed trios using a validated analysis pipeline. We evaluated the significance of the genetic findings by reanalyzing sequence data generated at Ambry Genetics, and from a number of additional case and control cohorts.
Results
In 54 previously undiagnosed trios, we identified two de novo missense variants in SCN8A in the highly expressed alternative exon 5 A—an exon only recently added to the Consensus Coding Sequence database. One additional undiagnosed epilepsy patient harboring a de novo variant in exon 5 A was found in the Ambry Genetics cohort. Missense variants in SCN8A exon 5 A are extremely rare in the population, further supporting the pathogenicity of the de novo alterations identified.
Conclusion
These results expand the range of SCN8A variants in epileptic encephalopathy patients and illustrate the necessity of ongoing reanalysis of negative exome sequences, as advances in the knowledge of disease genes and their annotations will permit new diagnoses to be made.
Keywords: alternative exon, epilepsy, epileptic encephalopathy, SCN8A, whole-exome sequencing
Introduction
The Epilepsy Genetics Initiative (EGI) is a signature program of Citizens United for Research in Epilepsy (http://www.cureepilepsy.org/egi/index.html). It was created to house and periodically reanalyze exome sequence data for patients with seizure disorders who have had clinical exome or genome sequencing performed as part of their medical care. The EGI reanalyzes data within the repository every 6 months with two broad goals: (i) to identify novel epilepsy genes through aggregate analyses of one of the largest sources of exome sequence data in patients with epilepsy and (ii) to reanalyze exome sequence data, using the most up-to-date knowledge, for missed genetic diagnoses in epilepsy patients who initially received an inconclusive result. Newly identified genetic diagnoses are returned to patients and families through their referring physician.
Through the work of the EGI, we report here the identification of three novel disease-causing variants in alternative exon 5 A of SCN8A in three unrelated patients with epilepsy. These diagnoses were missed by clinical exome sequencing because, at the time of analysis, exon 5 A was not recognized as protein coding in the consensus coding sequence database (CCDS; https://www.ncbi.nlm.nih.gov/projects/CCDS/).
The SCN8A gene encodes the sodium channel Nav1.6. Mutations cause SCN8A epileptic encephalopathy or early infantile epileptic encephalopathy type 13 (OMIM 614558), accounting for ~1% of epileptic encephalopathy cases.1 SCN8A encephalopathy is typically associated with seizures beginning in infancy, developmental delay and varying degrees of impaired speech and motor function. Most patients have multiple seizure types, and onset is generally not associated with fever or illness.
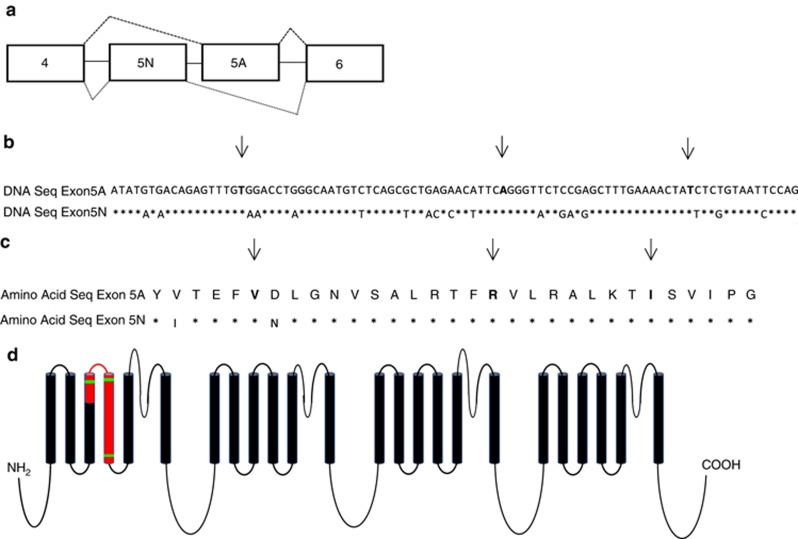
Nav1.6 consists of four homologous domains (DI–DIV), each of which contains six transmembrane segments (S1–S6).2 The SCN8A gene is comprised of 26 protein-coding exons.3 The majority of pathogenic variants in SCN8A are de novo missense variants or missense variants inherited from a mosaic parent.1 SCN8A contains two pairs of tandemly duplicated, mutually exclusive, alternatively spliced exons with a common evolutionary origin—exons 5 A/5 N and exons 18 A/18 N (Figure 1).4 Exons 5 and 18 encode portions of transmembrane segments S3 and S4 in domain I and domain III of Nav1.6, respectively. The 18 N transcript contains a conserved in-frame stop codon, which is predicted to result in protein truncation and is widely expressed at low levels in nonneuronal tissue.4 Exons 5 N (neonatal) and 5 A (adult) differ by two out of 31 amino acids (Figure 1).5 In humans and rodents, there is evidence that the expression of exon 5 A increases during development;5, 6 the expression of “neonatal” exon 5 N decreases over time, but it continues to be expressed at a low level in the adult brain. This is confirmed in exon-level data publically available in the BrainSpan database (http://www.brainspan.org/). Before this work, five disease-causing variants had been identified in exon 5 N of the SCN8A gene, and none had been described in exon 5A.7
Figure 1.
Exon 5 of SCN8A. (a) Exon 5 is encoded by two sequences in the genome (exons 5 N and 5 A). Only one of the two exons remains in the transcripts after splicing occurs. The nucleotide (b) and amino acid sequences (c) of exons 5 N and 5 A are nearly identical. Asterisks indicate the sites with the same sequence and arrows highlight the sites where a novel disease-causing SCN8A variant was identified in exon 5 A in this study. All disease-causing variants were found at sites where the sequence was identical between exons 5 A and 5 N. (d) Exon 5, highlighted in red, spans two transmembrane domains and a small extracellular region of Nav1.6. The three disease-causing variants, marked in green, were all located in the regions encoding transmembrane-spanning portions of the protein. (Adapted from ref. 4.)
Materials and methods
In this study, we analyzed exome sequence data from 54 unrelated probands and their unaffected parents (trios). Sequence data were generated at GeneDx, Ambry Genetics, University of California Los Angeles, Children’s Hospital of Philadelphia, the Laboratory of Personalized Genomic Medicine at Columbia University, the Broad Institute, and the Center for Advanced Studies, Research and Development in Sardinia. We also secondarily evaluated: (i) 13,448 unrelated control samples (not ascertained for neuropsychiatric conditions) sequenced at the Institute for Genomic Medicine, (ii) individuals with nonlesional focal epilepsy (n = 1,187), genetic generalized epilepsy (n = 640), and epileptic encephalopathy (n = 280) who were sequenced as part of the Epi4K Consortium,8, 9 and (iii) 3,693 individuals who underwent diagnostic sequencing through Ambry Genetics, 1,013 of whom had been diagnosed with a seizure disorder. The study was approved by the institutional review boards at Columbia University Medical Center, University of California San Francisco, Children’s Hospital of Philadelphia, Boston Children’s Hospital, New York University Langone Medical Center, Ann & Robert H. Lurie Children’s Hospital of Chicago, and Duke University as well as the Solutions Institutional Review Board (Ambry Genetics). After informed consent was obtained, exome data files (FASTQ or BAM) generated at diagnostic exome sequencing establishments were transferred to the EGI repository at the Institute for Genomic Medicine at the Columbia University Medical Center.
For individuals 1 and 2, trio data were analyzed with a pipeline based on the Genome Analysis Toolkit best-practices protocol, as reported by Zhu et al.10 Trio sequence data were analyzed using an updated version of our established trio sequencing framework. SureSelect XT2 All Exon V4 and SureSelect Human All Exon 50 Mb XT kits (Agilent Technologies; Santa Clara, CA) were used for the exome capture in individual 1 and individual 2, respectively.
Individual 3 was identified through reinterrogation of clinical exome sequence data by Ambry Genetics following the identification of the two candidate variants of exon 5 A in SCN8A in the EGI cohort. The capture kit used by Ambry Genetics was xGen Exome Research Panel version 1.0 (Integrated DNA Technologies; Coralville, IA). This individual was subsequently enrolled in the EGI to obtain phenotype information.
Variants in exon 5 A of SCN8A are annotated throughout the paper based on the RefSeq identifier XM_005269075.1.
Results
Fifty-four individuals were enrolled as trios through the EGI; most had severe epilepsies, particularly epileptic encephalopathies. Two were found to have a de novo variant in alternative exon 5 A of the SCN8A gene. The first individual found to carry a variant in exon 5 A of SCN8A (c.667A>G;p.Arg223Gly) had undergone exome sequencing in 2015. The clinical report was negative for causative variants in disease genes associated, or possibly associated, with the reported phenotype. The second individual had a different exon 5 A variant (c.632T>C;p.Val211Ala). This individual had undergone exome sequencing in 2014; while no diagnostic findings (pathogenic or likely pathogenic) had been reported, this SCN8A variant was reported as a de novo heterozygous intronic variant of uncertain significance. Clinical details of these two cases are shown in Table 1.
Table 1. The Epilepsy Genetics Initiative (EGI) participant phenotype and summary of cases previously described in the literature.
| Summary of SCN8A EE cases previously described in the literature1, 13, 14 | EGI individual 1 | EGI individual 2 | EGI individual 3 | |
|---|---|---|---|---|
| Variant | — | c.667A>G (p.Arg223Gly) | c.632T>C (p.Val211Ala) | c.692T>C (p.Ile231Thr) |
| Inheritance | — | De novo | De novo | De novo |
| Sex | — | Female | Male | Male |
| Ethnicity | — | Paternal–European Maternal–Chinese | Paternal–European Maternal–Filipino | Paternal–European Maternal–European |
| Age at seizure onset | 4–5 months on average | 6 months | 3 months | Febrile seizures at ~7–8 months; first unprovoked seizure at ~1 year of age |
| Seizure type(s) | Focal, tonic, clonic, myoclonic, and absence seizures reported; epileptic spasms; typically not associated with fever; convulsive or nonconvulsive status epilepticus in some | Clusters of head drops that evolved into extensor spasms | Tonic–clonic, then asymmetric supplementary motor seizures (left > right); tonic; focal dyscognitive | Febrile generalized tonic–clonic seizures, focal seizures, absence seizures |
| Development | Intellectual disability is common and may range from mild to severe; no speech for some; wheelchair dependence for some | Global developmental delay | Global developmental delay; never developed language | Global developmental delay; learning difficulty |
| Regression | In most cases, development is normal from birth to seizure onset; regression often slows or regresses following seizure onset | No | Yes; used to walk but regressed in setting of prolonged seizures | No |
| Electroencephalogram | Moderate to severe background slowing with focal or multifocal epileptiform discharges | Hypsarrhythmia with electroclinical spasms with electrodecrement | Left focal spikes, generalized spike/wave, background slowing and disorganization | Multifocal epileptiform discharges with background slowing |
| Magnetic resonance imaging | Variable degrees of generalized atrophy | Diffuse atrophy | Mild white matter volume loss; increased FLAIR hyperintensity in the left hemisphere in the acute setting of seizures | Normal |
| Other | Hypotonia, dystonia, ataxia, hyperreflexia, choreoathetosis | — | — | Hypotonia, ataxia |
| Diagnosis before exome sequencing | — | West syndrome of unknown cause | Epileptic encephalopathy of unknown cause | Epileptic encephalopathy of unknown cause |
| Response to treatment | Seizures refractory for many patients | Refractory to all anti- AEDs | Refractory to many AEDs; felbamate withdrawal precipitated convulsive status; addition of cannabidiol was reported to be helpful; once SCN8A diagnosis was made, phenytoin resulted in a >90% reduction in seizures | Responder to levetiracetam and topiramate; good seizure control since ~3 years of age; still has staring spells |
AED, antiepileptic drug; EE, epileptic encephalopathy; FLAIR, fluid-attenuated inversion recovery.
A review of 3,693 individuals who underwent diagnostic sequencing through Ambry Genetics revealed one additional de novo variant in SCN8A exon 5 A (c.692T>C;p.Ile231Thr). This finding was not identified at the time of the patient’s initial clinical sequencing. The exon is well sequenced on the Ambry Genetics sequencing platform. Clinical details of this case are shown in Table 1.
The XM_005269075.1 transcript contains exon 5 A of SCN8A. Exon 5 A was not included in CCDS 53794.1 or CCDS 44891.1 at the time the patients underwent sequencing, but it has since been included in the recent release 20 (CCDS81692.1). Since diagnostic companies often analyze only the secure annotations defined by CCDS, these variants were not called as pathogenic.
All three variants in exon 5 A are classified as probably damaging by PolyPhen-2,11 are absent from the Exome Aggregation Consortium and Genome Aggregation Database,12 and were not seen in any of the 13,448 in-house controls, despite sufficient sequence depth across both sets of controls at these sites to have called the variants (at least 90% of the 92 bases were sequenced at least 10-fold in more than 99% of the controls evaluated; average coverage across exon 42.3).
No missense variants were identified in exon 5 A of SCN8A in any of the other epilepsy cohorts we evaluated, including 280 individuals with epileptic encephalopathy, 640 individuals with genetic generalized epilepsy, and 1,187 individuals with nonlesional focal epilepsy. Each base of exon 5 A was sequenced at least 10-fold in these cohorts (average coverage across exon 55.6), so it is unlikely that a variant was missed.
Discussion
After reinterrogation of existing sequence data, we report three de novo variants in exon 5 A of SCN8A that were not reported as pathogenic during previous diagnostic exome interpretations. Despite evidence in the literature of the exon being present and expressed in the SCN8A transcript, it was not included in previous CCDS releases; these mutations were therefore either not reported or reported as variants of uncertain significance. Nonetheless, we believe these variants are pathogenic for the following reasons:
All three reported individuals experienced seizure onset in the first year of life and global developmental delay, which are phenotypic characteristics consistent with those previously described in SCN8A1, 13, 14 (Table 1).
According to the American College of Medical Genetics criteria, these three variants would be classified as likely pathogenic. All three are de novo variants (both maternity and paternity confirmed; PS2), absent from controls (PM2), and have multiple lines of computational evidence (PolyPhen and SIFT) supporting a deleterious effect (PP3).15
In the second case, while the variant was classified as a variant of uncertain significance by the clinical laboratory, the ordering physician felt that it may contribute to the patient’s phenotype and introduced phenytoin to the patient’s drug regimen based on previous reports of marked benefit in the SCN8A patient population.16 This resulted in a greater than 90% reduction in seizures in the patient, who was previously refractory to many antiepileptic drugs. While not conclusive evidence of pathogenicity, this pattern is similar to that reported previously for patients with disease-causing SCN8A variants.
Each base located in the genomic coordinates corresponding to exon 5 A of SCN8A (hg19, chr12:52082789-52082880) was sequenced at least 10-fold in all 54 probands studied and in 13,228 unrelated controls. We observed no missense variants in any of the 13,228 controls evaluated where exon 5 A was completely sequenced at least 10-fold, nor in any of the other 120 controls where the exon was only partially sequenced. In the Exome Aggregation Consortium and Genome Aggregation Database, with aggregate variant frequencies from the data of more than 138,000 exome- and genome-sequenced individuals, each base in the exon was sequenced 10-fold in more than 90% of individuals, and only two missense variants were reported (c.653C>T;p.Ala218Val and c.644A>G;p.Asn215Ser) out of a total of nine variant sites in the exon. Assuming that all base substitutions in exon 5 A can occur, we estimate the expected rate of nonsynonymous variation in the exon to be 0.704 by taking the sum of the estimated mutability associated with each possible nonsynonymous substitution considering its trinucleotide context8 and dividing it by the sum of the trinucleotide mutability for all possible base substitutions across the exon. Given that we expect 70.4% of all possible substitutions to result in a nonsynonymous amino acid change, an observation of only two of the nine (22%) variant sites is unlikely to occur by chance (P = 0.004, binomial exact test). This overall pattern indicates that this exon is probably under purifying selection, and suggests that protein-altering genetic variation may also be associated with a disease phenotype.
One of the amino acid substitutions in exon 5 A described here, p.Arg223Gly, was previously identified in exon 5 N (NM_014191.2: c.667A>G) in a patient with epileptic encephalopathy.17 Functional analysis identified altered activity of the channel.17 A second amino acid substitution in exon 5 A described here, p.Val211Ala (c.632T>C), is located adjacent to the pathogenic exon 5 N variant p.Phe210Leu (NM_014191.3: c.628T>C).18
Since the alternative splicing of exon 5 is conserved across several of the mammalian sodium channel alpha subunit genes, including two other known epilepsy genes, we should consider the potential for missed diagnoses in those genes as well. For SCN2A, it is unlikely that diagnoses have been missed because both forms of the exon are annotated in the CCDS. In fact, Nakamura et al.19 identified a genetic variant in the SCN2A transcript variant 3 (NM_001040143.1), which is the neonatal (6 N) isoform (CCDS 33313.1), as well as genetic variants in transcript variant 1 (NM_021007.2), which contains the 6 A exon (CCDS 33314.1). In contrast, although SCN1A also contains transcripts with both neonatal (5 N) and adult (5 A) exons,20 only the adult exon 5 A is included in the CCDS. This suggests that disease-causing variants in exon 5 N of SCN1A may also be missed.
Given the developmental regulation of SCN8A, we considered that variation in the alternate exon 5 A may translate to a different age of onset or presentation in affected individuals. While one individual reported here had a later than average age of onset and a milder course, no consistent pattern was identified in our cohort compared with the phenotype of individuals with variants elsewhere in SCN8A.
In conclusion, we report three novel epilepsy diagnoses in SCN8A, all of which were located in an alternate copy of exon 5 (exon 5 A) and overlooked by clinical exome sequencing. These findings motivate immediate re-evaluation of the genomic sequence defining exon 5 A of SCN8A, particularly in patients with severe refractory subtypes of epilepsy. In addition, they motivate re-evaluation of other well-defined alternative exons in known epilepsy genes, which may detect missed diagnoses in the epilepsy patient population. The results also provide a clear demonstration of the value of deep analysis and iterative interrogation of clinical exome sequence data that were initially found to be inconclusive. This is important not only because new disease genes and new phenotypes for known genes are still being routinely discovered, but because our knowledge of gene structures and important genomic regions continues to evolve.
Acknowledgments
The authors acknowledge Citizens United for Research in Epilepsy (339143) and the National Institute for Neurological Disorders and Stroke (U01-NS077303-04S1) for providing the funding to make this research possible. We thank the patients and families enrolled in the EGI for their participation in this research and dedication to furthering our understanding of the genetic causes of epilepsy. We also thank the external providers and clinicians who referred their patients and provided phenotype data.
Author contributions
The Steering Committee conceived, designed, and implemented the activities of the EGI. The Enrollment Group consisted of delegates at each site responsible for the enrollment of patients, phenotyping, and returning significant results to patients. Patients who took part specifically in this study were enrolled by the Columbia University Medical Center, University of California San Francisco, Children’s Hospital of Philadelphia, Boston Children’s Hospital, New York University Langone Medical Center, Ann & Robert H. Lurie Children’s Hospital of Chicago, and Duke University. Sequence and phenotypic data transfer was overseen by L.B., M.E.E., and T.M.A. In-depth phenotypic assessments of the patients were performed by J.S., I.E.S., and S.F.B. Data analysis and interpretation were performed by A.S.A., V.A., M.E.E., D.B.G., E.L.H., K.L.H., N.C.L., M.H.M., M.S.M., S.P., N.S., and S.T. All authors discussed the results and participated in the writing of the manuscript.
Footnotes
MEMBERS OF THE EPILEPSY GENETICS INITIATIVE
Samuel F. Berkovic, MD – Epilepsy Research Center, Department of Medicine, The University of Melbourne and Austin Health, Heidelberg, VIC 3084, Australia
Tracy Dixon-Salazar, PhD – Citizens United for Research in Epilepsy, Chicago, IL, 60654
David B. Goldstein, PhD – Institute for Genomic Medicine, Columbia University Medical Center, New York, NY, 10032
Erin L. Heinzen, PhD – Institute for Genomic Medicine, Columbia University Medical Center, New York, NY, 10032
Brandon L. Laughlin, MS – Citizens United for Research in Epilepsy, Chicago, IL, 60654
Daniel H. Lowenstein, MD – Department of Neurology, University of California, San Francisco, CA, 94143
Laura Lubbers, PhD – Citizens United for Research in Epilepsy, Chicago, IL, 60654
Julie Milder, PhD – Citizens United for Research in Epilepsy, Chicago, IL, 60654
Randall Stewart, PhD – National Institute for Neurological Disorders and Stroke, Bethesda, MD, 20892
Vicky Whittemore, PhD – National Institute for Neurological Disorders and Stroke, Bethesda, MD, 20892
Kaitlin Angione, MS – Department of Pediatrics, Section of Neurology, University of Colorado School of Medicine, Aurora, CO 80045
Carl W. Bazil, MD – Division of Epilepsy, The Neurological Institute, Columbia University Medical Center, New York, NY, 10032
Louise Bier, MS – Institute for Genomic Medicine, Columbia University Medical Center, New York, NY, 10032
Judith Bluvstein, MD – New York University Langone Medical Center, New York, NY, 10016
Elise Brimble, MSc, MS – Department of Neurology and Neurological Sciences, Stanford Medicine, Stanford, California, 94305
Colleen Campbell, PhD, MS – Iowa Institute of Human Genetics, University of Iowa, Carver College of Medicine, Iowa City, IA 52242
Chelsea Chambers, MS – Department of Neurology, University of Virginia, Charlottesville, VA, 22908
Hyunmi Choi, MD – Division of Epilepsy, The Neurological Institute, Columbia University Medical Center, New York, NY, 10032
Maria Roberta Cilio, MD – Departments of Neurology and Pediatrics, Benioff Children’s Hospital, University of California, San Francisco, San Francisco, CA, 94158
Michael Ciliberto, MD – Department of Pediatrics, University of Iowa Hospitals and Clinics, Iowa City, IA, 52242
Susannah Cornes, MD – Department of Neurology, University of California, San Francisco, CA, 94143
Norman Delanty, BSc, FRCPI – Department of Molecular and Cellular Therapeutics, Royal College of Surgeons in Ireland, Dublin 2, Ireland; Department of Neurology, Beaumont Hospital, Dublin 9, Ireland
Scott Demarest, MD – Department of Pediatrics, Section of Neurology, University of Colorado School of Medicine, Aurora, CO 80045
Orrin Devinsky, MD – New York University Langone Medical Center, New York, NY, 10016
Dennis Dlugos, MD – Division of Neurology, The Children’s Hospital of Philadelphia, Philadelphia, PA, 19104
Holly Dubbs, MS – Division of Neurology, The Children’s Hospital of Philadelphia, Philadelphia, PA, 19104
Patricia Dugan – New York University Langone Medical Center, New York, NY, 10016
Michelle E. Ernst, MS – Institute for Genomic Medicine, Columbia University Medical Center, New York, NY, 10032
William Gallentine, DO – Department of Pediatrics, Duke University Medical Center, Durham, NC, 27705
Melissa Gibbons, MS – Department of Pediatrics, Section of Neurology, University of Colorado School of Medicine, Aurora, CO 80045
Howard Goodkin, MD – Division of Pediatric Neurology, Department of Neurology, University of Virginia, Charlottesville, VA, 22908
Bronwyn Grinton – Epilepsy Research Center, Department of Medicine, The University of Melbourne and Austin Health, Heidelberg, VIC 3084, Australia
Ingo Helbig, MD – Division of Neurology, The Children’s Hospital of Philadelphia, Philadelphia, PA, 19104
Laura Jansen, MD – Division of Pediatric Neurology, Department of Neurology, University of Virginia, Charlottesville, VA, 22908
Kaleas Johnson – Department of Neurology, University of California, San Francisco, CA, 94143
Charuta Joshi, MD – Department of Pediatrics, Section of Neurology, University of Colorado School of Medicine, Aurora, CO 80045
Natalie C. Lippa, MS – Institute for Genomic Medicine, Columbia University Medical Center, New York, NY, 10032
Mohamad A. Makati, MD – Departments of Pediatrics and Neurobiology, Duke University Medical Center, Durham, NC, 27705
Eric Marsh, MD – Division of Neurology, The Children’s Hospital of Philadelphia, Philadelphia, PA, 19104; Departments of Neurology and Pediatrics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, 19104
Alejandro Martinez, MS – Division of Neurology, The Children’s Hospital of Philadelphia, Philadelphia, PA, 19104
John Millichap, MD – Division of Neurology, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL, 60611; and Departments of Pediatrics and Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL
Yuliya Moskovich – Division of Neurology, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL, 60611
Maureen S. Mulhern, MS – Institute for Genomic Medicine, Columbia University Medical Center, New York, NY, 10032
Adam Numis, MD – Departments of Neurology and Pediatrics, Benioff Children’s Hospital, University of California, San Francisco, San Francisco, CA, 94158
Kristen Park, MD – Department of Pediatrics, Section of Neurology, University of Colorado School of Medicine, Aurora, CO 80045
Annapurna Poduri, MD, MPH – Epilepsy Genetics Program, Boston Children’s Hospital, Boston, MA, 02115
Brenda Porter, MD – Department of Neurology and Neurological Science, Stanford School of Medicine, Palo Alto, California, 94304
Tristan T. Sands, MD – Division of Child Neurology, Morgan Stanley Children’s Hospital–New York Presbyterian, New York, NY, 10032
Ingrid E. Scheffer, MD – Epilepsy Research Centre and Department of Medicine, Austin Health, The University of Melbourne, Melbourne, VIC, Australia; The Florey Institute of Neuroscience and Mental Health, Melbourne, VIC, Australia; Departments of Paediatrics and Neurology, The Royal Children’s Hospital, The University of Melbourne, Melbourne, VIC, Australia
Beth Sheidley, MS – Epilepsy Genetics Program, Boston Children’s Hospital, Boston, MA, 02115
Nilika Singhal, MD – Departments of Neurology and Pediatrics, Benioff Children’s Hospital, University of California, San Francisco, San Francisco, CA, 94158
Lacey Smith, MS – Epilepsy Genetics Program, Boston Children’s Hospital, Boston, MA, 02115
Joseph Sullivan, MD – Departments of Neurology and Pediatrics, Benioff Children’s Hospital, University of California, San Francisco, San Francisco, CA, 94158
James J. Riviello Jr., MD – Division of Child Neurology, Morgan Stanley Children’s Hospital–New York Presbyterian, New York, NY, 10032
Alan Taylor, MS – Epilepsy Genetics Program, Boston Children’s Hospital, Boston, MA, 02115
Patricia Tolete – New York University Langone Medical Center, New York, NY, 10016.
OTHER CONTRIBUTORS
Tahseen M. Afgani – Institute for Genomic Medicine, Columbia University Medical Center, New York, NY, 10032
Vimla Aggarwal, MBBS – Institute for Genomic Medicine, Columbia University Medical Center, New York, NY, 10032; Department of Pathology and Cell Biology, Columbia University Medical Center, New York, NY 10032
Andrew S. Allen, PhD – Department of Biostatistics and Bioinformatics, Duke University, Durham, North Carolina, 27710
Rizwan Hamid, MD, PhD – Vanderbilt Medical Genetics and Genomic Medicine, Nashville, TN, 37232
Katherine L. Helbig, MS – Division of Clinical Genomics, Ambry Genetics, Aliso Viejo, CA, 92656
Miriam H. Meisler, PhD – Department of Human Genetics, University of Michigan, Ann Arbor, MI 48109
Slavé Petrovski, PhD – Department of Medicine, Royal Melbourne Hospital, The University of Melbourne, Victoria, 3050, Australia
Jean Pfotenhauer, MS – Vanderbilt Medical Genetics and Genomic Medicine, Nashville, TN, 37232
Nicholas Stong, PhD – Institute for Genomic Medicine, Columbia University Medical Center, New York, NY, 10032
Sha Tang, PhD – Division of Clinical Genomics, Ambry Genetics, Aliso Viejo, CA, 92656.
K.L.H. and S.T. are employed by Ambry Genetics. SCN8A sequencing in the setting of gene panel testing and whole-exome sequencing is among the tests commercially available at Ambry Genetics. S.P., O.D., and D.B.G. serve on the advisory board of Pairnomix. The other authors declare no conflict of interest.
References
- Meisler MH, Helman G, Hammer MF et al. SCN8A encephalopathy: research progress and prospects. Epilepsia 2016;57:1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Meisler MH. Recurrent and non-recurrent mutations of SCN8A in epileptic encephalopathy. Front Neurol 2015;6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer NW, Galt J, Jones JM et al. Exon organization, coding sequence, physical mapping, and polymorphic intragenic markers for the human neuronal sodium channel gene SCN8A. Genomics 1998;54:287–296. [DOI] [PubMed] [Google Scholar]
- Plummer NW, McBurney MW, Meisler MH. Alternative splicing of the sodium channel SCN8A predicts a truncated two-domain protein in fetal brain and non-neuronal cells. J Biol Chem 1997;272:24008–24015. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Castle J, Garrett-Engele P et al. Expression of alternatively spliced sodium channel alpha-subunit genes. Unique splicing patterns are observed in dorsal root ganglia. J Biol Chem 2004;279:46234–46241. [DOI] [PubMed] [Google Scholar]
- Gazina EV, Richards KL, Mokhtar MB, Thomas EA, Reid CA, Petrou S. Differential expression of exon 5 splice variants of sodium channel alpha subunit mRNAs in the developing mouse brain. Neuroscience 2010;166:195–200. [DOI] [PubMed] [Google Scholar]
- Butler KM, da Silva C, Shafir Y et al. De novo and inherited SCN8A epilepsy mutations detected by gene panel analysis. Epilepsy Res 2017;129:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome Project, Epi4k Consortium. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet 2014;95:360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi4K Consortium, Epilepsy Phenome/Genome Project. Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol 2017;16:135–143. [DOI] [PubMed] [Google Scholar]
- Zhu X, Petrovski S, Xie P et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med 2015;17:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J, Carvill GL, Gardella E et al. The phenotypic spectrum of SCN8A encephalopathy. Neurology 2015;84:480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally MA, Johnson J, Huisman TA et al. SCN8A epileptic encephalopathy: detection of fetal seizures guides multidisciplinary approach to diagnosis and treatment. Pediatr Neurol 2016;64:87–91. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerma RS, Braun KP, van den Broek MP et al. Remarkable phenytoin sensitivity in 4 children with SCN8A-related epilepsy: a molecular neuropharmacological approach. Neurotherapeutics 2016;13:192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kovel CG, Meisler MH, Brilstra EH et al. Characterization of a de novo SCN8A mutation in a patient with epileptic encephalopathy. Epilepsy Res 2014;108:1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercimek-Mahmutoglu S, Patel J, Cordeiro D et al. Diagnostic yield of genetic testing in epileptic encephalopathy in childhood. Epilepsia 2015;56:707–716. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kato M, Osaka H et al. Clinical spectrum of SCN2A mutations expanding to Ohtahara syndrome. Neurology 2013;81:992–998. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Yoon W, Tate SK et al. Nova2 interacts with a cis-acting polymorphism to influence the proportions of drug-responsive splice variants of SCN1A. Am J Hum Genet 2007;80:876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]