Abstract
Background
The number of targeted therapies utilized in precision medicine are rapidly increasing. Neuro-oncology offers a unique challenge due to the varying blood brain barrier (BBB) penetration of each agent. Neuro-oncologists face a difficult task weighing the growing number of potential targeted therapies and their likelihood of BBB penetration.
Methods
We developed the CNS TAP Working Group and performed an extensive literature review for the evidence-based creation of the CNS TAP tool, which was retrospectively validated by analyzing brain tumor patients who underwent therapy targeted based on genomic results from an academic sequencing study (MiOncoseq, n=17) or private molecular profiling (Foundation One, n=7).
Results
The CNS TAP tool scores relevant targeted agents by applying multiple variables (i.e., pre-clinical data, clinical data, BBB permeability) to patient specific genomic information and clinical trial availability. In the Michigan cohort, the CNS TAP tool predicted the selected agent 85.7% of the time. The CNS TAP tool predicted the agent independently selected by pediatric neuro-oncologists in the Colorado cohort 50% of the time. Patients with recurrent brain tumors treated with agents predicted by the CNS TAP tool demonstrated a median PFS of 4 months and four patients with recurrent high-grade glioma maintained ongoing partial responses of at least 6 months.
Conclusion
The CNS TAP tool is a formalized algorithm to assist clinicians select the optimal targeted therapy for neuro-oncology patients. The CNS TAP tool has relatively high concordance with selected therapies and clinical outcomes in patients receiving targeted therapy in this heterogeneous retrospective cohort were promising.
Keywords: precision medicine, neuro-oncology, algorithm, targeted therapy
INTRODUCTION
Precision medicine has become a crucial tool in the armamentarium of neuro-oncologists as clinically relevant molecular platforms become more efficient and available. With the ability to identify specific molecular alterations driving brain tumor proliferation and survival, precision medicine theoretically allows physicians to take advantage of targeted therapy, thus potentially avoiding systemic morbidity from more traditional cytotoxic chemotherapy agents.[1, 2] Precision medicine has already demonstrated efficacy in the field of neuro-oncology in multiple smaller studies, trials, and case reports.[3–7]
Concurrent with increased ease of molecular tumor profiling, the number of potential targeted therapies available has also continued to multiply. After a specific, potentially tumor-driving molecular aberration has been detected, there are often multiple biologically rational targeted therapies available. Selection of targeted therapies is influenced by evolving factors, such as Food and Drug Administration (FDA) approval, availability on a clinical trial, and physician/provider comfort level or familiarity with a particular agent.[8, 9] Additionally, given the rapidity of new published data and FDA approval of precision medicine therapies, many providers may not be aware of all possible agents that target a particular pathway. With the increasing availability of molecular information about each individual tumor, it is difficult for the treating clinician to fully interpret the likelihood of a tumor alteration driving tumor growth.[10, 11] Compounding these issues for brain tumor patients is that a majority of targeted therapies and chemotherapeutic compounds have little to no central nervous system (CNS) penetration.[9, 12] A large proportion of targeted therapies are substrates of efflux pumps, significantly limiting activity in brain tumors [13–18]. Thus, the ability to provide an objective, systematic tool to synthesize and quantify the above factors for available precision medicine agents is of critical importance to the treating neuro-oncologist.
To approach these challenges, we created a decision-making algorithm, the Central Nervous System Targeted Agent Prediction (CNS TAP) tool, to provide neuro-oncologists with a usable interface for scoring precision medicine therapies for brain tumor patients with personalized genomic data. This manuscript describes development of the CNS TAP tool and retrospectively explores the ability of the algorithm to predict the selected targeted therapy in previously sequenced and treated patients.
METHODS
Patient Selection and CNS TAP Development
Seventeen consecutive CNS tumor patients that were enrolled on a sequencing study (PEDS-MIONCOSEQ [age <21] or MIONCOSEQ [age>21]) at the University of Michigan that resulted in use of targeted therapies based on sequencing results were included in this retrospective cohort study. Additionally, seven patients with brain tumors from Children’s Hospital of Colorado/University of Colorado that underwent sequencing by Foundation One were retrospectively analyzed to provide a multicenter evaluation of the CNS TAP tool. The CNS TAP tool was developed by a multidisciplinary working group, including representatives from Neuro-Oncology, Pediatrics, Pharmacology, and Pathology. The tool’s design and weighting was further adjusted after obtaining consensus opinion with clinicians and researchers at multiple academic institutions that participated in the University of Michigan Precision Medicine Brain Tumor Conference.[9] To allow for dissemination of the tool to outside centers while maintaining its integrity, the CNS TAP Working Group has developed a web-based application for prospective use by clinicians and researchers.
MI-ONCOSEQ Study
MIONCOSEQ and PEDS-MIONCOSEQ are clinically-integrated sequencing studies were performed after obtaining informed consent (written assent if > 10 years old) from all patients or their parents/legal guardians, according to protocols approved by the Michigan Medicine Institutional Review Board (HUM00056496), following previously published methodology [19–22]. Briefly, sequencing involved targeted exome capture sequencing of paired tumor and germline DNA using a panel of 1707 genes (Oncoseq1700), transcriptome (tumor RNA) sequencing using whole exome capture, and genetic counseling. [19–21].
Results and treatment options from the PEDS-MIONCOSEQ study were discussed at a monthly multi-disciplinary, multi-center, teleconferenced University of Michigan Brain Tumor Precision Medicine Conference for each patient. Six different hospitals participate in this monthly conference.
Foundation One
For tumors profiled by Foundation One, sample collection and methodology was conducted per their established platform of commercial tumor profiling. Briefly, DNA was extracted from formalin-fixed paraffin-embedded tumor sections. Next generation sequencing was performed for exons of at least 331 cancer-related genes and select introns of 28 rearrangements using Illumina HiSeq 2000 (Illumina, Inc., San Diego, California). Testing was performed in a CLIA–certified, College of American Pathologists–accredited reference laboratory. The sample was evaluated for genomic alterations, including base pair substitutions, indels, copy number alterations, and rearrangements.[9]
MI-ONCOSEQ panel and Foundation One panel have 319 genes in common.
Retrospective Analysis
To test the validity of the CNS TAP tool, we performed a multi-institutional, retrospective cohort study. We analyzed the last 17 neuro-oncology patients enrolled in the PEDS-MIONCOSEQ or MIONCOSEQ studies, whose targeted therapies were chosen by the multi-disciplinary conference to determine if the CNS TAP tool was capable of accurately predicting the same drug regiments as this team of experts. We also validated the tool’s use with private molecular profiling with retrospective use in an additional seven patients profiled by Foundation One at the University of Colorado. Progression-free survival (PFS) was estimated using the Kaplan-Meier method and a median time to progression was reported with a 95% confidence interval. Time to progression was defined from the start of the treatment until disease progression, and survival data was censored at the last date the patient was known to be alive and free from progression. Statistical analyses were done using SAS 9.4 software (Cary, NC).
RESULTS
Description of Algorithm
The CNS TAP tool is designed to score relevant targeted agents by applying multiple variables to patient specific genomic information and clinical trial availability. Thirty-six targeted therapies were chosen based on common molecular aberrations detected in neuro-oncology patients and based on discussion from the prior three years at Brain Tumor Precision Medicine Conference at the University of Michigan. For each of the 36 targeted therapies evaluated, the CNS TAP Working Group assessed published data on 9 distinct categories to which a score was assigned. Scores were weighted based on consensus opinion from members of the Working Group and further adjusted by discussion in the conference (Table 1).
Table 1.
A summary of the distribution of points and the weights that were affixed to individual categories of the CNS TAP tool.
| CNS TAP Tool | |||
|---|---|---|---|
| Description of Points | Weight | Range of Points | |
| Pre-Clinical Data (in vitro) | 0 = No data 1 = Activity against any cell type 2 = Activity against primary CNS tumor cell type |
2 | 0 to 4 |
| Pre-Clinical Data (in vivo) | 0 = No data 1 = Activity against cells grafted into the flank 2 = Activity against orthotopic cells or cells grafted into the brain |
3 | 0 to 6 |
| Phase I Safety Data | 0 = No data 1 = Ongoing pediatric clinical trial 2 = Published phase I trial with phase I dosing |
3 | 0 to 6 |
| CNS Data with Response | −2 = Evidence of lack of positive response of primary CNS tumors −1 = Evidence of lack of positive response of brain metastasis 0 = No data 1 = Evidence of positive response of a brain metastasis 2 = Evidence of positive response of a primary CNS tumor |
5 | −10 to 10 |
| Brain Penetration | 0 = Insufficient CNS penetration 1 = < 5% CNS penetration but Cmax above IC50 2 = > 5% CNS penetration and Cmax above IC50 |
5 | 0 to 10 |
| FDA Approval | 0 = No FDA approval 1 = FDA approval |
10 | 0 to 10 |
| Clonality/variant allele fraction (%) | 0 = Pathway is non-dominant mutation 1 = Pathway is dominant mutation |
5 | 0 to 5 |
| Variant Tier Score | 0 = Tier IV 1 = Tier III 2 = Tier I or Tier II |
3 | 0 to 6 |
| Relevant Clinical Trial | 0 = No clinical trial considered 2 = Clinical trial is available that the patient is eligible for and willing to travel to |
10 | 0 to 20 |
Categories include: (1) pre-clinical data (in vitro), (2) pre-clinical data (in vivo), (3) clinical trial data, (4) human CNS data with response, (5) brain penetration, (6) FDA approval; as well as patient-specific variables, including (1) percentage of tumor with relevant alteration (dominant vs non-dominant), (2) variant tier score, and (3) relevant clinical trial availability.[10, 11] The final score for each agent is a weighted sum of scores assigned from published data. Each patient’s sequencing results and relevant clinical trials are then integrated to provide a final score, with scores of agents from relevant pathway(s) being listed on final CNS TAP report (Supplementary Figure 1). A description of each category follows below (Table 1):
Pre-Clinical Data (in vitro)
For each drug, an extensive literature review is conducted regarding any in vitro pre-clinical data of the agent demonstrating activity in cell lines harboring the designated pathway alteration. The main metric assessed is whether the drug in question demonstrates activity against cultured tumor cells confirmed to harbor the pathway alteration with a 50 percent inhibitory concentration (IC50) of less than micro-molar concentrations. Higher score is given if the data is in a primary brain tumor cell culture.
Pre-Clinical Data (in vivo)
This category is similar to the in vitro pre-clinical data except the agent is required to demonstrate activity (reduction in tumor growth or improved survival) against cancer cells with the designated pathway alteration in an animal model. Agents are scored higher if the animal model involves tumor cells implanted in the brain.
Phase I Safety Data
The phase I safety data category is included for patients less than 21 and examines whether there is any safety/toxicity data (pediatric phase I data) for the specific drug. The goal of this category is to give greater weight to agents for which dosing information has been determined for pediatric patients. This category is ignored for patients over 21 years old.
CNS Data with a Response
We performed an exhaustive literature search to identify any evidence that there have been positive, negative, or neutral responses to the agent against primary brain tumors or CNS metastases of non-CNS tumors. We define a positive response as a partial response (PR) or clinically significant stable disease (SD), as determined by the authors of the primary studies. Prospective clinical trials are prioritized in score assignment, but retrospective series and case reports are also considered. Responses in case reports or retrospective series in patients with concurrent or radiation therapy immediately preceding targeted therapy that may have impacted treatment response were not considered. Responses in CNS metastases in non-primary CNS tumors are given a lower score in comparison with responses in primary CNS tumors. For prospective clinical data demonstrating lack of efficacy in primary CNS tumors, a negative score is given.
Brain Penetration
Due to the unique nature of brain tumors, consideration of the ability of an agent to penetrate the BBB is imperative. To evaluate likelihood of CNS penetration, a compound’s molecular weight, lipophilicity, and polarity is assessed using Lipinski’s rule of five.[9] In addition, the percentage of protein binding and affinity for efflux pumps (e.g., p-glycoprotein and breast cancer resistance protein [BCRP]) is evaluated. Published data on CNS concentrations and penetration is evaluated in relation to the IC50 of the agent against the pathway and the predicted concentration of the agent within the brain, where available.
FDA approval
This category indicated whether the drug was FDA approved and commercially available for any indication.
Clonality/Variant Allele Fraction (%)
In the case of tumor sequencing harboring multiple potentially targetable alterations, the CNS TAP tool is adjusted, based on the percentage of tumor that expresses the pathway alteration (variant allele fraction), in order to give weight to the “dominant”, or more clonal, pathway. Clonal homozygous deletions and copy number amplifications are scored as 100%.
Variant tier score
The variant tier score is a metric that classifies the somatic variant in a tumor and allows physicians to gain diagnostic, prognostic, and therapeutic information.[10, 11] As previously published, this classification system gives a score of I–IV.[11] A somatic variant with either level A evidence or level B evidence are in tier I. Tier II somatic variants have either level C or level D evidence. Somatic variants without convincing published evidence are tier III. If there is no existing published evidence of any association with cancer, the somatic variant is assigned to tier IV.[10, 11]
Relevant Clinical Trial
The importance of promoting clinical trial enrollment and investigational therapies is paramount. An agent receives extra points if there is a clinical trial for which the patient may be eligible.
CNS TAP Tool Validation
Michigan Cohort
Seventeen neuro-oncology patients with MIONCOSEQ sequencing results received 21 different targeted therapies based on the University of Michigan Brain Tumor Precision Medicine Conference (Figure 1A and 1B). Of the 17 total patients, 11 (64.7%) had a single, dominant mutation that was treated with a single therapeutic agent (Table 2). For these patients with a single targetable lesion, the CNS TAP tool accurately predicted the precision medicine agent that was decided upon by the multi-disciplinary conference in all cases (11/11; 100%).
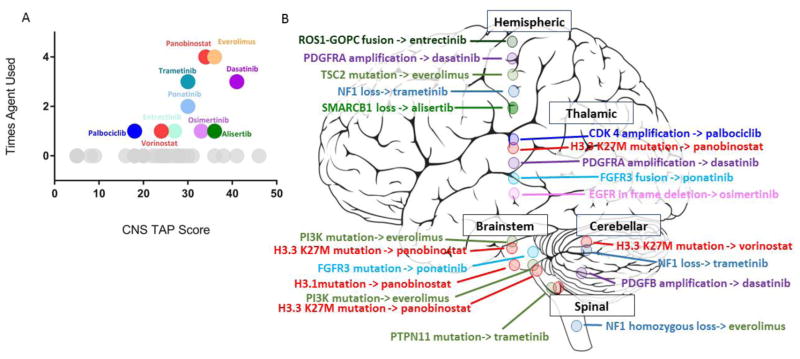
Fig 1.
A) Graphic representation of the base score given to the 36 targeted therapies currently evaluated by the CNS TAP tool by the number of times that targeted therapy was used in the retrospective University of Michigan cohort. Gray circles designate that the targeted therapy was not used in the cohort. Each specific color corresponds to a pathway (i.e. both panobinostat and vorinostat are red since they both inhibit the histone deacetylase [HDAC] pathway). B) Pictorial representation of where the CNS tumor was located in the CNS: hemispheric, thalamic, cerebellar, brainstem, or spinal. Each circle represents a major pathway mutation and each color represents a different pathway. (Abbreviations: ROS1-GOPC = Golgi-associated PDZ and coiled-coil domains-containing gene, PDGFRA = platelet-derived growth factor receptor, TSC = tuberous sclerosis complex, NF = neurofibromatosis, SMARCB = SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B, CDK = cyclin-dependent kinase, FGFR = fibroblast growth factor receptor, EGFR = epidermal growth factor receptor, PI3K = phosphatidylinositol 3-kinase, PTPN = protein-tyrosine phosphate nonreceptor).
Table 2.
For each of the 17 patients in the Michigan Cohort, a clinical description is provided along with the genomic pathway that was profiled for that specific patient. The potential drug options were listed along with the individual scoring for each drug. The targeted therapy that the CNS TAP tool predicted is juxtaposed next to the actual targeted therapy that was chosen to treat the specific patient.
| CNS TAP Tool: University of Michigan Patients | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient Number |
Age at Enrollment (years) |
Clinical diagnosis |
Genomic findings |
Drug | Pathway | Tumor line/ pre- clinical data (in vitro) |
Tumor line/ pre- clinical data (in vivo) |
Phase I safety data |
CNS Data with response |
Brain penetration |
FDA approval |
Clonality/ variant allele fraction (%) |
Variant tier score |
Relevant clinical trial |
Total Points |
Drug Predicted |
Drug Chosen |
| 1 | 1 | GBM | PDGFRA amplicafication | Dasatinib | PDGFR | 4 | 6 | 6 | 5 | 10 | 10 | 5 | 3 | 0 | 49 | Dasatanib | Dasatinib |
| Pazopanib | PDGFR | 4 | 6 | 6 | 5 | 5 | 10 | 5 | 3 | 0 | 44 | ||||||
| Sunitinib | PDGFR | 2 | 6 | 6 | 5 | 0 | 10 | 5 | 3 | 0 | 37 | ||||||
| Ponatinib | PDGFR | 4 | 3 | 0 | 0 | 10 | 10 | 5 | 3 | 0 | 35 | ||||||
| Sorafenib | PDGFR | 2 | 0 | 6 | 0 | 0 | 10 | 5 | 3 | 0 | 26 | ||||||
| Crenolanib | PDGFR | 2 | 0 | 3 | 0 | 0 | 0 | 5 | 3 | 0 | 13 | ||||||
| 2 | 6 | GBM | H3F3A point mutation | Panobinostat | HDAC | 2 | 6 | 6 | 10 | 0 | 10 | 0 | 3 | 0 | 37 | Panobinostat | Panobinostat |
| Vorinostat | HDAC | 2 | 6 | 6 | −10 | 10 | 10 | 0 | 3 | 0 | 27 | ||||||
| CDK4 amplification | Ribociclib | CDK | 2 | 0 | 6 | 0 | 10 | 10 | 5 | 3 | 0 | 36 | Ribociclib | Palbociclib | |||
| Abemaciclib | CDK | 2 | 6 | 0 | 10 | 5 | 0 | 5 | 3 | 0 | 31 | ||||||
| Palbociclib | CDK | 2 | 6 | 0 | 0 | 0 | 10 | 5 | 3 | 0 | 26 | ||||||
| 3 | 5 | DIPG | PIK3CA activating mutation | Everolimus | PI3K/mTOR | 2 | 3 | 6 | 10 | 5 | 10 | 0 | 6 | 0 | 42 | Everolimus | Everolimus |
| GDC-0084 | PI3K/mTOR | 4 | 0 | 6 | 10 | 10 | 0 | 0 | 6 | 0 | 36 | ||||||
| BKM120 | PI3K/mTOR | 4 | 0 | 0 | 0 | 5 | 0 | 0 | 6 | 0 | 15 | ||||||
| MK-2206 | AKT | 4 | 6 | 6 | 0 | 0 | 0 | 0 | 6 | 0 | 22 | ||||||
| Perifosine | AKT | 2 | 0 | 6 | 0 | 0 | 0 | 0 | 6 | 0 | 14 | ||||||
| HIST1H3B activating mutation | Panobinostat | HDAC | 2 | 6 | 6 | 10 | 0 | 10 | 5 | 3 | 0 | 42 | Panobinostat | Panobinostat | |||
| Vorinostat | HDAC | 2 | 6 | 6 | −10 | 10 | 10 | 5 | 3 | 0 | 32 | ||||||
| 4 | 1 | GBM | PDGFB amplification | Dasatinib | PDGFR | 4 | 6 | 6 | 5 | 10 | 10 | 5 | 3 | 0 | 49 | Dasatanib | Dasatinib |
| Pazopanib | PDGFR | 4 | 6 | 6 | 5 | 5 | 10 | 5 | 3 | 0 | 44 | ||||||
| Sunitinib | PDGFR | 2 | 6 | 6 | 5 | 0 | 10 | 5 | 3 | 0 | 37 | ||||||
| Ponatinib | PDGFR | 4 | 3 | 0 | 0 | 10 | 10 | 5 | 3 | 0 | 35 | ||||||
| Sorafenib | PDGFR | 2 | 0 | 6 | 0 | 0 | 10 | 5 | 3 | 0 | 26 | ||||||
| Crenolanib | PDGFR | 2 | 0 | 3 | 0 | 0 | 0 | 5 | 3 | 0 | 13 | ||||||
| 5 | 8 | Spinal anaplastic OD | FGFR activating mutation | Ponatinib | FGFR | 4 | 6 | 0 | 0 | 10 | 10 | 5 | 6 | 0 | 41 | Ponatinib | Everolimus |
| Pazopanib | FGFR | 2 | 0 | 6 | 5 | 5 | 10 | 5 | 6 | 0 | 39 | ||||||
| NF1 mutation | Trametinib | MEK | 2 | 0 | 3 | 5 | 10 | 10 | 0 | 3 | 0 | 33 | |||||
| Selumetinib | MEK | 4 | 0 | 0 | 0 | 10 | 10 | 0 | 3 | 0 | 27 | ||||||
| 6 | 13 | DIPG | FGFR3 activating mutation | Ponatinib | FGFR | 4 | 6 | 0 | 0 | 10 | 10 | 5 | 6 | 0 | 41 | Ponatinib | Ponatinib |
| Pazopanib | FGFR | 2 | 0 | 6 | 5 | 5 | 10 | 5 | 6 | 0 | 39 | ||||||
| H3.3 mutation | Panobinostat | HDAC | 2 | 6 | 6 | 10 | 0 | 10 | 0 | 3 | 0 | 37 | |||||
| Vorinostat | HDAC | 2 | 6 | 6 | −10 | 10 | 10 | 0 | 3 | 0 | 27 | ||||||
| 7 | 15 | GBM | TSC2 inactivating mutation | Everolimus | PI3K/mTOR | 2 | 3 | 6 | 10 | 5 | 10 | 5 | 3 | 0 | 44 | Everolimus | Everolimus |
| GDC-0084 | PI3K/mTOR | 4 | 0 | 6 | 10 | 10 | 0 | 5 | 3 | 0 | 38 | ||||||
| BKM120 | PI3K/mTOR | 4 | 0 | 0 | 0 | 5 | 0 | 5 | 3 | 0 | 17 | ||||||
| 8 | 17 | GBM | NF1 homozygous loss | Trametinib | MEK | 2 | 0 | 3 | 5 | 10 | 10 | 5 | 3 | 0 | 38 | Trametinib | Trametinib |
| Selumetinib | MEK | 4 | 0 | 0 | 0 | 10 | 10 | 5 | 3 | 0 | 32 | ||||||
| H3F3A point mutation | Panobinostat | HDAC | 2 | 6 | 6 | 10 | 0 | 10 | 0 | 3 | 0 | 37 | Panobinostat | Vorinostat | |||
| Vorinostat | HDAC | 2 | 6 | 6 | −10 | 10 | 10 | 0 | 3 | 0 | 27 | ||||||
| 9 | 17 | GBM | PDGFRA mutation | Dasatinib | PDGFR | 4 | 6 | 6 | 5 | 10 | 10 | 5 | 3 | 0 | 49 | Dasatanib | Dasatinib |
| Pazopanib | PDGFR | 4 | 6 | 6 | 5 | 5 | 10 | 5 | 3 | 0 | 44 | ||||||
| Sunitinib | PDGFR | 2 | 6 | 6 | 5 | 0 | 10 | 5 | 3 | 0 | 37 | ||||||
| Ponatinib | PDGFR | 4 | 3 | 0 | 0 | 10 | 10 | 5 | 3 | 0 | 35 | ||||||
| Sorafenib | PDGFR | 2 | 0 | 6 | 0 | 0 | 10 | 5 | 3 | 0 | 26 | ||||||
| Crenolanib | PDGFR | 2 | 0 | 3 | 0 | 0 | 0 | 5 | 3 | 0 | 13 | ||||||
| 10 | 11 | Anaplastic OD | FGFR3-PHGDH fusion | Ponatinib | FGFR | 4 | 6 | 0 | 0 | 10 | 10 | 5 | 6 | 0 | 41 | Ponatinib | Ponatinib |
| Pazopanib | FGFR | 2 | 0 | 6 | 5 | 5 | 10 | 5 | 6 | 0 | 39 | ||||||
| 11 | 11 | Choroid plexus carcinoma | NF1 frameshift insertion | Trametinib | MEK | 2 | 0 | 3 | 5 | 10 | 10 | 5 | 3 | 0 | 38 | Trametinib | Trametinib |
| Selumetinib | MEK | 4 | 0 | 0 | 0 | 10 | 10 | 5 | 3 | 0 | 32 | ||||||
| 12 | 8 | DIPG | PIK3CA activating mutation | Everolimus | PI3K | 2 | 3 | 6 | 10 | 5 | 10 | 0 | 6 | 0 | 42 | Panobinostat | Panobinostat |
| GDC-0084 | PI3K | 4 | 0 | 6 | 10 | 10 | 0 | 0 | 6 | 0 | 36 | ||||||
| BKM120 | PI3K | 4 | 0 | 0 | 0 | 5 | 0 | 0 | 6 | 0 | 15 | ||||||
| MK-2206 | AKT | 4 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | ||||||
| Perifosine | AKT | 2 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | ||||||
| H3F3A activating mutation | Panobinostat | HDAC | 2 | 6 | 6 | 10 | 0 | 10 | 5 | 3 | 0 | 42 | Everolimus | Everolimus | |||
| Vorinostat | HDAC | 2 | 6 | 6 | −10 | 10 | 10 | 5 | 3 | 0 | 32 | ||||||
| 13 | 29 | Anaplastic astroblastoma | SMARCB1homozygous deletion | Alisertib | INI1 | 4 | 6 | 6 | 10 | 10 | 0 | 5 | 3 | 0 | 44 | Alisertib | Alisertib |
| Tamoxifen | INI1 | 2 | 0 | 0 | 0 | 10 | 10 | 5 | 3 | 0 | 30 | ||||||
| Tazemetostat | INI1 | 2 | 0 | 3 | 0 | 0 | 0 | 5 | 3 | 0 | 13 | ||||||
| 14 | 11 | GBM | EGFR in-frame deletion | Osimertinib | EGFR | 2 | 6 | 0 | 5 | 10 | 10 | 5 | 3 | 0 | 41 | Osimertinib | Osimertinib |
| Pazopanib | EGFR | 2 | 0 | 6 | 5 | 5 | 10 | 5 | 3 | 0 | 36 | ||||||
| Erlotinib | EGFR | 4 | 0 | 6 | −10 | 10 | 10 | 5 | 3 | 0 | 28 | ||||||
| 15 | 3 | DIPG | PTPN11 mutation | Trametinib | MEK | 2 | 0 | 3 | 5 | 10 | 10 | 5 | 6 | 0 | 41 | Trametinib | Trametinib |
| Selumetinib | MEK | 4 | 0 | 0 | 0 | 10 | 10 | 5 | 6 | 0 | 35 | ||||||
| H3.1 mutation | Panobinostat | HDAC | 2 | 6 | 6 | 10 | 0 | 10 | 0 | 3 | 0 | 37 | |||||
| Vorinostat | HDAC | 2 | 6 | 6 | −10 | 10 | 10 | 0 | 3 | 0 | 27 | ||||||
| 16 | 4 | DIPG | H3.1 and ACVR1 mutation | Panobinostat | HDAC | 2 | 6 | 6 | 10 | 0 | 10 | 5 | 3 | 0 | 42 | Panobinostat | Panobinostat |
| Vorinostat | HDAC | 2 | 6 | 6 | −10 | 10 | 10 | 5 | 3 | 0 | 32 | ||||||
| 17 | 15 | Malignant glioneuronal tumor | ROS1-GOPC fusion | Entrectinib | ALK | 4 | 0 | 3 | 10 | 10 | 0 | 5 | 3 | 20 | 55 | Entrectinib | Entrectinib |
| Ceritinib | ALK | 2 | 0 | 6 | 0 | 10 | 10 | 5 | 3 | 0 | 36 | ||||||
| Alectanib | ALK | 2 | 0 | 3 | 0 | 10 | 10 | 5 | 3 | 0 | 33 | ||||||
Abbreviations: GBM = glioblastoma multiforme, PDGFRA = platelet-derived growth factor receptor, HDAC = histone deacetylase, CDK = cyclin-dependent kinase, DIPG = diffuse intrinsic pontine glioma, PI3K = phosphatidylinositol 3-kinase, mTOR = mechanistic target of rapamycin, AKT = protein kinase B, FGFR = fibroblast growth factor receptor, NF = neurofibromatosis, OD = oligodendroglioma, MEK = mitogen-activated protein kinase kinase, TSC = tuberous sclerosis complex, PHGDH = phosphoglycerate dehydrogenase, SMARCB = SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B, INI = integrase interactor, EGFR = epidermal growth factor receptor, PTPN = protein-tyrosine phosphate nonreceptor, ROS1-GOPC = Golgi-associated PDZ and coiled-coil domains-containing gene, ALK = anaplastic lymphoma kinase.
Of the 6 patients with two different mutations, 2 of the 6 (33.3%) were only given a single targeted therapy while 4 (66.7%) were given two agents, each targeting a separate pathway (Table 2). CNS TAP accurately predicted 1 of the 2 (50%) targeted therapies chosen for patients with two mutations but who only received a single drug. For the 4 patients who had two mutations and received two agents, CNS TAP accurately predicted at least one agent for all 4 patients. For two of the patients it predicted both drugs accurately while it only predicted 1 of the 2 drugs for the other two patients (6/8; 75%) (Table 2). In total, the CNS TAP tool correctly predicted 18 of the 21 (85.7%) drugs prescribed by the multi-disciplinary team of experts.
Within the limits of this small and heterogeneous group of patients, we performed an outcome analysis. Patients with newly diagnosed or recurrent brain tumors treated with agents predicted by the CNS TAP tool demonstrated a median PFS of 9 months and 4 months, respectively (Figures 2). Four pediatric patients with recurrent high-grade glioma maintained ongoing partial responses of at least 6 months.
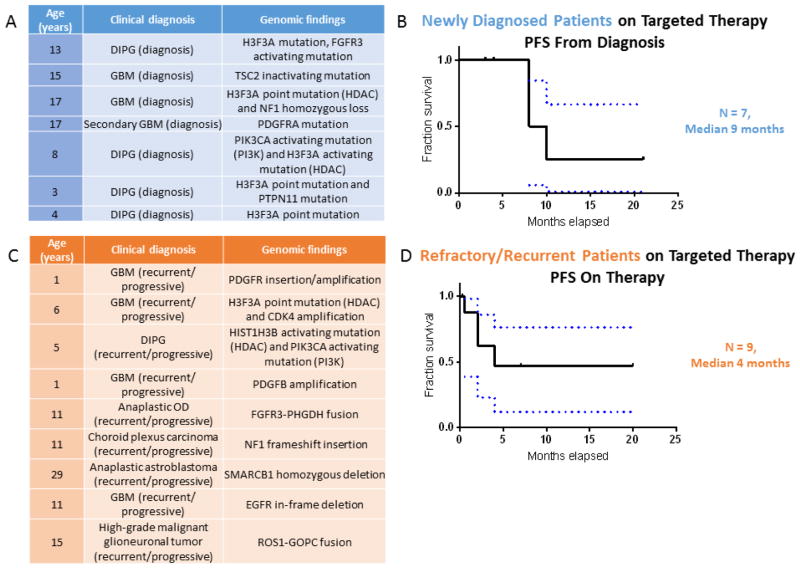
Fig 2.
A) Clinical characteristics of patients from the Michigan Cohort taking a targeted therapy that was congruent with the targeted therapy selected by the CNS TAP tool with a new diagnosis of a CNS tumor. B) A Kaplan Meier curve for the patients described in 2A demonstrating a median progression free survival (PFS) of 9 months. C) Clinical characteristics of patients from the Michigan Cohort taking a targeted therapy that was congruent with the targeted therapy selected by the CNS TAP tool with a refractory or recurrent CNS tumor. D) A Kaplan Meier curve for the patients described in 2C demonstrating a median PFS of 4 months. (Abbreviations: DIPG = diffuse intrinsic pontine glioma, FGFR = fibroblast growth factor receptor, GBM = glioblastoma multiforme, TSC = tuberous sclerosis complex, HDAC = histone deacetylase, NF = neurofibromatosis, PDGFRA = platelet-derived growth factor receptor, PI3K = phosphatidylinositol 3-kinase, PTPN = protein-tyrosine phosphate nonreceptor, CDK = cyclin-dependent kinase, OD = oligodendroglioma, PHGDH = phosphoglycerate dehydrogenase, SMARCB = SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B, EGFR = epidermal growth factor receptor, ROS1-GOPC = Golgi-associated PDZ and coiled-coil domains-containing gene, PFS = progression free survival).
Colorado Cohort
Seven neuro-oncology patients received 8 targeted therapies. Three of the 7 (42.9%) patients had a single mutation with a single targeted therapy given, 3 (42.9%) had two presumed pathogenic mutations with a single agent given, and 1 (14.2%) patient had two presumed pathogenic mutations with two drugs given (Table 3).
Table 3.
For each of the 7 patients in the Colorado Cohort, a clinical description is provided along with the genomic pathway that was profiled for that specific patient. The potential drug options were listed along with the individual scoring for each drug. The targeted therapy that the CNS TAP tool predicted is juxtaposed next to the actual targeted therapy that was chosen to treat the specific patient.
| CNS-TAP Tool: University of Colorado Patients | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient Number |
Age at Enrollment (years) |
Clinical diagnosis | Genomic findings |
Drug | Pathway | Tumor line/ pre- clinical data (in vitro) |
Tumor line/ pre- clinical data (in vivo) |
Phase I safety data |
CNS Data with response |
Brain penetration |
FDA approval |
Clonality/ variant allele fraction (%) |
Variant tier score |
Relevant clinical trial |
Total Points |
Drug Predicted |
Drug Chosen |
| 1 | 2 | Spinal pilomyxoid astrocytoma | FGFR1-TACC1 fusion | Ponatinib | FGFR | 4 | 6 | 0 | 0 | 10 | 10 | 5 | 6 | 0 | 41 | Ponatinib | AZD4547 |
| Pazopanib | FGFR | 2 | 0 | 6 | 5 | 5 | 10 | 5 | 6 | 0 | 39 | ||||||
| 2 | 11 | GBM | H3F3A K28M mutation | Entinostat | HDAC | 4 | 6 | 3 | 0 | 5 | 0 | 5 | 3 | 20 | 46 | Entinostat | Entinostat |
| Panobinostat | HDAC | 2 | 6 | 6 | 6 | 0 | 10 | 5 | 3 | 0 | 38 | ||||||
| Vorinostat | HDAC | 2 | 6 | 6 | −6 | 10 | 10 | 5 | 3 | 0 | 36 | ||||||
| PDGFRA mutation | Dasatinib | PDGFR | 4 | 6 | 6 | 3 | 10 | 10 | 0 | 3 | 0 | 42 | |||||
| Pazopanib | PDGFR | 4 | 6 | 6 | 3 | 5 | 10 | 0 | 3 | 0 | 37 | ||||||
| Ponatinib | PDGFR | 4 | 3 | 0 | 0 | 10 | 10 | 0 | 3 | 0 | 30 | ||||||
| Sunitinib | PDGFR | 2 | 6 | 6 | 3 | 0 | 10 | 0 | 3 | 0 | 30 | ||||||
| Sorafenib | PDGFR | 2 | 0 | 6 | 0 | 0 | 10 | 0 | 3 | 0 | 21 | ||||||
| Crenolanib | PDGFR | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 8 | ||||||
| 3 | 18 | GBM | NF1 mutation | Trametinib | MEK | 2 | 0 | 3 | 5 | 10 | 10 | 5 | 3 | 0 | 38 | Trametinib | Trametinib |
| Selumetinib | MEK | 4 | 0 | 0 | 0 | 10 | 10 | 5 | 3 | 0 | 32 | ||||||
| 4 | 0 | Metastatic clival chordoma | TSC1 loss | Everolimus | PI3K/mTOR | 2 | 3 | 6 | 10 | 5 | 10 | 5 | 6 | 0 | 47 | Everolimus | Temsirolimus |
| GDC-0084 | PI3K/mTOR | 4 | 0 | 6 | 10 | 10 | 0 | 5 | 6 | 0 | 41 | ||||||
| Temsirolimus | PI3K/mTOR | 4 | 6 | 6 | −5 | 5 | 10 | 5 | 6 | 0 | 37 | ||||||
| BKM120 | PI3K/mTOR | 4 | 0 | 0 | 0 | 5 | 0 | 5 | 6 | 0 | 20 | ||||||
| 5 | 12 | Anaplastic astrocytoma | CDKN2A deletion | Abemaciclib | CDK | 2 | 6 | 0 | 10 | 5 | 0 | 0 | 3 | 20 | 46 | Abemaciclib | Abemaciclib |
| Ribociclib | CDK | 2 | 0 | 6 | 0 | 10 | 10 | 0 | 3 | 0 | 31 | ||||||
| Palbociclib | CDK | 2 | 6 | 0 | 0 | 0 | 10 | 0 | 3 | 0 | 21 | ||||||
| EGFR mutation | Erlotinib | EGFR | 4 | 0 | 6 | −10 | 10 | 10 | 5 | 3 | 0 | 28 | Erlotinib | Cetuximab | |||
| Cetuximab | EGFR | 4 | 6 | 6 | −10 | 0 | 10 | 5 | 3 | 0 | 24 | ||||||
| 6 | 15 | GBM | CDKN2A deletion | Abemaciclib | CDK | 2 | 6 | 0 | 10 | 5 | 0 | 5 | 3 | 20 | 51 | Abemaciclib | Abemaciclib |
| Ribociclib | CDK | 2 | 0 | 6 | 0 | 10 | 10 | 5 | 3 | 0 | 36 | ||||||
| Palbociclib | CDK | 2 | 6 | 0 | 0 | 0 | 10 | 5 | 3 | 0 | 26 | ||||||
| PI3KCA E542K mutation | Everolimus | PI3K/mTOR | 2 | 3 | 6 | 10 | 5 | 10 | 0 | 6 | 0 | 42 | |||||
| GDC-0084 | PI3K/mTOR | 4 | 0 | 6 | 10 | 10 | 0 | 0 | 6 | 0 | 36 | ||||||
| Temsirolimus | PI3K/mTOR | 4 | 6 | 6 | −5 | 5 | 10 | 0 | 6 | 0 | 32 | ||||||
| BKM120 | PI3K/mTOR | 4 | 0 | 0 | 0 | 5 | 0 | 0 | 6 | 0 | 15 | ||||||
| MK-2206 | AKT | 4 | 6 | 6 | 0 | 0 | 0 | 0 | 6 | 0 | 22 | ||||||
| Perifosine | AKT | 2 | 0 | 6 | 0 | 0 | 0 | 0 | 6 | 0 | 14 | ||||||
| 7 | 10 | Anaplastic pleomorphic xanthoastrocytoma | BRAF V600E mutation | Dabrafenib | BRAF | 4 | 6 | 6 | 10 | 5 | 10 | 5 | 3 | 0 | 49 | Dabrafenib | Vemurafinib |
| Vemurafenib | BRAF | 2 | 0 | 0 | 10 | 5 | 10 | 5 | 3 | 0 | 35 | ||||||
| CDKN2A deletion | Ribociclib | CDK | 2 | 0 | 6 | 0 | 10 | 10 | 0 | 3 | 0 | 31 | |||||
| Abemaciclib | CDK | 2 | 6 | 0 | 10 | 5 | 0 | 0 | 3 | 0 | 26 | ||||||
| Palbociclib | CDK | 2 | 6 | 0 | 0 | 0 | 10 | 0 | 3 | 0 | 21 | ||||||
Abbreviations: FGFR = fibroblast growth factor receptor, TACC = transformic acidic coiled-coil-containing gene, GBM = glioblastoma multiforme, HDAC = histone deacetylase, PDGFRA = platelet-derived growth factor receptor, NF = neurofibromatosis, MEK = mitogen-activated protein kinase kinase, TSC = tuberous sclerosis complex, PI3K = phosphatidylinositol 3-kinase, mTOR = mechanistic target of rapamycin, CDK = cyclin-dependent kinase, EGFR = epidermal growth factor receptor, AKT = protein kinase B.
The CNS TAP tool accurately predicted the administered drug in only 1 of the 3 patients with a single mutation (33.3%), while administered drugs were accurately predicted for 2 of the 3 patients with two significant mutations (66.7%). For the single patient who had two mutations and two drugs administered, the CSN-TAP tool accurately predicted one of the two drugs (50%).
In total, the CNS TAP tool predicted 4 of the 8 (50%) targeted therapies that were chosen by the neuro-oncology team at the University of Colorado.
DISCUSSION
Precision medicine is an expanding field which allows physicians to provide individualized and unique targeted therapies to patients to treat their tumor-specific alterations. By understanding the molecular alterations unique to each brain tumor, physicians can select therapeutic agents targeting the inferred tumor-driving pathways. Perhaps equally important in neuro-oncology is the likelihood that the drug will achieve adequate CNS penetration after traversing (or efflux from) the BBB. As in many other cancer types, neuro-oncologists do not have a formalized method for prioritization of precision medicine therapies, either on or off trial. Neuro-oncologists are increasingly integrating molecular information in the management of their patients with higher risk of refractory brain tumors, and there are a growing number of targeted agents available. By creating a formalized, objective method of evaluating targeted therapies, the process of selecting precision medicine therapies can be standardized, potentially improving care for patients.
The CNS TAP tool is designed to be adaptive to the continued addition of new relevant agents and published data. The CNS TAP Working Group will re-visit the tool on a quarterly basis to add agents and adjust scores accordingly. Due to the rapid advances in the field, we plan to eventually integrate proteomic measurements into the CNS TAP tool to utilize biomarker monitoring to diagnose specific aspects of a disease as well as determine disease severity.[23] Additionally, as immuno-oncology continues to revolutionize the treatment of cancer patients, we will include well-studied immuno-therapies to the CNS TAP tool to ensure an algorithm that suggests the most effective, up-to-date therapies. We have developed a web-based application to allow access to clinicians and researchers at outside institutions for prospective validation.
Our preliminary retrospective data demonstrates that the CNS TAP tool was effective in selecting the same therapeutic agents that clinicians had selected for patients after consensus opinion in the University of Michigan Brain Tumor Precision Medicine Conference. The tool accurately predicted the correct agent each time when the patient had a single, dominant alteration. The CNS TAP tool was less consistent (accurate 50%) when compared to therapies selected by clinicians at the University of Colorado for patients who underwent private molecular profiling. This discrepancy may be partially accounted for by differences in the brain tumor programs at each institution. University of Michigan has a combined adult and pediatric brain tumor board and a subsequently older patient cohort. As well, Colorado’s Pediatric Neuro-Oncology program had a larger portfolio of consortium-based clinical trials and providers reported frequent prioritization of trial enrollment over off-trial targeted treatment. This difference in CNS TAP tool concordance highlights the different thought processes and practice patterns of institutions and underscores the need for a standardized tool to assist in the clinical decision making of physicians. In addition, Colorado’s practice is more driven by clinical trials as there is a greater accessibility to clinical trials compared to Michigan and their providers occasionally prioritized trial drugs over targeting the pathway with precision medicine agents.
PFS for patients with recurrent brain tumors was favorable compared to historical outcomes in recent therapeutic studies of pediatric patients with refractory or relapsed high-grade is glioma (the predominant patient population in our study), which demonstrate median PFS of 2.5–3 months.[24, 25] Given the retrospective, non-randomized nature of this analysis, it is pre-mature to draw conclusions regarding the efficacy of targeted therapies chosen by the CNS TAP algorithm. Ultimately, the Working Group plans to use the CNS TAP to aid in selection of targeted therapies for patients and then prospectively monitor responses to the agents selected by CNS TAP and compare outcomes to historical controls.
The design and use of our tool brings up several important points. Our data primarily included pediatric patients, which may represent the age group most likely to benefit from precision medicine. Precision medicine therapies may be more effective in tumors with one or few targetable pathways.[8] Recent surveys of pediatric tumors have shown that most are driven by relatively few mutational events, many of which may be targetable with precision medicine therapies.[26, 27] Accordingly, relatively few mutational events were observed in our pediatric cohort.
While extensive literature review and consensus opinion have been performed to provide the basis for this tool, the weights given to each category were based on the subjective judgement of a group of neuro-oncologists and clinical pharmacists. This limitation to the CNS TAP tool is unavoidable at this point due to the limited data and heterogeneous nature of the patient population. When enough data is available, a model utilizing regression coefficients may be utilized to strengthen and optimize the CNS TAP tool. We attempted to address this limitation by assessing a second cohort of patients from an outside institution. However, without a multicenter, comparative, prospective trial assessing clinical outcomes of patients, there is no way to determine whether the agent recommended by CNS TAP algorithm or the agents chosen by the multi-disciplinary conference represent the “correct” agent. Also, given the retrospective nature of the study, the heterogeneity of the patient population, and the small patient numbers, we are unable to compare outcomes in patients who received therapies which would have been recommended by the CNS TAP tool to patients who received alternative therapies with adequate statistical power.
Our group discussed in detail whether agents should receive a higher score based on logistical metrics, including the availability of clinical trials and FDA approval. Our consensus opinion was to continue to prioritize clinical trial enrollment to advance the field. As well, the ability to acquire and utilize therapies with FDA approval (most frequently for a non-brain tumor indication) outside of clinical trial allows for the prioritization of agents with more extensive safety analysis and avoidance of the logistical constraints of compassionate use programs.
Future prospective trials incorporating the CNS TAP tool will ultimately shed light on its full utility. Additionally, new computational methodologies may be considered for incorporation into the tool. Integration of tumor proteomics may improve treatment selection algorithms, as groups have shown alterations in protein signaling can be very powerful predictors of targeted therapy response in pre-clinical models of GBM.[28] As well, genomic analysis of mutational burden and “immuno-phenotype” of tumor and tumor infiltrating immune cells could be incorporated in our algorithm to prioritize immune-based therapies.[29]
CONCLUSION
The CNS TAP tool is a formalized algorithm to assist clinicians select the optimal targeted therapy for neuro-oncology patients based on drug properties, clinical and pre-clinical data, and patient-specific sequencing data. In retrospective analysis, CNS TAP selected targeted therapies that were consistent with clinician/precision medicine conference choice in most cases. Clinical outcomes in patients receiving targeted therapy in this heterogeneous cohort were promising compared with historical outcomes.
Supplementary Material
A finalized copy of a report that is currently returned to a clinician summarizing the data from the CNS TAP tool. (Abbreviations: DIPG = diffuse intrinsic pontine glioma, ACVR = activin receptor, HDAC = histone deacetylase).
Acknowledgments
Funding: C.K. is supported by NIH/NINDS K08-NS099427-01, the University of Michigan Pediatric Brain Cancer Research Initiative. The PEDS-MIONCOSEQ study was supported by grant 1UM1HG006508 from the National Institutes of Health Clinical Sequencing Exploratory Research Award (PI: Arul Chinnaiyan).
The authors thank the patients and their families. Additionally, the authors thank the Michigan Center for Translational Pathology for whole exome and transcriptome tumor sequencing analysis through the PEDS-MIONCOSEQ program.
Footnotes
Conflict of Interest: The authors report no disclosures or conflicts of interests.
References
- 1.Gupta S, Smith TR, Broekman ML. Ethical considerations of neuro-oncology trial design in the era of precision medicine. J Neurooncol. 2017 doi: 10.1007/s11060-017-2502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 3.Brastianos PK, Shankar GM, Gill CM, Taylor-Weiner A, Nayyar N, Panka DJ, Sullivan RJ, Frederick DT, Abedalthagafi M, Jones PS, Dunn IF, Nahed BV, Romero JM, Louis DN, Getz G, Cahill DP, Santagata S, Curry WT, Barker FG. Dramatic Response of BRAF V600E Mutant Papillary Craniopharyngioma to Targeted Therapy. J Natl Cancer Inst. 2016:108. doi: 10.1093/jnci/djv310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norden AD, Drappatz J, Wen PY. Targeted drug therapy for meningiomas. Neurosurg Focus. 2007;23:E12. doi: 10.3171/FOC-07/10/E12. [DOI] [PubMed] [Google Scholar]
- 5.Wang P, Xiao P, Ye Y, Liu P, Han L, Dong L, She C, Yu J. Rapid response of brain metastasis to crizotinib in a patient with KLC1-ALK fusion and MET gene amplification positive non-small cell lung cancer: a case report. Cancer Biol Med. 2017;14:183–186. doi: 10.20892/j.issn.2095-3941.2017.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, Kabuubi P, Ayers-Ringler J, Rabbitt J, Page M, Fedoroff A, Sneed PK, Berger MS, McDermott MW, Parsa AT, Vandenberg S, James CD, Lamborn KR, Stokoe D, Haas-Kogan DA. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:579–584. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lasorella A, Sanson M, Iavarone A. FGFR-TACC gene fusions in human glioma. Neuro Oncol. 2017;19:475–483. doi: 10.1093/neuonc/now240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prados MD, Byron SA, Tran NL, Phillips JJ, Molinaro AM, Ligon KL, Wen PY, Kuhn JG, Mellinghoff IK, de Groot JF, Colman H, Cloughesy TF, Chang SM, Ryken TC, Tembe WD, Kiefer JA, Berens ME, Craig DW, Carpten JD, Trent JM. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro Oncol. 2015;17:1051–1063. doi: 10.1093/neuonc/nov031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marini BL, Benitez LL, Zureick AH, Salloum R, Gauthier AC, Brown J, Wu YM, Robinson DR, Kumar C, Lonigro R, Vats P, Cao X, Kasaian K, Anderson B, Mullan B, Chandler B, Linzey JR, Camelo-Piragua SI, Venneti S, Mc Keever PE, McFadden KA, Lieberman AP, Brown N, Shao L, Leonard MAS, Junck L, McKean E, Maher CO, Garton HJL, Muraszko KM, Hervey-Jumper S, Mulcahy-Levy JM, Green A, Hoffman LM, Dorris K, Vitanza NA, Wang J, Schwartz J, Lulla R, Smiley NP, Bornhorst M, Haas-Kogan DA, Robertson PL, Chinnaiyan AM, Mody R, Koschmann C. Blood-brain barrier-adapted precision medicine therapy for pediatric brain tumors. Translational research: the journal of laboratory and clinical medicine. 2017 doi: 10.1016/j.trsl.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons DW, Roy A, Yang Y, Wang T, Scollon S, Bergstrom K, Kerstein RA, Gutierrez S, Petersen AK, Bavle A, Lin FY, López-Terrada DH, Monzon FA, Hicks MJ, Eldin KW, Quintanilla NM, Adesina AM, Mohila CA, Whitehead W, Jea A, Vasudevan SA, Nuchtern JG, Ramamurthy U, McGuire AL, Hilsenbeck SG, Reid JG, Muzny DM, Wheeler DA, Berg SL, Chintagumpala MM, Eng CM, Gibbs RA, Plon SE. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2015.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, Nikiforova MN. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardridge WM. Molecular Trojan horses for blood-brain barrier drug delivery. Discov Med. 2006;6:139–143. [PubMed] [Google Scholar]
- 13.de Gooijer MC, Zhang P, Thota N, Mayayo-Peralta I, Buil LC, Beijnen JH, van Tellingen O. P-glycoprotein and breast cancer resistance protein restrict the brain penetration of the CDK4/6 inhibitor palbociclib. Invest New Drugs. 2015;33:1012–1019. doi: 10.1007/s10637-015-0266-y. [DOI] [PubMed] [Google Scholar]
- 14.Minocha M, Khurana V, Qin B, Pal D, Mitra AK. Co-administration strategy to enhance brain accumulation of vandetanib by modulating P-glycoprotein (P-gp/Abcb1) and breast cancer resistance protein (Bcrp1/Abcg2) mediated efflux with m-TOR inhibitors. International journal of pharmaceutics. 2012;434:306–314. doi: 10.1016/j.ijpharm.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minocha M, Khurana V, Qin B, Pal D, Mitra AK. Enhanced brain accumulation of pazopanib by modulating P-gp and Bcrp1 mediated efflux with canertinib or erlotinib. International journal of pharmaceutics. 2012;436:127–134. doi: 10.1016/j.ijpharm.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell LK, Scaduto M, Sharp W, Dufton L, Van Slyke D, Whitlock JA, Compas B. A meta-analysis of the neurocognitive sequelae of treatment for childhood acute lymphocytic leukemia. Pediatric blood & cancer. 2007;49:65–73. doi: 10.1002/pbc.20860. [DOI] [PubMed] [Google Scholar]
- 17.Sasongko L, Link JM, Muzi M, Mankoff DA, Yang X, Collier AC, Shoner SC, Unadkat JD. Imaging P-glycoprotein transport activity at the human blood-brain barrier with positron emission tomography. Clin Pharmacol Ther. 2005;77:503–514. doi: 10.1016/j.clpt.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Agarwal S, Shaik NM, Chen C, Yang Z, Elmquist WF. P-glycoprotein and breast cancer resistance protein influence brain distribution of dasatinib. J Pharmacol Exp Ther. 2009;330:956–963. doi: 10.1124/jpet.109.154781. [DOI] [PubMed] [Google Scholar]
- 19.Mody RJ, Wu YM, Lonigro RJ, Cao X, Roychowdhury S, Vats P, Frank KM, Prensner JR, Asangani I, Palanisamy N, Dillman JR, Rabah RM, Kunju LP, Everett J, Raymond VM, Ning Y, Su F, Wang R, Stoffel EM, Innis JW, Roberts JS, Robertson PL, Yanik G, Chamdin A, Connelly JA, Choi S, Harris AC, Kitko C, Rao RJ, Levine JE, Castle VP, Hutchinson RJ, Talpaz M, Robinson DR, Chinnaiyan AM. Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. Jama. 2015;314:913–925. doi: 10.1001/jama.2015.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, Gursky A, Siddiqui J, Tomlins SA, Roychowdhury S, Pienta KJ, Kim SY, Roberts JS, Rae JM, Van Poznak CH, Hayes DF, Chugh R, Kunju LP, Talpaz M, Schott AF, Chinnaiyan AM. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nature genetics. 2013;45:1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, Cheng AJ, Kunju LP, Siddiqui J, Tomlins SA, Wyngaard P, Sadis S, Roychowdhury S, Hussain MH, Feng FY, Zalupski MM, Talpaz M, Pienta KJ, Rhodes DR, Robinson DR, Chinnaiyan AM. Identification of targetable FGFR gene fusions in diverse cancers. Cancer discovery. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson DR, Wu YM, Lonigro RJ, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, Schuetze S, Alva A, Siddiqui J, Chugh R, Worden F, Zalupski MM, Innis J, Mody RJ, Tomlins SA, Lucas D, Baker LH, Ramnath N, Schott AF, Hayes DF, Vijai J, Offit K, Stoffel EM, Roberts JS, Smith DC, Kunju LP, Talpaz M, Cieślik M, Chinnaiyan AM. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duarte TT, Spencer CT. Personalized Proteomics: The Future of Precision Medicine. Proteomes. 2016:4. doi: 10.3390/proteomes4040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruggiero A, Cefalo G, Garre ML, Massimino M, Colosimo C, Attina G, Lazzareschi I, Maurizi P, Ridola V, Mazzarella G, Caldarelli M, Di Rocco C, Madon E, Abate ME, Clerico A, Sandri A, Riccardi R. Phase II trial of temozolomide in children with recurrent high-grade glioma. J Neurooncol. 2006;77:89–94. doi: 10.1007/s11060-005-9011-2. [DOI] [PubMed] [Google Scholar]
- 25.Wetmore C, Daryani VM, Billups CA, Boyett JM, Leary S, Tanos R, Goldsmith KC, Stewart CF, Blaney SM, Gajjar A. Phase II evaluation of sunitinib in the treatment of recurrent or refractory high-grade glioma or ependymoma in children: a children’s Oncology Group Study ACNS1021. Cancer Med. 2016;5:1416–1424. doi: 10.1002/cam4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janeway KA, Place AE, Kieran MW, Harris MH. Future of clinical genomics in pediatric oncology. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:1893–1903. doi: 10.1200/JCO.2012.46.8470. [DOI] [PubMed] [Google Scholar]
- 27.Gajjar A, Pfister SM, Taylor MD, Gilbertson RJ. Molecular insights into pediatric brain tumors have the potential to transform therapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:5630–5640. doi: 10.1158/1078-0432.CCR-14-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei W, Shin YS, Xue M, Matsutani T, Masui K, Yang H, Ikegami S, Gu Y, Herrmann K, Johnson D, Ding X, Hwang K, Kim J, Zhou J, Su Y, Li X, Bonetti B, Chopra R, James CD, Cavenee WK, Cloughesy TF, Mischel PS, Heath JR, Gini B. Single-Cell Phosphoproteomics Resolves Adaptive Signaling Dynamics and Informs Targeted Combination Therapy in Glioblastoma. Cancer Cell. 2016;29:563–573. doi: 10.1016/j.ccell.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, Durno C, Krueger J, Cabric V, Ramaswamy V, Zhukova N, Mason G, Farah R, Afzal S, Yalon M, Rechavi G, Magimairajan V, Walsh MF, Constantini S, Dvir R, Elhasid R, Reddy A, Osborn M, Sullivan M, Hansford J, Dodgshun A, Klauber-Demore N, Peterson L, Patel S, Lindhorst S, Atkinson J, Cohen Z, Laframboise R, Dirks P, Taylor M, Malkin D, Albrecht S, Dudley RW, Jabado N, Hawkins CE, Shlien A, Tabori U. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol. 2016;34:2206–2211. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A finalized copy of a report that is currently returned to a clinician summarizing the data from the CNS TAP tool. (Abbreviations: DIPG = diffuse intrinsic pontine glioma, ACVR = activin receptor, HDAC = histone deacetylase).