Summary
Neuronal maturation requires dramatic morphological and functional changes, but the molecular mechanisms governing this process are not well understood. Here, we studied the role of Rbfox1-3 proteins, a family of tissue-specific splicing regulators mutated in multiple neurodevelopmental disorders. We generated Rbfox triple knockout (tKO) ventral spinal neurons to define a comprehensive network of alternative exons under Rbfox regulation and to investigate their functional importance in the developing neurons. Rbfox tKO neurons exhibit defects in alternative splicing of many cytoskeletal, membrane, and synaptic proteins, and display immature electrophysiological activity. The axon initial segment (AIS), a subcellular structure important for action potential initiation, is diminished upon Rbfox depletion. We identified an Rbfox-regulated splicing switch in ankyrin G, the AIS “interaction hub” protein, that regulates ankyrin G-beta spectrin affinity and AIS assembly. Our data show that the Rbfox-regulated splicing program plays a crucial role in structural and functional maturation of postmitotic neurons.
Keywords: alternative splicing, neuronal maturation, Rbfox, actin cytoskeleton, axon initial segment, AnkG
eTOC Blurb
Jacko et al. identified a comprehensive neuronal Rbfox splicing program, enriched for cytoskeletal, membrane, and synaptic genes. Rbfox1/2/3 triple knockout motor neurons exhibit defects in excitability and axon initial segment assembly, regulated by a developmental splicing switch in Ankyrin G.
Introduction
Newborn neurons in the developing mammalian central nervous system (CNS) undergo a complex process of maturation characterized by the establishment of axo-dendritic polarity, neurite outgrowth, axon initial segment (AIS) assembly, synaptogenesis, and the formation of intricate neural circuits (Barnes and Polleux, 2009; Rasband, 2010). The maturation process needs to be tightly regulated to ensure correct expression of cell-type and cell-stage specific protein repertoire. Alternative splicing (AS), a molecular process that allows the generation of multiple transcript and protein isoforms from a single gene, is increasingly being recognized as a driver of molecular diversity in the CNS (Raj and Blencowe, 2015; Vuong et al., 2016). Transcriptomic studies based on deep RNA sequencing (RNA-seq) demonstrated that usage of alternative exons changes dramatically during brain development (Dillman et al., 2013; Weyn-Vanhentenryck et al., 2018). While the functional significance of the majority of AS events remains unclear, striking examples of individual alternatively spliced exons were shown to control neuronal migration, axon guidance, or synapse formation (Vuong et al., 2016).
To obtain a deeper molecular understanding of how AS regulation contributes to neuronal development and maturation, we focused on the Rbfox family of RNA-binding proteins (RBPs). This family consists of three highly conserved members, Rbfox1 (A2bp1), Rbfox2 (Rbm9) and Rbfox3 (NeuN), specifically enriched in neurons, heart, and muscle (Kuroyanagi, 2009). Mutations in RBFOX genes were reported in patients with autism, schizophrenia, and epilepsy (Barnby et al., 2005; Bhalla et al., 2004; Martin et al., 2007; Sebat et al., 2007; Xu et al., 2008). At the molecular level, Rbfox proteins are known to regulate AS of neuronal transcripts by binding a specific (U)GCAUG sequence element (Jin et al., 2003; Ponthier et al., 2006). Based on the presence of conserved motif sites and the biochemical binding footprints of Rbfox proteins in the mouse brain mapped by crosslinking and immunoprecipitation followed by high-throughput sequencing (HITS-CLIP) (Licatalosi et al., 2008), we predicted over a thousand exons as potential Rbfox targets, many in transcripts with known functions in the adult neurons and muscle (Weyn-Vanhentenryck et al., 2014; Weyn-Vanhentenryck et al., 2018; Zhang et al., 2008).
The function of Rbfox proteins in the nervous system has been studied using genetically engineered mouse models. Rbfox1 knockout (KO) mice are more susceptible to seizures (Gehman et al., 2011), and Rbfox2 KO mice show cerebellar defects and ataxia (Gehman et al., 2012). Yet, despite the large number of predicted targets and biochemically mapped binding sites, only a few dozen transcripts were reported to undergo splicing changes in these single KO models. Even double knockdown of Rbfox1 and Rbfox3 in primary hippocampal neuronal culture resulted in relatively moderate splicing changes, and no cellular phenotypes were reported (Lee et al., 2016). These moderate changes are most likely due to the functional redundancy between Rbfox family members, as all three members are co-expressed in most subtypes of postmitotic neurons (Gehman et al., 2011; Kuroyanagi, 2009), and they bind to the same (U)GCAUG sequence motif in a largely overlapping set of RNAs (Weyn-Vanhentenryck et al., 2014). Thus, the full extent of the Rbfox-regulated transcripts and their impact on neuronal development, function, and disease remain elusive.
The axon initial segment (AIS) is a neuron-specific cytoskeleton-based structure in the proximal part of the axon in vertebrate neurons. This structure is assembled in early post-mitotic neurons in a cell autonomous manner, but the molecular mechanism is incompletely understood. The high density of ion channels clustered in the AIS facilitates the initiation of action potentials, and thereby, the AIS modulates neuronal excitability and plasticity (Rasband, 2010). Recent super-resolution imaging studies suggest that the backbone of the AIS is formed by a periodic cytoskeletal structure composed of actin, βIV-spectrin and ankyrin G (AnkG, encoded by Ank3), with a well-defined 180–190 nm periodicity (Leterrier et al., 2015; Xu et al., 2013; Zhong et al., 2014). The AIS lattice replaces the existing lattice composed of the actin, βII/αII-spectrin and ankyrin B (AnkB, encoded by Ank2) that initially forms along the entire axon. The AIS-specific AnkG protein is a crucial “interaction hub” protein that recruits many of the AIS-localized ion channels and cell adhesion molecules, and its accumulation in the AIS is essential for establishment of the structure. However, the mechanism underlying recruitment and accumulation of AnkG at the AIS is still not well understood.
In this study, we investigated the function of Rbfox proteins and their target transcripts in neurons lacking all three Rbfox genes (triple knockout or tKO). This was achieved by combining CRISPR/Cas9 genome engineering of mouse embryonic stem cells (ESCs) (Cong et al., 2013; Mali et al., 2013) with directed in vitro differentiation of ventral spinal neurons enriched in motor neurons (Wichterle et al., 2002; Wichterle and Peljto, 2008). This system, which mirrors in vivo spinal cord development, allowed us to define the comprehensive Rbfox splicing target network by mapping Rbfox binding sites and profiling splicing perturbations in tKO neurons. Our analysis demonstrated that loss of Rbfox leads to retention of part of the embryonic splicing program, accompanied by immature electrophysiological activity and severe defects in AIS assembly. We discovered that Rbfox-mediated exclusion of an alternative exon in Ank3 transcript increases AnkG affinity to βII- and βIV-spectrins. Constitutive expression of the embryonic (inclusion) splice isoform severely disrupts AnkG accumulation in the proximal axon resulting in a defective AIS. Our study supports a model in which AnkG-β spectrin interaction is a critical step in the AIS assembly and reveals a novel regulatory mechanism relying on an AS event of AIS components.
Results
An in vitro maturation system that mirrors in vivo neuronal development
As pan-neuronal Rbfox1/2 double null mice are not viable (Gehman et al., 2012), we elected to study the function of Rbfox proteins in an in vitro mouse ESC differentiation system that mirrors ventral spinal cord development (Wichterle et al., 2002; Wichterle and Peljto, 2008). To examine neuronal maturation, we cultured differentiated neural cells in the presence of neurotrophic factors, to promote neuronal maturation, and mitotic inhibitors, to prevent expansion of glial progenitors. Neurons rapidly elaborated branched processes and increased their soma size, and by day 5 (D5) after plating most cells fired trains of action potentials upon current injection in patch-clamp experiments (Figure 1A, B), indicative of morphological and electrophysiological maturation (Miles et al., 2004).
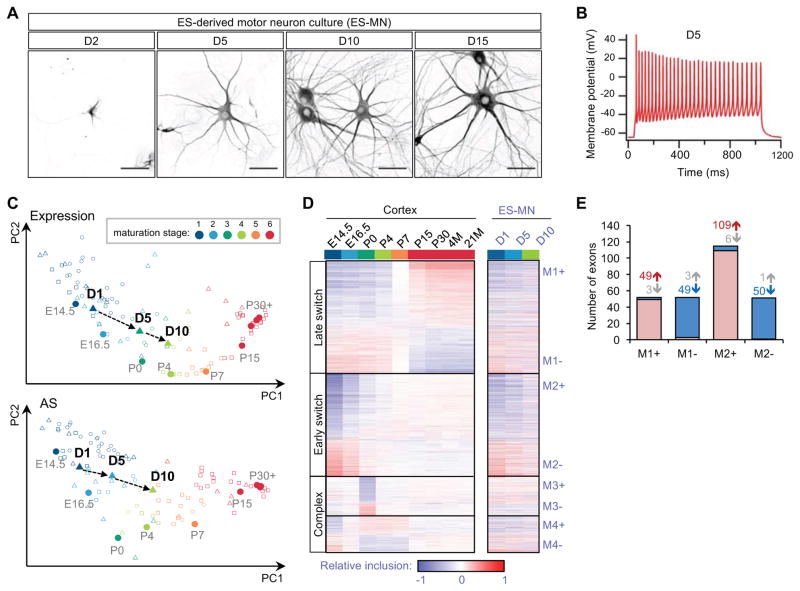
Figure 1. In vitro maturing ventral spinal neurons derived from ESCs undergo developmental splicing changes paralleling those observed in mouse cortex.
(A) Map2 staining shows morphological changes of motor neurons from day 2 (D2) to day 15 (D15) of in vitro culture after plating. Scale bar: 50 μm.
(B) A representative trace from whole-cell patch-clamp recording of maturing motor neuron on day 5 after current injection.
(C) Maturation of ventral spinal neurons is staged by their gene expression (top) or splicing (bottom) profiles using cortex samples as a reference. Transition of maturation stages for motor neurons on day 1, day 5 and day 10 are indicated. Additonal samples derived from different types of neurons or neuronal tissues are also included for comparison.
(D) Four modules of exons show distinct temporal patterns of splicing in developing cortex (left) and spinal neurons (right)(Weyn-Vanhentenryck et al, 2018). These exons were identified and ordered based on their developmental splicing changes in the cortex and their splicing profiles in spinal neurons are shown in the same order. Exons in each module are further divided into two groups (e.g., M1+ and M1-) based on the direction of splicing changes
E) Overlap between exons showing developmental splicing changes in the cortex and exons showing splicing changes in developing spinal neurons. Exons with increased or decreased inclusion in each module and direction are shown separately. The number of exons in each group is indicated.
To characterize the functional maturation process at the molecular level, we performed RNA-seq of the cultured neurons at three time points (day 1, 5 and 10 after dissociation and plating), representing distinct maturation stages based on neuronal morphology and electrophysiology (Table S2). We analyzed the RNA-seq data using Splicescope, a computational tool we recently developed to stage neuronal maturation based on gene expression and splicing profiles, using the developing primary cortex as a reference (Weyn-Vanhentenryck et al., 2018). We found that ESC-derived spinal neurons progressed from stage 1 on day 1 to stage 2 on day 5 and stage 4 on day 10, roughly equivalent to E14.5, E16.5 and P4 mouse cortex (Figure 1C).
Using stringent thresholds (|ΔΨ|≥0.2 and FDR≤0.05), we identified 719 cassette exons with monotonic changes (either increasing or decreasing) in exon inclusion during maturation. We compared these data with developmentally regulated exons identified in the cortex that were clustered into four modules (M1–M4) with distinct temporal profiles (Figure 1D and Table S2; (Weyn-Vanhentenryck et al., 2018)). In total, 270 exons show significant monotonic increase or decrease in exon inclusion in both systems (odds ratio=19.4, p<1.4×10−187, Fisher’s exact test; Figure 1E). Importantly, 95% of these exons exhibit developmental changes in the same direction, with the most pronounced changes observed for exons showing early splicing switches in module M2. Together, these observations suggest that ESC-derived ventral spinal neurons undergo in vitro maturation that parallels the normal development of neuronal tissue in vivo.
Complete depletion of Rbfox in neurons results in dramatic splicing changes with minor impact on the steady-state mRNA level
Taking advantage of CRISPR/Cas9-mediated genome engineering, we targeted ESCs to generate a Rbfox1/2/3 triple knockout (tKO) cell line (Figure 2A and Figure S1A), thus overcoming challenges associated with the functional redundancy of Rbfox proteins. The tKO line showed no apparent defects in expression of the pluripotency marker Oct4 and only a minor reduction in ESC proliferation rate (Figure S1B, C). The Rbfox tKO ESC line differentiated into neurons lacking Rbfox proteins (Figure 2A and Figure S1D) with similar efficiency as the parental wild-type (WT) line (Figure 2B and Figure S1E).
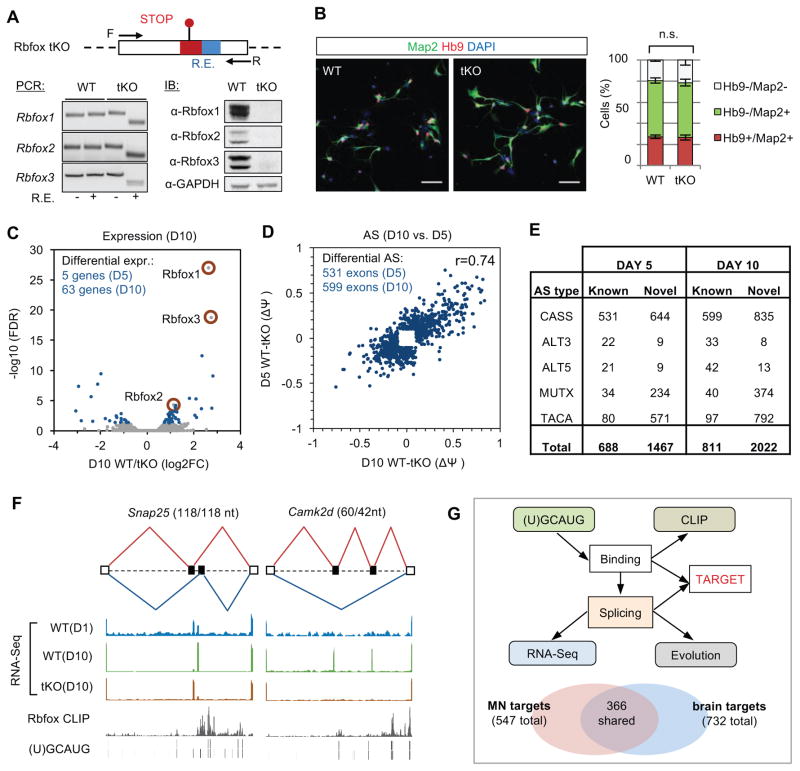
Figure 2. Depletion of Rbfox proteins in ventral spinal neurons perturbs the global splicing program with only minor changes at the steady-state mRNA level.
(A) Schematic illustration of the Rbfox triple knockout (tKO) mouse ES cell line generated by CRISPR/Cas9-mediated genome engineering. Each Rbfox gene is disrupted by an 11-nt insertion that includes in-frame stop codons and a restriction enzyme (RE) digestion site for genotyping. The triple knockout was confirmed by PCR genotyping of ESCs using primers flanking the mutated site and restriction digestion. Depletion of Rbfox proteins was confirmed by immunoblot analysis in in vitro differentiated neurons; GAPDH is a loading control.
(B) Comparison of motor neuron differentiation efficiency from day 2 WT and Rbfox tKO motor neuron culture. Representative immunostaining images are shown on the left. Map2 and Hb9 are pan-neuronal and motor neuron markers, respectively. Scale bar: 50 μm. The result of quantification is shown on the right (two-way ANOVA with post hoc Bonferroni’s multiple test correction, n.s., p>0.05, n=4); error bars represent standard error of the mean (S.E.M.).
(C) Differential gene expression analysis of WT versus Rbfox tKO neurons. Genes with significant changes on day 10 are shown in blue (fold change >1.5, FDR<0.05). Rbfox1, 2 and 3 genes are highlighted.
(D) Differential splicing analysis of annotated cassette exons in WT versus Rbfox tKO neurons. The scatter plot shows the magnitude of splicing changes (ΔΨ) at the two time points. Only exons with significant splicing changes (|ΔΨ|≥0.1 and FDR≤0.05) on day 5 or day 10 are shown. The correlation of splicing changes between the two time points is indicated.
(E) The total number of differentially spliced AS events in WT versus Rbfox tKO neurons (|ΔΨ|≥0.1 and FDR≤0.05). Novel AS events were identified in the mouse cortex transcriptome. CASS, cassette exon; TACA, tandem cassette exons; MUTX, mutually exclusive exons; ALT5, alternative 5′ splice sites; ALT3, alternative 3′ splice sites.
(F) Two examples of Rbfox-regulated alternative exons. RNA-seq, Rbfox1–3 pooled CLIP data and the Rbfox binding (U)GCAUG motif sites are shown as separate tracks in each panel.
(G) Integrative modeling defines direct Rbfox targets. Schematic of the integrative modeling framework is shown at the top. The comparison of Rbfox targets defined using motor neuron culture data and those defined using data derived from mouse brain and other sources (without including motor neuron data) is shown at the bottom.
See also Figures S1–S3.
To experimentally identify transcripts regulated by Rbfox, we performed RNA-seq profiling of WT and tKO neurons matured for 5 and 10 days. It has been reported that Rbfox proteins regulate mRNA stability (Lee et al., 2016), however, we found only a small number of genes (5 on day 5 and 63 on day 10) with significant transcript changes in tKO neurons (>1.5 fold change, FDR<0.05; Figure 2C). In contrast, we detected dramatic perturbation of AS upon Rbfox depletion. We initially focused on annotated cassette exons, the most common type of AS in mammals. Among these, 531 exons on day 5 and 599 exons on day 10 showed significant, well correlated changes (|ΔΨ|≥0.1 and FDR≤0.05; Figure 2D, F and Table S3). Both the number of affected exons and the magnitude of splicing changes observed in tKO neurons are much larger than those observed in recently reported Rbfox1/3 double knockdown hippocampal neurons (Lee et al., 2016) (Figure S2; 548 exons in tKO neurons on day 10 vs. 84 exons upon Rbfox1/3 double knockdown; only exons with sufficient read coverage in both datasets were included), or in Rbfox1 or Rbfox2 single KO mouse brains (Gehman et al., 2012; Gehman et al., 2011). When we extended our analysis to other types of AS events, including the novel ones we identified in the mouse cortex (Yan et al., 2015), we found 2155 events on day 5 and 2833 events on day 10 that show Rbfox-dependent splicing changes (Figure 2E and Table S3), in total affecting over 1100 genes. These data demonstrate the importance of complete Rbfox depletion to uncover the full complement of AS events they regulate. The list of Rbfox-regulated exons thus provides a valuable resource to understand the function of Rbfox proteins and specific AS events in development and disease.
Rbfox proteins can regulate splicing by either directly binding to the (U)GCAUG elements or by being recruited to their target transcripts as a part of a large multiprotein complex (Damianov et al., 2016). To distinguish between the two possibilities, we mapped Rbfox1, 2 and 3 binding sites in day 10 WT neurons using HITS-CLIP. Similar to our previous analysis of mouse brains (Weyn-Vanhentenryck et al., 2014), the binding profiles of the three Rbfox proteins were highly similar to each other (data not shown), allowing us to pool the CLIP data in subsequent analyses. Regulation of exon inclusion or skipping by Rbfox depends on the position of its binding sites relative to the alternative exon (Underwood et al., 2005; Zhang et al., 2008). By overlaying (U)GCAUG motif sites or CLIP tags with Rbfox-dependent exons identified in tKO neurons, we found a substantial enrichment of Rbfox binding sites in the downstream intron of exons with Rbfox-dependent inclusion (Figure S3; see examples in Figure 2F), consistent with direct regulation. Interestingly, the enrichment for Rbfox binding sites in the upstream intron of skipped exons is less pronounced, suggesting that many of these events might be controlled by Rbfox indirectly or as part of a large complex.
To define the list of direct Rbfox target exons with high confidence, we performed integrative modeling of RNA-seq and CLIP data, both derived from the same spinal neuron culture, using a Bayesian network approach (Weyn-Vanhentenryck et al., 2014; Zhang et al., 2010) (Figure 2G). Using a stringent threshold (FDR<0.01), we identified 547 annotated cassette exons as direct Rbfox targets. Among them, about one-third (181 exons) are ventral spinal neuron-specific targets, not predicted in the brain or other examined cell types (Figure 2G bottom and Table S4; (Weyn-Vanhentenryck et al., 2018)). Altogether, the combination of experimental evidence from WT and Rbfox tKO neurons allowed us, for the first time, to define a high-confidence set of alternative exons directly regulated by Rbfox factors.
Rbfox tKO neurons retain an embryonic splicing program and exhibit immature electrophysiological properties
To assess the impact of Rbfox depletion on neuronal maturation at the splicing level, we compared the predicted maturation stages of Rbfox tKO neurons and their WT counterparts using Splicescope analysis. While day 10 WT neurons reached maturation stage 4, Rbfox tKO neurons remained at stage 2 (Figure 3A). In contrast, no changes in maturation stages were inferred from the analysis of steady-state transcript levels, which is consistent with the small number of genes showing expression changes upon Rbfox depletion (Figure 3B).
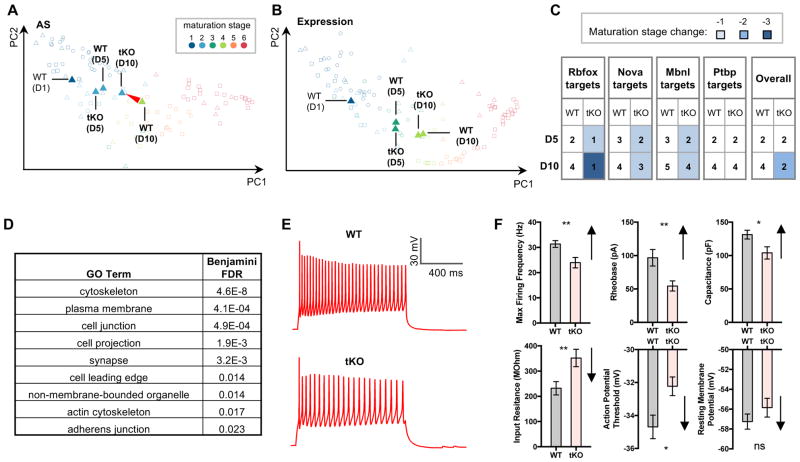
Figure 3. Rbfox tKO neurons retain an embryonic-like splicing profile and immature electrophysiological properties.
(A, B) Comparison of maturation stages of WT and Rbfox tKO neurons based on splicing profiles (A) and gene expression profiles (B).
(C) Maturation of WT and Rbfox tKO neurons is staged using splicing profiles of Rbfox targets. Results from target exons of several other RBPs (Nova, Mbnl and Ptbp) are shown for comparison.
(D) Representative gene ontology (GO) terms enriched in Rbfox target transcripts defined by Bayesian network analysis. See also Table S5 for the complete list.
(E) Whole-cell patch-clamp recording of maturing motor neuron on day 5. Representative traces of action potential firing in WT and tKO motor neurons in response to current injection are shown.
(F) Quantification of whole-cell patch-clamp measurements. For each parameter, the direction of change during neuronal maturation is indicated by the arrow on the right of the bar plot (** p<0.01, * p<0.05, t-test, N=34 in each group). Error bars represent S.E.M.
See also Figure S4.
Because Rbfox proteins regulate only a subset of alternative exons, the impact of Rbfox on the developmental splicing switches of these exons might be diluted in the analysis of the overall splicing profile. Therefore, we also predicted neuronal maturation stages of WT and Rbfox tKO neurons using only direct Rbfox target exons. Using this subset of exons, both day 5 and day 10 tKO neurons were assigned to stage 1, which is more immature compared to the stage inferred from the overall splicing profile, supporting the instrumental role of Rbfox in promoting the mature pattern of the developmental splicing program (Figure 3C). We note that a larger shift in maturation stages inferred from Rbfox targets is not a trivial observation, because this will not occur unless exons are regulated by Rbfox in the same developmental direction (i.e. Rbfox systematically promotes the mature splicing pattern). As a control, when known targets of several other RBPs were used for maturation staging, the difference between WT and tKO neurons diminished, especially after removal of exons co-regulated by Rbfox (Figure S4A). Taken together, these data provide direct experimental evidence that Rbfox proteins are instrumental in establishing the mature RNA splicing program in post-mitotic neurons.
In order to gain insight into the functional consequences of Rbfox-mediated AS, we examined gene ontology (GO) of Rbfox target genes. The target genes were overrepresented in the “cytoskeleton” and “plasma membrane” categories (Figure 3D and Table S5), consistent with the notion that neuronal morphogenesis and electrophysiological maturation involve substantial cytoskeletal and membrane remodeling. Whole-cell patch-clamp recordings confirmed a functional delay in maturation of tKO neurons (Figure 3E, F and Figure S4 B, C), including lower rheobase (a depolarized action potential firing threshold), and a reduction in maximum firing frequency (Ziskind-Conhaim, 1988). Additionally, Rbfox tKO motor neurons exhibited a decrease in cell body growth (Figure S4 D, E), consistent with the observed reduction in capacitance and increase in input resistance.
Rbfox tKO neurons have impaired axon initial segment (AIS) assembly due to disorganization of AnkG
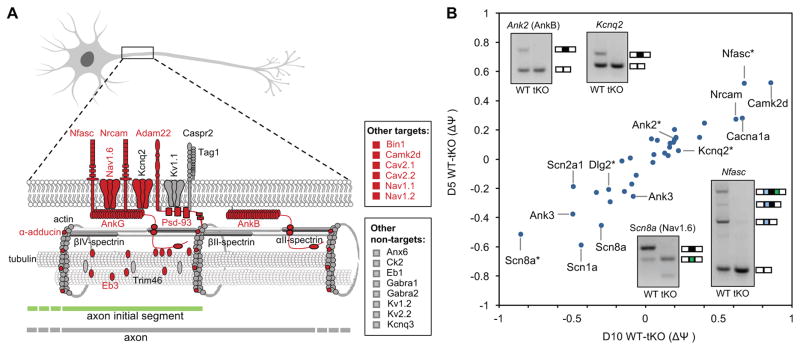
Given the impairment of molecular and electrophysiological maturation of Rbfox tKO neurons, we examined genes involved in development of specific subcellular compartments, such as axons and dendrites. Strikingly, we found that 16 of 32 (50%) genes encoding AIS-enriched proteins contain at least one Rbfox-dependent alternative exon (p<3.3×10−6; Binomial test; Figure 4A, B and Table S6). These genes include the core AIS components Ank3 (AnkG), sodium and potassium channels (Nav1.6/Scn8a, Nav1.2/Scn2a1, Kcnq2), adhesion molecules (neurofascin/Nfasc, Nrcam, Adam22), and other components (AnkB/Ank2, Camk2d, and Ppp3ca).
Figure 4. Rbfox-regulated exons are enriched in genes encoding regulators and components of the AIS.
(A) Genes encoding defined AIS components are shown in the schematic diagram. Genes with Rbfox-regulated exons as defined by Bayesian network and RNA-seq are indicated in red. Additional AIS genes reported in the literature are also shown on the right.
(B) Magnitude of Rbfox-dependent splicing changes in AIS genes as measured by RNA-seq and RT-PCR validation. Two deregulated alternative exons of Scn8a and Ank3 are shown in the scatter plot separately. Exons with asterisks in the scatter plot correspond to those tested by RT-PCR.
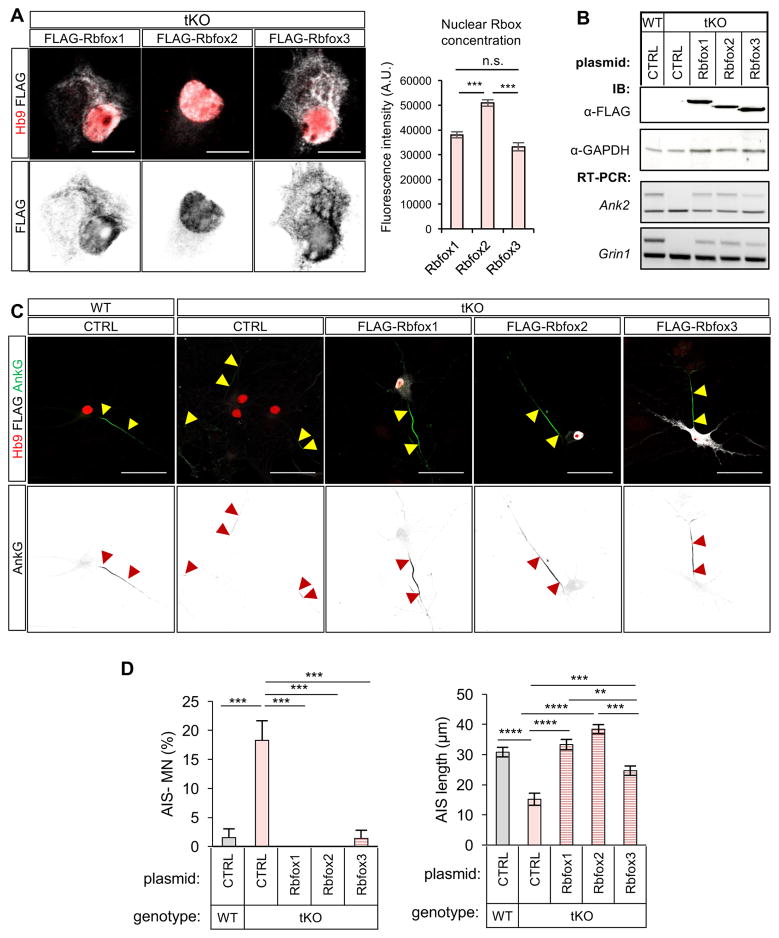
Intrigued by this observation, we tested whether Rbfox tKO neurons exhibit defects in AIS formation by examining the AnkG localization in WT and tKO neurons. We initially focused on motor neurons, distinguished from the other spinal neurons by expression of the Hb9 (Mnx1) transcription factor (Wichterle et al., 2002), to avoid potential differences in the maturation of individual neuronal subtypes. Compared to WT controls, AnkG immunostaining in tKO motor neurons is more heterogeneous (Figure 5A). Only 56% of tKO motor neurons on day 2 and 65% on day 5 have AnkG positive segments, as compared to 78% and 95% for WT motor neurons at the same time points (p<0.001, t-test; Figure 5B left panel). Moreover, even when we limited our analysis to motor neurons with detectable AIS (AnkG staining >5μm), the length of AIS in Rbfox tKO neurons was significantly reduced at all examined time points (p<0.001, t-test; Figure 5B right panel). The deficit in AnkG accumulation in AIS is not due to a global decrease in AnkG protein expression (Figure S5A) or an impairment of axon specification (as revealed by Tau-1 immunostaining; Figure 5C and Figure S5B) in tKO motor neurons. In addition to AnkG, we examined two additional AIS markers, βIV-spectrin and the potassium channel Kcnq3, which do not contain Rbfox-regulated alternative exons yet show a reduction in tKO motor neurons, indicating that the entire AIS structure is impaired (Figure 5D and Figure S5C, D). The AIS defect is not limited to motor neurons, as similar analysis performed on Hb9-negative ventral spinal interneurons revealed a higher proportion of tKO interneurons lacking AnkG staining (25% of tKO vs. 5% of WT interneurons; p<0.01, t-test) and interneurons containing AIS exhibited a significantly shorter AnkG positive segment (16.5 μm in tKO vs. 24.6 μm in WT interneurons; p<0.001, t-test; Figure S5E, F). These results suggest that most, if not all, neurons rely on Rbfox proteins for effective AIS assembly.
Figure 5. Rbfox tKO motor neurons show severe pertubation of AIS and AnkG organization.
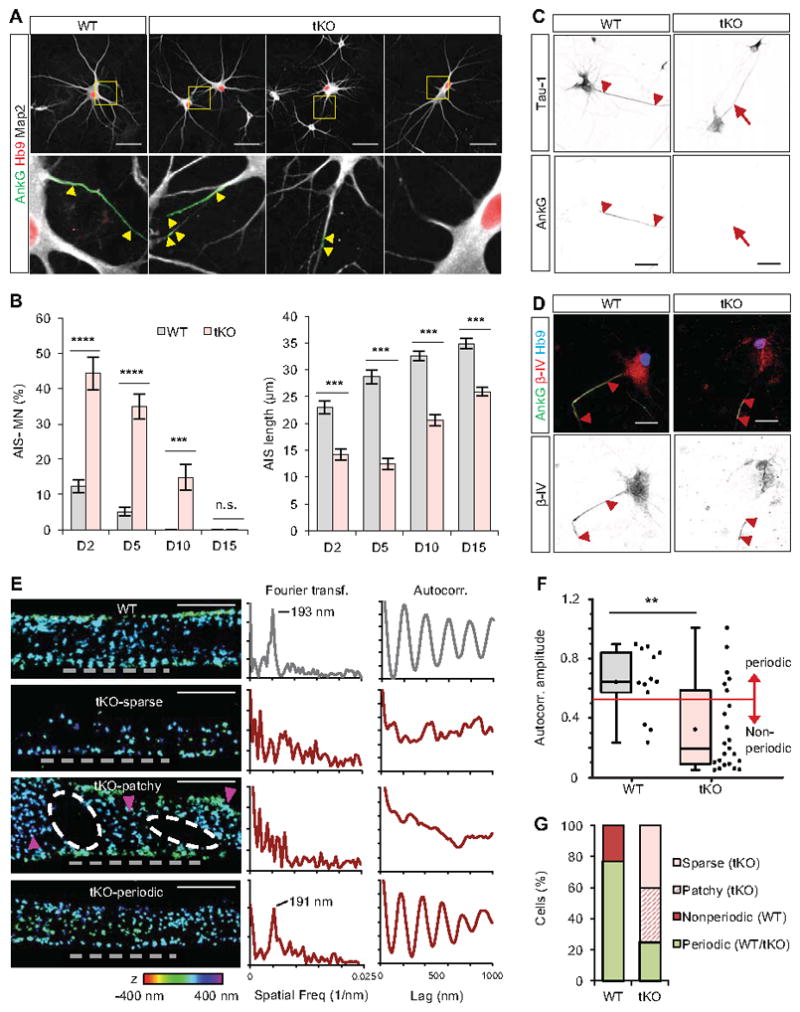
(A) Immunostaining analysis showing AIS morphology in day 5 WT and Rbfox tKO motor neurons. Motor neurons are identified by Hb9 staining. The arrowheads mark the beginning and the end of the AIS. Scale bar: 50μm.
(B) Quantification of WT and Rbfox tKO motor neurons that lack AnkG acumulation in the proximal axon (left) or have reduced AIS length based on AnkG staining (right) during maturation (two-way repeated measures ANOVA, post hoc Bonferroni’s multiple comparisons test; ****p<0.0001, ***p<0.001, **p<0.01, n.s. p>0.05; N ranges from 45 to 91 for each group). Error bars represent S.E.M.
(C, D) Immunostaining of axonal marker Tau-1 (C) and βIV-spectrin (D) on day 5 WT and tKO motor neurons. Scale bar: 50 μm.
(E) Representative 3D-STORM images and quantitative analysis of AnkG in the AIS in day 10 WT and tKO neurons. Left: 3D-STORM images of immunolabeled AnkG in AIS. tKO results are classified into sparse, patchy and periodic patterns. Alternation of low and high AnkG density regions in the patchy pattern is indicated by the white ovals and magenta arrowheads, respectively. The color scale used to indicate depth in z is shown at the bottom. Scale bars: 1 μm. Middle and right: Fourier transform (middle) and autocorrelation analyses (right) of AnkG distribution in the indicated regions of AIS in the left panel (grey dashed lines).
(F) Box plot of average autocorrelation amplitude of AnkG in all WT and tKO AIS (p=0.005, Wilcoxon-Mann-Whitney test, N=13 AIS for WT and N=23 for tKO neurons). The red line indicates the threshold used to classify periodic vs. non-periodic AIS.
(G) Stacked column graph showing fractions of AnkG patterns out of all analysed AIS.
See also Figures S5 and S6.
Rbfox-regulated genes include actin cytoskeleton stability regulators α-adducin (Add1) and tropomyosin alpha-1 chain (Tpm1) (Figure 4A and Tables S3 and S4), raising the possibility that the defect in AnkG recruitment might be secondary to a global disruption in the axonal actin-spectrin cytoskeletal lattice. We used three-dimensional stochastic optical reconstruction microscopy (3D-STORM) (Huang et al., 2008; Rust et al., 2006) to visualize and quantify the distribution of individual AnkG and actin molecules with ~20 nm spatial resolution. 3D-STORM imaging of day 10 motor neurons revealed that the periodic arrangement of actin is intact in the AIS region of tKO motor neurons and comparable to its WT counterparts (Figure S6A, B). In sharp contrast, the AnkG localization pattern in tKO motor neurons was severely disrupted, with only 25% of neurons showing AnkG periodicity as compared to 77% of WT motor neurons. 40% of tKO motor neurons exhibited substantially sparser distribution of AnkG without local periodicity, and 35% exhibited alternating patterns of locally high and low AnkG density, which we denote as “patchy” patterns that were also observed previously and might be related to clustering of Kv2.1 channels (King et al., 2014; Leterrier et al., 2015) (Figure 5E–G and Figure S6C). To quantify the orderliness of periodicity, we performed a Fourier transform (Xu et al., 2013) and autocorrelation analysis (Zhong et al., 2014) to determine the presence and degree of periodicity, respectively. For most WT motor neurons and a small fraction of “periodic” tKO neurons, Fourier transform showed well-defined peaks around the spatial frequency corresponding to 190 nm spacing, a value similar to previous reports (Leterrier et al., 2015; Zhong et al., 2014) (Figure 5E and Figure S6D). Overall, WT neurons had a significantly higher degree of AnkG periodicity than tKO neurons, even with the “periodic” tKO subtype included (P<0.005, Wilcoxon-Mann-Whitney test; Figure 5F). Taken together, our data suggest that the observed AIS defect caused by Rbfox depletion is not due to a global disruption of the axonal cytoskeleton, but rather due to a specific impairment of AnkG integration.
AIS defects in Rbfox tKO neurons is due to loss of the nuclear function of Rbfox
While half of the genes encoding AIS components contain Rbfox-dependent AS exons, Rbfox has been also shown to control mRNA stability and translation (Carreira-Rosario et al., 2016; Lee et al., 2016). To investigate which Rbfox function accounts for the AIS phenotype, we took advantage of the fact that Rbfox2 is localized strictly to the nucleus while Rbfox1 and Rbfox3 are also present in the cytoplasm of neurons (Lee et al., 2016). We overexpressed a 3×FLAG-tagged version of Rbfox1, Rbfox2, or Rbfox3 in tKO neurons and characterized the neurons 5 days after transfection. Quantification of immunofluorescence intensity confirmed localization of Rbfox2 to the nucleus (Figure 6A, B). While all Rbfox factors rescued AIS assembly, the level of rescue correlated with the degree of nuclear localization (Figure 6A, B). The AIS length was rescued most prominently upon overexpression of the nuclear Rbfox2 (Figure 6C, D), or by overexpression of an Rbfox1 splice isoform with a bias towards nuclear vs. cytoplasmic localization (Lee et al., 2009) (Figure S7). These experiments indicate that the nuclear function of Rbfox is more important for proper AIS assembly than its cytosolic function.
Figure 6. Rescue of AIS defects in Rbfox tKO neurons by overexpression of individual Rbfox protein.
(A) Subcelullar localization of the exogenously expressed Rbfox proteins in motor neurons. Representative immunostaining images are shown on the left (scale bar: 10 μm) and quantification of nuclear protein concentration based on fluorescence intensity is shown on the right (***p<0.001, t-test, N ranges from 60 to 63 for each group).
(B) Immunoblot analysis to validate expression of FLAG-tagged Rbfox proteins after plasmid transfection (top). Rescue of splicing of two representative Rbfox target exons is also shown at the bottom.
(C) Representative images for AIS analysis upon overexpression of Rbfox1, Rbfox2 and Rbfox3 in Rbfox tKO motor neurons, as well as WT and tKO control. Scale bar: 50 μm.
(D) AIS quantification in control WT and Rbfox tKO motor neurons, and Rbfox tKO motor neurons upon overexpression of individual FLAG-tagged Rbfox protein (one-way ANOVA with post hoc Tukey’s multiple test correction; ****p<0.0001, ***p<0.001, **p<0.01; N ranges from 60 to 63 for each group). Error bars represent S.E.M.
See also Figure S7.
An Rbfox-dependent developmental splicing switch in AnkG is critical for normal AIS assembly
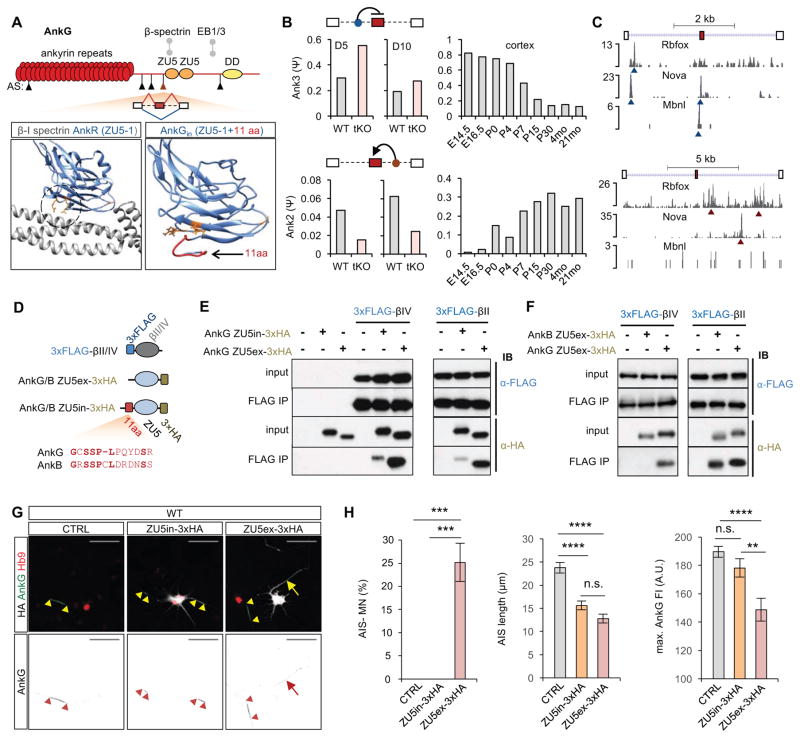
We noted that Ank3 (AnkG) itself contains five Rbfox-dependent alternative exons (Figure 7A), raising the possibility that these exons might contribute to AnkG recruitment to the AIS. A 33-nt cassette exon encoding a peptide immediately upstream of the first (N-terminal) ZU5 domain (ZU5-1) was particularly interesting. This exon showed progressive exclusion in both the maturing cortex and ESC-derived ventral spinal neurons. Rbfox and two other tissue-specific splicing factors Nova and Mbnl bind the upstream intron, consistent with the observed skipping of the exon at the time of the AIS formation (Figure 7B, C). The ZU5-1 domain downstream of the alternatively spliced exon is required for anchoring ankyrin to the underlying actin cytoskeleton via interaction with β-spectrin (Ackerman et al., 1997; Leonardo et al., 1997). Structural modeling predicted that the peptide encoded by the alternative exon is located proximally to the two crucial arginines engaged in the interaction with spectrin repeat 14 in β-spectrins (Figure 7A, bottom). Interestingly, despite a substantial sequence divergence, a homologous cassette exon is also present in Ank2 (AnkB), but in contrast to the exon in Ank3, this exon shows progressive inclusion during development (Figure 7B–D).
Figure 7. Inclusion of a developmentally regulated alternative exon upstream of AnkG ZU5-1 domain negatively affects interaction of AnkG with βIV- and βII-spectrins.
(A) Top: A schematic illustration of AnkG protein domains. Arrowheads indicate five alternative exons differentially spliced in Rbfox tKO neurons on day 5 or day 10. The candidate 33-nt cassette exon immediately upstream of the ZU5-1 domain is highlighted in red. Bottom: Structural modelling of the ZU5-1/β-spectrin complex. A published structural model of βI-spectrin repeats 13–15 (gray) and AnkR ZU5-1 (blue) domain (PDB ID: 3KBT) is shown on the left and structural prediction of AnkG ZU5-1 (blue) including the segment encoded by the candidate alternative exon (red) is shown on the right. The three crucial interaction interface residues (two arginines at the bottom and one alanine on the right) are highlighted in orange in both models.
(B) RNA-seq quantification of the candidate cassette exon in Ank3 (top) and its homolog in Ank2 (bottom) in WT and Rbfox tKO neurons on day 5 and day 10 (left) and in developing mouse cortex (right).
(C) CLIP tags indicating positions of Rbfox, Mbnl2 and Nova binding sites in Ank3 (top) and Ank2 (bottom) in the alternatively spliced region are shown and the strongest peaks are indicated by blue and red arrowheads, depending on their positions relative to the alternative exon.
(D) Shematics of constructs for co-immunoprecipitation of AnkG/AnkB ZU5–1 domains with βII-/βIV-spectrin repeats 13–15 overexpressed in NIH/3T3 cells. The peptide encoded by the candidate alternative exon in AnkG and its homolog in AnkB is also shown.
(E, F) Immunoblot analysis of the co-immunoprecipitation experiments. The + and – signs denote the combination of the transfected plasmid constructs. In all experiments, anti-FLAG antibodies were used for immunoprecipitation. (E) Comparison of AnkG inclusion and exclusion isoforms for β-spectrin binding. (F) Comparison of AnkG and AnkB exclusion isoforms for β-spectrin binding.
(G) Immunostaining analysis showing AIS in WT motor neurons transfected with AnkG ZU5in-3×HA or AnkG ZU5ex-3×HA plasmid vector. AIS is stained using AnkG antibody that detects only the endogenous protein, and the overexpressed ZU5-1 domain isoforms are detected by anti-HA antibody. The arrowheads mark the beginning and the end of the AIS. The arrow indicates an HA-positive motor neuron showing weak and distributed AnkG staining.
(H) Quantification of AIS presence and length based on AnkG staining (one-way ANOVA with post hoc Tukey’s multiple test correction; ****p<0.0001, ***p<0.001, **p<0.01, n.s. p>0.05, N ranges from 47 to 70 for each group). Error bars represent S.E.M. Scale bar: 50 μm.
These observations raised the possibility that retention of the 33-nt exon in tKO neurons might play a role in the AIS assembly by interfering with the interaction between AnkG and β-spectrins. To test the role of the alternatively spliced exon, we co-expressed HA-tagged AnkG ZU5-1 domain containing the 33-nt exon (ZU5in-3×HA) or the mature isoform lacking the exon (ZU5ex-3×HA) together with FLAG-tagged βII/IV-spectrin repeats 13–15 (3×FLAG-βII and 3×FLAG-βIV) in NIH/3T3 cells (Figure 7D). Co-immunoprecipitation analysis revealed that the mature AnkG ZU5ex isoform co-immunoprecipitated with βII- and βIV-spectrins (both of which are present in the AIS (Figure S6E–G) (Zhong et al., 2014)) but AnkB ZU5-1 co-immunoprecipitated only with βII-spectrin (Figure 7E, F). Importantly, inclusion of the alternatively spliced exon resulted in a significantly weaker interaction of the immature AnkG ZU5in isoform with both βII- and βIV-spectrin, consistent with the prediction that the exon weakens the ZU5-1 dependent spectrin interaction (Figure 7E).
To probe the relevance of the ZU-5-1 domain for AnkG recruitment and AIS assembly, we overexpressed the two AnkG ZU5-1 domain isoforms in motor neurons. The ZU5-1 domain is the minimal functional unit sufficient for interaction with β-spectrins, and it does not include any additional known functional domains ((Freal et al., 2016; He et al., 2012); also see Discussion). Therefore, overexpression of the ZU5-1 domains alone was expected to act in a dominant negative fashion by competing for β-spectrin binding with the endogenous AnkG, while minimizing additional perturbations. Four days after transfection, we observed a significant increase in the number of neurons lacking AIS and in neurons with shorter AIS characterized by weaker AnkG staining intensity (Figure 7G, H). Importantly, the overexpression of ZU5ex-3×HA isoform, which interacts more strongly with β-spectrins, resulted in a more severe phenotype with 25% of motor neurons showing a complete loss of AnkG staining in the AIS, while ZU5in-3×HA expressing motor neurons all had detectable AnkG staining (Figure 7H). These findings are consistent with the notion that ZU5-1 mediated interaction of AnkG with β-spectrins is important for accumulation of AnkG in the AIS of maturing neurons.
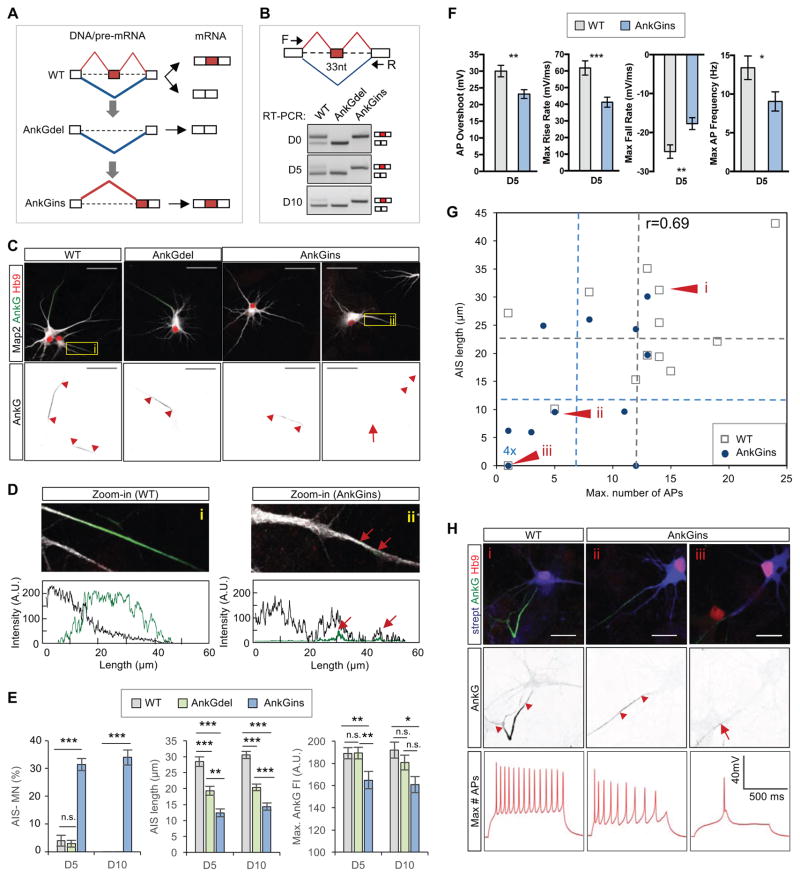
To directly test the importance of this developmentally regulated exon in AnkG, we used CRISPR/Cas9 to delete the exon in ESCs (denoted AnkGdel) and to re-insert it next to the downstream constitutive exon (denoted AnkGins), which allowed us to force the expression of the adult or embryonic isoform during motor neuron maturation (Figure 8A, B and Figure S8A, B). Constitutive expression of the embryonic AnkGins resulted in an AIS phenotype that was even stronger than the one observed in Rbfox tKO neurons. Over 30% of AnkGins motor neurons lacked AnkG accumulation in the proximal axon, and the remaining cells displayed significant reduction in AIS length and in AnkG staining intensity (Figure 8C–E and Figure S8C). In addition, the AnkGins mutant neurons without AIS displayed distributed puncta of low-intensity AnkG staining along the axon and exhibited an impairment of Map2 restriction in the somato-dendritic compartment (Figure 8D). Together our findings indicate that a defect in splicing of a single exon in AnkG is sufficient to disrupt the assembly of AIS, recapitulating a key phenotype observed in Rbfox tKO neurons.
Figure 8. Deregulation of the alternative exon upstream of AnkG ZU5-1 domain negatively affects AIS assembly.
(A) A schematic shows generation of ESC lines for constitutive expression of the AnkG exclusion (AnkGdel) or AnkG inclusion (AnkGins) isoform by deletion and subsequent re-insertion of the candidate alternative exon in the Ank3 (AnkG) gene.
(B) RT-PCR analysis of the candidate exon inclusion in WT, AnkGdel and AnkGins maturing neurons.
(C) Immunostaining analysis showing AIS in day 5 WT, AnkGdel and AnkGins motor neurons. The red arrowheads mark the beginning and the end of AIS. The arrow shows an axon with weak and distributed AnkG staining. Scale bar: 50 μm.
(D) Map2 restriction in somatodendritic compartment is perturbed in AnkGins motor neurons without AIS. Zoom-in views of the boxed area in panel (C) are shown at the top and respective axonal tracing fluorescence intensity profiles for Map2 and AnkG are at the bottom. The red arrows indicate AnkG puncta in the axon.
(E) Quantification of AIS presence and length in WT, AnkGdel and AnkGins motor neurons on day 5 and day 10 based on AnkG staining (two-way ANOVA with post hoc Tukey’s multiple test correction; ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05; N equals 66 for both groups). Error bars represent S.E.M.
(F) Quantification of significantly changed electrophysiological characteristics from whole-cell patch-clamp measurements of WT and AnkGins motor neurons on day 5. (t-test, ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05; N ranges from 25 to 28 per group). Error bars represent S.E.M.
(G) Correlation of action potential firing and AIS length in WT and AnkGins motor neurons. Post hoc quantification of AIS was performed in motor neurons based on AnkG staining after whole-cell patch-clamp recording. The gray and the blue datapoints represent individual WT and AnkGins motor neurons, respectively with the dotted lines indicating the average values.
(H) Immunostaining and whole-cell patch clamp recordings showing the maximum number of fired action potentials after current injection for representative motor neurons, denoted i, ii and iii in panel (G).
See also Figures S9 and S10.
See also Figure S8.
To determine whether defects associated with aberrant splicing of Ank3 alone are sufficient to disrupt firing properties of maturing neurons, we performed whole-cell patch-clamp recordings of AnkGins motor neurons followed by post hoc immunostaining analysis of AnkG and Hb9. AnkGins neurons fired fewer action potentials, the action potential overshoot was reduced, and the duration was increased as a result of reduction in both the action potential rise and fall rates (Figure 8F and Figure S8D), indicative of a reduction in the AIS ion channel densities. Furthermore, the level of the AIS perturbation as measured by AIS length correlated well with the degree of the firing reduction (Pearson r=0.69, p<3.8×10−5, correlation test; Figure 8G, H and Figure S8E). These data demonstrate that the developmental splicing switch in AnkG is required for the establishment of mature neuronal firing properties.
Immunoprecipitation studies established that ZU5-1 domain lacking the alternative exon interacts strongly with β-spectrins, raising the possibility that forced exclusion of the 33-nt exon might lead to the formation of a more robust AIS. In contrast to the severe AIS defects observed in AnkGins motor neurons, almost all motor neurons with constitutive expression of the mature AnkGdel isoform in the WT background have detectable AIS, and the intensity of AnkG staining in the proximal AIS region is also comparable to that of WT control (Figure 8C, E). Interestingly, AnkGdel motor neurons exhibited a shortening of the AIS (Figure 8E), indicating that the formation of normal AIS depends on a developmentally regulated balance between the immature and mature isoforms of alternatively spliced AnkG. Finally, we examined whether expression of the mature AnkG isoform would be sufficient to rescue AIS defects observed in Rbfox tKO neurons. Constitutive expression of AnkGdel significantly increased AnkG staining intensity in the AIS (p<0.001, one-way ANOVA; Figure S8F, G), and resulted in a slight increase in the number of neurons with AIS and in the length of AIS, although these differences did not reach statistical significance. Together these results suggest that deregulation of a single 33-nt exon in AnkG is an important, but not the only, contributor to the AIS phenotype observed in Rbfox tKO neurons.
Discussion
Here we studied the functional role of Rbfox proteins in neuronal maturation at the molecular and cellular levels. Rbfox proteins are well established splicing factors that have been extensively studied in different cellular contexts, including brain, cardiac and skeletal muscle, stem cells, and cancer (see reviews by (Conboy, 2017; Kuroyanagi, 2009)). However, Rbfox-regulated transcripts in neurons are largely undetermined and their function, especially at the cellular level, is poorly defined. This is due in large part to the co-expression of three redundant family members in neurons, which makes it challenging to perform experimental perturbation in a physiologically relevant cellular context.
We overcame this challenge by using CRISPR/Cas9 mediated genome engineering in ESCs in combination with directed differentiation of ESCs to defined neuronal cell populations, which allowed us to generate Rbfox tKO neurons for the first time. We observed much more dramatic splicing changes in tKO neurons, compared to mouse brain tissue lacking individual Rbfox family members (Gehman et al., 2012; Gehman et al., 2011) or primary neuronal culture with simultaneous knockdown of Rbfox1 and Rbfox3 (Lee et al., 2016) (Figure S2). Using this system, we identified Rbfox-dependent splicing in over 1,100 genes using stringent criteria, including 1,175 known and novel cassette exons in day 5 neurons and 1,434 cassette exons in day 10 neurons (Figure 2E). Further integrative modeling of RNA-Seq data, CLIP data and Rbfox-binding motif sites allowed us to define 547 previously annotated cassette exons as direct Rbfox targets, and a comparable number of direct Rbfox targets among novel cassette exons (Weyn-Vanhentenryck et al. unpublished data). We note that another study knocked down Rbfox1 during differentiation of human neural stem cells and reported alteration of over 900 cassette exons (Fogel et al., 2012). However, these exons were identified using less stringent criteria (P<0.05 without multiple test correction or filtering on ΔΨ) and none of the changes remained statistically significant after multiple test correction. Therefore, the list of Rbfox-regulated exons reported in this study represents the most comprehensive and robust set so far identified in the neuronal contexts, which will be a valuable resource for studying the physiological function of Rbfox proteins and their downstream targets in the nervous system and other tissues.
We found that Rbfox-regulated exons are enriched in genes involved in cytoskeletal remodeling, neurite outgrowth, and synaptic formation or function. Rbfox tKO neurons offer a unique opportunity to elucidate the functional consequences caused by disruption of these alternative exons during neuronal maturation. We determined that Rbfox tKO neurons exhibit morphological and electrophysiological features that resemble immature neurons, which is consistent with the failure to properly execute part of a developmental splicing program. Further analysis of Rbfox-regulated alternative exons led to an unexpected discovery that Rbfox factors control AS of about half of characterized AIS components and play a crucial role in its assembly and function. Super-resolution microscopy imaging of AIS components revealed that the AIS defects observed in Rbfox tKO neurons are not due to general disruption of the periodic actin cytoskeletal lattice, but result from the failure of AnkG to integrate into the lattice. The AIS defects are consistent with our observation that Rbfox tKO neurons are less excitable compared to WT controls. Interestingly, neuronal excitability was also reported to be reduced in Purkinje cells upon simultaneous cell-type-specific depletion of Rbfox1 and Rbfox2 (Purkinje cells do not express Rbfox3) (Gehman et al., 2012), but the cause of this defect has not been determined. Our study suggests a novel AS-based mechanism to modulate neuronal excitability by regulating the AIS assembly.
We pursued a detailed analysis of an Ank3 (AnkG) alternative exon based on its developmental splicing pattern and its predicted role in modulating ankyrin-spectrin interaction. This alternative exon has also diverged from a homologous exon in Ank2 (AnkB) in terms of both sequence and the direction of splicing regulation (maturation-dependent exclusion in Ank3 vs. inclusion in Ank2), suggesting that the AnkG exon might have evolved a unique role in modulating AnkG insertion into the cytoskeleton and its function in maturing neurons. Indeed, the importance of this exon was supported by multiple lines of evidence demonstrating its role in the modulation of AnkG affinity to βII- and βIV-spectrins. Most strikingly, the AIS defects and disruption of action potential firing were largely recapitulated upon forced expression of the immature AnkG inclusion isoform in WT neurons. Unexpectedly, some of the defects observed in tKO neurons were corrected in more mature neurons. Even the splicing change of the AnkG exon is larger on day 5 than on day 10, indicating compensatory splicing-regulatory mechanisms that might be mediated by other splicing factors, like Nova or Mbnl. Indeed, the constitutive inclusion of the alternatively spliced exon in AnkGin neurons resulted in more severe AIS defects than the one observed in Rbfox tKO neurons, providing further support to the notion that the described AIS phenotypes can be to large extent attributed to the defective splicing of AnkG.
Combining what is known from previous studies and the current work, we propose the following model of AIS assembly. The AIS assembly is preceded by establishment of the axonal membrane cytoskeleton consisting of the AnkB-βII-spectrin-actin periodic lattice (Zhong et al., 2014) (Figure S6F–G). This is followed by recruitment and accumulation of AnkG in the proximal axon. Multiple mechanisms likely contribute to this process, including direct AnkG interaction with βII-spectrin (as observed in this study and reported in the lateral membrane of epithelial cells (He et al., 2014)), with the microtubule end-binding proteins EB1/3 (Freal et al., 2016; Jenkins et al., 2015; Leterrier et al., 2011), and with the plasma membrane through its N-terminal palmitoylation (He et al., 2012). Subsequently, AnkG recruits ion channels, cell adhesion molecules, and βIV-spectrin that partially replaces βII-spectrin in the proximal axon, thus forming the mature AIS (Hedstrom et al., 2008). This model is consistent with the observation that recruitment of AnkG and AIS assembly are disrupted by depletion of βII spectrin (Galiano et al., 2012), but not by depletion of βIV-spectrin (Hedstrom et al., 2007; Yang et al., 2007). Genetic ablation of AnkG impairs the recruitment of its binding partners and leads to failure of AIS establishment (Hedstrom et al., 2007). Previous studies established the importance of the interaction between AnkG ZU5-1 domain and βIV-spectrin for the long-term stability of AIS (Komada and Soriano, 2002; Uemoto et al., 2007). Our data extend the existing model of AIS assembly by showing that AnkG is able to bind to both βIV- and βII-spectrin via the ZU5-1 domain, and importantly, that the interaction affinity is developmentally modulated by the Rbfox-regulated alternative exon in AnkG. Skipping of the exon during early stages of neuronal maturation allows high-affinity interaction of AnkG with βII-spectrin that likely contributes to initial recruitment and stabilization of AnkG in the proximal axon by its tethering to the actin-spectrin cytoskeletal lattice. Therefore, Rbfox-proteins play an important role in the establishment of AIS, demonstrating the importance of AS as a powerful mechanism to modulate the dynamic, highly regulated process of neuronal maturation.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-Rbfox1 (anti-Fox1) D8F8 | EMD Millipore | MABE159 |
| rabbit anti-Rbfox2 (anti-Rbm9) | Bethyl Laboratories | A300-864A |
| mouse anti-Rbfox3 (anti-NeuN) A60 | EMD Millipore | MAB377 |
| mouse anti-Map2 AP-20 | EMD Millipore | MAB3418 |
| rabbit anti-AnkG H-215 | Santa Cruz Biotech | Sc-28561 |
| mouse anti-AnkG 463 | Santa Cruz Biotech | Sc-12719 |
| mouse anti-bIV-spectrin | Neuromabs | N393/76 |
| guinea pig Hb9 | Gift from Thomas M. Jessell (Columbia U.) | N/A |
| Rabit anti-Kcnq3 | Alomone | APC-051 |
| mouse anti-tau-1 PC1C6 | EMD Millipore | MAB3420 |
| mouse anti-FLAG M2 | Sigma-Aldrich | F3165 |
| goat anti-Oct4 | Abcam | ab27985 |
| rabbit anti-GAPDH FL-335 | Santa Cruz Biotech | Sc-25778 |
| Alexa Fluor 647 goat anti-mouse | Life Technologies | A21236 |
| Alexa Fluor 555 goat anti-guinea pig | Life Technologies | A21435 |
| custom-labeled Alexa Fluor 488 donkey anti-guinea pig | Jackson ImmunoResearch | N/A |
| Alexa 488-conjugated, Cy3-conjugated and Cy5-conjugated donkey secondary antibodies | Life Technologies | - |
| NeutrAvidin, Texas Red conjugate | ThermoFisher Scientific | A2665 |
| HA-Tag (6E2) Mouse mAb (HRP Conjugate) | Cell Signaling Technology | #2999 |
| Monoclonal ANTI-FLAG® M2-Peroxidase (HRP) antibody produced in mouse | Sigma-Aldrich | A8592 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Human BDNF | PeproTech | 450-02 |
| Recombinant human GDNF | R&D Systems | 212-GD-050/CF |
| TRO 19622, Tocris | Thermo Fischer Scientific | 29-061-0 |
| Critical Commercial Assays | ||
| Mouse Neural Stem Cell Nucleofector® Kit | Lonza | VPG-1004 |
| Deposited Data | ||
| RNA-seq data from WT and Rbfox tKO motor neuron culture | Short Reads Archive | SRP128054 |
| Rbfox1-3 CLIP data from day 10 motor neuron culture | Short Reads Archive | SRP128054 |
| Experimental Models: Cell Lines | ||
| mES Hb9::CD2-IRES-GFP WT | Ikiz et al. (2015) | |
| mES Hb9::CD2-IRES-GFP Rbfox tKO | This paper | |
| mES Hb9::CD2-IRES-GFP AnkGins | This paper | |
| mES Hb9::CD2-IRES-GFP AnkGdel | This paper | |
| mES Hb9::CD2-IRES-GFP Rbfox tKO+AnkGdel | This paper | |
| NIH3T3 cells | - | - |
| Oligonucleotides (source of primers: this paper) | ||
| A full list of oligonucleotides is presented in Table S1. | NA | NA |
| Recombinant DNA | ||
| pCAGGS-Cas9-mCherry | This paper | |
| pgRNA empty | Addgene | #41824 |
| pgRNA Rbfox1 KO | This paper | |
| pgRNA Rbfox2 KO | This paper | |
| pgRNA Rbfox3 KO | This paper | |
| pgRNA1 AnkG del | This paper | |
| pgRNA2 AnkG del | This paper | |
| pgRNA1 AnkG ins | This paper | |
| pgRNA2 AnkG ins | This paper | |
| pCAGGS-3xFLAG-Rbfox1 | This paper | |
| pCAGGS-3xFLAG-Rbfox2 | This paper | |
| pCAGGS-3xFLAG-Rbfox3 | This paper | |
| pCAGGS-3xFLAG-bIV spectrin (repeats 13–15) | This paper | |
| pCAGGS-3xFLAG-bII spectrin (repeats 13–15) | This paper | |
| pCAGGS-AnkG ZU5in-3xHA | This paper | |
| pCAGGS-AnkG ZU5ex-3xHA | This paper | |
| pCAGGS-AnkB ZU5ex-3xHA | This paper | |
| Software and Algorithms | ||
| AIS quantification algorithm (Matlab) | (Grubb and Burrone, 2010) | (ais_z3.m) http://grubblab.org/resources/ |
| Fiji | (Schindelin et al., 2012) | https://fiji.sc/ |
| RNA-seq analysis by Quantas | (Yan et al., 2015) | http://zhanglab.c2b2.columbia.edu/index.php/Quantas |
| CLIP data analysis by CTK | (Shah et al., 2017) | http://zhanglab.c2b2.columbia.edu/index.php/CTK |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture
Mouse embryonic stem cells (ESCs) were cultured in EmbryoMax DMEM (EMD Millipore) supplemented with 15% embryonic stem cell screened fetal bovine serum (HyClone), 2mM L-glutamine (Life Technologies), 1x non-essential amino acids EmbryoMax MEM (EMD Millipore), 1x EmbryoMax nucleosides (EMD Millipore), 0.1mM β-mercapthoethanol (Sigma-Aldrich), 100U/ml penicillin-streptomycin (Life Technologies), 1000U/ml ESGRO Leukemia inhibitory factor (EMD Millipore), 2.5μM GSK-3 inhibitor XVI (EMD Millipore) and 50nM FGF receptor antagonist PD173074 (Tocris). NIH3T3 cells were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific), 2mM glutamine (Life Technologies), and 100U/ml penicillin-streptomycin (Life Technologies). All cells were determined to be negative for mycoplasma using the Venor GeM Mycoplasma detection kit (Sigma-Aldrich).
METHOD DETAILS
CRISPR/Cas9 genome engineering
We generated a plasmid construct to express the Cas9 enzyme with a self-cleavable mCherry reporter (Cas-p2A-mCherry), which was used in combination with a gRNA plasmid previously generated (Mali et al., 2013). Mouse embryonic stem cells (ESCs) were transfected with these plasmids, together with an ssDNA oligo used as a donor for homologous recombination. The ssDNA used to generate Rbfox1-3 null mutants consists of two homologous arms designed based on the region of interest, two consecutive stop codons (6 nt), and a single SpeI restriction site (6 nt) for homozygous clone selection. The first nucleotide of the restriction site is shared with the last stop codon, which results in a downstream frame-shift due to the 11-nt insertion. The stop codons and the additional frameshift were designed to disrupt Rbfox translation upstream of its RNA-binding domains (Figure S1A). After 24 hours of expression, the cells were harvested and subject to FACS analysis using the mCherry reporter. A portion of the mCherry+ cells was used for estimating the number of mutant cells in population (typically 20–40%) by PCR and restriction digestion analysis. The rest of the cells were cultured further at low density to pick individual clones that were expanded for genotyping and further culturing. The knockout clones were identified using two rounds of PCR genotyping. The first round of genotyping using F1+R primers identified clones with insertion mutation. The positive clones were subjected to the second round (F2+R) of genotyping combined with restriction digestion of the PCR product to identify homozygous mutant clones. PCR products of homozygous clones were sequenced by Sanger sequencing to assure that there are no additional mutations in adjacent regions. Rbfox1-3 triple knock out (tKO) was generated by serial mutation of each individual Rbfox gene. AnkG mutant mES cell lines were generated similarly. Exon deletion was achieved by cleavage of two gRNAs flanking the targeted region followed by non-homologous end joining. Exon insertion was achieved by homologous recombination as described above. We generated 3–4 ESC knockout clones for each of the genotypes. Each biological replicate of subsequent experiments was performed using a different knockout clone. All experiments described this study were performed using at least three biological replicates unless specified explicitly.
Motor neuron differentiation and culture
ESCs were differentiated into spinal neurons in embryoid bodies following an established protocol (Wichterle et al., 2002; Wichterle and Peljto, 2008). After 6 days of differentiation, embryoid bodies were dissociated and neurons were plated on polyornithine/laminin coated plates in maturation media (Advanced DMEM/F12 (Life Technologies) + Neurobasal medium (Life Technologies) (1:1), 2mM L-glutamine (Life Technologies), 1x B-27 Supplement (serum free) (Life Technologies) containing neurotrophic factors (10ng/ml BDNF (Peprotech), 5ng/ml GDNF (R&D Systems), 7μM TRO1962 (Thermo Fisher Scientific), and 1μM 5′-fluoro-2′-deoxyuridine (Sigma-Aldrich) to inhibit proliferation of the undifferentiated cells). Half of the media was periodically replaced every two days starting from one day after plating. Non-neuronal cells were depleted by day 5 of maturation and no contamination with oligodendrocytes or astrocytes was detected based on marker expression in RNA-seq profiling (e.g., Olig2 and GFAP). This pure neuronal culture contains on average 40% of motor neurons.
Motor neuron survival analysis
We performed a comparative survival analysis taking advantage of the motor neuron-specific GFP reporter (Hb9::GFP) in the wild type (WT) and Rbfox tKO ESC lines used in this study. We mixed Rbfox tKO GFP-positive motor neurons with an equal number of WT motor neurons with a Hb9::RFP reporter. Quantification of GFP- and RFP-positive motor neurons on day 5 and day 10 revealed no discernable difference in the survival rate of tKO and control motor neurons (Figure S1E).
Purification of motor neurons
To avoid contamination with non-neuronal cells on day 1, the motor neurons were FACS sorted before plating based on transiently expressed Hb9::GFP reporter. The GFP fluorescence was at a background level after the first day in culture and it did not interfere with subsequent immunostainings. The sorted motor neuron culture contained on average 75–80% motor neurons. The sorted WT and tKO neurons used for electrophysiological measurements were cultured with primary mouse cortical astrocytes prepared as described previously (Miles et al., 2004). The maturation media contained neurotrophic factors (10ng/ml BDNF, 5ng/ml GDNF), and 1μM 5′-fluoro-2′-deoxyuridine (during the first 5 days).
RT-PCR validation of splicing quantification
To measure exon inclusion, 2 μg of total RNA was reverse transcribed using SuperScript III Reverse transcriptase (Invitrogen) in 20 μl reaction with oligodT primer (f.c. 0.5 μM) and random hexamer primers (f.c. 0.5 ng/μl), the cDNA was PCR amplified for 34 cycles and resolved by electrophoresis in 2% agarose gel with 1x Gelred (Biotium) for visualization. PCR primers are listed in Table S1.
RNA-seq library preparation
For each genotype, two or three independent sets of parallel differentiations (biological replicates) were performed to collect RNA which was subject to paired-end RNA-seq analysis (except for one day 1 WT motor neuron sample for which single-end sequencing was performed). RNA-seq libraries were prepared following the standard Illumina TruSeq poly-dT library preparation protocol and sequenced on the Illumina HiSeq 2000 platform (101-nt reads). We are in the process of depositing this dataset to NCBI Short Read Archive (SRA).
HITS-CLIP library preparation
Rbfox HITS-CLIP experiments were performed following the BrdU-CLIP protocol as described previously (Weyn-Vanhentenryck et al., 2014). For each library ~10 million cells were used. The resulting PCR amplified cDNA libraries were sequenced on the Illumina HiSeq 2000 platform (single-end 101-nt reads). We are in the process of depositing this dataset to NCBI SRA.
Analysis of RNA-seq and HITS-CLIP data
RNA-seq data were mapped by OLego v1.1.2 (Wu et al., 2013) to the reference genome (mm10) and a comprehensive database of exon junctions was provided for read mapping. Gene expression and splicing of known and novel AS events were quantified using the Quantas pipeline (http://zhanglab.c2b2.columbia.edu/index.php/Quantas), as we described previously (Yan et al., 2015). Differential gene expression was detected using edgeR as part of the Quantas pipeline (>1.5 fold change, FDR<0.05 and RPKM greater than median in WT or tKO).
To identify exons with differential splicing in two compared conditions, a Fisher’s exact test was used to evaluate the statistical significance of splicing changes using both exonic and junction reads that support each of the two splice isoforms. The false discovery rate (FDR) was estimated by the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995). An AS event was called differentially spliced in the two compared conditions with the following criteria: coverage≥20, Benjamini FDR≤0.05, and |ΔΨ |≥0.1 or 0.2 as indicated.
Rbfox HITS-CLIP data were analyzed using the CTK package, as described previously (Shah et al., 2017; Zhang and Darnell, 2011) and more details can be found on our supporting website (http://zhanglab.c2b2.columbia.edu/index.php/CTK). Due to long (101-nt) reads in these experiments, we removed 3′ adapters before collapsing exact PCR duplicates. Filtered reads were mapped by bwa (Li and Durbin, 2009), followed by a second step of duplicate collapsing to identify unique CLIP tags using a model based approach. CLIP tag clusters were identified by grouping overlapping CLIP tags in combination with a valley seeking algorithm, and the statistical significance of CLIP tag cluster peak height was evaluated as described previously (Shah et al., 2017).
Rbfox splicing-regulatory network in motor neurons
We used an integrative modeling approach to define direct Rbfox target exons in motor neurons, as described previously (Weyn-Vanhentenryck et al., 2014; Zhang et al., 2010). Rbfox-dependent splicing in both day 5 and day 10 motor neurons were used as evidence of splicing changes in the Bayesian network analysis. Rbfox1-3 pooled CLIP tag cluster scores in motor neurons were used as evidence of Rbfox binding. The use of the other datasets, model training, cross validation and prediction were the same as we described previously (Weyn-Vanhentenryck et al., 2014). In total, 547 cassette exons were predicted as direct Rbfox targets in motor neurons with FDR≤0.01. Among these, we were able to determine the direction of splicing regulation for 534 exons with probability of activation or repression ≥0.7.
Gene ontology (GO) analysis
GO enrichment analysis was performed using the online tool DAVID (Dennis et al., 2003). Genes with cassette exons regulated by Rbfox, as defined by Bayesian network analysis, were compared with all genes having at least one cassette exons with sufficient read coverage (≥20).
Evaluation of neuronal maturation based on the splicing or gene expression profile
We defined 6 distinct maturation stages from the mouse cortex data E14.5, E16.5, P0, P4, P7, and P15 or older, which were represented by stages 1–6. P15 or older were grouped as one stage because of high correlation between samples after P15. For each motor neuron sample, we obtained the splicing profile of 1,909 annotated module cassette exons defined in the cortex reference, which was used to assign the sample to a specific maturation stage by comparison to the cortex reference. To evaluate the contribution of the four specific RBP families we focused on to neuronal maturation, we also predicted neuronal maturation stages based on the splicing profile of their target exons. More detail of the method was described in (Weyn-Vanhentenryck et al., 2018).
Immunofluorescence analysis
The standard immunolabeling procedure started with paraformaldehyde fixation (4% paraformaldehyde in 1x PBS) for 10 min at room temperature. Coverslips were washed three times with 1xPBS and permeabilized/blocked for 30 minutes with Blocking buffer (1x PBS, 10% horse serum, 0.2% Triton X-100, 0.05% sodium azide). The primary antibodies were diluted in Antibody buffer (1x PBS, 5% horse serum, 0.2% Triton X-100, 0.05% sodium azide) and applied on coverslips for overnight incubation at 4°C in humidified chamber. After three washing steps with 1xPBS, the secondary antibodies in Antibody buffer were applied and the coverslips were incubated for 2 hours at 4°C. Next, the coverslips were washed three times with 1x PBS and mounted on slides with Aqua Poly/Mount (Polysciences, Inc).
Whole cell current clamp
Excitability was assessed using conventional whole cell current clamp technique. Briefly, astrocytes were prepared as previously described (Miles et al., 2004) and plated on 15-mm diameter coverslips at a density of 100,000 cells per well in a 24-well plate. 4–10 days following astrocyte plating, either FACS sorted motor neurons in the WT versus tKO experiment, or non-sorted motor neurons in the WT versus AnkGins experiment were added to the wells at a density of 50,000 cells per well. Cultures were maintained for 5 days before recording. Membrane potential recordings were performed using a Multiclamp 700B amplifier and a Digidata 1550 digital-to-analog converter. Signals were recorded at a 10-kHz sample rate using pClamp 10 software (all equipment from Molecular Devices). Patch pipettes were fabricated with a P-97 pipette puller (Sutter Instruments) using 1.5 mm outer diameter, 1.28 mm inner diameter filamented capillary glass (World Precision Instruments). Pipette resistance was 2–5 MΩ when filled with the pipette solution. The external recording solution contained 145 mM NaCl, 5 mM KCl, 10 mM HEPES, 10 mM glucose, 2 mM CaCl2 and 2 mM MgCl2. The pH was adjusted to 7.3 using NaOH and the osmolality adjusted to 325 mOsm with sucrose. The pipette solution contained 130 mM CH3KO3S, 10 mM CH3NaO3S, 1 mM CaCl2, 10 mM EGTA, 10 mM HEPES, 5 mMMgATP and 0.5 mM Na2GTP (pH 7.3, 305 mOsm). Experiments were performed at room temperature (21–23 °C). During recordings, current was injected to hold the cells at −60 mV. The liquid junction potential between pipette and external solutions was calculated empirically, and the correction applied before the experiment. Resting membrane potential was measured immediately following establishment of the whole-cell configuration. Membrane resistance and capacitance were calculated from the membrane potential changes in response to 1 s duration hyperpolarizing current steps that increased incrementally by 5 pA. Action potentials were evoked and rheobase obtained using 1 s duration depolarizing current steps that increased incrementally by 5 pA.
An action potential was defined as a transient depolarization of the membrane which had a minimum rise rate > 10 mV/ms and reached a peak amplitude > 0 mV. Action potential characteristics were measured from the first action potential at rheobase. The threshold potential was measured at the point where the voltage increases at a rate greater than 10 ms/mV. The duration was calculated from the full width at the half maximum voltage. For this calculation, the amplitude was measured from the threshold potential to the maximum potential. The maximum number of action potentials was measured from a 1 s current step. The amplitude of the step was dependent on the individual cell. Quantification was carried out using custom written scripts for Igor Pro v. 6 (Wavemetrics, USA) and R v. 3 (www.R-project.org). Outliers within Rheobase, Input Resistance, and Capacitance, that would indicate a poor seal, were identified using the ROUT method (Q = 0.5%) in GraphPad Prism version 7.0a for Mac, GraphPad Software, La Jolla California USA (www.graphpad.com). For each outlier detected, data from the entire neuron was removed from the analysis. For the WT and tKO, recording data from 3 and 7 neurons respectively were removed and for the WT and AnkGins, recording data from 3 neurons was removed from each group. Statistical comparisons were made using unpaired Student’s t-test in GraphPad Prism. P-values < 0.05 were considered significant.
Axon initial segment analysis
Motor neurons plated on primary cortical astrocytes were imaged using a laser-scanning confocal microscope (LSM 800, Zeiss) using a 40x oil-immersion objective. The settings were adjusted to prevent signal saturation and the images were taken in z-stacks with 0.49 μm steps. Z-stack images were projected into a single plane using maximum intensity projections and imported into MATLAB computer software (Mathworks) for AIS analysis using a custom program as previously described (Grubb and Burrone, 2010). Briefly, the software automatically determines AIS length based on AnkG fluorescence intensity profile along a semi-automatically traced path. The cutoff intensity for AIS start/end was set to 1/3 of the maximum intensity as recommended in the original study. The motor neurons with no detectable AnkG immunostaining or with immunostaining shorter than 5 μm (puncta) were categorized as AIS(−) motor neurons. It should be noted that the majority of AIS(−) neurons showed weak AnkG puncta throughout the axon (Figure S5B).
STORM imaging
10 days matured neurons plated on primary cortical astrocytes were washed with 1x PBS and immediately fixed. For AnkG STORM imaging, the cells were fixed with 4% paraformaldehyde for 20 minutes and washed with 1xPBS. For actin filament imaging, the cells were first fixed/permeabilized with 0.3% (v/v) glutaraldehyde and 0.25% (v/v) Triton X-100 in cytoskeleton buffer (10mM MES, pH6.1, 150mM NaCl, 5mM EGTA, 5mM glucose and 5mM MgCl2) for 1min, and then post-fixed for 25min in 2% (v/v) glutaraldehyde in cytoskeleton buffer. The glutaraldehyde-fixed samples were treated with freshly prepared 0.1% (w/v) sodium borohydride in 1xPBS for 2x 5min. All coverlips were stored in 1x PBS with 20mM sodium azide and stained within 2 weeks. The fixed coverslips were incubated with STORM blocking buffer (3% bovine serum albumin (BSA), 0.1% Triton X-100 in 1x PBS) and stained with corresponding primary antibodies at 4°C overnight. After washing with 0.3% BSA and 0.01% Triton X-100 in 1xPBS, secondary antibody (2.5 μg/ml) labeling was performed at room temperature for 1 hour. For actin labeling, samples were incubated with Alexa Fluor-647 conjugated phalloidin (0.4 μM) (Life Technologies, A22287) for 45 minutes at room temperature, and briefly washed with 1x PBS before imaging.
3D-STORM imaging was performed on a homebuilt setup based on a Nikon Eclipse Ti-U inverted optical microscope with an oil immersion objective (Nikon CFI Plan Apochromat λ 100x, NA 1.45). Lasers at 647 (MPB Communications) and 405 (Coherent) were coupled into an optical fiber after an acousto-optic tunable filter and then introduced into the sample from the back focal plane of the microscope. The laser beams were shifted toward the edge of the objective with a translation stage, making incidence angles slightly smaller than the critical angle of the glass-water interface. Continuous illumination of 647 nm laser (~2kW cm−2) was used to excite Alexa Fluor 647 molecules and switch them into the dark state. Illumination of the 405 nm laser (0–1 W cm−2) was tuned during image acquisition so that any given instant, only a small fraction of fluorophores in the sample were in the emitting state, which is optically resolvable. For z position determination, a cylindrical lens was inserted between the electron-multiplying charge-coupled device (EMCCD) camera (iXon Ultra 897, Andor) and the microscope, so that images of single molecules were elongated in x and y for molecules proximal and distal sides of the focal plane (relative to the objective), respectively. Imaging buffer used was 100 mM Tris-HCl (pH7.5) containing 140 mM cysteamine, 5% glucose, 0.8 mg.ml−1 glucose oxidase and 40 μg.ml−1 catalase.
Recorded STORM movies were analyzed according to previously described methods (Huang et al., 2008; Rust et al., 2006). Spatial localization of molecules was determined from the centroids of 2D-Gaussian fit (for x and y) and ellipticity (for z) of single fluorescent molecules. Molecules in single frames were added up and drift-corrected by cross-correlation analysis of every ~200 frames. Fourier transform analysis was performed with a custom-written MATLAB program (Xu et al., 2013). Fluorescence intensity along the axial direction of the ROI was measured and subject to 1D-Fast Fourier Transform (FFT). The spatial frequency at the peak of FFT curve was taken as the frequency with maximal probability, whose reciprocal gave the period. Autocorrelation curves were plotted with a custom-written MATLAB program, where the autocorrelation function of fluorescence intensity was computed by shifting the data along the axon and quantifying the extent of correlation. The autocorrelation amplitude was defined as the difference between the first peak and the average of two neighboring valleys (Zhong et al., 2014).
Structural modeling of AnkG ZU5–1 and βIV-spectrin
We used the previously solved structure of the homologous AnkR ZU5–1 domain together with the βI-spectrin repeats 13–15 (PDB ID: 3KBT) (Ipsaro and Mondragon, 2010) and UCSF Chimera (v1.11) (Pettersen et al., 2004) was used for visualization. This structure suggested that two arginines and an alanine of ZU5–1 are engaged in interaction with β-spectrin (shown in red in Figure 7A). We used the I-TASSER protein structure prediction software (Yang et al., 2015) to model the structure of βIV-spectrin and AnkG ZU5–1 with alternative exon inclusion (shown in orange in Figure 7A) and found that the peptide encoded by the alternative exon is predicted to localize proximally to the two crucial arginines at the interface of protein-protein interaction.
Co-immunoprecipitation of AnkG ZU5–1 isoforms and βIV-spectrin
AnkG ZU5–1 domain isoforms were cloned into a pCAGGS-GOI-3xHA vector. βIV-spectrin repeats 13–15 were cloned into pCAGGS-3xFLAG-GOI vector from mouse cortex cDNA using primers indicated in Table S1. 4mu;g of plasmid DNA for each construct were used for transfection (Lipofectamine 3000, Invitrogen,) of a single 80% confluent 6cm dish of NIH/3T3 cells. 24hrs after transfection, the plates were carefully washed twice with ice-cold 1x PBS and the cells were lysed directly on the plate by addition of 400mu;l of Lysis buffer. The lysate was collected using a plastic cell scraper, transferred into a microcentrifuge tube, further resuspended by passing 8x through a 26G needle attached to a 1ml TB syringe and incubated on ice for 20 minutes.
The soluble fraction of the cell lysate was separated by centrifugation (14,000rpm, 20minutes, 4°C) and transferred into a new microcentrifuge tube. 28μl of the soluble fraction from each sample was aliquoted and mixed with 10μl of 4xLDS buffer and 2μl of 1M DTT. The remaining lysate was mixed with the magnetic beads and incubated 1hr on a rotating device at 4°C. The beads were then washed 2x with Wash buffer, transferred into a new tube and washed two more times with Wash buffer. After removal of the last wash, the beads were collected and the bottom of the tube by a brief spin in microcentrifuge (3 seconds) and the remaining liquid was removed. The protein was eluted by addition of 28μl water, 10μl of 4xLDS buffer and 2μl of 1M DTT and boiling for 5 minutes at 90°C. 10μl of each sample was separated on 15-well 10% Bis-Tris PAGE gel (Thermo Fisher Scientific), transferred onto nitrocellulose membrane using X-cell blot wet transfer set up and the protein was detected using standard western blot protocol with anti-FLAG HRP (1:3000, or anti-HA HRP (1:3000) antibodies.
Immunoprecipitation of βII and βIV-spectrin was performed using Dynabeads Protein G (Thermo Fisher Scientific, 10004D) and anti-FLAG M2 antibodies (Sigma-Aldrich). 25μl of magnetic beads per sample were washed 2x with Binding buffer (1x PBS, 0.05% Tween 20), mixed with 2.5μg antibodies diluted in Binding buffer, and incubated overnight or at least 2hrs on a rotating device at 4°C. After the incubation, the beads were washed 2x with Wash buffer (50mM HEPES pH7.5, 150mM NaCl, 1% NP-40, 1mM EDTA) and 1x with Lysis buffer (50mM HEPES pH7.5, 100mM NaCl, 1% NP-40, 1mM EDTA, cOmplete Protease Inhibitor Cocktail (Sigma-Aldrich)).
Gene overexpression in maturing motor neurons
To overexpress proteins in motor neurons, we trypsinized differentiating embryoid bodies on day 4 of in vitro differentiation (motor neuron progenitors) and nucleofected 5×106 cells with 5μg of plasmid DNA using Amaxa mouse NSC nucleofector kit (nucleofector setting B-16) (Lonza). For Rbfox rescue experiments, we used full-length Rbfox1, Rbfox2 or Rbfox3 cloned into pCAGGS-3xFLAG-GOI vector. For the competition with the dominant negative ankyrin G ZU5–1 isoforms, we used constructs described above. The empty vector was used as control in these experiments.
The transfected cells were plated on polyornithine/laminin coated plates and cultured in motor neuron maturation media with neurotrophic factors (10ng/ml BDNF, 5ng/ml GDNF) and 5μM DAPT to improve differentiation efficiency of the dissociated cells. After the first day, the maturation media was completely removed and replaced with fresh media without DAPT and with addition of 1μM 5′-fluoro-2′-deoxyuridine to prevent proliferation of undifferentiated cells. Typically, the transgene overexpression was observed in 30–40% of motor neurons. The AIS was analyzed using confocal microscopy as described above.
Supplementary Material
Table S1: List of oligonucleotide sequences used in this study. Related to Methods.
Table S2: Splicing profile of developmentally regulated cassette exons used to stage maturation of motor neurons. Related to Figure 1.
Table S3: List of Rbfox-dependent AS events in day 5 and day 10 motor neurons. Related to Figure 2.
Table S4: Motor neuron Rbfox targets from Bayesian network analysis. Related to Figure 2.
Table S5: Gene Ontology (GO) of Rbfox targets defined by Bayesian network analysis. Genes with cassette exons with coverage ≥20 reads on day 5 or day 10 were used as control. Related to Figure 3.
Table S6: Rbfox target transcripts encoding AIS genes. Related to Figure 4.
Highlights.
Rbfox proteins control splicing of cytoskeletal, membrane, and synaptic genes
Rbfox tKO neurons retain an immature splicing program and electrophysiology
The AIS is perturbed in Rbfox tKO neurons due to defects in AnkG localization
A developmental splicing switch in AnkG is critical for AIS assembly
Acknowledgments
We thank Michael Closser and other members of the Zhang and Wichterle laboratories for helpful discussions during the project. We also thank Columbia Genome Center for sequencing of HITS-CLIP and RNA-seq libraries and James Caicedo (Serge Przedborski laboratory, Columbia University) for providing primary mouse cortical astrocytes. This study was supported by grants from the National Institutes of Health (NIH) (R00GM095713, R21NS098172, and R03HG009528 to C.Z., R01NS089676 to C.Z. and H.W., and R01NS078097 to H.W.), the Simons Foundation Autism Research Initiative (307711 to C.Z.) and Project ALS Foundation (to H.W.). M.J. was in part supported by the Bakala Foundation. H.F. was in part supported by a Scholarship from the China Scholarship Council. R.Y. and K.X. acknowledge support from the Beckman Young Investigator Program and the Packard Fellowships for Science and Engineering. High-performance computation was supported by NIH grants S10OD012351 and S10OD021764.
Footnotes
Author Contributions
MJ, HW and CZ conceived the project. MJ performed most experiments with assistance from JP. SMW, HF and CZ performed bioinformatics analysis. JWS and DJW performed whole cell patch clamp recordings. RY and KX performed STORM imaging. MJ, HW and CZ wrote the paper with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackerman SL, Kozak LP, Przyborski SA, Rund LA, Boyer BB, Knowles BB. The mouse rostral cerebellar malformation gene encodes an UNC-5-like protein. Nature. 1997;386:838–842. doi: 10.1038/386838a0. [DOI] [PubMed] [Google Scholar]
- Barnby G, Abbott A, Sykes N, Morris A, Weeks DE, Mott R, Lamb J, Bailey AJ, Monaco AP. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. Am J Hum Genet. 2005;76:950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc B. 1995;57:289–300. [Google Scholar]
- Bhalla K, Phillips HA, Crawford J, McKenzie OLD, Mulley JC, Eyre H, Gardner AE, Kremmidiotis G, Callen DF. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- Carreira-Rosario A, Bhargava V, Hillebrand J, Kollipara RK, Ramaswami M, Buszczak M. Repression of pumilio protein expression by Rbfox1 promotes germ cell differentiation. Dev Cell. 2016;36:562–571. doi: 10.1016/j.devcel.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy JG. Developmental regulation of RNA processing by Rbfox proteins. Wiley Interdiscip Rev RNA. 2017;8 doi: 10.1002/wrna.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Ying Y, Lin CH, Lee JA, Tran D, Vashisht AA, Bahrami-Samani E, Xing Y, Martin KC, Wohlschlegel JA, et al. Rbfox proteins regulate splicing as part of a large multiprotein complex LASR. Cell. 2016;165:606–619. doi: 10.1016/j.cell.2016.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane H, Lempicki R. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- Dillman AA, Hauser DN, Gibbs JR, Nalls MA, McCoy MK, Rudenko IN, Galter D, Cookson MR. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat Neurosci. 2013;16:499–506. doi: 10.1038/nn.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Parikshak N, Konopka G, Geschwind DH. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21:4171–4186. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freal A, Fassier C, Le Bras B, Bullier E, De Gois S, Hazan J, Hoogenraad CC, Couraud F. Cooperative interactions between 480 kDa Ankyrin-G and EB proteins assemble the axon initial segment. J Neurosci. 2016;36:4421–4433. doi: 10.1523/JNEUROSCI.3219-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano MR, Jha S, Ho TSY, Zhang CS, Ogawa Y, Chang KJ, Stankewich MC, Mohler PJ, Rasband MN. A distal axonal cytoskeleton forms an intra-axonal boundary that controls axon initial segment assembly. Cell. 2012;149:1125–1139. doi: 10.1016/j.cell.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Meera P, Stoilov P, Shiue L, O’Brien JE, Meisler MH, Ares M, Otis TS, Black DL. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb MS, Burrone J. Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature. 2010;465:1070–U1131. doi: 10.1038/nature09160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Abdi KM, Bennett V. Ankyrin-G palmitoylation and betaII-spectrin binding to phosphoinositide lipids drive lateral membrane assembly. J Cell Biol. 2014;206:273–288. doi: 10.1083/jcb.201401016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Jenkins P, Bennett V. Cysteine 70 of ankyrin-G is S-palmitoylated and is required for function of ankyrin-G in membrane domain assembly. J Biol Chem. 2012;287:43995–44005. doi: 10.1074/jbc.M112.417501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom KL, Ogawa Y, Rasband MN. AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J Cell Biol. 2008;183:635–640. doi: 10.1083/jcb.200806112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom KL, Xu X, Ogawa Y, Frischknecht R, Seidenbecher CI, Shrager P, Rasband MN. Neurofascin assembles a specialized extracellular matrix at the axon initial segment. J Cell Biol. 2007;178:875–886. doi: 10.1083/jcb.200705119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Mondragon A. Structural basis for spectrin recognition by ankyrin. Blood. 2010;115:4093–4101. doi: 10.1182/blood-2009-11-255604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins PM, Kim N, Jones SL, Tseng WC, Svitkina TM, Yin HH, Bennett V. Giant ankyrin-G: a critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc Natl Acad Sci U S A. 2015;112:957–964. doi: 10.1073/pnas.1416544112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AN, Manning CF, Trimmer JS. A unique ion channel clustering domain on the axon initial segment of mammalian neurons. J Comp Neurol. 2014;522:2594–2608. doi: 10.1002/cne.23551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M, Soriano P. Beta IV-spectrin regulates sodium channel clustering through ankyrin-G at axon initial segments and nodes of Ranvier. J Cell Biol. 2002;156:337–348. doi: 10.1083/jcb.200110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Damianov A, Lin CH, Fontes M, Parikshak NN, Anderson ES, Geschwind DH, Black DL, Martin KC. Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron. 2016;89:113–128. doi: 10.1016/j.neuron.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo ED, Hinck L, Masu M, Keino-Masu K, Ackerman SL, Tessier-Lavigne M. Vertebrate homologues of C. elegans UNC-5 are candidate netrin receptors. Nature. 1997;386:833–838. doi: 10.1038/386833a0. [DOI] [PubMed] [Google Scholar]
- Leterrier C, Potier J, Caillol G, Debarnot C, Rueda Boroni F, Dargent B. Nanoscale architecture of the axon initial segment reveals an organized and robust scaffold. Cell Rep. 2015;13:2781–2793. doi: 10.1016/j.celrep.2015.11.051. [DOI] [PubMed] [Google Scholar]
- Leterrier C, Vacher H, Fache MP, d’Ortoli SA, Castets F, Autillo-Touati A, Dargent B. End-binding proteins EB3 and EB1 link microtubules to ankyrin G in the axon initial segment. Proc Natl Acad Sci U S A. 2011;108:8826–8831. doi: 10.1073/pnas.1018671108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Duvall J, Ilkin Y, Simon J, Arreaza M, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell C, Rao K, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- Miles GB, Yohn DC, Wichterle H, Jessell TM, Rafuse VF, Brownstone RM. Functional properties of motoneurons derived from mouse embryonic stem cells. J Neurosci. 2004;24:7848–7858. doi: 10.1523/JNEUROSCI.1972-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, Winkelmann JC, et al. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Rasband MN. The axon initial segment and the maintenance of neuronal polarity. Nat Rev Neurosci. 2010;11:552–562. doi: 10.1038/nrn2852. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics. 2017;33:566–567. doi: 10.1093/bioinformatics/btw653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemoto Y, Suzuki S, Terada N, Ohno N, Ohno S, Yamanaka S, Komada M. Specific role of the truncated betaIV-spectrin Sigma6 in sodium channel clustering at axon initial segments and nodes of ranvier. J Biol Chem. 2007;282:6548–6555. doi: 10.1074/jbc.M609223200. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong CK, Black DL, Zheng SK. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17:265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck S, Mele A, Sun S, Yan Q, Farny N, Zhang Z, Xue C, Silver PA, Zhang MQ, Krainer AR, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Feng H, Ustianenko D, Duffié R, Yan Q, Jacko M, Martinez JC, Goodwin M, Zhang X, Hengst U, et al. Precise temporal regulation of alternative splicing during neural development. bioRxiv. 2018 doi: 10.1038/s41467-018-04559-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Peljto M. Differentiation of mouse embryonic stem cells to spinal motor neurons. Curr Protoc Stem Cell Biol. 2008:Unit 1H 1 1–1H 1 9. doi: 10.1002/9780470151808.sc01h01s5. [DOI] [PubMed] [Google Scholar]
- Wu J, Anczukow O, Krainer AR, Zhang MQ, Zhang C. OLego: Fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic Acids Res. 2013;41:5149–5163. doi: 10.1093/nar/gkt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Xu K, Zhong GS, Zhuang XW. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Weyn-Vanhentenryck SM, Wu J, Sloan SA, Zhang Y, Chen K, Wu JQ, Barres BA, Zhang C. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci U S A. 2015;112:3445–3350. doi: 10.1073/pnas.1502849112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ogawa Y, Hedstrom KL, Rasband MN. BetaIV spectrin is recruited to axon initial segments and nodes of Ranvier by ankyrinG. J Cell Biol. 2007;176:509–519. doi: 10.1083/jcb.200610128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotech. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong GS, He J, Zhou RB, Lorenzo D, Babcock HP, Bennett V, Zhuang XW. Developmental mechanism of the periodic membrane skeleton in axons. Elife. 2014;3:e04581. doi: 10.7554/eLife.04581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziskind-Conhaim L. Electrical properties of motoneurons in the spinal cord of rat embryos. Dev Biol. 1988;128:21–29. doi: 10.1016/0012-1606(88)90262-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: List of oligonucleotide sequences used in this study. Related to Methods.
Table S2: Splicing profile of developmentally regulated cassette exons used to stage maturation of motor neurons. Related to Figure 1.
Table S3: List of Rbfox-dependent AS events in day 5 and day 10 motor neurons. Related to Figure 2.
Table S4: Motor neuron Rbfox targets from Bayesian network analysis. Related to Figure 2.
Table S5: Gene Ontology (GO) of Rbfox targets defined by Bayesian network analysis. Genes with cassette exons with coverage ≥20 reads on day 5 or day 10 were used as control. Related to Figure 3.
Table S6: Rbfox target transcripts encoding AIS genes. Related to Figure 4.