Abstract
Backcrossing together with simple sequence repeat marker strategy was adopted to improve popular Malaysian chilli Kulai (Capsicum annuum L.) for heat tolerance. The use of molecular markers in backcross breeding and selection contributes significantly to overcoming the main drawbacks such as increase linkage drag and time consumption, in the ancient manual breeding approach (conventional), and speeds up the genome recovery of the recurrent parent. The strategy was adopted to introgress heat shock protein gene(s) from AVPP0702 (C. annuum L.), which are heat-tolerant, into the genetic profile of Kulai, a popular high-yielding chilli but which is heat sensitive. The parents were grown on seed trays, and parental screening was carried out with 252 simple sequence repeat markers. The selected parents were crossed and backcrossed to generate F1 hybrids and backcross generations. Sixty-eight markers appeared to be polymorphic and were used to assess the backcross generation; BC1F1, BC2F1 and BC3F1. The average recipient allele of the selected four BC1F1 plants was 80.75% which were used to produce the BC2F1 generation. BC1-P7 was the best BC1F1 plant because it had the highest recovery at 83.40% and was positive to Hsp-linked markers (Hsp70-u2 and AGi42). After three successive generations of backcrossing, the average genome recovery of the recurrent parent in the selected plants in BC3F1 was 95.37%. Hsp gene expression analysis was carried out on BC1F1, BC2F1 and BC3F1 selected lines. The Hsp genes were found to be up-regulated when exposed to heat treatment. The pattern of Hsp expression in the backcross generations was similar to that of the donor parent. This confirms the successful introgression of a stress-responsive gene (Hsp) into a Kulai chilli pepper variety. Furthermore, the yield performance viz. plant height, number of fruits, fruit length and weight and total yield of the improved plant were similar with the recurrent parent except that the plant height was significantly lower than the Kulai (recurrent) parent.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-017-0836-3) contains supplementary material, which is available to authorized users.
Keywords: Backcrossing, Marker-assisted selection, Chilli, Heat stress, Heat shock protein 70, Simple sequence repeat (SSR) markers
Introduction
In Malaysia, the estimated annual production of chilli for 2014 was 40,520 Mt planted over 3581.80 ha of cultivated land area (DOA 2014). Due to high demand for chilli and insufficient supplies, Malaysia imports chilli to meet the rising demand. In 2005, Malaysia imported 45,900 t of chilli, while in 2007, import of peppers declined to 35,500 t. Recently, chilli output in Malaysia plunged due to El Nino, leading to a shortage of supply. El Nino is a complex series of climatic changes that occurs irregularly and affects sea surface temperature in most tropics and subtropics. With a population of about 30 million, Malaysia is the 26th largest greenhouse gas emitter in the world (Alam et al. 2011) which is projected to increase its average temperature by 0.3 to 4.5 °C with a subsequent rise in sea level of above 95 cm over a hundred years. This will lead to fluctuations in rainfall from about −30% to +30% and will ultimately affect crop yield negatively (Chong and Mathews 2001).
Heat stress is an increase in temperature above the critical value for periods long enough to pose serious harm or damage to the growth and development of plants (Larkindale and Knight 2002). Crop plants, including chilli peppers, are exposed to heat stress at different stages of their lifecycle. High temperature is the main environmental factor negatively affecting plant growth in tropical and sub-tropical areas (Usman et al. 2014). As soon as cells are exposed to various stress conditions such as heat stress, heat shock factors (HSFs) residing in the cytosol dissociate from Hsps, become activated and undergo trimerization. These HSF trimers react with phosphorus and are moved to the nucleus where they bind to the heat shock elements located in the promoter region of the Hsp genes. Hsp mRNA is then transcribed and translated which leads to increased levels of Hsps in the aqueous component of the cytoplasm. Hence, these factors act as chaperones for misformed proteins and help in the movement and/or degradation of already damaged proteins (Young 2010). To whatever degree, all organisms show the expression of Hsps that belong to the family Hsp70 with the molecular weight of the family falling between 68 and 78 kDa (Lindquist 1986). Heat-responsive gene (Hsp70) is a stress-inducible protein present in living organisms throughout evolution and is highly conserved. They act as a molecular chaperone and are important for allowing cells to cope with acute stressor insults, especially those affecting the protein machinery. Hsp70 proteins give protection to cells against heat stress. Regardless of their function in preventing aggregation of proteins and in helping again in the folding of proteins that are not native in environments that are not favourable, several Hsp70s also play a crucial role in housekeeping activities under favourable conditions (Usman et al. 2017; Tompa and Kovacs 2010).
The process involving molecular markers for the indirect selection of a genetic determinant(s) of a desirable trait could be referred to as marker-assisted selection (e.g. heat tolerance). It is a breeding selection technique for a desirable gene via molecular marker analysis with markers closely associated with the target gene. The selections are considered with high reliability and genotype selection could be performed in early generation, thus speeding up the breeding process (Chen et al. 2005). Precise transfer of genes from wild species was found effective by gene introgression assisted by marker selection with less linkage drag (Lang et al. 2015) and could identify genotypes carrying the desired gene in the early backcross generations even if it is subdued in a genetic background (Hurni et al. 2014). The success of the incorporation relies on the ability of the gene(s) of interest to showcase the expected outcome once incorporated into the genetic background of the recipient parent. The main advantages of this breeding approach are precise selection for the desired locus, effective genetic profiling of the recipient parent and reduced linkage drag adjacent to the incorporated locus. It is also an efficient way to manipulate the genetic make-up of plants to develop new genotypes with favourable characters (Miah et al. 2015).
Lecomte et al. (2004) through marker-assisted backcrossing (MAB) incorporated five quantitative trait loci (QTLs) highly linked to tomato fruit quality into three different recipient lines. Lawson et al. (1997) observed that the level of resistance in individuals incorporated for five pest-resistant QTLs through MAB was less when compared with the interspecific F1 hybrids. Another experimental study was reported by Thabuis et al. (2004) in which through marker-assisted backcrossing, four QTL-resistant alleles were incorporated from small fruited pepper into bell pepper recipient line in just three backcross generations. Further, the successful introgression and recovery of the recipient parent was confirmed following phenotypic evaluations of each backcross generation which showed an increased level of resistance and an efficient return to the recipient phenotype for the fruit weight. Through marker-assisted backcrossing, Babu et al. (2005) reported that high lysine opaque2 gene was incorporated to phi057 and umc1066, which are located within the opaque2 gene itself. Successful introgression of two chromosomal regions containing six QTLs (RM3586, RM160, RM3735, RM3471, RM3687 and RM3536) for heat tolerance from N22 Dular rice variety through marker-assisted backcrossing was reported (Lang et al. 2015). From the available literature, development of heat-tolerant vegetable (including chilli) varieties through marker-assisted backcrossing was not reported. However, the AVRDC–The World Vegetable Center has made significant contributions to the development of heat-tolerant tomato, peppers and Chinese cabbage lines and the subsequent release of adapted, tropical varieties worldwide (De la Peña and Hughes 2007). The objective of the present study was to introgress Hsp70 gene into widely cultivated high-yielding Kulai variety, to identify polymorphic molecular markers for heat-tolerant characteristics and background recoveries and to validate the backcross progenies for heat tolerance (Hsp loci).
Materials and methods
Plant materials
The chilli genotype AVPP0702 (Capsicum annuum L.), a heat-tolerant breeding line from the AVRDC—The World Vegetable Center, was used as the donor parent. Its yield is 2.5–3 t/ha under heat conditions in Taiwan (AVRDC 2001). The recipient pepper variety Kulai is widely grown in Malaysia (especially in Peninsular Malaysia). Kulai was developed by the Malaysian Agricultural Research and Development Institute (MARDI 2009). The yield potential of this variety is 15–20 t/ha under normal growth conditions (27–30 °C) but can decline by 50% under heat conditions (above 35 °C) (Usman et al. 2015). AVPP0702 was used as heat-tolerant donor parents, and the cultivated chilli variety Kulai, sensitive to heat stress, served as the recurrent parent. The Kulai variety was crossed with AVPP0702 to produce the F1 seed generation. The plants in F1 generation (as a female parent) were then backcrossed with the recurrent parent to form the first backcross generation, BC1F1. Selection for the desired alleles (plants carrying the target genes) and background selection for the recovery of the recurrent parent were performed using microsatellite markers distributed across the 12 chilli chromosomes. Progenies with the desired allele and maximum recovery of the recurrent parents were again backcrossed with Kulai to produce the BC2F1 generation. This backcross breeding program proceeds until BC3F1 generation. Backcross selection was conducted at each backcross generation to determine the percentage recovery of the recurrent parent.
Molecular marker analysis
The pepper simple sequence repeat markers closely linked to the Hsp gene were used to select backcross plants possessing the target gene. Marker screening for polymorphism was carried out between the two selected parents using the tightly-linked (Hsp70-u2 and AGi42) markers, which have been found to be related to the Hsp genes (Magaji et al. 2016; Ince et al. 2010), and 250 paired SSR markers spread across the 12 chilli pepper chromosomes (Magaji et al. 2016). A minimum of four polymorphic SSR background markers per chromosome were used for recurrent parent genome recovery analysis.
Precision of introgression line screening for heat tolerance
Heat tolerance screening was carried out in the Laboratory of Climate-Smart Food Crop Production, Institute of Tropical Agriculture and Food Security, Universiti Putra Malaysia. Seeds of the BC generations (BC1F1, BC2F1 and BC3F1) including parents (AVPP0702 and Kulai) were germinated in plastic cups with three replications. The plants were exposed to a gradual temperature increase (10 min for every 5° increase) from 25 to 35 °C using an experimental plant growth chamber (GC-101C; Daeyang ETS, Hwasung-si, Kyunggi-do, South Korea). After the temperature reached 30 °C, the plants were kept at 35 °C for 2 and 4 h, and the same method of stress treatment was applied for extreme heat stress (45 °C) for 2 and 4 h duration. After each heat treatment, leaf samples were excised and immediately suspended in liquid nitrogen for subsequent analyses.
RNA isolation and quantitative real-time PCR
Hsp gene expression analysis was performed using qRT-PCR to confirm the introgression of the heat-responsive gene in BC1F1, BC2F1 and BC3F1 in response to the different levels of the heat stress. Total RNA was extracted following TRIzol. The integrity and purity of the total RNA were evaluated with NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and 1.0% agarose gel. The genomic DNA of total RNA was removed by DNaseI. First-strand cDNA was synthesized using Thermo RT First Strand cDNA Synthesis Kit according to the manufacturer’s instruction. The quantitative real-time PCR (qRT-PCR) was performed on CFX96 Real-Time PCR system (Bio-Rad, Mississauga, Ontario, Canada). The primer3Plus software was used to design Hsp70. The UBI-3 and EF1-α were used as an internal control. Each qRT-PCR reaction was performed with Power 2× SYBR real-time PCR premixture (BioTek). The reactions were subjected to 95 °C for 2 min followed by 40 cycles at 95 °C for 20 s, 60 °C for 20 s and 72 °C for 60 s. The primers of EF1-α were forward primer, 5′ TGAAGAATGGTGATGCTGGC 3′; reverse primer, 5′ GACAACACCAACAGCAACAG 3′, and those of UBI-3 were forward primer, 5′ TGTCCATCTGCTCTCTGTTG 3′; reverse primer, 5′ CACCCCAAGCACAATAAGAC 3′. The primers of Hsp70-u2 were forward primer, 5′ ACGAAGGGTGTCTCAGCAAG 3′; reverse primer, 5′ GGAAGATTTGCCAGTGAAGG 3′, and the primers of OsHsp24 were forward primer, 5′ TTCCAGGTCAACGTCGAGT 3′; reverse primer, 5′ GCACGGTTCTTCCGC TTCA 3′. Each reaction was done in duplicate, and two non-template controls were included.
Selection for phenotypic resemblance
Plants carrying the heat-resistant gene(s) with a maximum phenotypic resemblance to the recipient parent Kulai were selected during the growing period. Phenotypic selection was carried out over the entire population of BC1F1, BC2F1 and BC3F1 generations after foreground selection. The phenotypic parameters considered are plant height, number of fruits, fruit length, fruit weight and yield per plant. Individuals at each backcross generation having the highest phenotypic resemblance with the recipient Kulai as well as carrying the Hsp70-linked gene were selected for the next generation.
Extraction of DNA, PCR conditions and electrophoresis
Fresh leaves of approximately 4-week-old plants from all parents and the backcross generations were excised for total genomic DNA extraction using cetyltrimethylammonium bromide (CTAB) method as described by Doyle and Doyle (1990). The quality and size of genomic DNA of the DNA samples were checked using NanoDrop spectrophotometry (ND1000 spectrophotometer) and evaluated on gel electrophoresis at 1%. Samples within the range of 1.8–2.0 260/280 were selected for the polymerase chain reaction. NanoDrop indicates the presence of protein and organic acid contamination. A 260/280 ratio is generally used to determine protein contamination of a nucleic acid sample, and a 260/230 ratio indicates the presence of organic contaminants. A 260/280 ratio of ~1.8 is generally accepted as “pure” for DNA and a 260/230 close to 2.0 generally accepted as “pure” DNA. A single high-molecular-weight band was considered good DNA, and smeared DNA band was of poor quality. The stock DNA samples were diluted with 1× TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0) to make a final concentration of 70 ng/μl and kept in −20 °C refrigerator for further analysis.
The PCR amplification was performed in 1 μl 70 ng template DNA, 1.0 μM of each forward and reverse primer, 7.4 μl DreamTaq Green PCR Master Mix (2×) (Thermo Scientific) and 4.6 μl nuclease-free water per 15 μl using thermocycler (T100TM, Bio-Rad). The PCR condition was followed using a touchdown PCR program with the following profile: 94 °C for 5 min followed by 38 cycles at 94 °C for 1 min, 45–62 °C (depending on annealing temperature of the primer pair) for 1 min, then 72 °C for 2 min, and a final extension for 10 min at 72 °C followed by rapid cooling to 4 °C prior to analysis both for foreground and background markers. A total of 2.5% Metaphor™ agarose (Lonza) gel was prepared comprising of 1 μl Midori green in 1× TBE buffer (0.05 M Tris, 0.05 M boric acid, 1 mM EDTA, pH 8.0) for electrophoresis. The gel was run at a constant voltage of 80 V for 80 min, and the amplified products were visualized with the aid of a Molecular Imager® (GelDoc™ XR, Bio-Rad Laboratories Inc., USA). The molecular weights of the different alleles were measured using the software provided by Bio-Rad and attached to the gel documentation system.
Statistical analysis
The primers’ banding pattern was scored with reference to two parents. In the foreground selection, the homozygous recipient allele, homozygous dominant allele and heterozygous allele were scored as “A”, “K” and “H”. With respect to the background selection, the marker data was analysed using the Graphical Genotyper (GGT 2.0) software (Van Berloo 2008). The proportion of recurrent parent (% K), the percent donor alleles (% A) and heterozygous segments (% H) were calculated. The Chi-square (χ 2) analysis for the sensitive and tolerant ratio was calculated by using the formula, χ 2 = (O − E)2/E, where O is the observed value, and E is the expected value. An independent t test and Tukey’s HSD test using SAS 9.4 program were done to separate the mean difference of the growth and yield between the parental lines and heat-tolerant improved lines. Relative gene expression analysis was calculated following the 2−ΔΔCT method (Pfaffl 2001) using the software provided by Bio-Rad. UBI-3 and EF1-α were used as housekeeping gene for normalization.
Results and discussion
Improving abiotic stress tolerance of plants involves breeding plants that can survive under extreme temperature range beyond what exists in the current germplasm. The molecular breeding approach is a potential alternative to conventional breeding. This is because it is less time-consuming, labour-saving and cost-effective. Various researchers have reported that known gene(s) can be manipulated using indirect (linked) markers such as in wheat (AnLi et al. 2005) and tomato (Barone et al. 2005). However, from the available literature, development of heat-tolerant vegetable varieties through MAB was not reported.
Parental SSR polymorphism screening
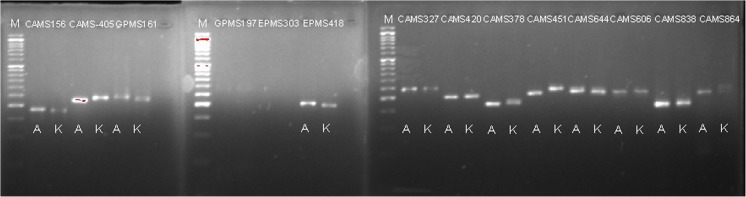
A total of 252 SSR markers were used for the parental polymorphism screening. Of these, 68 polymorphic markers between the two parents (Magaji et al. 2016) spreading across all 12 chilli pepper chromosomes were used to assess the BC1F1, BC2F1 and BC3F1 generations for background analysis. All selected SSR markers showed clear polymorphisms between the parental lines. The ratio of polymorphic markers on the parental survey was approximately 26.98% (Magaji et al. 2016). Figure 1 showed the parental screening of polymorphic markers used in this study. Our results showed different alleles between the contrasting parental lines using the screened SSR markers, indicating the existence of polymorphism of the same gene in the same chilli population. Similar results were reported by Chamikara et al. (2015) and Ince et al. (2010). Hence, the markers can be applicable for marker-assisted selection in each backcross generation.
Fig. 1.
Screening of parental lines (A AVPP0702 and K Kulai) for polymorphism using some of the SSR marker. Running on 2.5% metaphor agarose gel stained with midori green. M 50 bp ladder
Genotyping F1 generation
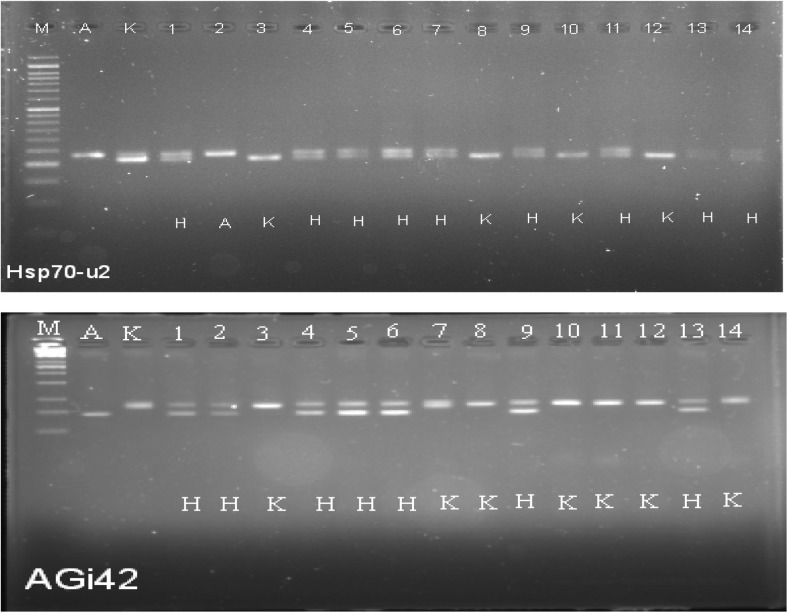
The F1 seeds were produced from the cross between Kulai and AVPP0702, and F1 plants were tested for both characters of the parents using the Hsp70-u2- and AGi42-linked markers. Almost all the tested individuals were heterozygous (carrying both characters of the two parents) using the foreground markers (Fig. 2).
Fig. 2.
Genotyping F1 (derived from K Kulai × A AVPP0702) using foreground markers Hsp70-u2 (14 individuals) and AGi42 (26 individuals) plus the two parents. H indicates heterozygous individuals. Running on 2.5% metaphor agarose gel stained with midori green. M 50 bp ladder
Genotyping BC1F1 generation
Foreground selection
The best F1 plants carrying the two tightly linked markers were backcrossed with Kulai to generate 60 BC1F1 plants. Forty-six individuals were heterozygous for the Hsp70-u2 and AGi42 markers tightly linked to Hsp genes (Fig. 3). The plants with the “H” score for the Hsp-linked marker were subjected to background selection with unlinked markers. The non-heterozygous condition of the other F1 individuals indicates that due to backcrossing, the Hsp gene disappeared.
Fig. 3.
Genotyping with markers Hsp70-u2 and AGi42 linked to Hsp genes in BC1F1 population of chilli derived from K Kulai × A AVPP0702. H indicates heterozygous individuals. Running on 2.5% metaphor agarose gel stained with midori green, only 14 samples plus the two parents for each marker are shown (M 50 bp ladder)
Genetic background selection
Sixty-eight polymorphic markers were initially used for selection in BC1F1 generation. The distribution of the SSR polymorphic markers on the 12 chromosomes varied, ranging from four (chromosome 2, 7 and 9) to seven on chromosomes 1, 3, 11 and 12. Individuals with fixed donor alleles were excluded from selection in the next generation. The recurrent parent genome recovery percentage was from 70.30 to 83.40% in the BC1F1 generation. The mean recurrent parent genome recovery of four favoured BC1F1 plants carrying the Hsp-linked genes was 80.75%. The recovery percentage and heterozygous segment of chosen plants in BC1F1 population were summarized and presented (Table 1). The Hsp gene from the donor parent is located on chromosome numbers 3 and 8. The best individual in BC1F1 generation was plant BC1-P7 with the highest recovery percentage (83.40%) and the lowest heterozygous regions with little or no linkage drag. Based on the selection for the Hsp linked and unlinked markers, four BC1F1 plants were chosen and used to produce BC2F1 populations. This finding was in conformity with the works of Cuc et al. (2012) who reported recovery percentage from 80.00 to 89.01% in BC1F1 populations. MAB approach for screening BC1F1 plants provides information regarding the physical map of the chosen plant; this is very important and makes selection for next generation easy. The use of exact primers located in the Hsp70 segment absolutely leads to reduced incorporation size of the Hsp in Kulai variety. It also lessens the chiasma between the gene regions, which decreased the false-positive results in selection for the target gene.
Table 1.
Recurrent parent genome recovery in percentage and heterozygous segment of selected individuals in BC1F1 population
| Progenies | K (recurrent parent) | H | A (donor parent) | Total (cM) |
|---|---|---|---|---|
| P1 | 76.3 | 13.3 | 10.4 | 1576.3 |
| P4 | 83.4 | 9.1 | 7.5 | 1576.3 |
| P6 | 82.6 | 11.6 | 5.8 | 1576.3 |
| P7 | 80.7 | 16.8 | 2.5 | 1576.3 |
Genotyping BC2F1 generation
Foreground selection
Two markers Hsp70-u2 and AGi42 tightly linked to Hsp genes for heat tolerance on chromosomes 3 and 8 (Magaji et al. 2016) were screened to select BC1F1 individuals carrying Hsp alleles and from which BC2F1 were generated. Out of 50 plants of BC2F1 generation, introgression of the heat-resistant gene was confirmed in 17 plants using the foreground markers. The proportion of sensitive and tolerant progenies in BC2F1 generation using the linked marker is shown in Table 2. The markers showed a good fit to the expected test cross ratio (1:1) for a single gene in BC2F1 population indicating an association with Hsp genes for heat tolerance in chilli pepper, which shows non-significance at 5% probability level. Target genes are now conveniently transferred through marker-assisted foreground selection, which confirms the target alleles in the individuals produced either by selfing of a cross or F1 crossed between two parental lines (Allard 1999). Foreground markers and the target gene are close together (tightly linked) on the same chromosome, hence carried together onto the next generation.
Table 2.
Proportion of sensitive and tolerant individuals in BC2F1 generation of the foreground marker using Chi-square test of association
| Foreground marker | Chromosome | Marker analysis | Chi square | Prob < 0.05 | |
|---|---|---|---|---|---|
| aa | Aa | ||||
| Hsp70-u2 | 3 | 46 | 54 | 0.64 | 0.42 |
| AGi42 | 8 | 48 | 52 | 0.16 | 0.69 |
Background selection
Sixty-eight polymorphic markers were used to screen among BC2F1 plants from a selection of plants possessing the target gene. The extent of recurrent parent genome recovery percentage was from 86.30 to 90.10% in BC2F1 generation. The four best plants having maximum recurrent parent genome introgression and possessing the desired gene were coupled favourably with phenotypic resemblance with the recipient parent. The mean recovery percentage of the individuals selected was 87.76%. The recovery percentage and heterozygous segment of chosen plants in BC2F1 population were summarized and presented in Table 3. The best individual carrying both linked markers in BC2F1 generation was plant BC1-P7-P10 with the highest recovery percentage (89.80%).
Table 3.
Recurrent parent genome recovery in percentage and heterozygous segment of selected individuals in BC2F1 population
| Progenies | K (recurrent parent) | H | A (donor parent) | Total (cM) |
|---|---|---|---|---|
| P7-9 | 88.6 | 5.7 | 5.8 | 1575.9 |
| P7-10 | 83.7 | 10.7 | 5.6 | 1575.9 |
| P1-3 | 89.8 | 10.2 | 0 | 1575.9 |
| P1-5 | 90.1 | 6.8 | 3.1 | 1575.9 |
| P1-10 | 86.6 | 4.9 | 4.9 | 1575.9 |
From our investigation, the backcross program was successful, having individuals satisfying the strongest conditions thereby making selection easier and more efficient. Based on this, the selected plant was used to generate the BC3F1 population. Using background analysis, several other reports confirm these findings including the works of Singh et al. (2013) who found the recurrent parent genome recovery at 91.6% in BC2F1, Cuc et al. (2012) at 93.75% and Khanh et al. (2013) who found recurrent parent genome recovery at 89.8%.
Genotyping BC3F1 generation
Foreground selection
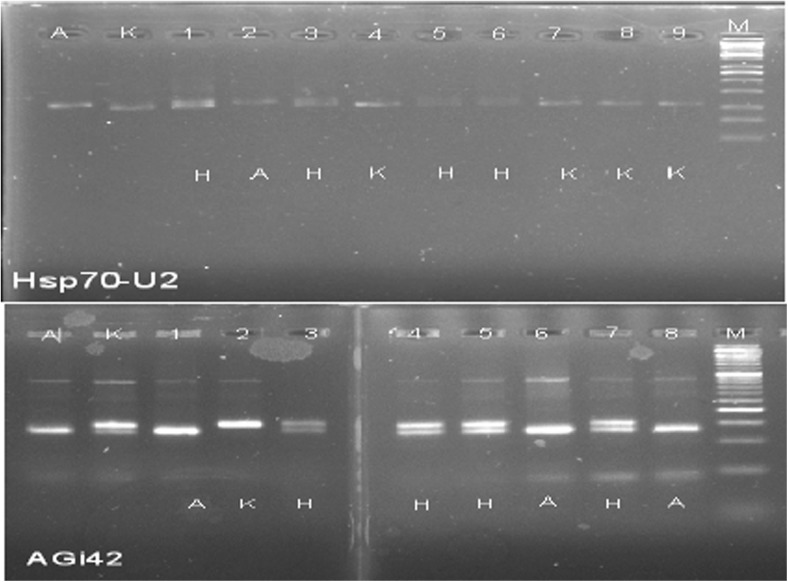
Hsp-tightly linked markers were used to confirm the introgression of Hsp genes in BC3F1 generation. A total of 20 BC3F1 plants confirmed the introgression of Hsp using Hsp70-u2 and AGi42 (Fig. 4). Both markers showed a goodness-of-fit to the expected test cross ratio (1:1) for a single gene in the BC3F1 population indicating an association with Hsp genes for heat tolerance in chilli pepper. Table 4 showed the proportion of sensitive and tolerant plants in BC3F1. This is confirmed by the works of Iftekharuddaula (2008) who reported 1:1 ratio in the BC generation.
Fig. 4.
BC3F1 confirmation using foreground markers Hsp70-u2 (9 individuals) and AGi42 (8 individuals). A AVPP0702 and K Kulai; H heterozygous region. Only eight individual samples were shown plus the two parents. M 50 bp ladder
Table 4.
Proportion of sensitive and tolerant individuals in BC3F1 generation of the foreground marker using Chi-square test of association
| Foreground marker | Chromosome | Marker analysis | Chi square | Prob < 0.05 | |
|---|---|---|---|---|---|
| aa | Aa | ||||
| Hsp70-u2 | 3 | 49 | 51 | 0.04 | 0.84 |
| AGi42 | 8 | 45 | 55 | 0.82 | 0.37 |
Though not reported in chilli peppers, incorporation of heat shock proteins has shown considerable success in the improvement of crop plants against heat stress. Ristic et al. (1998) reported that Hsp 45 kDa was observed in maize F2 plants that displayed an increased ability to recover from heat stress. Using MAB in our study, SSR markers closely linked to Hsp genes were used for foreground selection in the backcross generations. This accelerates the breeding process and reduces linkage drag.
Background selection
The recurrent genome recovery of the BC3F1 generation was also analysed from GGT2.0 software. The recurrent parent genome recovery percentage was from 94.60 to 96.50% in BC3F1 generation. The three best plants having maximum recurrent parent genome introgression and possessing the desired gene were coupled favourably with phenotypic resemblance with the recipient parent to generate the homozygous (carrying homozygous Hsp alleles) improved lines through selfing. The mean recovery percentage of the selected plants was 95.37% (Table 5). From this, the best plant BC1-P7-P10-P3 carrying the Hsp genes had a recovery percentage of 94.60%. Chromosome-wise recovery percentage of BC1-P7-P10-P3 plant was shown (Supplementary I).
Table 5.
Recurrent parent genome recovery in percentage and heterozygous segment of selected individuals in BC3F1 population
| Progenies | K (recurrent parent) | H | A (donor parent) | Total (cM) |
|---|---|---|---|---|
| P10-1 | 95.0 | 1.8 | 3.1 | 1575.9 |
| P10-3 | 94.6 | 1.9 | 3.4 | 1575.9 |
| P10-4 | 96.5 | 1.1 | 2.5 | 1575.9 |
Marker selection for maximum introgression recovery of the recipient parent speeds up the recovery percentage of the recurrent parent and delimits the number of backcross generations (Hasan et al. 2015; Miah et al. 2013; Hospital and Charcosset 1997). Apparently, the recipient genome is fully recovered on all chromosomes with some few donor and homozygous regions which can be eliminated when selfed to produce the BC3F2 homozygous lines, as reported (Basavara et al. 2010). With advanced generations, the recurrent parent genome recovery may increase due to the attachment of recipient parent allele from the heterozygous alleles. Young and Tanksley (1989) reported that selection against genetic drag can save ten generations using molecular markers in selection for unlinked markers, which accelerates the recovery of the recipient genome by 2 to 3 cycles. Factors that determine the number of backcross generations in backcross breeding program include the genetic distance between the recipient and non-recipient parents, the breeder’s preference and first-hand assessment in the early backcross generations on the phenotypic and genotypic selections. In our study, the backcross generation was extended to the third generation because the genome recovery of the recipient parent was relatively slow.
Agro-morphological performance of the BC3F1 generation
To confirm the successful recovery of the recurrent parent genome in the improved BC3F1 lines, the agronomical and morphological performances of the improved lines (carrying the heat-responsive Hsp genes) together with the adaptable recurrent parent were compared using both Tukey’s HSD test and independent t test procedures. Introgression of target genes (such as Hsp genes) and recovery of the recipient parent must be confirmed by phenotyping the cross-generation usually at the individual level.
The t test was satisfied via Folded F’s test, F(19) = 1.53 to 53.21, p = 0.037 to 0.790 (Table 6). This indicates the equality of variances was not significant for all the dependent variables except the number of fruits, so a pooled variance method was used to calculate t (Table 6). The independent-sampled t test was associated with no statistically significant effect, t(3) = 0.98 to 2.610, p = 0.079 to 0.586 for all dependent variables except for plant height t(3) = 9.350, p = 0.0013 (Table 6). This means that the growth and yield performance of the recurrent (recipient) parent and improved BC3F1 plants are similar, indicating a successful recovery of the recipient genome through marker-assisted backcross breeding after successful introgression of heat-tolerant Hsp gene. Hence, the results confirm the null hypothesis that performance of improved BC3F1 plants was recovered when compared to recipient plants.
Table 6.
Descriptive t test and homogeneity variance test statistics for the mean comparison between the recurrent (recipient) parent and BC3F1 lines using independent t test
| Genotype | Plant height | Number of fruits | Fruit length | Fruit weight | Yield (g/plant) |
|---|---|---|---|---|---|
| BC3F1 (improved lines) | 47.00b | 57.67 | 14.00 | 14.66 | 618.16 |
| Kulai | 72.17a | 64.00 | 17.23 | 17.68 | 818.10 |
| t test | 9.350 | 0.590 | 1.52 | 0.98 | 2.61 |
| p < 0.05 | 0.001 | 0.586 | 0.250 | 0.385 | 0.079 |
| Folded F test | 2.11 | 53.21 | 12.55 | 1.53 | 3.58 |
| p < 0.05 | 0.644 | 0.037 | 0.147 | 0.790 | 0.437 |
Means followed by different letters within the same column are significantly different at 5% level of probability
Consistently, the Tukey’s HSD test demonstrated similar phenotypic characteristics between the improved lines and the recipient parent except in the plant height (Table 7). It appears that the growth performance of the improved lines was maintained statistically as compared with the recipient from this study. Similar observations were reported by Lau et al. (2017). Phenotypic performance is an index of a good genotype if the genes have a major effect on the phenotype, and the error of the phenotype is minimal (Ye and Smith 2008).
Table 7.
Morphological performance of the recipient (recurrent) parent and improved BC3F1 lines using Tukey’s HSD test
| Variables | Kulai (recurrent parent) | BC3F1 (improved lines) |
|---|---|---|
| Plant height | 72.17a | 47b |
| Number of fruits | 64.00 | 57.67 |
| Fruit length | 17.23 | 14.00 |
| Fruit weight | 17.68 | 14.66 |
| Yield (g/plant) | 818.10 | 618.16 |
Means followed by different letters across the row are significantly different at 5% level of probability
Hsp gene expression in BC1F1, BC2F1 and BC3F1 backcross generations
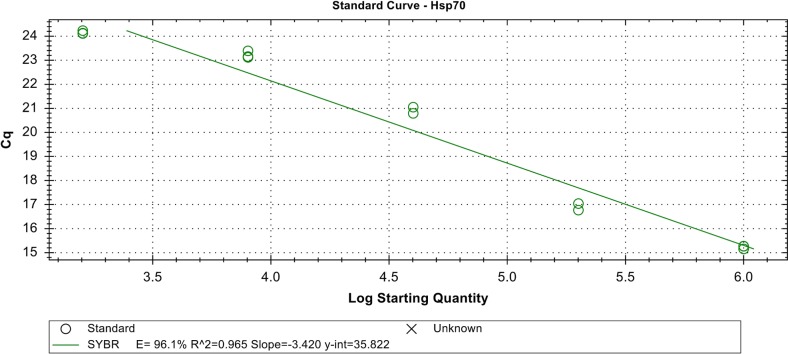
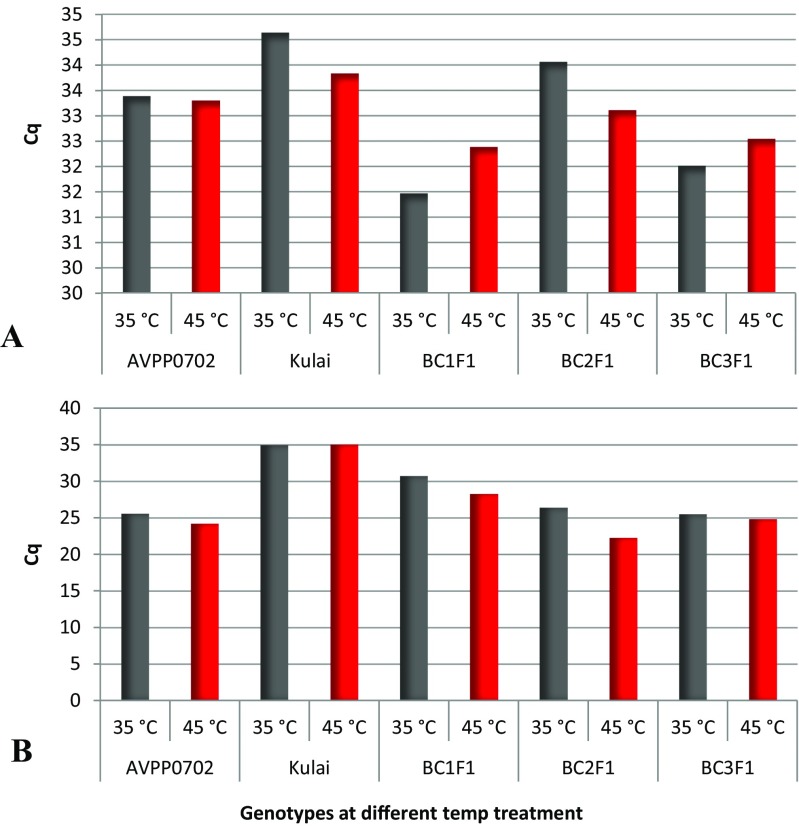
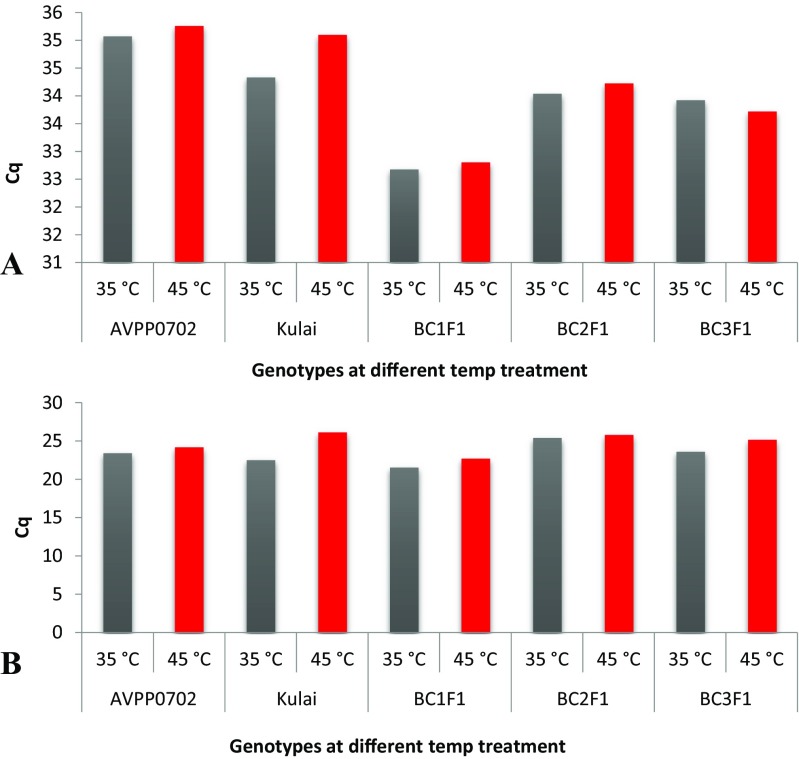
To assess the efficiency, accuracy, reliability and specificity of the gene of interest, a standard curve was generated using a 5-fold serial dilution of cDNA template amplified on the thermal cycler real-time system. Each dilution was assayed in duplicate. Figure 5 showed the amplification curves of the 5-point dilution series of Hsp70-u2 and standard curve with the CT plotted against the log of the starting quantity of template for each dilution. The results of quantitative RT-PCR showed that at 2-h exposure, the accumulation of target Hsp70-u2 gene was moderate across all the genotypes and weak in the recurrent parent (Fig. 6). Strong positive threshold cycle (Cq < 29) values were observed in the backcross generations at 4 h after heat exposure at 35 and 45 °C which is similar to the donor parent, indicative of abundant target nucleic acid in the samples. While Kulai, the recurrent parent, showed a weak reaction (Cq of 38–40) indicative of a minimal amount of the target nucleic acid in the sample. Similarly, OsHsp24 at 4-h heat treatment amplified early with lower Cq values, indicating an abundance of the nucleic acid transcript (Fig. 7).
Fig. 5.
Standard curve with the C q plotted against the log of the starting quantity of cDNA template for Hsp70-u2
Fig. 6.
Amplification levels of candidate target Hsp70-u2 gene in the different parents and backcross generations under differential heat treatment and exposure time (A 2 h and B 4 h), C q threshold cycle (triplicate) indicates the abundance of the target nucleic acid; the lower the C q level, the higher the nucleic acid (in this case, Hsp70-u2)
Fig. 7.
Accumulation of candidate target OsHsp24 gene in the different parents and backcross generations under differential heat treatment and exposure time (A 2 h and B 4 h), C q threshold cycle (triplicate) indicates the abundance of the target nucleic acid; the lower the C q level, the higher the nucleic acid (in this case, OsHsp24)
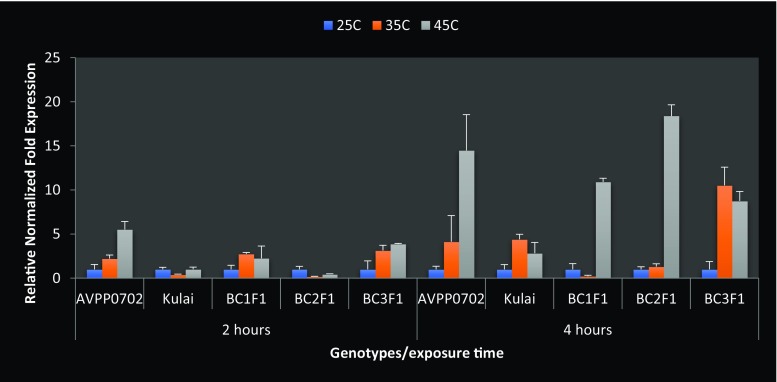
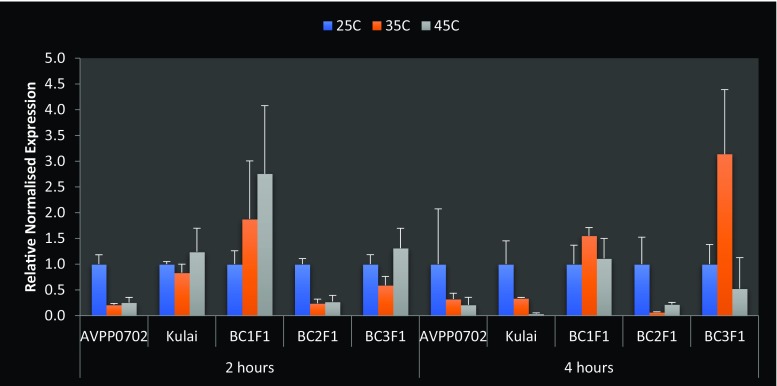
The results of quantitative RT-PCR showed that OsHsp24 and Hsp70-u2 were expressed at different heat stress treatments in the backcross generations including the parents; the expression of Hsp70-u2 in BC1F1, BC2F1 and BC3F1 was higher than that of the parents when compared with the control, and the optimal stress-inducing temperature was 45 °C. The expression peak in BC3F1 was noted when compared with the donor parent at 2 h after heat stress, and all the backcross generations consistently showed the response to heat treatment by expressing the incorporated heat-induced gene (Hsp70-u2) at 4 h exposure time (Fig. 8). This was true for the backcross individuals which were significantly up-regulated (Table 8) by more than 10.9-, 18.4- and 8.8-fold for BC1F1, BC2F1 and BC3F1, respectively (Fig. 8), like the expression level of the AVPP0702 (donor parent). The highest expression was given at BC2F1 generation with 18.37-fold increase in Hsp70-u2 (Fig. 8). Hsp70-u2 exhibit high transcript levels in the leaves indicating that this gene may play key roles in maintaining the normal leaf functions, such as respiration and photosynthesis. Kulai (the recurrent parent) was either down-regulated or less expressed in response to heat stress. Table 8 shows the regulation and significance of the gene expression at each heat treatment. However, as shown in Fig. 9, the OsHsp24 was expressed at different times under heat treatment. The mRNA of OsHsp24 was induced by heat stress but showed low-level expression compared to that of Hsp70-u2. The transcripts increased 2 h after the treatment at 45 °C and began to decrease at 4 h after treatment and gradually back to the control or beyond with slight increase at 35 °C in BC3F1 generation. Since this gene mainly increases expression within the first 2 h of heat treatment, they may function mainly at the early stage of heat stress response. From our investigation, after recovery for 24 h, the transcript levels of the OsHsp24 and Hsp70-u2 genes decreased to the level prior to stress treatment. This indicated that the Hsp genes were not destroyed compared with the control except that the Hsp70-u2 showed a slight decrease. This was however similar with the results of Ye et al. (2012).
Fig. 8.
Changes in the expression level of Hsp70-u2 gene in AVPP0702 (tolerant), “Kulai” (sensitive) BC1F1, BC2F1 and BC3F1 Capsicum annuum genotypes under heat shock treatment of 25, 35 and 45 °C for 2 and 4 h. UBI-3 and EF1-α were used as endogenous control. Error bars indicate SE (n = 3)
Table 8.
Gene regulation and probability value of the analysis of Hsp70 and OsHsp24 under differential heat stress condition among the parents and backcross generations
| 2 h | 4 h | ||||
|---|---|---|---|---|---|
| Population | Temp | Regulation | Sig (5%) | Regulation | Sig (5%) |
| Hsp70 | |||||
| AVPP0702 | 35.00 | Up-regulated | 0.251 | Up-regulated | 0.020 |
| 45.00 | Up-regulated | 0.026 | Up regulated | 0.023 | |
| Kulai | 35.00 | Down-regulated | 0.071 | Up-regulated | 0.050 |
| 45.00 | No change | 0.987 | Down-regulated | 0.030 | |
| BC1F1 | 35.00 | Up-regulated | 0.057 | Down-regulated | 0.303 |
| 45.00 | Up-regulated | 0.337 | No change | 0.819 | |
| BC2F1 | 35.00 | Down-regulated | 0.117 | No change | – |
| 45.00 | No change | 0.209 | Up-regulated | 0.001 | |
| BC3F1 | 35.00 | Up-regulated | 0.026 | Up-regulated | 0.027 |
| 45.00 | Up-regulated | 0.013 | Up-regulated | 0.075 | |
| OsHsp24 | |||||
| AVPP0702 | 35.00 | No change | 0.007 | No change | 0.423 |
| 45.00 | No change | 0.022 | Down-regulated | 0.385 | |
| Kulai | 35.00 | No change | 0.419 | No change | 0.240 |
| 45.00 | No change | 0.542 | Down-regulated | 0.147 | |
| BC1F1 | 35.00 | No change | 0.418 | No change | 0.528 |
| 45.00 | Up-regulated | 0.144 | No change | 0.857 | |
| BC2F1 | 35.00 | No change | 0.013 | Down-regulated | 0.170 |
| 45.00 | No change | 0.032 | Down-regulated | 0.215 | |
| BC3F1 | 35.00 | No change | 0.204 | No change | 0.437 |
| 45.00 | No change | 0.446 | No change | 0.898 | |
Fig. 9.
Changes in the expression level of OsHsp24 gene in AVPP0702 (tolerant), “Kulai” (sensitive), BC1F1, BC2F1 and BC3F1 Capsicum annuum genotypes under heat shock treatment of 25, 35 and 45 °C for 2 and 4 h. UBI-3 and EF1-α were used as endogenous control. Error bars indicate SE (n = 3)
Hsp70-u2 and OsHsp24 were found to be heat-inducible, suggesting that these genes contribute to the heat stress response via some mechanisms. A plant synthesized Hsps (stress-related proteins) when exposed to high temperature and signals from the cells, resulting from changing the expression of genes and accumulation of transcripts at the molecular level (Guo et al. 2014). In this study, individuals selected from BC1F1, BC2F1 and BC3F1 along with the parents were exposed to heat treatments to confirm the introgression of the Hsp gene(s) in the backcross generation for further selection. The heat stress induction was not distinct at first from the donor parent, while the accumulation of Hsp70 significantly increased after 4 h (about 18.36-fold) compared with the control and recurrent parent. The BC1F1, BC2F1 and BC3F1 showed almost similar expression pattern with the donor parent with the maximum level of expression at 45 °C treatment. Our data revealed that these genes have different specific expression profiles and different time exposure responses to heat stress, implying their differential function in abiotic stress response. High-temperature stress is detrimental throughout the growth period of pepper; for example, the extreme temperature can disturb pollination and fertilization, resulting in a decline in production and quality of pepper fruits (Guo et al. 2014).
Conclusion
Marker-assisted backcrossing strategy highlighted in this study was used successfully to incorporate Hsp genes into Kulai. In chilli pepper breeding programs, MAB was not fully utilized compared to cereals such as rice and wheat. The recovery percentage of the recipient genome was hastened indicating the potentiality of MAB approach to recover the genome of the recurrent parent in chillies. The improved heat-tolerant Kulai chilli carrying Hsp genes could serve as donors for heat tolerance in chilli pepper breeding programs, and this could become increasingly important as other desirable genes are introduced into the heat-tolerant Kulai chilli pepper. This approach could be further exploited to introgress not only Hsp genes in important varieties of chilli with a least possible introgression region and in the shortest possible time. Incorporated Hsp genes were found differentially expressed in the BC generations at different time exposures either more than or like the donor parent. It is expected that the newly developed heat-tolerant lines will be able to increase chilli pepper production to enhance and sustain future livelihoods and food security in Malaysia and other heat-prone areas in the context of climate change.
Electronic supplementary material
(DOCX 135 kb)
Acknowledgements
We are grateful to the Ministry of Education, Malaysia, for adequate research funding through the Fundamental Research Grant Scheme (FRGS/1/2012/STWN03/UPM/02/2: 07-01-13-1240FR). We also acknowledged the contribution of the Universiti Putra Malaysia to the success of the research through the Geran Putra (GP—IBT/2013/9421000).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-017-0836-3) contains supplementary material, which is available to authorized users.
References
- Alam M, Siwar C, Murad W, Toriman ME. Farm level assessment of climate change, agriculture and food security issues in Malaysia. World Appl Sci J. 2011;14(3):431–442. [Google Scholar]
- Allard RW. Principles of plant breeding. 2. New York: Wiley; 1999. [Google Scholar]
- AnLi G, HuaGang H, QuanZhan C, ShouZhong Z, PeiDu C (2005) Pyramiding wheat powdery mildew resistance genes Pm2, Pm4a and Pm21 by molecular marker-assisted selection. Acta Agronom Sinica 31:1400–1405
- AVRDC, IPGRI, CATE . Descriptors for Capsicum (Capsicum spp) Rome: International Plant Genetic Resources Institute; 2001. [Google Scholar]
- Babu R, Nair SK, Kumar A, Venkatesh S, Sekhar JC, Singh NN, Srinivasan G, Gupta HS. Two-generation marker-aided backcrossing for rapid conversion of normal maize lines to quality protein maize (QPM) Theor Appl Genet. 2005;111:888–897. doi: 10.1007/s00122-005-0011-6. [DOI] [PubMed] [Google Scholar]
- Barone A, Ercolano MR, Langella R, Monti L, Frusciante L. Molecular marker-assisted selection for pyramiding resistance genes in tomato. Adv Hortic Sci. 2005;19:147–152. [Google Scholar]
- Basavara SH, Singh VK, Singh AT, Singh AS, Singh AN, Anand D, Yadav S, Ellur RK, Singh D, Gopalakrishnan S, Nagarajan M, Mohapatra T, Prabhu KV, Singh AK. Marker assisted improvement of bacterial blight resistance in parallel lines of PusaRH10, a superfine grain aromatic rice hybrid. Mol Breed. 2010;26:293–305. doi: 10.1007/s11032-010-9407-3. [DOI] [Google Scholar]
- Chamikara MDM, Ishan M, Karunadasa SS, Perera MKDI, Rajapaksha PI, Lelwala RV, Sooriyapathirana SDSS (2015) Morphological and microsatellite marker analysis of fruit size and shape in selected accessions and commercial cultivars of capsicum species in Sri Lanka. Intl J of multidisciplinary Studies 2(1):11–32
- Chen ZW, Guan HZ, Wu WR, Zhou YC, Han QD. The screening of molecular markers closely linked to rice blast-resistant gene Pi-1 and their application. J Fujian Agric For Univ. 2005;34:74–77. [Google Scholar]
- Chong AL, Mathews P (2001) National Response Strategies to Climate Change. In. Putrajaya: Ministry of Science, Technology and the Environment pp 389–422. MOSTE publisher, Putrajaya Malaysia
- Cuc IM, Huyen LTN, Hien PTM, Hang VTT, Dam N, Mui PT, Quang VD, Ismail AM, Ham LH. Application of marker assisted backcrossing to introgress the submergence tolerance QTL SUB1 into the Vietnam elite rice variety—AS996. Am J Plant Sci. 2012;3:528–536. doi: 10.4236/ajps.2012.34063. [DOI] [Google Scholar]
- De la Peña R, Hughes J. Improving vegetable productivity in a variable and changing climate. J SAT Agric Res. 2007;4(1):1–22. [Google Scholar]
- DoA (Department of Agriculture) (2014) Ministry of Agriculture. Malaysia: Kuantiti (MT) dan Nilai (RM) Export Sayur sayuran mengikut Negara dituju@Destinasi baagi tahun 2014
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus (San Franc) 1990;12:13–15. [Google Scholar]
- Guo M, Zhai YF, Lu JP, Chai L, Chai WG, Gong ZH, Lu MH. Characterization of CaHsp70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L. Int J Mol Sci. 2014;15:19741–19759. doi: 10.3390/ijms151119741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MM, Rafii MY, Ismail MR, Mahmood M, Rahim HA, Alam MA, Ashkani S, Malek MA, Latif MA. Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol Biotechnol Equip. 2015;29(2):237–254. doi: 10.1080/13102818.2014.995920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F, Charcosset A. Marker-assisted introgression of quantitative trait loci. Genetics. 1997;147:1469–1485. doi: 10.1093/genetics/147.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurni S, Brunner S, Stirnweis D, Herren G, Peditto D, McIntosh RA. The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J. 2014;79:904–913. doi: 10.1111/tpj.12593. [DOI] [PubMed] [Google Scholar]
- Iftekharuddaula KM (2008) Comparison of new selection strategies or marker assisted backcrossing for a submergence tolerant gene rice. Doctoral dissertation, Bangladesh Agricultural University Mymensingh, Bangladesh, pp 1–205
- Ince AG, Karaca M, Onus AN. Polymorphic microsatellite markers transferable across Capsicum species. Plant Mol Biol Report. 2010;28(2):285–291. doi: 10.1007/s11105-009-0151-y. [DOI] [Google Scholar]
- Khanh TD, Linh LH, Linh TH, Ham IH, Xuan TD. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into the Vietnamese elite rice variety. J Plant Breed Crop Sci. 2013;5:26–33. doi: 10.5897/JPBCS12.052. [DOI] [Google Scholar]
- Lang NT, Ha PTT, Tru PC, Toan TB, Buu BC, Cho YC. Breeding for heat tolerance rice based on marker-assisted backcrossing in Vietnam. Plant Breed Biotechnol. 2015;3(3):274–281. doi: 10.9787/PBB.2015.3.3.274. [DOI] [Google Scholar]
- Larkindale J, Knight MR. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 2002;128:682–695. doi: 10.1104/pp.010320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WCP, Rafii MY, Ismail MR, Puteh A, Latif MA, Asfaliza R, Miah G. Development of advanced fragrant rice lines from MR269 3 × Basmati 370 through marker-assisted backcrossing. Euphytica. 2017;213:11. doi: 10.1007/s10681-016-1794-z. [DOI] [Google Scholar]
- Lawson DM, Lunde CF, Mutschler MA. Marker-assisted transfer of acylsugar-mediated pest resistance from the wild tomato Lycopersicon pennellii to the cultivated tomato Lycopersicon esculentum. Mol Breed. 1997;3:307–317. doi: 10.1023/A:1009677412902. [DOI] [Google Scholar]
- Lecomte L, Duffé P, Buret M, Servin B, Hospital F, Causse M. Marker-assisted introgression of five QTLs controlling fruit quality traits into three tomato lines revealed interactions between QTLs and genetic backgrounds. Theor Appl Genet. 2004;109:658–668. doi: 10.1007/s00122-004-1674-0. [DOI] [PubMed] [Google Scholar]
- Lindquist S (1986) The heat-shock response. Annu Rev Biochem 55:1151–1191 [DOI] [PubMed]
- Magaji UG, Rafii MY, Martini MY, Yusuff OA, Ismail MR. Polymorphism between popular Kulai and heat tolerant donor AVPP0702 chilli peppers revealed through SSR markers. Trans Persatuan Genet Malays. 2016;3:39–44. [Google Scholar]
- Malaysian Agriculture Research and Development Institute—MARDI (2009) Ingin Meningkatkan Hasil Dan Kualiti Produk Pertanian Anda? MARDI publisher, Serdang, Selangor, Malaysia
- Miah G, Rafii MY, Ismail MR, Puteh AB, Rahim HA, Islam KN, Latif MA. A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int J Mol Sci. 2013;14(11):22499–22528. doi: 10.3390/ijms141122499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miah G, Rafii MY, Ismail MR, Puteh AB, Rahim HA, Latif MA. Recurrent parent genome recovery analysis in a marker-assisted backcrossing program of rice (Oryza sativa L.) C R Biol. 2015;338(2):83–94. doi: 10.1016/j.crvi.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45–e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic Z, Yang G, Martin B, Fullerton S. Evidence of association between specific heat-shock protein (s) and the drought and heat tolerance phenotype in maize. J Plant Physiol. 1998;153(30):497–505. doi: 10.1016/S0176-1617(98)80180-6. [DOI] [Google Scholar]
- Singh VK, Singh A, Singh SP, Ellur RK, Singh D, Krihnan SG, Bhowmick PK, Nagarajan M, Vinod KK, Singh UD, Mohapatra T, Prabhu KV, Singh AK (2013) Marker assisted simultaneous but stepwise backcross breeding for pyramiding blast resistance genes Piz5 and Piz54 into an elite Basmati rice restorer line ‘PRR78’. Plant Breed (132):486–495
- Thabuis A, Palloix A, Servin B, Daubeze AM, Signoret P, Lefebvre V. Marker-assisted introgression of 4 Phytophthora capsici resistance QTL alleles into a bell pepper line: validation of additive and epistatic effects. Mol Breed. 2004;14(1):9–20. doi: 10.1023/B:MOLB.0000037991.38278.82. [DOI] [Google Scholar]
- Tompa P, Kovacs D (2010) Intrinsically disordered chaperones in plants and animals. This paper is one of a selection of papers published in this special issue entitled-Canadian Society of Biochemistry, Molecular & Cellular Biology 52nd Annual Meeting-Protein Folding: Principles a Biochem Cell Biol 88: 167–174 [DOI] [PubMed]
- Usman MG, Rafii MY, Ismail MR, Malek MA, Latif MA (2014) Heritability and genetic advance among chili pepper genotypes for heat tolerance and morphophysiological characteristics. ScientificWorldJournal 308042. doi:10.1155/2014/308042 [DOI] [PMC free article] [PubMed]
- Usman MG, Rafii MY, Ismail MR, Malek MA, Latif MA. Expression of target gene Hsp70 and membrane stability determine heat tolerance in chili pepper. J Am Soc Hortic Sci. 2015;140:144–150. [Google Scholar]
- Usman MG, Rafii MY, Ismail MR, Martini MY, Yusuff OA, Miah G (2017) Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnol Genet Eng Rev. doi:10.1080/02648725.2017.1340546 [DOI] [PubMed]
- Van Berloo R. GGT 2.0: versatile software for visualization and analysis of genetic data. J Heredity. 2008;99(2):232–236. doi: 10.1093/jhered/esm109. [DOI] [PubMed] [Google Scholar]
- Ye G, Smith KF. Marker-assisted gene pyramiding for inbred line development: basic principles and practical guidelines. Int J Plant Breed. 2008;2(1):1–10. doi: 10.3923/ijpbg.2008.1.8. [DOI] [Google Scholar]
- Ye S, Yu S, Shu L, Wu J, Wu A, Luo L. Expression profile analysis of 9 heat shock protein genes throughout the life cycle and under abiotic stress in rice. Chin Sci Bull. 2012;57:336–343. doi: 10.1007/s11434-011-4863-7. [DOI] [Google Scholar]
- Young JC. Mechanisms of the Hsp70 chaperone system. Biochem Cell Biol. 2010;88:291–300. doi: 10.1139/O09-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Tanksley SD. Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor Appl Genet. 1989;77:95–101. doi: 10.1007/BF00292322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 135 kb)