Abstract
microRNA (miR)-612 shows anticancer activity in several types of cancers, yet its function in melanoma is still unclear. This study was undertaken to investigate the expression of miR-612 and its biological relevance in melanoma cell growth, invasion, and tumorigenesis. The expression and prognostic significance of miR-612 in melanoma were examined. The effects of miR-612 overexpression on cell proliferation, colony formation, tumorigenesis, and invasion were determined. Rescue experiments were conducted to identify the functional target gene(s) of miR-612. miR-612 was significantly downregulated in melanoma tissues compared to adjacent normal tissues. Low miR-612 expression was significantly associated with melanoma thickness, lymph node metastasis, and shorter overall, and disease-free survival of patients. Overexpression of miR-612 significantly decreased cell proliferation, colony formation, and invasion of SK-MEL-28 and A375 melanoma cells. In vivo tumorigenic studies confirmed that miR-612 overexpression retarded the growth of A375 xenograft tumors, which was coupled with a decline in the percentage of Ki-67-positive proliferating cells. Mechanistically, miR-612 targeted Espin in melanoma cells. Overexpression of Espin counteracted the suppressive effects of miR-612 on melanoma cell proliferation, invasion, and tumorigenesis. A significant inverse correlation (r = −0.376, P = 0.018) was observed between miR-612 and Espin protein expression in melanoma tissues. In addition, overexpression of miR-612 and knockdown of Espin significantly increased the sensitivity of melanoma cells to doxorubicin. Collectively, miR-612 suppresses the aggressive phenotype of melanoma cells through downregulation of Espin. Delivery of miR-612 may represent a novel therapeutic strategy against melanoma.
Keywords: aggressiveness, downregulation, metastasis, microRNA, target
INTRODUCTION
Melanoma is one of the most lethal malignancies worldwide, accounting for 80% of skin cancer-related deaths (Kircher et al., 2016; van Akkooi et al., 2016). It has a high metastatic potential and poor prognosis. The median survival for advanced melanoma is less than 1 year (Pal et al., 2016; Tas, 2012). Deciphering the mechanism governing the aggressiveness of melanoma is critical for the development of effective therapeutic strategies against this malignancy.
Espin encodes an actin-binding protein and is implicated in actin cytoskeleton remodeling. Mutations in Espin cause deafness in both humans and mice (Donaudy et al., 2006; Zheng et al., 2000). It contains one actin-bundling module, which is necessary for F-actin bundling activity (Sekerková et al., 2006). Espin has exhibited the ability to modulate cell growth, migration, and invasion (Taura et al., 2016; Wang et al., 2012). A previous study has demonstrated that Espin expression is increased in human primary and metastatic melanomas and that depletion of Espin significantly impairs the invasion capacity of melanoma cells (Yanagishita et al., 2014). This study provides evidence for the oncogenic role of Espin in melanoma.
microRNAs (miRs) are a large family of small non-coding RNA molecules that negatively regulate gene expression on the post-transcriptional level, typically through binding to the 3′-untranslated region (UTR) of target mRNAs (Cheerla and Gevaert, 2017). Although thousands of miRs have been detected in cancers (Shu et al., 2017; Zhang et al., 2016), the functions of most of them in tumor progression are not elucidated. Several miRs have been shown to contribute to the aggressive phenotype of melanoma cells (Komina et al., 2016; Xu et al., 2016). For instance, inhibition of miR-4286 exerts antiproliferative and pro-apoptotic effects on melanoma cells (Komina et al., 2016). It was found that miR-9 can suppress the growth and invasion of malignant melanoma cells (Xu et al., 2016).
miR-612 is one less characterized miR. Recent studies have reported that miR-612 acts as a tumor suppressor in colorectal cancer (Sheng et al., 2015) and liver cancer (Tang et al., 2014). Overexpression of miR-612 can inhibit the epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma (Tao et al., 2013). However, its expression and function in melanoma is still elusive. The aim of this study is to determine the expression and clinical relevance of miR-612 in melanoma and uncover its biological role in melanoma cell growth, invasion, and tumorigenesis.
MATERIALS AND METHODS
Tissue specimens
In this study, 89 cases of melanoma tissues (37 primary melanomas and 52 metastatic melanomas) and matched adjacent normal tissues were collected from melanoma patients who underwent surgical resection at Qilu Hospital (China). This cohort included 56 males and 33 females, with a median age of 51 years (range 39–78 years). The patients receiving any anticancer treatment before operation were excluded. Tissue specimens were snap-frozen in liquid nitrogen immediately after surgery and stored at −80°C until use. This study was approved by the Ethics Committee of Shandong University (China), and written informed consent for research purposes was obtained from all patients.
Cell lines
Human melanoma cell lines (SK-MEL-28, SK-MEL-3, A375, HT-144, and Hs294T) were purchased from the American Type Culture Collection (ATCC, USA) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactive fetal bovine serum (FBS; Gibco/Thermo Fisher Scientific, USA) in a CO2 incubator at 37°C. Human epidermal melanocytes were purchased from Scien-Cell Research Laboratories (USA) and cultured in Melanocyte Medium (ScienCell Research Laboratories) containing 10% FBS. HEK293T cells were purchased from the Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences (China) and cultured in DMEM with 10% FBS.
Doxorubicin treatment
Melanoma cell lines were plated in 96-well plates (1 × 104 cells/well) and exposed to different concentration of doxorubicin (0, 1, 2, 5, and 10 μM; Sigma-Aldrich, USA) for 24 h. The cell culture was added with 0.5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich). After incubation for 4 h at 37°C, dimethyl sulfoxide was added. Absorbance was measured at 570 nm. The half maximal inhibitory concentration (IC50) for doxorubicin was calculated.
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from tissue samples and cell lines using TRIzol reagent (Invitrogen, USA). The expression level of miR-612 was measured by qRT-PCR using the TaqMan MicroRNA Assay Kit (Applied Biosystems, USA). RNU6B was used as an endogenous control.
Plasmids
To generate a miR-612-expressing plasmid, a DNA fragment containing human miR-612 precursor and its flanking sequence was amplified by PCR and inserted into pcDNA3.1(+) vector. Human Espin-encoding cDNA (Origene Technologies, USA) lacking the 3′-UTR was amplified by PCR and cloned into pBABE-puro vector. For luciferase reporter assays, the entire Espin 3′-UTR was obtained by PCR and inserted into pLUC firefly luciferase vector. The mutated Espin 3′-UTR with disruption of the miR-612 binding site was generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Germany) according to the manufacturer’s protocol. All the plasmids were validated by sequencing.
Cell transfection
Transfection of the plasmids into cells was performed using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s instructions. To generate miR-612 stable clones, melanoma cells were transfected with the miR-612-expressing plasmid and after 24 h, selected in the presence of 800 μg/ml G418 (Sigma-Aldrich) for 2 weeks. For co-expression of miR-612 and Espin, miR-612 stable cell lines were re-transfected with the pBABE-Espin plasmid and then selected in the presence of 800 μg/ml G418 and 2 μg/ml puromycin (Sigma-Aldrich) for 1 week. For knockdown of Espin, specific small interfering RNAs (siRNAs) targeting Espin (Santa Cruz Biotechnology, USA) were transfected at a concentration of 40 nM.
Cell proliferation assay
Cell proliferation was measured using the Cell Proliferation ELISA, BrdU (colorimetric) Kit (Roche Applied Science, USA), as described previously [12]. Absorbance was measured at 370 nm.
Colony formation assay
Colony formation assay was done as described previously [13]. Transfected cells (500 cells per well) were seeded onto 6-well plates and cultured for 10 days. Colonies were stained with 0.5% crystal violet (Sigma-Aldrich) solution and counted.
Xenograft tumor studies in mice
In vivo tumorigenic studies in mice were performed as described previously (Alaoui-Jamali et al., 2009). In brief, A375 melanoma cells stably expressing miR-612 or together with Espin (1 × 106 cells per mouse) were subcutaneously implanted into the flanks of nude mice (5-week old; Shanghai Laboratory Animal Center, Chinese Academy of Sciences, China). Tumor volume was calculated weekly for 4 weeks, and growth curves were plotted. After the last measurement, the animals were killed. Xenograft tumors were fixed, sectioned, and stained with anti-Ki-67 antibody (ab15580, Abcam, Cambridge, UK; 1:200 dilution), as described previously (Waters et al., 2015). Animal experiments were approved by the Animal Care and Use Committee of Shandong University.
In vitro wound-healing assay
Cells were seeded in 6-wells plates (3 × 106 cells/well) and allowed to grow to confluence and incubated with 10 μg/mL mitomycin C (Sigma-Aldrich) for 2 h. A scratch wound was made with a 200-μl pipette tip. Cells were further cultured in DMEM medium containing 10% FBS. Pictures were taken at 0 and 48 h after scratching. The percentage of wound closure was determined from three independent experiments.
Transwell invasion assay
Cancer cell invasive ability was assessed using Transwell invasion assay (Li et al., 2016). In brief, 2 × 104 cells suspended in serum-free medium were seeded in the upper chamber of 24-well Transwell plates, which were precoated with Matrigel (BD Biosciences, USA). The lower chamber was filled with the cell culture medium containing 10% FBS. After incubation for 48 h, cells that had invaded through Matrigel membrane were fixed, stained with 0.1% crystal violet, and counted.
Luciferase reporter assay
HEK293T cells were co-transfected with the Espin 3′-UTR reporter constructs together with the miR-612-expressing plasmid or vector using Lipofectamine 2000. The pRL-TK vector (Promega, USA) that encodes Renilla luciferase was also transfected to control for transfection efficiency. Luciferase activities were measured with the Dual-Luciferase Reporter System (Promega) 48 h after transfection.
Western blot analysis
Tissue or cell lystates were prepared using radioimmunoprecipitation assay (RIPA) buffer (Beyotime, China) supplemented with protease and phosphatase inhibitors (Sigma-Aldrich). Protein samples were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. After blocking non-specific binding sites, membranes were incubated with anti-Espin (sc-393469, Santa Cruz Biotechnology; 1:500 dilution), anti-E-cadherin (ab15148, Abcam, Abcam, USA; 1:800 dilution), anti-vimentin (ab8978, Abcam; 1:500 dilution), and anti-β-actin (Sigma-Aldrich; 1:2000 dilution) antibodies, followed by incubation with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). Protein bands were visualized using an enhanced chemiluminescence detection system (Thermo Fisher Scientific, USA). Densitometric analysis was carried out using the Quantity One software (Bio-Rad Laboratories, USA).
Statistical analysis
Data are expressed as mean ± standard deviation and analyzed by the Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s Post-hoc test. Associations of miR-612 expression with demographic and clinicopathological characteristics of patients were evaluated by the chi-square test. Kaplan-Meier and log-rank tests were conducted to determine the correlation between miR-612 expression and overall and disease-free survival of melanoma patients. The correlation between miR-612 and Espin protein expression was determined by Pearson correlation analysis. The P values less than 0.05 were considered significant.
RESULTS
Clinical significance of miR-612 expression in melanoma
We first examined the levels of miR-612 in a set of melanoma tissues and their matched noncancerous normal tissues. As illustrated in Fig. 1A, the miR-612 transcript level was significantly lower in melanoma tissues than in paired adjacent normal tissues (P = 0.0154). According to the median level of miR-612, the series of 89 melanoma specimens were divided into two groups, low (n = 46) and high (n = 43) miR-612 expression. We then analyzed the relationship between miR-612 expression and clinical features of melanoma patients. Low miR-612 expression was significantly associated with melanoma thickness (P = 0.0013) and lymph node metastasis (P = 0.0169; Table 1), suggesting that miR-612 may modulate melanoma progression. We then examined the association of miR-612 expression with patient survival. The Kaplan-Meier survival analysis revealed that low miR-612 expression was significantly associated with shorter overall (P = 0.0138) and disease-free survival (P = 0.0022) in melanoma patients (Fig. 1B).
Fig. 1. Downregulation of miR-612 predicts poor prognosis in melanoma patients (n = 89).
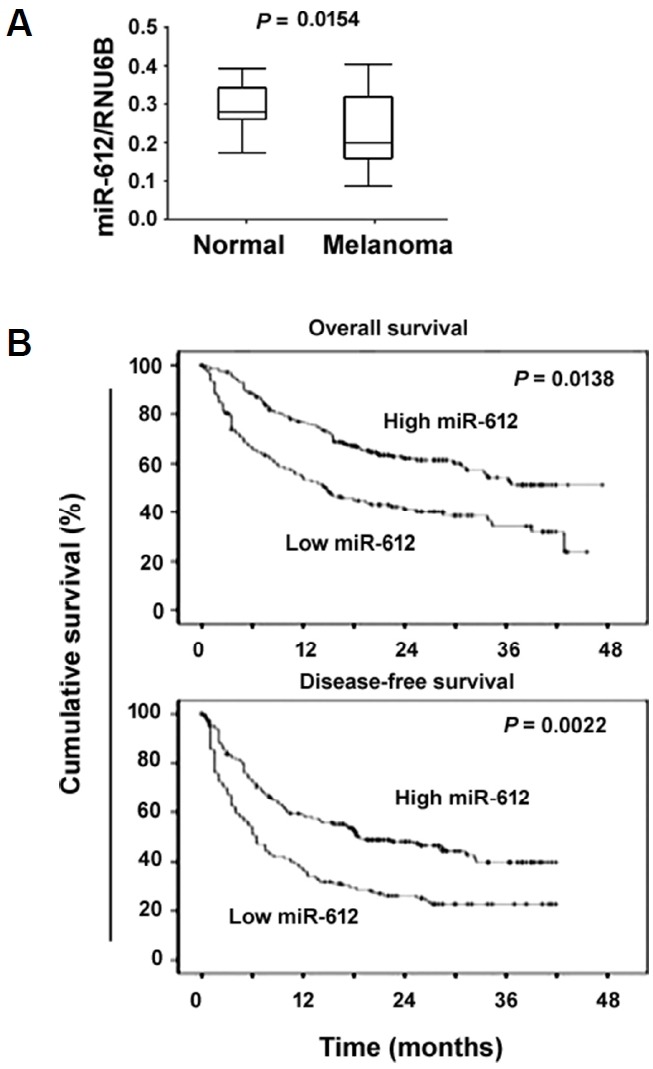
(A) qRT-PCR analysis of miR-612 levels in 89 pairs of melanoma tissues and adjacent normal tissues. (B) Kaplan-Meier analysis of the correlation between miR-612 expression and melanoma patient survival. Significance was determined by the log-rank test.
Overexpression of miR-612 reduces melanoma growth in vitro and in vivo
Next, we explored the function of miR-612 in melanoma progression. Compared to non-malignant melanocytes, miR-612 was commonly downregulated in melanoma cell lines (Fig. 2A). Overexpression of miR-612 (Fig. 2B) significantly reduced proliferation in both SK-MEL-28 and A375 melanoma cells, as determined by BrdU incorporation assays (P < 0.05; Fig. 2C). Colony-forming assays revealed that miR-612-overexpressing melanoma cells formed a significantly lower number of colonies than control cells (P < 0.05; Fig. 2D). In vivo xenograft studies confirmed that tumors derived from miR-612-overexpressing A375 melanoma cells grew significantly slower than control tumors (Fig. 2E). The tumor volume obtained at the last measurement in the miR-612 group was 28% of control values (P < 0.05). The tumors were then stained for Ki-67, a proliferation marker. A significant decline in the percentage of Ki-67-positive proliferating cells was noted in the miR-612 overexpression group relative to the control group (20.5 ± 4.8% vs. 53.2 ± 6.5%, P < 0.05; Fig. 2F).
Fig. 2. Restoration of miR-612 reduces proliferation and tumorigenesis.
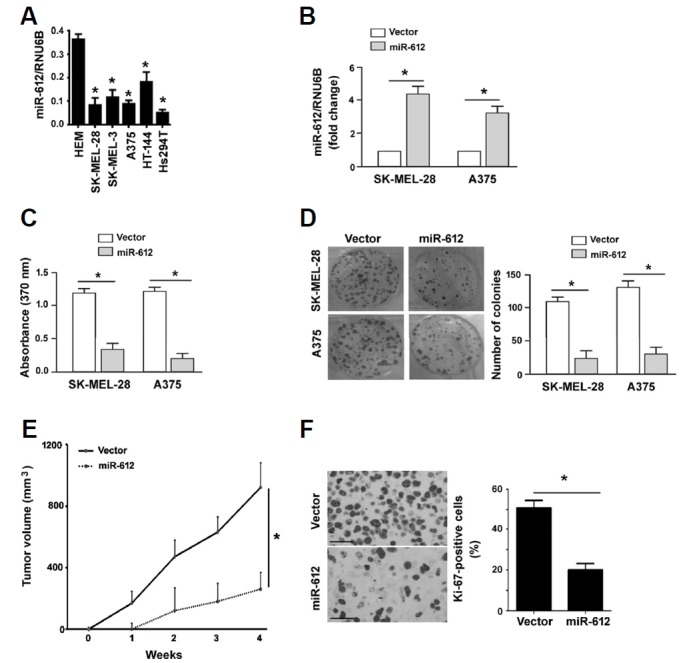
(A) Measurement of miR-612 levels in human epidermal melanocytes (HEM) and melanoma cell lines. *P < 0.05 relative to HEM. (B) qRT-PCR analysis of miR-612 expression in melanoma cells transfected with vector or miR-612-expressing plasmid. *P < 0.05. (C) BrdU incorporation assays were performed to assess the proliferation of melanoma cells transfected with vector or miR-612-expressing plasmids. (D) Colony formation assay. Melanoma cells stably transfected with vector or miR-612-expressing plasmids were cultured for 10 days to allow to form colonies. Representative images of colonies are shown in left panels. (E) Xenograft tumor studies. Subcutaneous tumors derived from miR-612-overexpressing A375 melanoma cells grew significantly slower than control tumors (n = 4). (F) Ki-67 immunohistochemistry. Sections of xenograft tumors were stained with anti-Ki-67 antibody. Right, quantification of Ki-67 immunostaining. Scale bar = 50 μm. *P < 0.05.
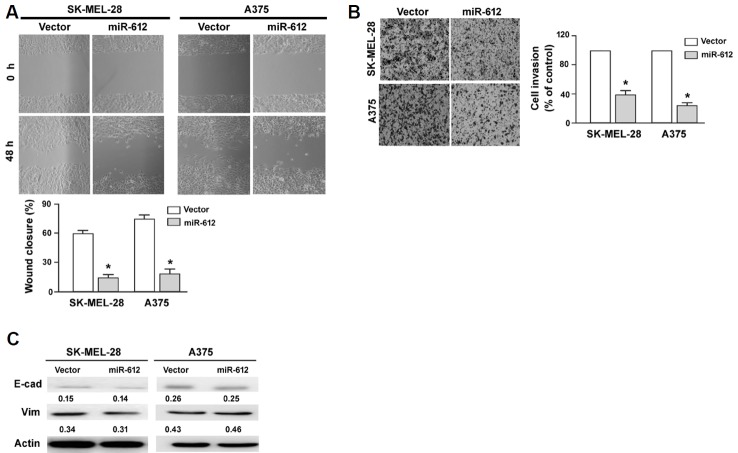
Overexpression of miR-612 suppresses melanoma cell migration and invasion
Next, we investigated whether miR-612 participated in the regulation of migration and invasion of melanoma cells. In vitro wound-healing assay demonstrated that miR-612-overexpressing SK-MEL-28 and A375 cells had a significantly lower migration capacity than corresponding controls (P < 0.05; Fig. 3A). Transwell invasion assays showed that ectopic expression of miR-612 led to a 60% and 72% reduction of invasion in SK-MEL-28 and A375 cells, respectively (P < 0.05 vs. control cells; Fig. 3B). Western blot analysis of epithelial-mesenchymal transition (EMT) markers showed that the expression of E-cadherin and vimentin remained unchanged after miR-612 overexpression. Collectively, these data indicate that miR-612 has suppressive activity against melanoma.
Fig. 3. miR-612 inhibits melanoma cell migration and invasion.
(A) In vitro wound healing assays demonstrated that miR-612 overexpression significantly inhibited the migration of SK-MEL-28 and A375 cells. (B) Transwell invasion assay. Ectopic expression of miR-612 significantly suppressed the invasion of SK-MEL-28 and A375 cells. Top, representative images of invaded cells. Bottom, quantification of cell invasion. *P < 0.05 relative to vector-transfected cells. (C) Western blot analysis of E-cadherin (E-cad) and vimentin (Vim) protein levels in SK-MEL-28 and A375 cells transfected with indicated constructs. Numbers indicate normalized protein levels, which were determined from three independent experiments.
miR-612 targets Espin in melanoma cells
To gain more insight into the role of miR-612 in melanoma, we attempted to identify functionally relevant targets. Using the TargetScan algorithm (http://www.targetscan.org/vert_71/), we found that the Espin gene harbored a putative miR-612 responsive site in its 3′-UTR (Fig. 4A). Luciferase reporter assays demonstrated that miR-612 overexpression significantly suppressed the expression of the reporter gene with wild-type but not mutant 3′-UTR of Espin (Fig. 4B). Consistently, enforced expression of miR-612 caused a significant reduction of Espin protein expression in both SK-MEL-28 and A375 cells, compared to vector-transfected cells (P < 0.05; Fig. 4C). Clinical evidence further supported that there was a significant inverse correlation between miR-612 and Espin protein expression in melanoma tissues (r = −0.376, P = 0.018; Fig. 4D).
Fig. 4. miR-612 targets Espin in melanoma cells.
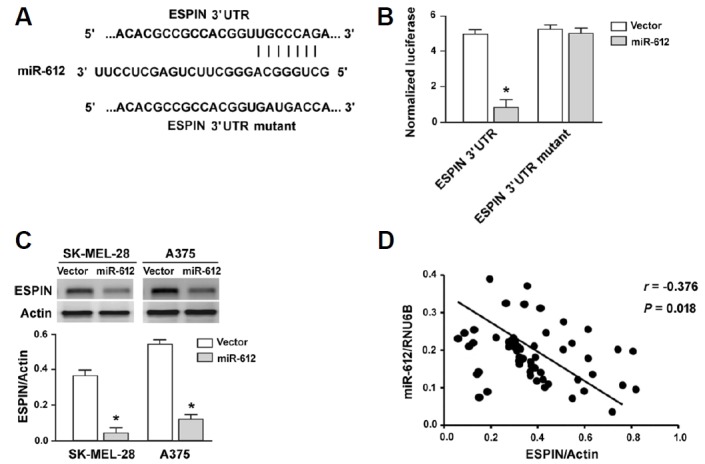
(A) prediction of a putative miR-612 responsive site in the 3′-UTR of Espin based on the TargetScan algorithm (http://www.targetscan.org/vert_71/). (B) Luciferase reporter assay showing that miR-612 overexpression significantly suppressed the expression of the reporter gene with wild-type Espin 3′-UTR reporters. (C) Western blot analysis of ESPIN protein levels in melanoma cells transfected with vector or miR-612-expressing plasmids. *P < 0.05 relative to vector-transfected cells. (D) Pearson correlation analysis was performed to determine the correlation between miR-612 and Espin protein expression in 75 cases of melanoma tissues.
Overexpression of Espin rescues melanoma cells from miR-612-mediated anticancer activity
Next, we conducted rescue experiments to validate the involvement of Espin in the activity of miR-612 in melanoma. As determined by Western blot analysis, co-transfection with the Espin-expressing plasmid resulted in abundant expression of Espin in miR-612-overexpressing A375 cells (Fig. 5A). In vitro studies confirmed that enforced expression of Espin rendered melanoma cells more resistant to miR-612-mediated suppression of growth (Fig. 5B) and invasion (Fig. 5C). Moreover, overexpression of Espin partially restored the growth of xenograft tumors formed by miR-612-overexpressing A375 cells (Fig. 5D).
Fig. 5. Overexpression of Espin rescues melanoma cells from miR-612-mediated anticancer activity.
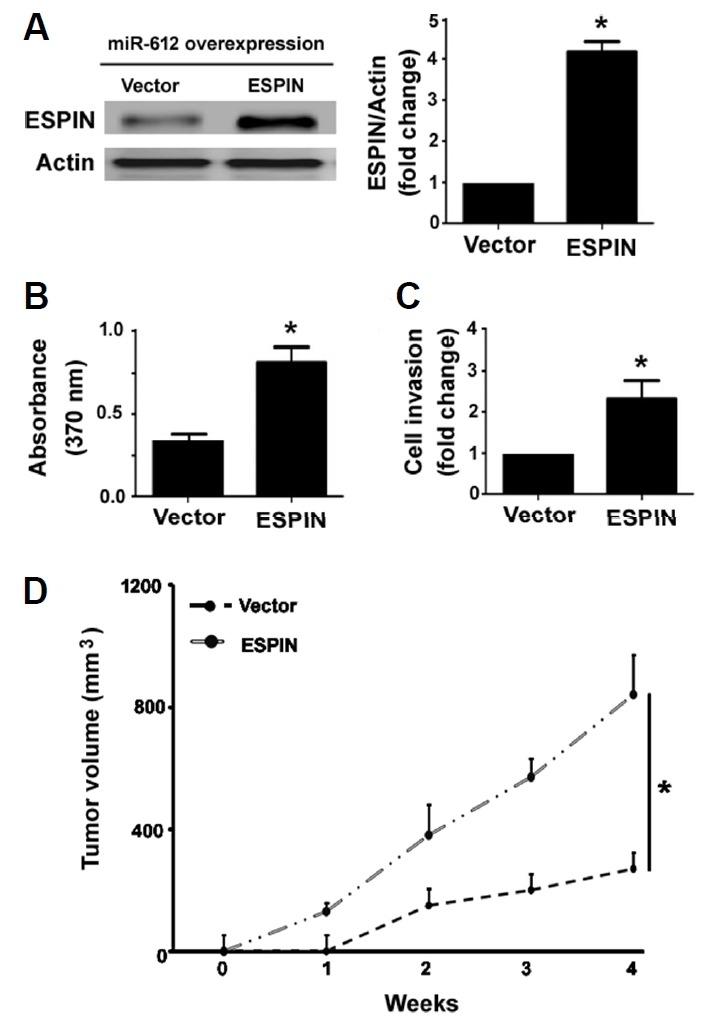
miR-612-overexpressing A375 cells were re-transfected with vector or Espin-expressing plasmid and tested for gene expression, proliferation, invasion, and tumorigenesis. (A) Western blot analysis of ESPIN protein expression. (B) Cell proliferation assessed by BrdU incorporation assay. (C) Cell invasion determined by Transwell invasion assay. (D) miR-612-overexpressing A375 cells were re-transfected with vector or Espin-expressing plasmid were subcutaneously injected to mice and tumor growth was measured. *P < 0.05 vs. the vector group.
miR-612 overexpression and Espin knockdown enhances the sensitivity of melanoma cells to doxorubicin
Finally, we checked the role of miR-612/Espin axis in the regulation of drug sensitivity in melanoma cells. As illustrated in Fig. 6A, the IC50 for doxorubicin was reduced by 2- and 4-fold in miR-612-overexpressing SK-MEL-28 and A375 cells, respectively. Similarly, Espin knockdown led to a significant decline in the IC50 for doxorubicin in both SK-MEL-28 and A375 cells (Fig. 6B). However, the sensitivity of melanoma cells to paclitaxel was not affected by either miR-612 overexpression or Espin knockdown (data not shown).
Fig. 6. miR-612 overexpression and Espin knockdown enhances the sensitivity of melanoma cells to doxorubicin.
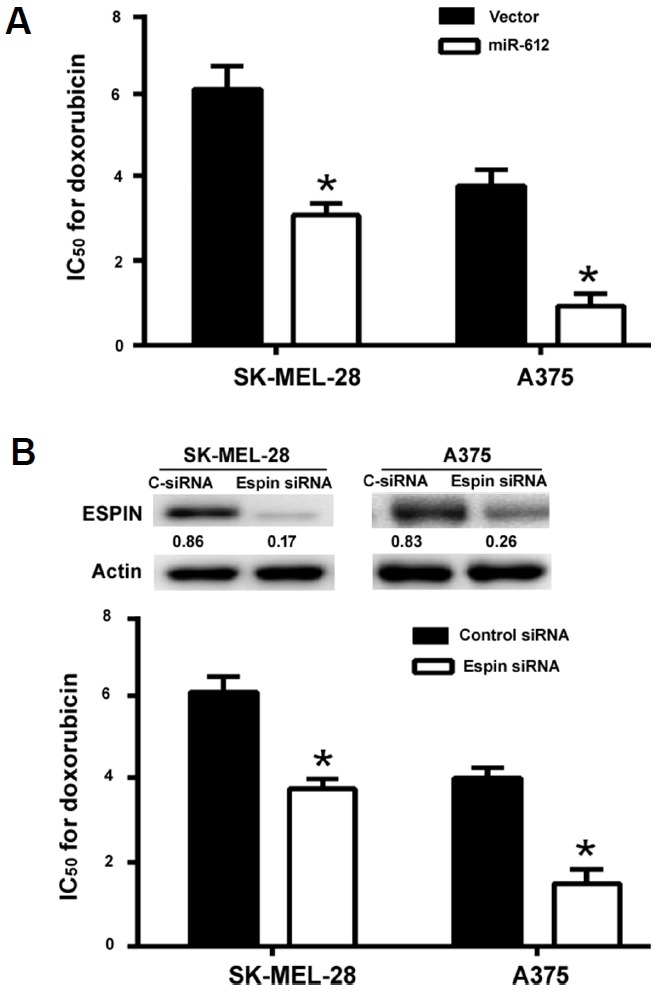
(A) SK-MEL-28 and A375 cells were transfected with empty vector or miR-612-expressing plasmid and 24 h post-transfection, exposed to different concentrations of doxorubicin for additional 24 h. The IC50 for doxorubicin was significantly reduced by miR-612 overexpression. (B) Measurement of the IC50 for doxorubicin in SK-MEL-28 and A375 cells transfected with Espin-targeting or negative control siRNAs. Inserts show representative Western blots for Espin protein. *P < 0.05.
DISCUSSION
It has been documented that miR-612 is downregulated in lung metastatic lesions of hepatocellular carcinoma (HCC) relative to primary HCC tissues (Tao et al., 2013). Another study demonstrated that miR-612 expression is significantly reduced in colorectal cancer (CRC) tissues, in particular metastatic CRC lesions (Sheng et al., 2015). miR-612 is dysregulated in pancreatic cancer (Mao et al., 2017). Consistently, our data showed that melanoma tissues had significantly decreased levels of miR-612, compared to paired adjacent normal tissues. Furthermore, low miR-612 expression correlated with aggressive parameters and predicted poor prognosis in melanoma patients. The clinical data suggest a link between miR-612 dysregulation and melanoma progression. To confirm the downregulation of miR-612 in tumor cells, we also examined the expression of miR-612 in a panel of melanoma cell lines. In line with clinical findings, miR-612 was generally downregulated in melanoma cells relative to normal human melanocytes.
Functionally, it was observed that ectopic expression of miR-612 restrained melanoma cell proliferation, colony formation, an invasion in vitro and tumorigenesis in vivo. These findings point toward a suppressive role for miR-612 in melanoma progression, which may explain the downregulation of miR-612 in melanoma specimens. In accordance with our results, it has been reported that miR-612 overexpression inhibits CRC cell growth and metastasis (Sheng et al., 2015) and HCC cell invasion and metastasis (Tao et al., 2013). miR-612 overexpression can suppress tumor growth and stemness in HCC (Liu et al.,2016). Therefore, miR-612 may serve as a tumor suppressor in a broad range of cancers.
Mechanistically, it was found that miR-612 negatively regulated the expression of Espin in melanoma cells. Luciferase reporter assays confirmed that miR-612 targeted the 3′-UTR of Espin mRNA. Most importantly, a significant inverse correlation between miR-612 and Espin expression was noted in melanoma tissues. These findings suggest the possibility that Espin acts as a functional target gene of miR-612 in melanoma. In support of this hypothesis, rescue experiments demonstrated that enforced expression of Espin abolished the suppression of proliferation, invasion, and tumorigenesis induced by miR-612. Moreover, overexpression of miR-612 and depletion of Espin elicited a similar effect on the sensitivity of melanoma cells to doxorubicin. Espin encodes an actin-binding protein and is implicated in actin cytoskeleton remodeling, thus affecting cell growth, migration, and invasion (Taura et al., 2016; Wang et al., 2012). A previous study has demonstrated that Espin expression is increased in human primary and metastatic melanomas and that depletion of Espin significantly impairs the invasion capacity of melanoma cells (Yanagishita et al., 2014). Therefore, it is suggested that repression of Espin accounts for miR-612-mediated anticancer activity in melanoma cells. Apart from Espin, miR-612 can also target other genes such as AKT2 (Sheng et al., 2015; Tang et al., 2014). However, AKT2 may not be a direct mediator of miR-612 action in melanoma, as overexpression of AKT2 failed to rescue the suppressive effect of miR-612 on melanoma cells (data not shown). Microarray-based profiling of the genes modulated by miR-612 would provide more insight into the mechanisms by which miR-612 suppresses melanoma growth and metastasis.
In conclusion, miR-612 is downregulated and acts as a tumor suppression in melanoma. Targeting Espin represents an important mechanism for miR-612-mediated inhibition of aggressive phenotype of melanoma cells. Restoration of miR-612 could be a potential therapeutic strategy against melanoma.
REFERENCES
- Alaoui-Jamali M.A., Bismar T.A., Gupta A., Szarek W.A., Su J., Song W., Xu Y., Xu B., Liu G., Vlahakis J.Z., et al. A novel experimental heme oxygenase-1-targeted therapy for hormone-refractory prostate cancer. Cancer Res. 2009;69:8017–8024. doi: 10.1158/0008-5472.CAN-09-0419. [DOI] [PubMed] [Google Scholar]
- Deacu E., Mori Y., Sato F., Yin J., Olaru A., Sterian A., Xu Y., Wang S., Schulmann K., Berki A., et al. Activin type II receptor restoration in ACVR2-deficient colon cancer cells induces transforming growth factor-beta response pathway genes. Cancer Res. 2004;64:7690–7696. doi: 10.1158/0008-5472.CAN-04-2082. [DOI] [PubMed] [Google Scholar]
- Donaudy F., Zheng L., Ficarella R., Ballana E., Carella M., Melchionda S., Estivill X., Bartles J.R., Gasparini P. Espin gene (ESPN) mutations associated with autosomal dominant hearing loss cause defects in microvillar elongation or organisation. J Med Genet. 2006;43:157–161. doi: 10.1136/jmg.2005.032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerla N., Gevaert O. MicroRNA based Pan-Cancer diagnosis and treatment recommendation. BMC Bioinformatics. 2017;18:32. doi: 10.1186/s12859-016-1421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrioğlu U., Dodurga Y., Elmas L., Seçme M. Ferulic acid decreases cell viability and colony formation while inhibiting migration of MIA PaCa-2 human pancreatic cancer cells in vitro. Gene. 2016;576:476–482. doi: 10.1016/j.gene.2015.10.061. [DOI] [PubMed] [Google Scholar]
- Li B., Xie Z., Li Z., Chen S., Li B. MicroRNA-613 targets FMNL2 and suppresses progression of colorectal cancer. Am J Transl Res. 2016;8:5475–5484. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu Y., Liu D.L., Dong L.L., Wen D., Shi D.M., Zhou J., Fan J., Wu W.Z. miR-612 suppresses stem cell-like property of hepatocellular carcinoma cells by modulating Sp1/Nanog signaling. Cell Death Dis. 2016;7:e2377. doi: 10.1038/cddis.2016.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher D.A., Silvis M.R., Cho J.H., Holmen S.L. Melanoma Brain Metastasis: Mechanisms, Models, and Medicine. Int J Mol Sci. 2016;17(9) doi: 10.3390/ijms17091468. pii: E1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komina A., Palkina N., Aksenenko M., Tsyrenzhapova S., Ruksha T. Antiproliferative and Pro-Apoptotic Effects of MiR-4286 Inhibition in Melanoma Cells. PLoS One. 2016;11:e0168229. doi: 10.1371/journal.pone.0168229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Shen J., Lu Y., Lin K., Wang H., Li Y., Chang P., Walker M.G., Li D. RNA sequencing analyses reveal novel differentially expressed genes and pathways in pancreatic cancer. Oncotarget. 2017;8:42537–42547. doi: 10.18632/oncotarget.16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal H.C., Hunt K.M., Diamond A., Elmets C.A., Afaq F. Phytochemicals for the Management of Melanoma. Mini Rev Med Chem. 2016;16:953–979. doi: 10.2174/1389557516666160211120157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekerková G., Zheng L., Loomis P.A., Mugnaini E., Bartles J.R. Espins and the actin cytoskeleton of hair cell stereocilia and sensory cell microvilli. Cell Mol Life Sci. 2006;63:2329–2341. doi: 10.1007/s00018-006-6148-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng L., He P., Yang X., Zhou M., Feng Q. miR-612 negatively regulates colorectal cancer growth and metastasis by targeting AKT2. Cell Death Dis. 2015;6:e1808. doi: 10.1038/cddis.2015.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X., Hildebrandt M.A., Gu J., Tannir N.M., Matin S.F., Karam J.A., Wood C.G., Wu X. MicroRNA profiling in clear cell renal cell carcinoma tissues potentially links tumorigenesis and recurrence with obesity. Br J Cancer. 2017;116:77–84. doi: 10.1038/bjc.2016.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Tao Z.H., Wen D., Wan J.L., Liu D.L., Zhang S., Cui J.F., Sun H.C., et al. MiR-612 suppresses the stemness of liver cancer via Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2014;447:210–215. doi: 10.1016/j.bbrc.2014.03.135. [DOI] [PubMed] [Google Scholar]
- Tao Z.H., Wan J.L., Zeng L.Y., Xie L., Sun H.C., Qin L.X., Wang L., Zhou J., Ren Z.G., Li Y.X., et al. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210:789–803. doi: 10.1084/jem.20120153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas F. Metastatic behavior in melanoma: timing, pattern, survival, and influencing factors. J Oncol. 2012;2012:647684. doi: 10.1155/2012/647684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura A., Taura K., Koyama Y., Yamamoto N., Nakagawa T., Ito J., Ryan A.F. Hair cell stereociliary bundle regeneration by espin gene transduction after aminoglycoside damage and hair cell induction by Notch inhibition. Gene Ther. 2016;23:415–423. doi: 10.1038/gt.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Akkooi A.C., Atkins M.B., Agarwala S.S., Lorigan P. Surgical Management and Adjuvant Therapy for High-Risk and Metastatic Melanoma. Am Soc Clin Oncol Educ Book. 2016;35:e505–e514. doi: 10.1200/EDBK_159087. [DOI] [PubMed] [Google Scholar]
- Wang L., Zou J., Shen Z., Song E., Yang J. Whirlin interacts with espin and modulates its actin-regulatory function: an insight into the mechanism of Usher syndrome type II. Hum Mol Genet. 2012;21:692–710. doi: 10.1093/hmg/ddr503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A.M., Stewart J.E., Atigadda V.R., Mroczek-Musulman E., Muccio D.D., Grubbs C.J., Beierle E.A. Preclinical Evaluation of a Novel RXR Agonist for the Treatment of Neuroblastoma. Mol Cancer Ther. 2015;14:1559–1569. doi: 10.1158/1535-7163.MCT-14-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Chen X., He Q., Luo C. MicroRNA-9 suppresses the growth, migration, and invasion of malignant melanoma cells via targeting NRP1. Onco Targets Ther. 2016;9:7047–7057. doi: 10.2147/OTT.S107235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagishita T., Yajima I., Kumasaka M., Kawamoto Y., Tsuzuki T., Matsumoto Y., Watanabe D., Kato M. Actin-binding protein, Espin: a novel metastatic regulator for melanoma. Mol Cancer Res. 2014;12:440–446. doi: 10.1158/1541-7786.MCR-13-0468-T. [DOI] [PubMed] [Google Scholar]
- Zhang K., Wu X., Wang J., Lopez J., Zhou W., Yang L., Wang S.E., Raz D.J., Kim J.Y. Circulating miRNA profile in esophageal adenocarcinoma. Am J Cancer Res. 2016;6:2713–2721. [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Sekerková G., Vranich K., Tilney L.G., Mugnaini E., Bartles J.R. The deaf jerker mouse has a mutation in the gene encoding the espin actin-bundling proteins of hair cell stereocilia and lacks espins. Cell. 2000;102:377–385. doi: 10.1016/s0092-8674(00)00042-8. [DOI] [PMC free article] [PubMed] [Google Scholar]