Abstract
Here, we report isolation of multiple long non-coding RNAs (lncRNAs) expressed tissue-specifically during murine embryogenesis. One of these, subsequently came to be known as Redrum, is expressed in erythropoietic cells in fetal liver and adult bone marrow. Redrum transcription is also detected during pregnancy in the spleen where extramedullary hematopoiesis takes place. In order to examine the function of Redrum in vivo, we generated a gene-targeted murine model and analyzed its embryonic and adult erythropoiesis. The homozygous mutant embryo showed no apparent deficiency or defect in erythropoiesis. Adult erythropoiesis in bone marrow and in the spleen during pregnancy likewise showed no detectable phenotype as red blood cells matured in normal fashion. The phenotype is in contrast to the reported function of Redrum in vitro, and our observation implies that Redrum plays in vivo an accessory or supplementary role whose loss is compatible with normal erythropoiesis.
Keywords: erythropoiesis, hematopoiesis, long non-coding RNA, Redrum
INTRODUCTION
It has been well-established that most of the mammalian genome is transcribed but only 1~2% code for proteins (Djebali et al., 2012; Pertea, 2012). This implies that most of the RNA species are non-coding and that functional assignment has yet to be made for a vast number of genes. The so-called long non-coding RNAs (lncRNAs) featuring mRNA-like structure but without any apparent open reading frame have been shown to play critical functions in various cellular processes including proliferation, differentiation, stress response, senescence, and apoptosis through diverse molecular mechanisms (Perry and Ulitsky, 2016; Schmitz et al., 2016).
LncRNAs also likely regulate erythropoiesis, the process through which red blood cells are produced (Paralkar and Weiss, 2013). Specifically, several recent studies reported isolation and functional analyses of lncRNAs expressed in maturing erythroblasts and involved in erythropoiesis (Alvarez-Dominguez et al., 2014; Hu et al., 2011; Paralkar et al., 2014). These studies used high-throughput sequencing and RNA interference-based functional tests. Specifically, Alvarez-Dominguez and coworkers reported identification of lncRNAs expressed in early erythroid cells from fetal liver of mice using RNA-seq and functionally validated 12 candidates for their role in terminal maturation using specific siR-NAs (Alvarez-Dominguez et al., 2014). Similarly, Paralkar and coworkers performed RNA-seq to define specific lncRNAs in three distinct cell-types, erythroblasts, cultured megakaryocytes derived from mouse fetal liver and megakaryocyte-erythroid precursors from adult bone marrow (Paralkar et al., 2014). Subsequent RNAi-based screen led to multiple candidates with potential role in terminal erythroid differentiation. One of the lncRNAs that emerged from both studies is Redrum. Depletion of Redrum in vitro resulted in stunted expression of terminal markers of erythropoiesis (Alvarez-Dominguez et al., 2014; Paralkar et al., 2014).
Here, we describe independent isolation of Redrum, expression in various erythropoietic tissues and stages during differentiation and the effect of gene targeting in vivo. We show that Redrum is not just expressed in fetal liver but also in adult bone marrow and in the spleen during pregnancy. We also show that the peak of Redrum expression is near the terminal stage of red blood cell (RBC) differentiation. Surprisingly, deletion of the highly conserved 3rd exon of Redrum which accounts for more than 80% of the transcript had little effect in viability or in erythropoiesis.
MATERIALS AND METHODS
RNA-seq data acquisition and analysis
Total RNA was extracted from whole body and telencephalon of mouse fetuses at embryonic day 12.5 (E12.5) using the RNeasy Mini kit (Qiagen). RNA sequencing data sets for whole body and telencephalon were generated using TruSeq RNA Library Preparation kit v2 and Illumina HiSeq 2000 for 101 bp-long pair-end reads. Raw reads were successfully trimmed using Trimmomatic-0.3. Alignment, removing duplication and indexing were sequentially performed using Tophat v2.0.21 and Picard v1.127 based on the reference sequence of mouse ENSEMBL version78 in the GRCm38 assembly. LncRNAs were identified from ENSEMBL biotype status definition. Differentially expressed genes (DEG) between whole body and telencephalon were determined using Cuffdiff v2.2.1, and LncRNAs with differential expression test P-value < 0.05, gene expression in one of two tissues (FPKM) > 1.5, and log2 fold change > 2 were selected for further examination. The RNA-seq data were deposited in the Gene Express Omnibus (GEO) database [GEO: GSE103986].
RNA in situ hybridization
RNA in situ hybridization was performed essentially as previously described using paraformaldehyde–fixed and OCT-embedded mouse embryo sections (Ma et al., 1998). For whole-mount in situ hybridization, E10.5 mouse embryo was used. Plasmid templates for anti-sense RNA probes were either obtained from EST library (Open Biosystems) or generated by amplifying portions of target cDNA by PCR and ligating into pGEM-Teasy Vector plasmids (Promega). The EST IDs and oligonucleotide primers are provided in Supplementary Table S1. RNA probes were generated by in vitro transcription with T7, T3 or SP6 RNA polymerase and incorporated with digoxigenin (DIG)-labeled UTP using the labeling mix (Roche). After hybridization, DIG-labeled RNA probes were detected by anti-DIG antibody conjugated with alkaline phosphatase (Roche) which yields purple precipitates from Nitro blue tetrazolium (NBT) and 5-Bromo-4-chloro-3-indoly phosphate (BCIP).
Reverse transcription and PCR amplification
Total RNA was isolated from indicated tissues of wild type and Redrum KO mouse. Reverse transcription was carried out with 1 μg of RNA from each sample using QuantiTect Reverse Transcription Kit (QIAGEN) according to manufacturer’s instructions. Standard PCR was performed on Gene-Amp PCR system2700 (Applied Biosystems) using i-StarTaq DNA polymerase (iNtRON Biotechnology). Real-time PCR for quantification was carried out using Power SYBR Green PCR Master Mix (Applied Biosystems) and CFX96™ Real Time PCR Detection System (BIO-RAD). The following oligonucleotide primers were used for conventional PCR and real-time PCR: Redrum: (forward) 5′-AGACTGCTTTCTCAAGTGGCTTA G-3′, (reverse) 5′-AGCCAATGAATAGTCACTGTAGGG-3′; GAPDH: (forward) 5′-CAAGAAGGTGGTGAAGCAGG-3′, (reverse) 5′-CCGTATTCATTGTCATACCAGG-3′.
Gene targeting
Gene trap vector was designed for homologous recombination in the 2nd intron in Redrum genomic locus. The vector contains a splicing acceptor (SA) sequence in front of a reporter gene consisted in order of an internal ribosome entry site, nuclear localization sequence, enhanced green fluorescence protein (eGFP) cDNA sequence and poly A signal sequence. Genomic DNA fragments, each 3 kb in length, 5′ and 3′ to the targeted site were amplified from a BAC clone (RP23-37K18) by PCR and ligated to each end of the reporter cassette. The sequences of oligonucleotide primers used in vector construction are listed in Supplementary Table S2. The details of targeting vector construction are available upon request. A 20-bp guide sequence (5′-AGAACGGAA CTGACAGCCGT-3′) targeting DNA within the second intron of Redrum was selected for induction of homologous recombination. The target specific sgRNA and Cas9 mRNA were purchased from Toolgen, Inc. Pronuclear injection was performed on B6D2F1 eggs, and homologous recombinant line was established via Southern blotting among transgene positive pups. For Southern blotting, genomic DNA was extracted from tails, cut with Kpn I and subsequently processed following standard protocols. The probe was amplified from genomic region immediately upstream to 5′ arm of the targeting vector. Subsequent genotyping was carried out by PCR amplification of genomic DNA. Detailed protocols are available upon request. Oligonucleotide primers used for probe generation and genotyping are provided in Supplementary Table S3.
Flow cytometric analysis
Cells were stained with APC-conjugated Rat anti-mouse TER119 (BD Pharmingen, 557909) and PE-conjugated Rat anti-mouse CD71 (BD Pharmingen, 553267) and analyzed by flow cytometry using BD LSRFortessa (BD Bioscience) and BD FACSDiva Software (BD Bioscience). Cell sorting was carried out using BD FACSAria after labeling with anti-mouse TER119 and anti-mouse CD71. The nucleic acid content was examined using BD LSRFortessa after labeling cells with thiazole orange (TO; Sigma Aldrich) or with Retic Count (BD Bioscience). Forward scatter (FSC) analysis was carried out to determine the cell size likewise using BD LSRFortessa.
Research ethics
The plan for this study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Ewha Womans University.
RESULTS AND DISCUSSION
We extracted RNA from whole mouse embryo and from brain at E12.5 and generated RNA-seq data. Based on subsequent DEG analysis, we selected highly differentially expressed lncRNAs from each of the two sources relative to the other leading to 19 tissue-specifically expressed candidates that may function during embryogenesis (Supplementary Table S1). Gene names had already been assigned for some of the lncRNAs such as H19 and Miat while for one of the lncRNAs the gene name Redrum was assigned subsequent to initiation of our study. EST clones were available for most of them, and for those unavailable, cDNA fragments ranging from 500 to 1000 base pairs in length were cloned (Supplementary Table S1). The plasmid templates were used for generation of anti-sense probes for RNA in situ hybridization assay. We were able to detect significant tissue-specific expression from at least eight of them (Fig. 1). Of note, H19 showed strong expression in non-neural tissues consistent with a previous report (Poirier et al., 1991). Miat, AI854517, and A930011O12Rik were mostly expressed in the ventricular or subventricular zone of the developing central nervous system (CNS) where neural stem cells proliferate and migrate laterally as they mature suggesting possible roles in neurogenesis. Rian and Mirg showed limited expression in the CNS but more prominent expression in developing skeletal muscles suggestive of potential roles in musculogenesis. Two lncRNAs, Redrum and 2700038G22Rik, showed strong expression in the embryonic liver. The latter also showed low-level expression in the CNS and in lung buds.
Fig. 1. Expression of lncRNAs in E12.5 mouse embryo.
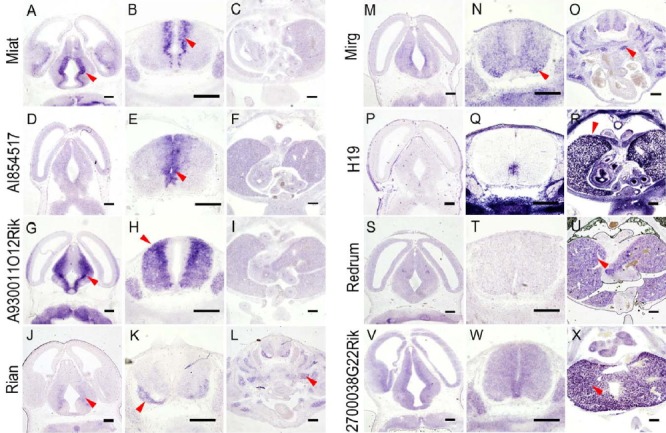
Eight of the 19 differentially expressed lncRNAs showed tissue-specific expression by RNA in situ hybridization. For each lncRNA, three sections through the brain, neural tube and trunk are shown. Note expression of Miat, AI854517, and A930011O12Rik in the CNS (arrowheads in A, B, E, G, H). Rian and Mirg are expressed both in the CNS and in muscular tissues (arrowheads in J, K, L, N, O). H19 is strongly expressed mainly outside the CNS (arrowhead in R) while Redrum and 2700038G22Rik mainly in the liver (arrowheads in U, X). (Scale bars: 200 μm.)
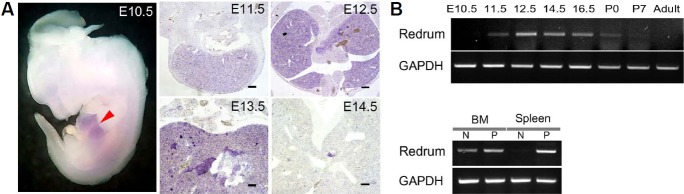
We chose Redrum for detailed analyses of expression and function. Two independent studies reported expression of Redrum during murine embryonic erythropoiesis which occurs in the developing liver (Alvarez-Dominguez et al., 2014; Paralkar et al., 2014). Consistently, Redrum expression was seen in the embryonic liver as early as E10.5 (Fig. 2A). The expression in the liver peaks in E12.5 and then declines through the embryogenesis largely coinciding with the level of fetal erythropoiesis (Figs. 2A and 2B). We also saw expression in the adult bone marrow, the primary tissue of hematopoiesis (Fig. 2C). Interestingly, Redrum expression was strongly induced during pregnancy in the spleen, the site of extramedullary erythropoiesis which occurs in response to physiological needs arising from pregnancy (Fig. 2C).
Fig. 2. Expression of Redrum in hematopoietic tissues.
Whole mount RNA in situ hybridization show expression of Redrum in developing liver (arrowhead in A). Transverse sections from embryos with indicated days show expression in the liver (A). Note the peak of expression in E12.5 (A) which was also the case in the RT-PCR analysis (B) carried out with liver tissues isolated from indicated stages. In adult mice, liver was negative for Redrum (B) while bone marrow was positive during both pregnant and non-pregnant states (C). Strong expression in the spleen was seen only during pregnancy (C). (BM, bone marrow; P, pregnant; N, non-pregnant)
Next, we sought to determine the stage of erythropoiesis during which Redrum is expressed and is therefore likely to function. Erythropoiesis can be divided into several stages based on the expression of two surface markers, CD71 and Ter119 (Zhang et al., 2003). Cells in the early stage of the differentiation are double negative (CD71−, Ter119−). Subsequently differentiating cells express CD71 (CD71+, Ter119−), then express Ter119 and become double positive (CD71+, Ter119+) and finally lose CD71 expression during terminal differentiation (CD71−, Ter119+). Real-time PCR analysis performed on E12.5 liver cells divided into four groups (R1–R4) based on the expression of CD71 and Ter119 indicated that Redrum expression is highest in the erythroblast stage (CD71+ TER119+, R3) which is marked by the initiation of enucleation process (Figs. 3A and 3B). Consistent with this, it has been reported that RNA interference targeting Redrum inhibits maturation of red blood cells through this stage and results in compromised enucleation in vitro (Alvarez-Dominguez et al., 2014; Paralkar et al., 2014). In order to examine the function of Redrum in vivo, we generated a mouse model in which the 3rd exon of the gene is deleted by homologous recombination. The 3rd exon accounts for >80% of the Redrum transcript and shows higher degree of conservation among rodent species than the 1st or 2nd exon (Supplementary Fig. S1). A gene trap vector, comprised of a splice acceptor and green fluorescent protein (GFP) gene connected to two 3kb genomic DNA fragments derived from 5′ and 3′ regions to the 3rd exon of the Redrum, was constructed (Fig. 4A). The trap vector was injected into fertilized eggs along with CRISPR9-based targeting sgRNA and Cas9 mRNA. The successful targeting was confirmed by Southern blotting and PCR amplification using genomic DNA prepared from progeny mice (Figs. 4B and 4C). RT-PCR analysis confirmed that no transcript containing the 3rd exon is generated in the homozygous mutant (data not shown). We failed to observe expression of GFP in the fetal liver for an unknown reason.
Fig. 3. Expression of Redrum in erythroblasts.
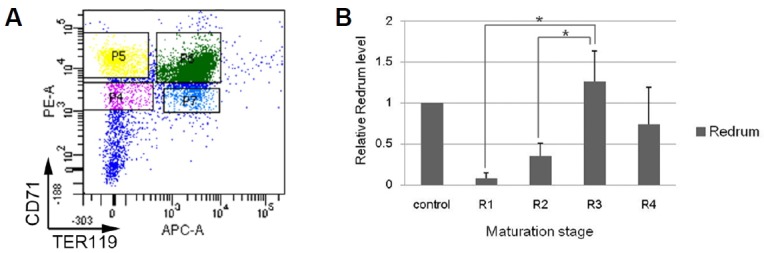
Cells were harvested from E12.5 liver, labelled with CD71 and TER119 antibodies and isolated by fluorescent activated cell sorting technique (A). RT-PCR analysis shows that Redrum expression is highest in the CD71+ Ter119+ erythroblast stage (B). Results represent mean ± SD of three independent assays. Statistical significance was confirmed by the Student’s t-test (* indicates P-value of < 0.05).
Fig. 4. Generation of Redrum-targeted mouse model.
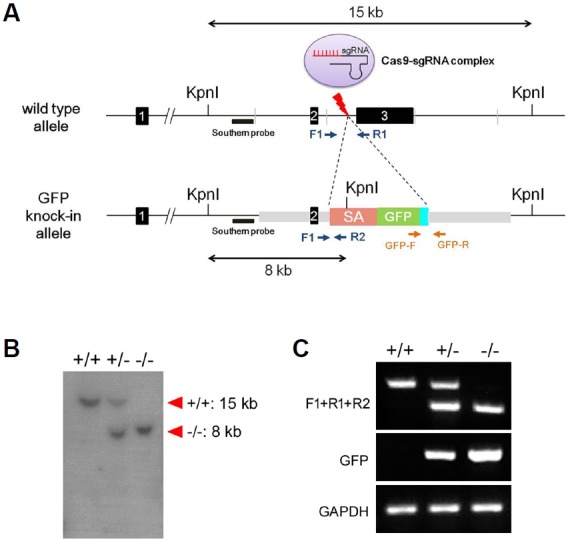
Map of the wild type Redrum locus is shown at the top, and the locus after homologous recombination is shown below (A). A Kpn I site is introduced by homologous recombination and would result in 8 kb fragment upon Kpn1 digestion. Locations of the probe for Southern blotting and primers (F1, R1, R2, GFP-F, and GFP-R) for PCR amplification are indicated (A). Southern blotting with genomic DNA derived from wild type, heterozygous and homozygous mice shows results consistent with the scheme of the homologous recombination (B). Likewise, PCR amplification resulted in the predicted pattern (C). (SA, splice acceptor; GFP, green fluorescent protein; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase)
The homozygous mutant mice were born at the Mendelian ratio, grossly normal, and fertile. Cells isolated from fetal livers were examined by flow cytometry to examine erythropoiesis. Compositions of sub-populations representing various stages erythropoiesis based on expression of two surface markers, CD71 and TER119, were virtually identical among embryos of different genotypes. The results indicated that erythropoiesis was largely normal in Redrum homozygous mutant (Fig. 5). This was also the case for erythropoiesis in the adult bone marrow and for extramedullary erythropoiesis in the spleen (Supplementary Fig. S2). A critical event that takes place in erythroblast stage when Redrum expression peaks is enucleation. Both cell size and nucleotide content undergo dramatic decrease during this process (Palis, 2008; Tsiftsoglou et al., 2009). CD71+ Ter119+ erythroblasts were thus isolated during embryonic, adult and extramedullary erythropoiesis and examined by flow cytometry for cell size and nucleotide content. We were particularly interested in extramedullary erythropoiesis during which Redrum was strongly induced. However, virtually no difference was noted between wild type and mutant spleens of pregnant mice in terms of cell size and nucleotide content of erythroblasts (Fig. 6). Likewise, cell size and nucleotide content of erythroblasts were virtually identical in fetal livers and adult bone marrows regardless of genotypes (Supplementary Fig. S3).
Fig. 5. Flow cytometric analysis of embryonic erythropoiesis.
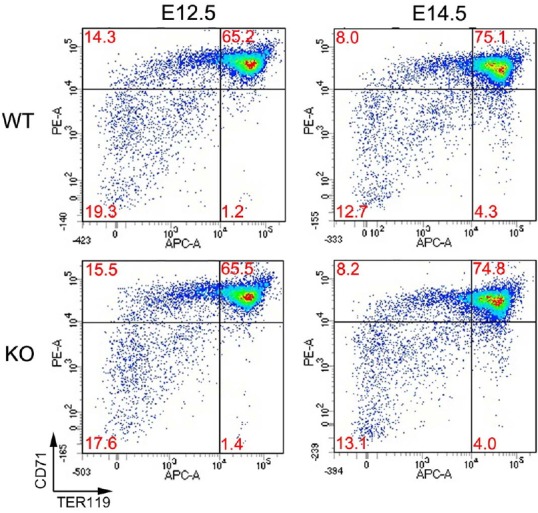
Cells were labelled with APC-conjugated anti-mouse TER119 and PE-conjugated CD71. Each quadrant represents distinct stages of development. Note the increase in proportions of relatively mature populations (CD71+ TER119+ and CD71− TER119+) in E14.5 cells compared to E12.5 cells. Note the lack of difference between wild type and homozygous mutant mice (E12.5, n = 6 & E14.5, n = 3).
Fig. 6. Flow cytometric analysis of erythroblast maturation.
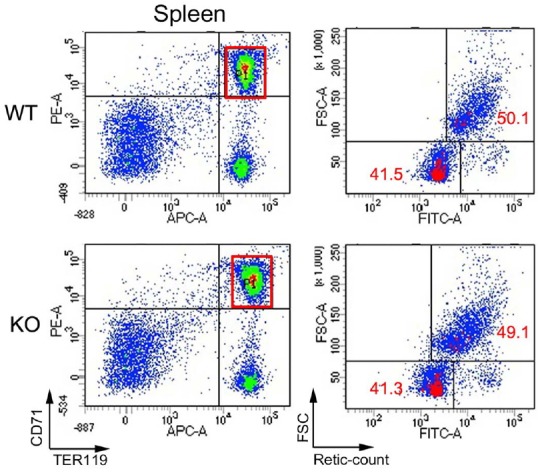
CD71+ TER119+ erythroblasts were isolated and stained with Retic Count. FSChigh and Retic Counthigh cells represent uncondensed cells prior to enucleation, and FSClow Retic Countlow cells represent condensed and enucleated cells. Note the similar proportions of the two populations in wild type and homozygous mutant mice (n = 7).
We report detailed embryonic expression patterns of multiple lncRNAs in this study. Our RNA in situ hybridization data in particular should provide bases for testable hypotheses regarding function of lncRNAs during embryonic development. Detailed study was subsequently focused on Redrum which was specifically expressed in the erythropoietic lineage. We report for the first time that Redrum expression peaks at the erythroblast stage during which enucleation takes place. We also are the first to note that Redrum expression is induced in the spleen during pregnancy likely in response to added demand for oxygen supply. Curiously, we were not able to find a deficiency associated with deletion of Redrum. This is despite that fact that depletion of this gene in vitro leads to retarded enucleation. The role of this lncRNA therefore is likely to be extremely subtle, or its deficiency may be compensated by other proteins or lncRNAs in vivo, thus rendering phenotypic consequences largely invisible. A more biochemically oriented analyses using Redrum gene products, such as identifying interacting partners, may be required for further elaboration of the gene function.
Supplementary Information
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation of Korea (NRF-2015K1A4A3047851 and 2013M3C7A1056562) funded by the Ministry of Science, ICT, and Future Planning, Republic of Korea.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Alvarez-Dominguez J.R., Hu W., Yuan B., Shi J., Park S.S., Gromatzky A.A., van Oudenaarden A., Lodish H.F. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123:570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Yuan B., Flygare J., Lodish H.F. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Chen Z., del Barco Barrantes I., de la Pompa J.L., Anderson D.J. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Palis J. Ontogeny of erythropoiesis. Curr Opin Hematol. 2008;15:155–161. doi: 10.1097/MOH.0b013e3282f97ae1. [DOI] [PubMed] [Google Scholar]
- Paralkar V.R., Weiss M.J. Long noncoding RNAs in biology and hematopoiesis. Blood. 2013;121:4842–4846. doi: 10.1182/blood-2013-03-456111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paralkar V.R., Mishra T., Luan J., Yao Y., Kossenkov A.V., Anderson S.M., Dunagin M., Pimkin M., Gore M., Sun D., et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123:1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.B., Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development. 2016;143:3882–3894. doi: 10.1242/dev.140962. [DOI] [PubMed] [Google Scholar]
- Pertea M. The human transcriptome: an unfinished story. Genes. 2012;3:344–360. doi: 10.3390/genes3030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier F., Chan C.T., Timmons P.M., Robertson E.J., Evans M.J., Rigby P.W. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development. 1991;113:1105–1114. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- Schmitz S.U., Grote P., Herrmann B.G. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiftsoglou A.S., Vizirianakis I.S., Strouboulis J. Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life. 2009;61:800–830. doi: 10.1002/iub.226. [DOI] [PubMed] [Google Scholar]
- Zhang J., Socolovsky M., Gross A.W., Lodish H.F. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.